Indel Diversity
Reading Config File:
knitr::opts_chunk$set(echo = TRUE, warning = FALSE, message = FALSE)
suppressMessages(suppressWarnings(library(knitr)))
suppressMessages(suppressWarnings(library(ggplot2)))
suppressMessages(suppressWarnings(library(ggrepel)))
suppressMessages(suppressWarnings(library(data.table)))
suppressMessages(suppressWarnings(library(rtracklayer)))
suppressMessages(suppressWarnings(library(plotly)))
suppressMessages(suppressWarnings(library(GenomicRanges)))
suppressMessages(suppressWarnings(library(parallel)))
suppressMessages(suppressWarnings(library(pbapply)))
suppressMessages(suppressWarnings(library(DT)))
targetName <- params$target_name
config <- yaml::read_yaml('./config.yml')
sampleSheet <- data.table::fread(config$sample_sheet)
pipeline_output_dir <- config$pipeline_output_dir
cut_sites <- rtracklayer::import.bed(config$cut_sites_file)
sampleComparisons <- data.table::fread(config$comparisons_file)Declare some common functions
importSampleBigWig <- function(pipeline_output_dir, samples, suffix = '.alnCoverage.bigwig') {
sapply(simplify = F, USE.NAMES = T,
X = unique(as.character(samples)),
FUN = function(s) {
f <- file.path(pipeline_output_dir, 'indels', s, paste0(s, suffix))
if(file.exists(f)) {
rtracklayer::import.bw(f, as = 'RleList')
} else {
stop("Can't find bigwig file for sample: ",s," at: ",
"\n",f,"\n")
}})
}
subsetRleListByRange <- function(input.rle, input.gr) {
as.vector(input.rle[[seqnames(input.gr)]])[start(input.gr):end(input.gr)]
}
getReadsWithIndels <- function(pipeline_output_dir, samples) {
readsWithIndels <- lapply(samples, function(sample) {
dt <- data.table::fread(file.path(pipeline_output_dir,
'indels',
sample,
paste0(sample, ".reads_with_indels.tsv")))
})
names(readsWithIndels) <- samples
return(readsWithIndels)
}
getIndels <- function(pipeline_output_dir, samples) {
indels <- sapply(simplify = FALSE, samples, function(s) {
f <- file.path(pipeline_output_dir,
'indels',
s,
paste0(s, ".indels.tsv"))
if(file.exists(f)) {
dt <- data.table::fread(f)
dt$sample <- s
return(dt)
} else {
stop("Can't open indels.tsv file for sample",s,
"at",f,"\n")
}})
return(indels)
}Subset sample sheet for those that match the target region of interest
targetName <- params$target_name
sampleSheet <- sampleSheet[target_name == targetName]
targetRegion <- as(sampleSheet[target_name == targetName]$target_region[1], 'GRanges')
#get the list of all guides used for the target region
#get sample-specific cut sites at the target region
sgRNAs <- unlist(strsplit(x = sampleSheet[sampleSheet$target_name == targetName,]$sgRNA_ids,
split = ':'))
cutSites <- cut_sites[cut_sites$name %in% sgRNAs]#import deletion coordinates
deletions <- do.call(rbind, lapply(getIndels(pipeline_output_dir, sampleSheet$sample_name),
function(dt) {
dt[indelType == 'D']
}))
sampleGuides <- lapply(sampleSheet$sample_name, function(s) {
sgRNAs <- unlist(strsplit(x = sampleSheet[sampleSheet$sample_name == s,]$sgRNA_ids,
split = ':'))
if(sgRNAs[1] == 'none') {
sgRNAs <- setdiff(unique(unlist(strsplit(x = sampleSheet[target_name == targetName,]$sgRNA_ids, split = ':'))), 'none')
}
return(sgRNAs)
})
names(sampleGuides) <- as.character(sampleSheet$sample_name)#find cut sites overlapping with the indels
# indels: a data.table object with minimal columns: start, end,
# cutSites: a GRanges object of cut site coordinates
# return: data.frame (nrow = nrow(indels), columns are sgRNA ids,
# values are 1 if indel overlaps cutsite, otherwise 0.
overlapCutSites <- function(indels, cutSites, extend = 5) {
cutSites_ext <- flank(cutSites, width = extend, both = TRUE)
#check if indel overlaps with the cut site
query <- GenomicRanges::makeGRangesFromDataFrame(indels)
overlaps <- as.data.table(findOverlaps(query, cutSites_ext, type = 'any', ignore.strand = TRUE))
M <- matrix(data = rep(0, nrow(indels) * length(cutSites)),
nrow = nrow(indels), ncol = length(cutSites))
colnames(M) <- cutSites$name
M[as.matrix(overlaps)] <- 1
return(M)
}
#get deletions within the target region
deletions <- as.data.table(subsetByOverlaps(GRanges(deletions), targetRegion, ignore.strand = TRUE))
#define deletion frequency: read support / coverage
deletions$freq <- deletions$ReadSupport/deletions$coverage
#find overlaps with cut sites (only considering guides used in the corresponding sample)
deletionCutsiteOverlaps <- cbind(deletions, overlapCutSites(deletions, cutSites))
deletionCutsiteOverlaps <- do.call(rbind, lapply(unique(deletionCutsiteOverlaps$sample), function(sampleName) {
dt <- deletionCutsiteOverlaps[sample == sampleName]
sgRNAs <- sampleGuides[[sampleName]]
dt$atCutSite <- apply(subset(dt, select = sgRNAs), 1, function(x) sum(x > 0) > 0)
return(dt)
}))# cs: data.frame with cut site coordinates. minimual columns: start, end, name
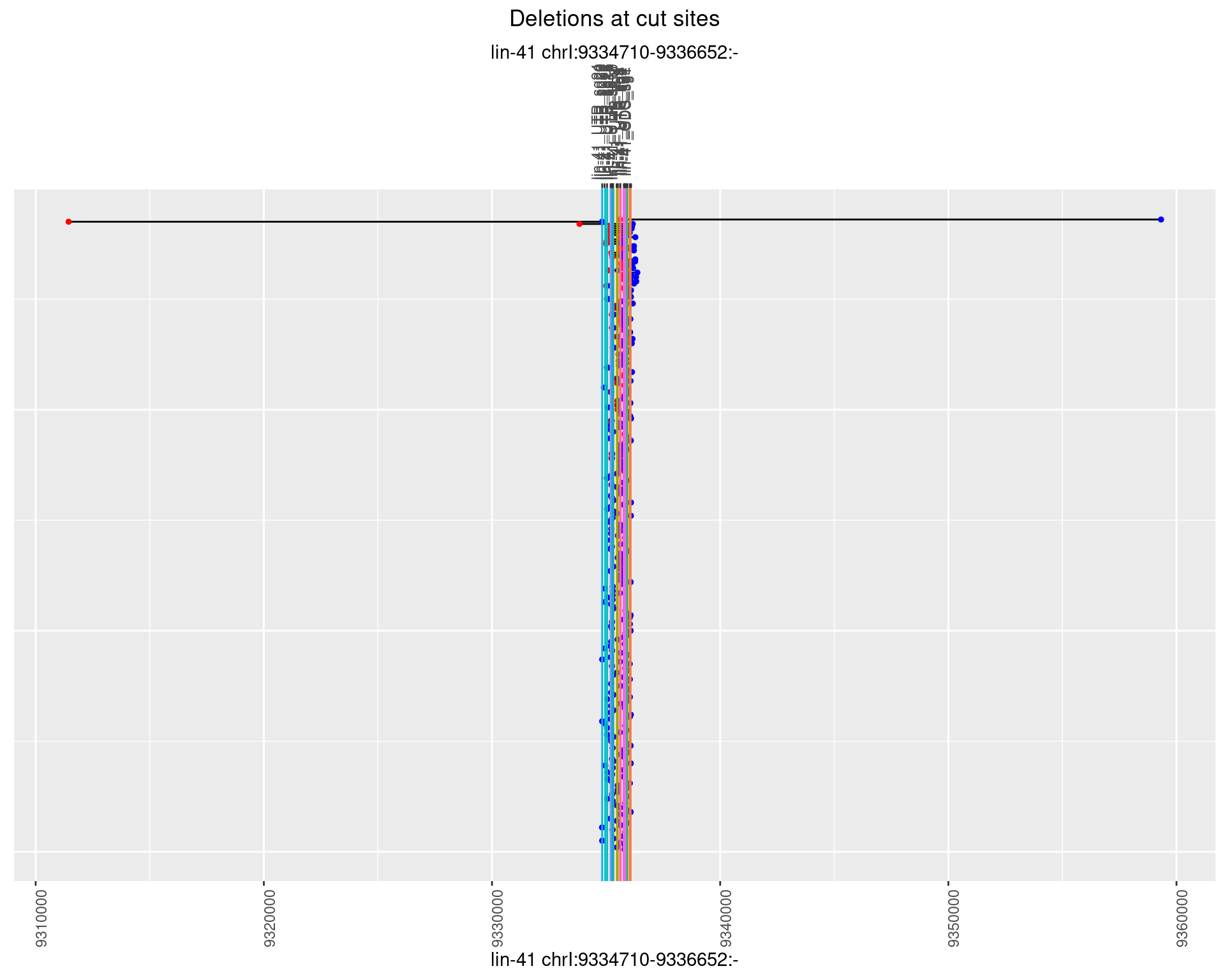
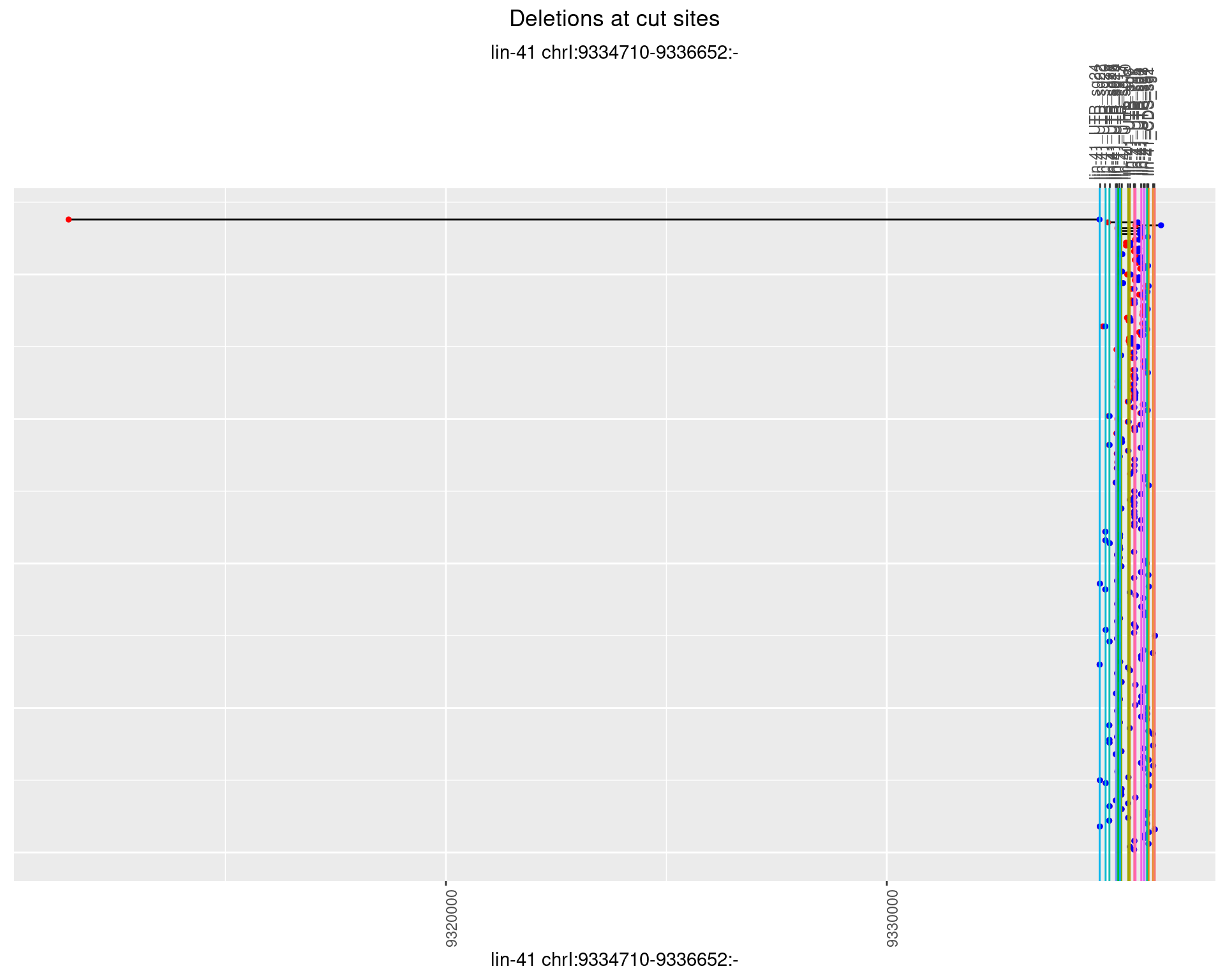
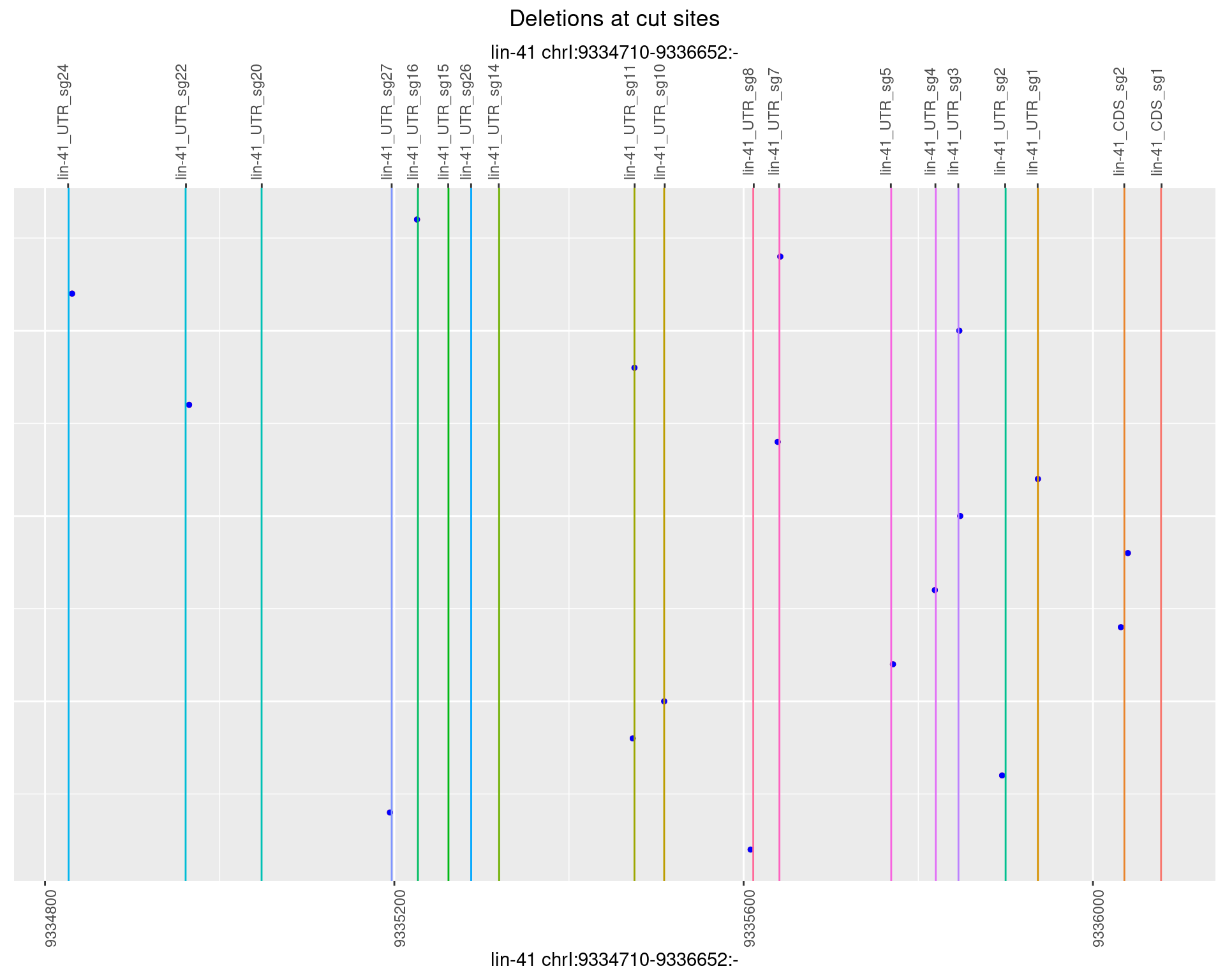
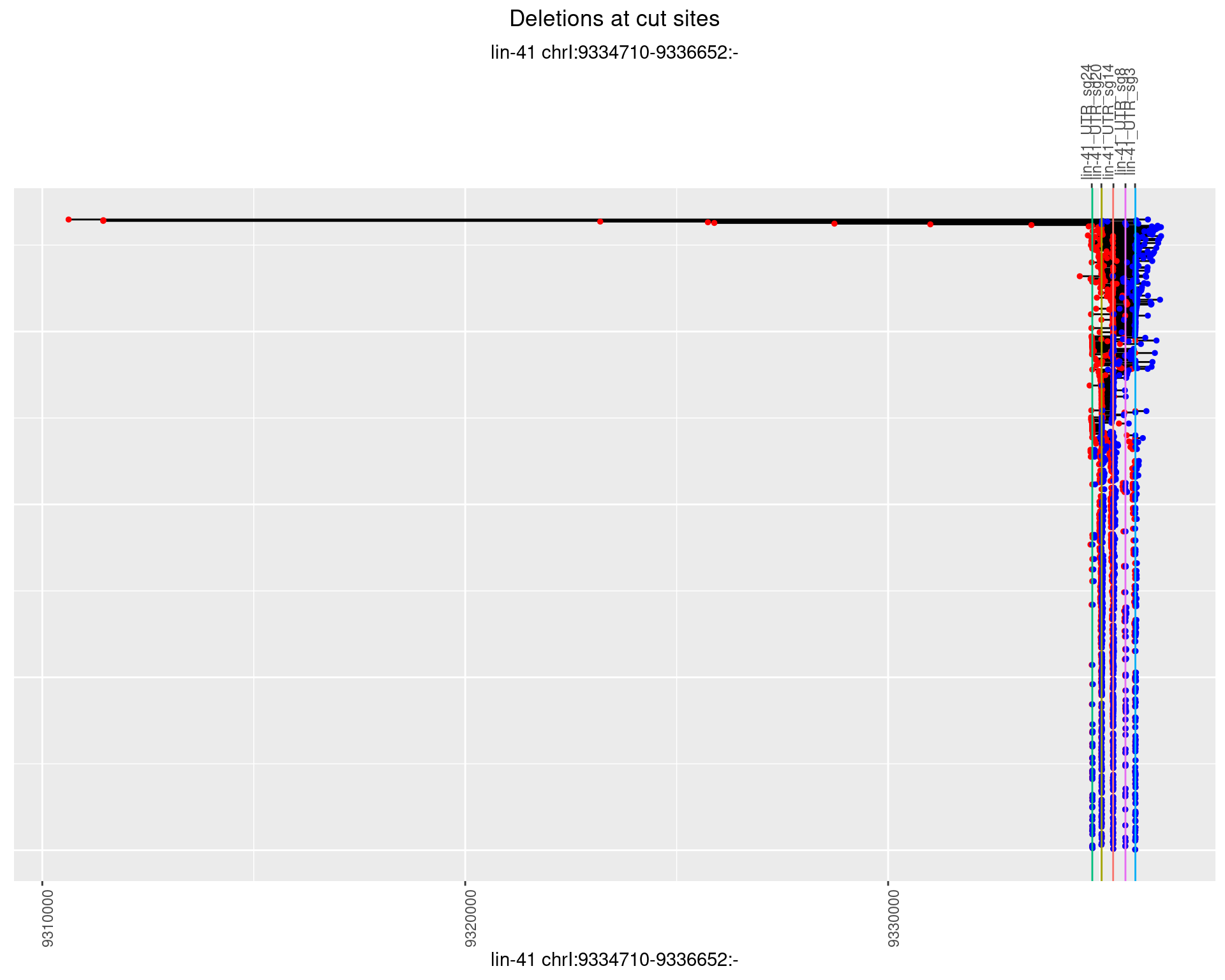
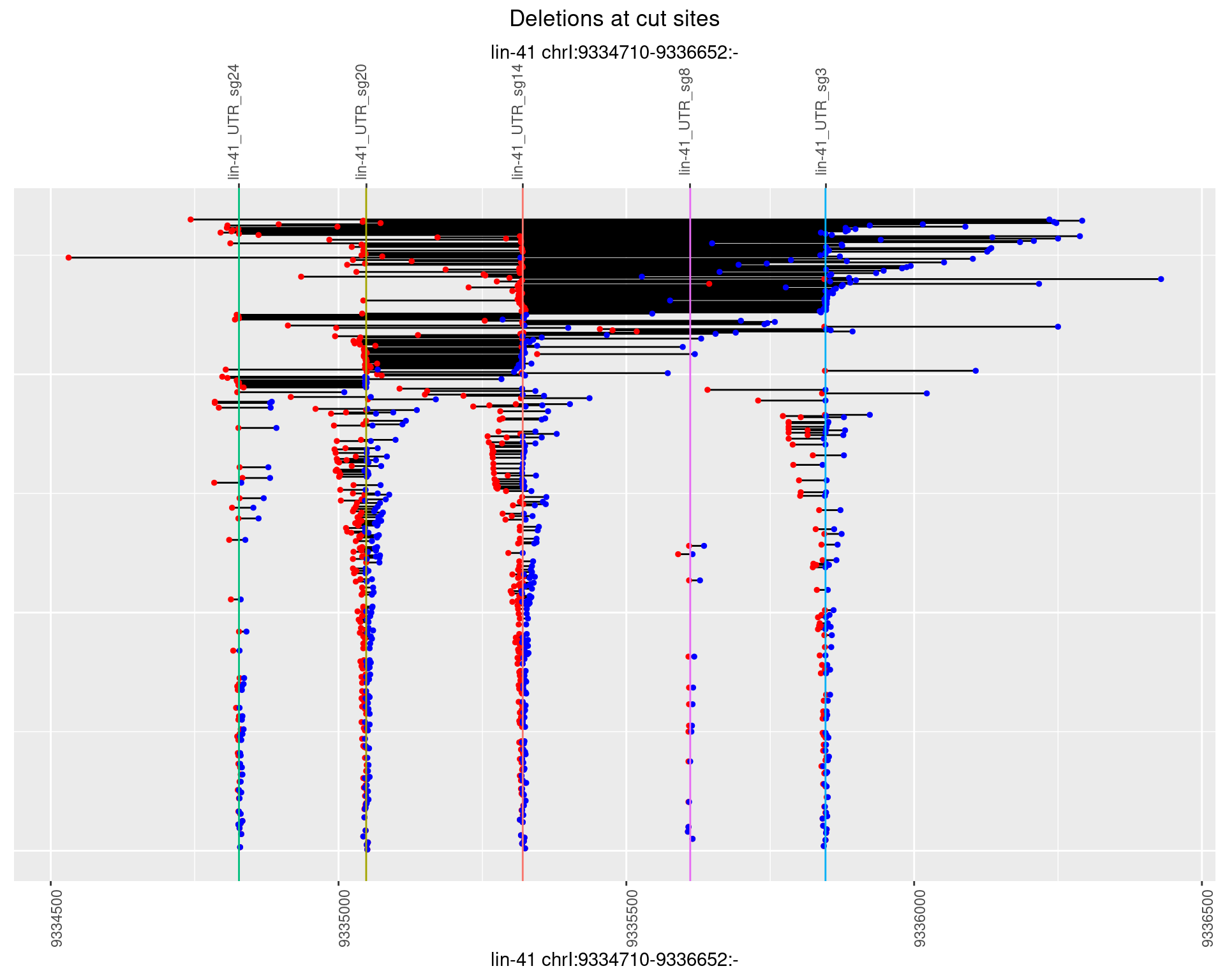
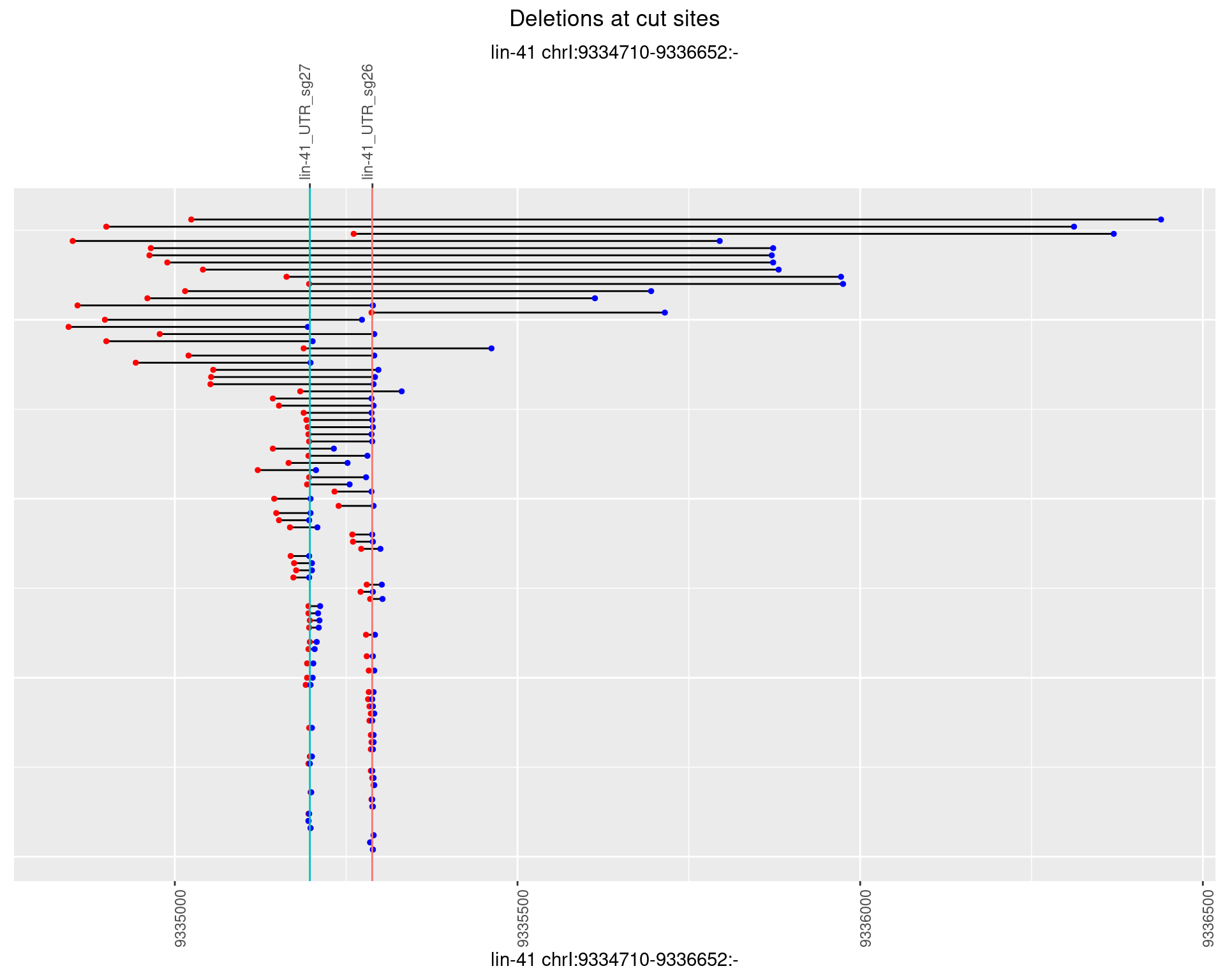
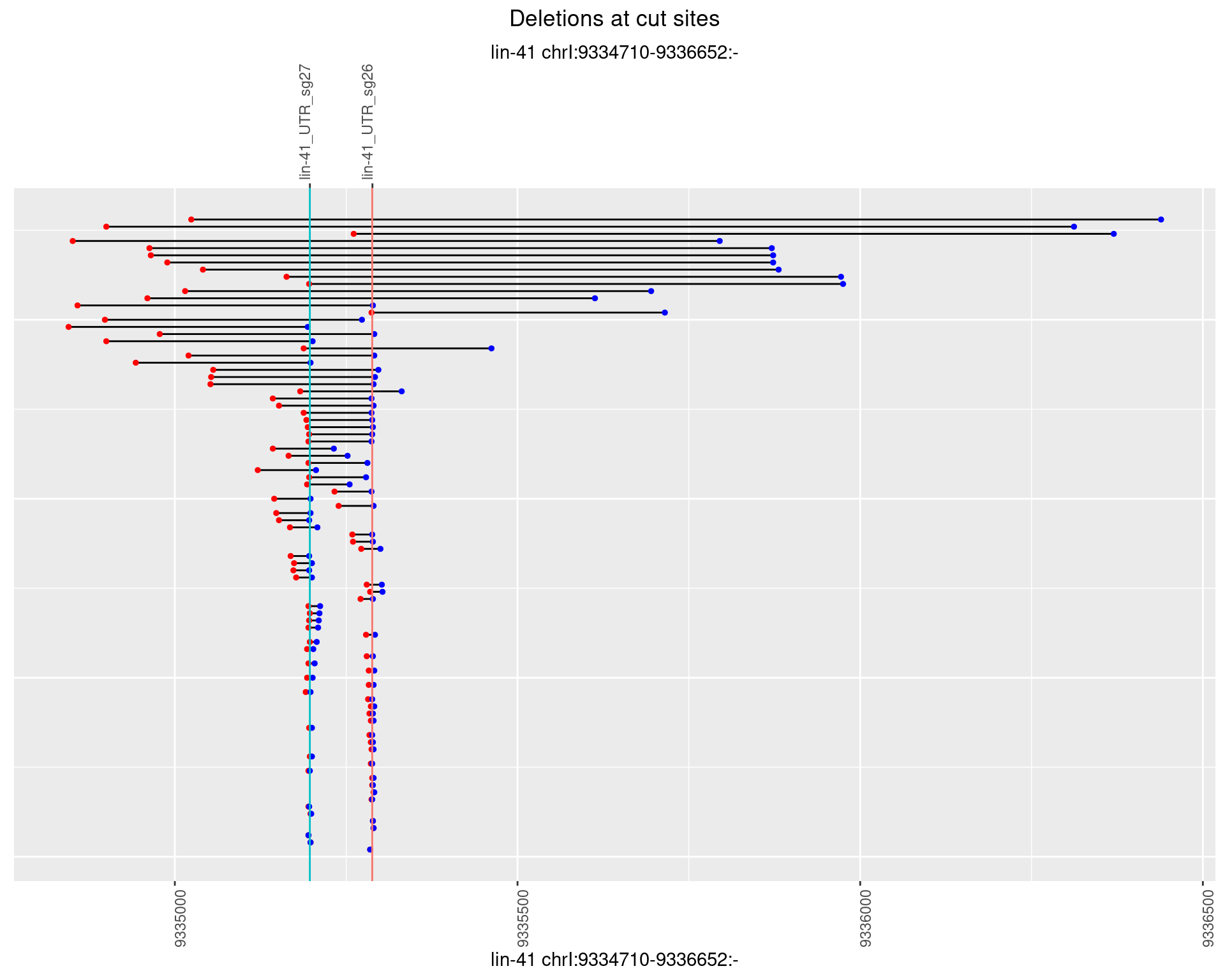
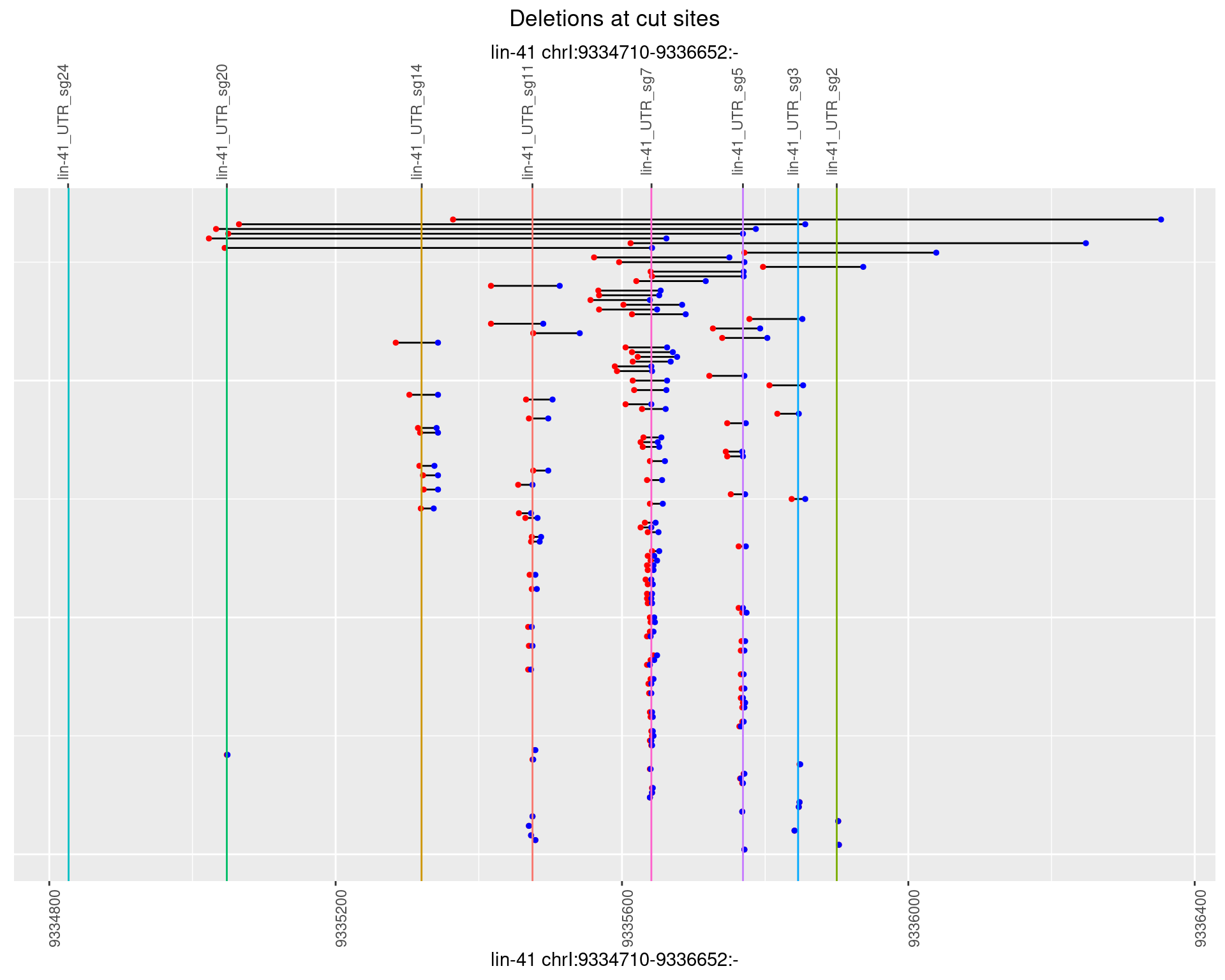
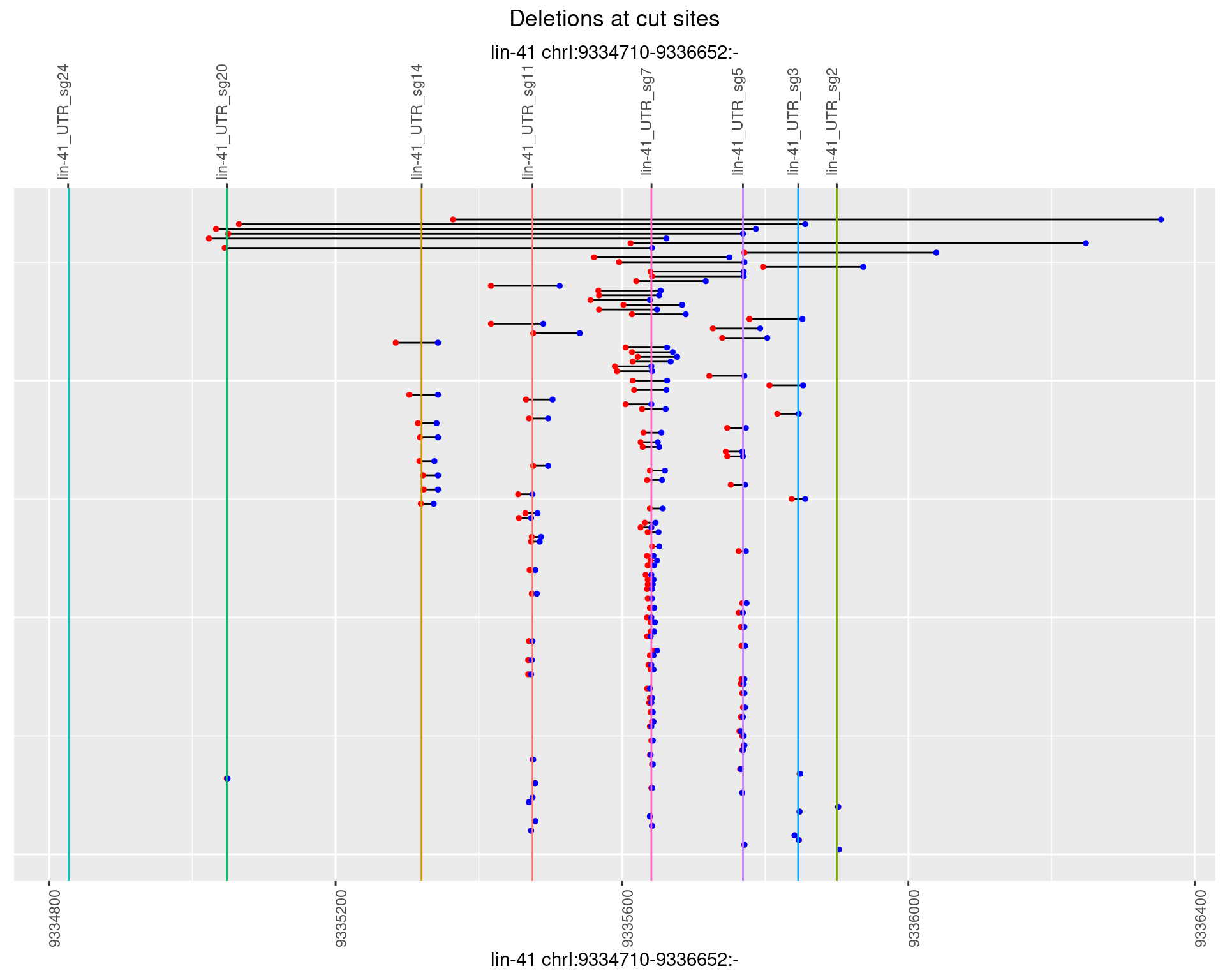
plotSegments <- function(dt, cs, readSupportThreshold = 0, freqThreshold = 0) {
dt <- dt[ReadSupport >= readSupportThreshold & freq >= freqThreshold]
if(nrow(dt) == 0){
return(NULL)
}
#first randomize the order (to avoid sorting by start position)
dt <- dt[sample(1:nrow(dt), nrow(dt))]
dt <- dt[order(end - start)]
dt$linePos <- 1:nrow(dt)
ggplot2::ggplot(dt, aes(x = linePos, ymin = start, ymax = end)) +
geom_linerange(size = 0.5) +
labs(title = "Deletions at cut sites") +
geom_point(data = dt, aes(x = linePos, y = start), size = 1, color = 'red') +
geom_point(data = dt, aes(x = linePos, y = end), size = 1, color = 'blue') +
geom_hline(data = cs,
aes(yintercept = start, color = name), show.legend = FALSE) +
theme(axis.text.y = element_blank(),
axis.title.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.x = element_text(angle = 90),
plot.title = element_text(hjust = 0.5)) +
scale_y_continuous(sec.axis = dup_axis(breaks = cs$start,
labels = cs$name)) +
coord_flip()
}
plots <- lapply(unique(deletionCutsiteOverlaps$sample), function(s) {
dt <- deletionCutsiteOverlaps[sample == s]
if(nrow(dt) == 0) {
return(NULL)
}
#segment plots with varying frequency thresholds
freqThresholds <- c(0, 0.00001, 0.0001, 0.001, 0.01, 0.1)
plots <- lapply(freqThresholds, function(t) {
p <- plotSegments(dt = dt[atCutSite == TRUE], #only plot those at cut sites
cs = as.data.frame(cutSites[cutSites$name %in% sampleGuides[[s]]]),
freqThreshold = t)
if(!is.null(p)) {
p <- p + labs(y = paste(targetName, targetRegion))
}
return(p)
})
names(plots) <- freqThresholds
return(plots)
})
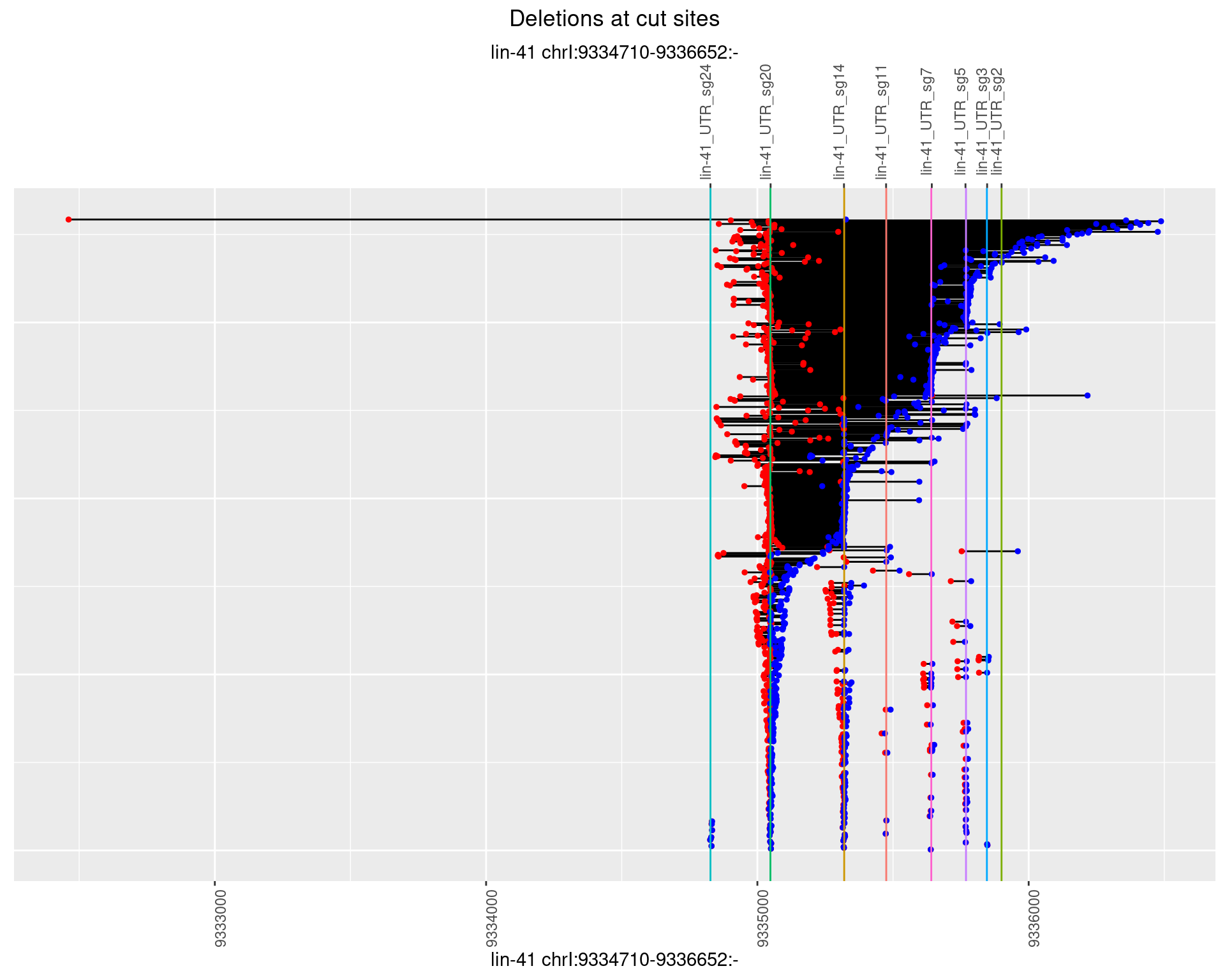
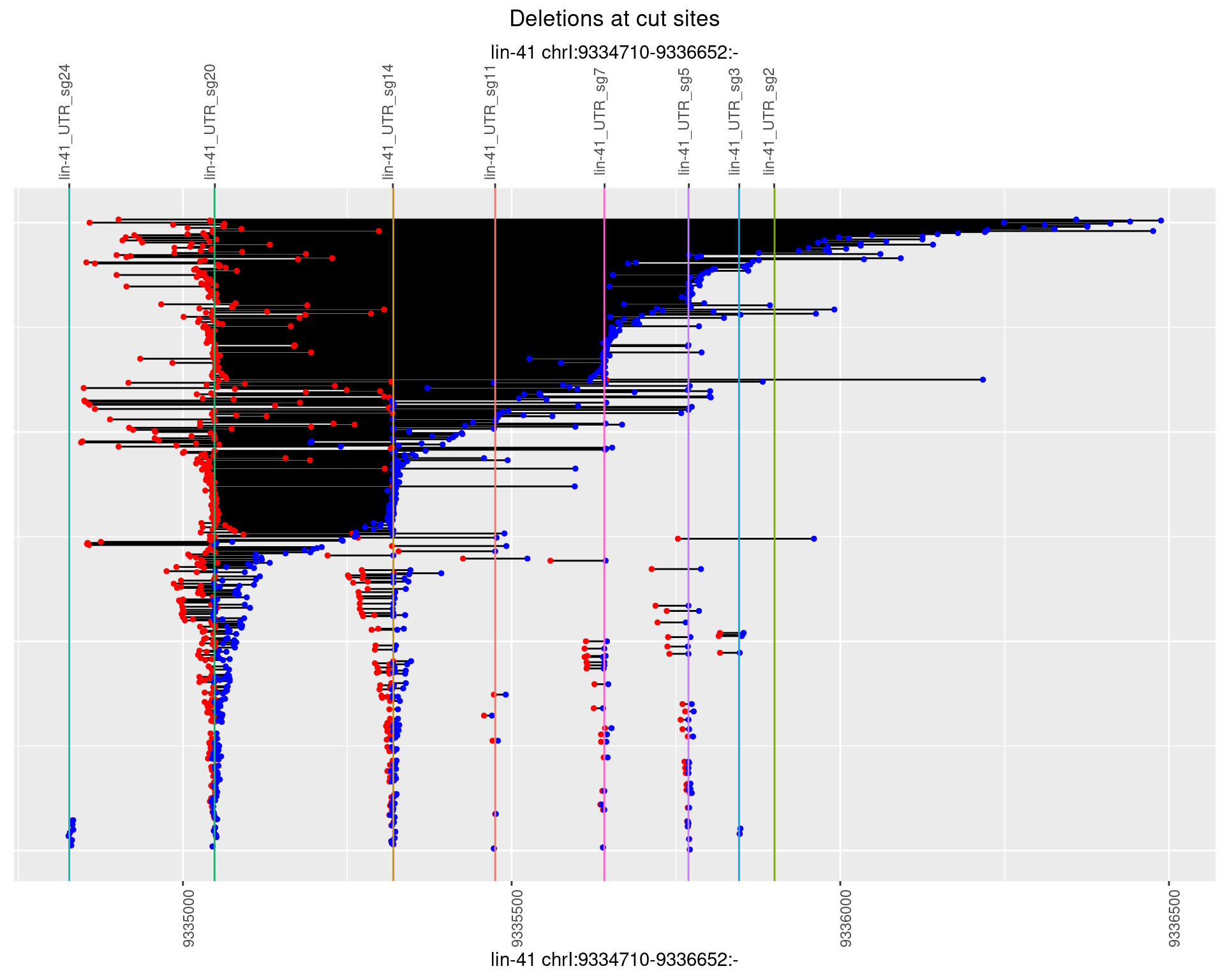
names(plots) <- unique(deletionCutsiteOverlaps$sample)1 Deletion diversity at cut sites
# folder to save pdf versions of the segment plots
dirPath <- paste(targetName, 'Indel_Diversity.deletion_segment_plots', sep = '.')
if(!dir.exists(dirPath)) {
dir.create(dirPath)
}
for (sample in names(plots)) {
cat('## ',sample,'{.tabset .tabset-fade .tabset-pills}\n\n')
for(i in names(plots[[sample]])) {
cat('### Freq:',i,'\n\n')
p <- plots[[sample]][[i]]
if(!is.null(p)) {
print(p)
ggsave(filename = file.path(dirPath, paste(sample, 'freq', i, 'pdf', sep = '.')),
plot = p, width = 10, height = 8, units = 'in')
} else {
cat("No plot to show\n\n")
}
cat("\n\n")
}
cat("\n\n")
}1.1 gen_1624C_F1_lin-41_N2
1.1.1 Freq: 0
1.1.2 Freq: 1e-05
1.1.3 Freq: 1e-04
1.1.4 Freq: 0.001
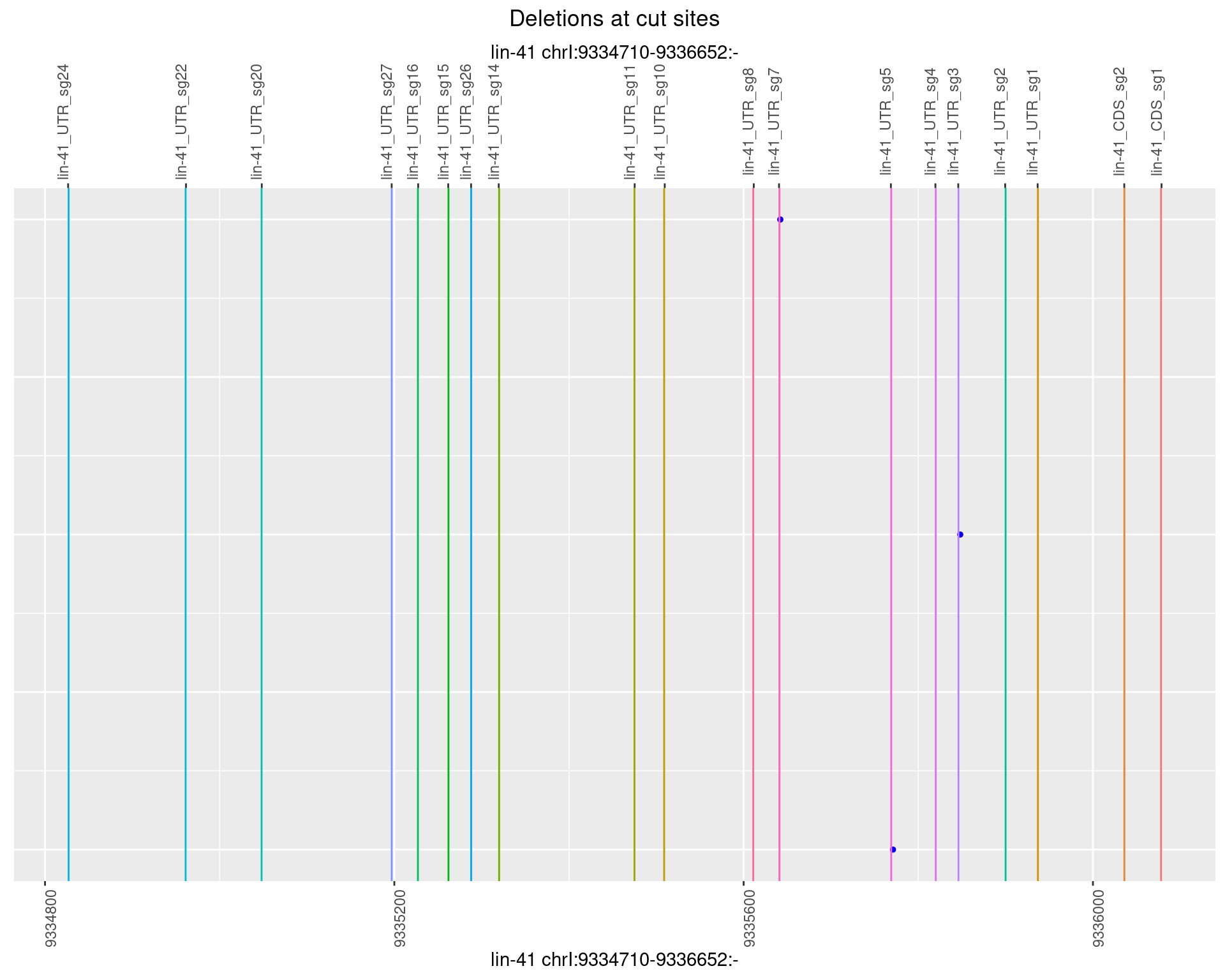
1.1.5 Freq: 0.01
No plot to show
1.1.6 Freq: 0.1
No plot to show
1.2 gen_1624C_F1_lin-41_sg15sg16
1.2.1 Freq: 0
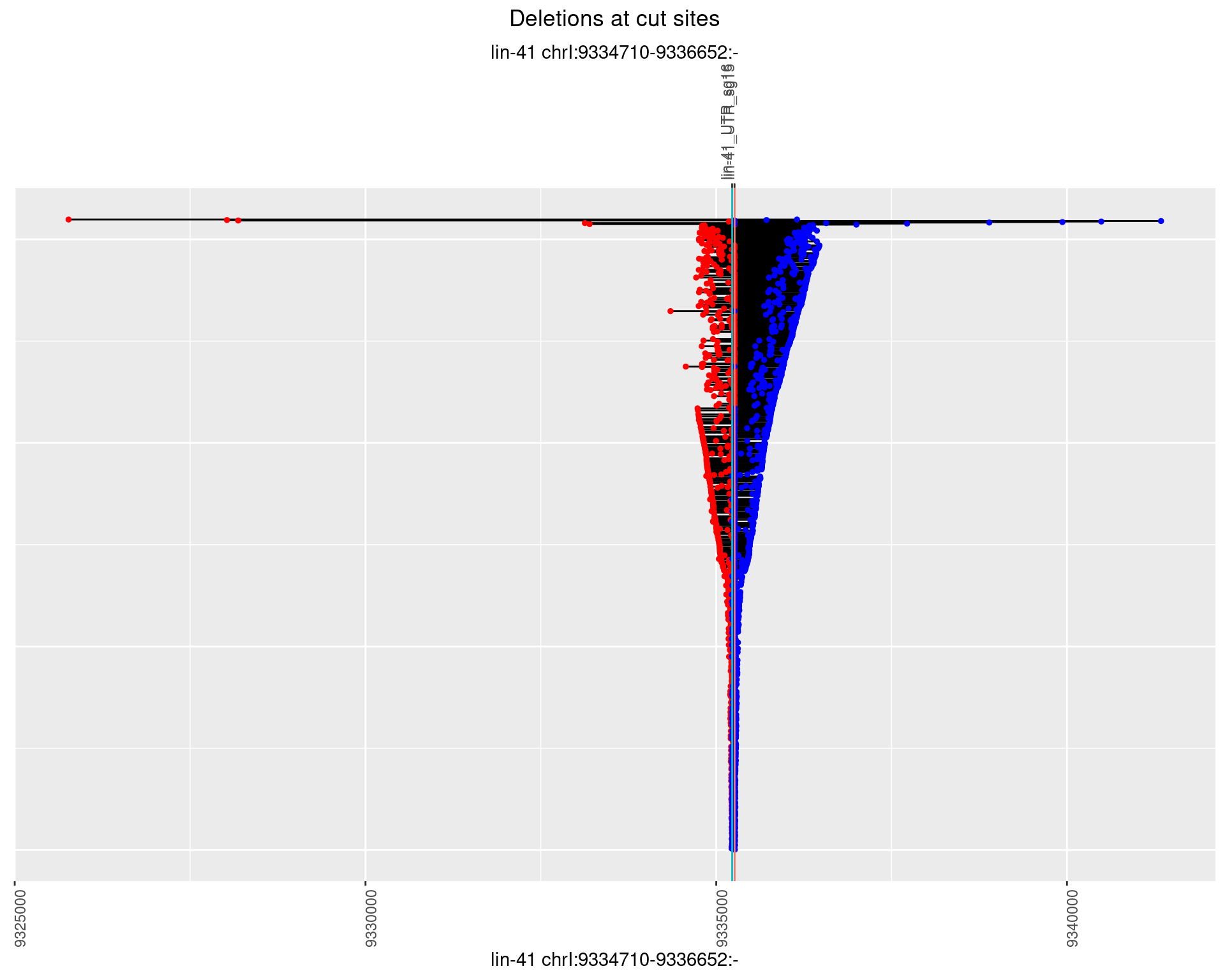
1.2.2 Freq: 1e-05
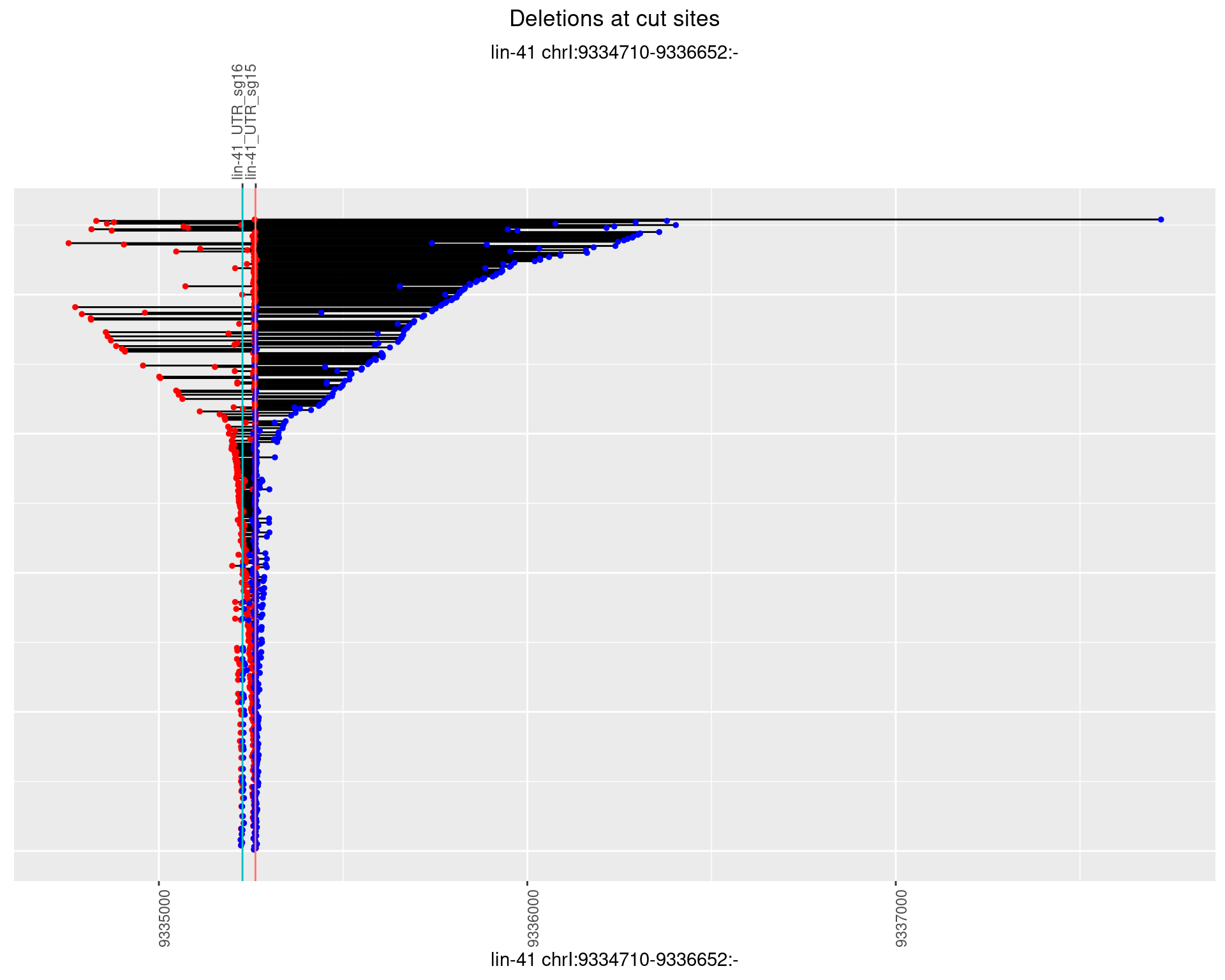
1.2.3 Freq: 1e-04
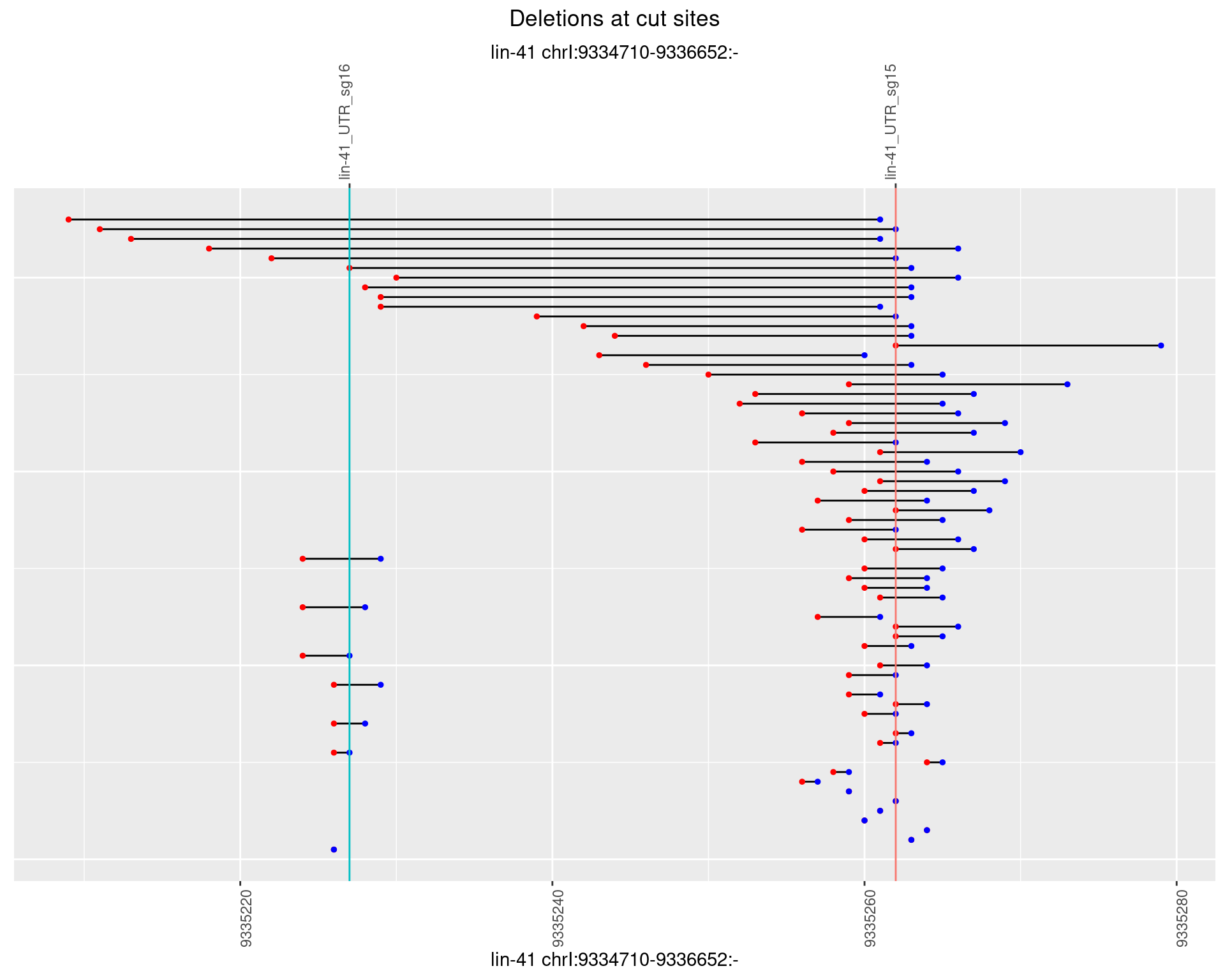
1.2.4 Freq: 0.001
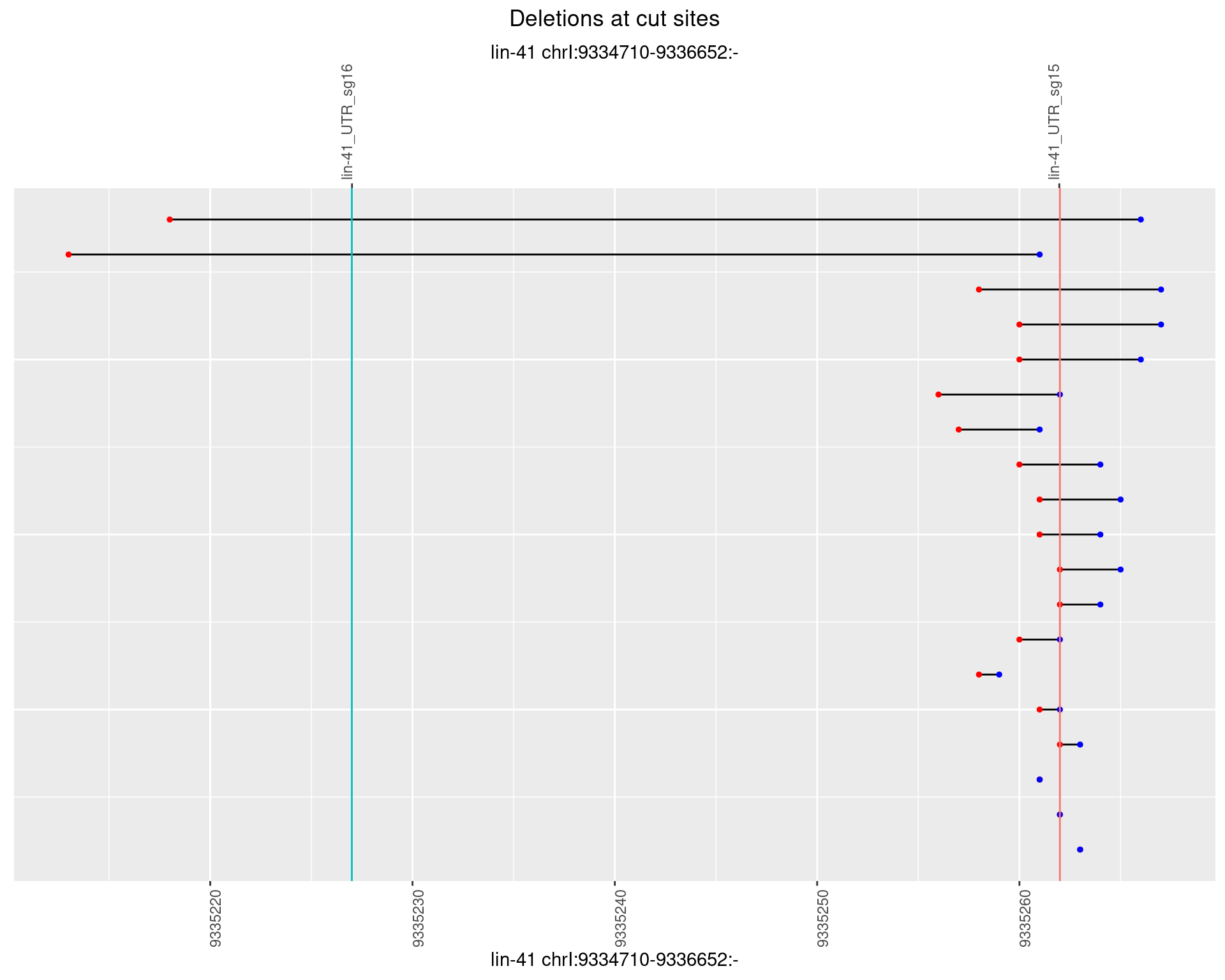
1.2.5 Freq: 0.01
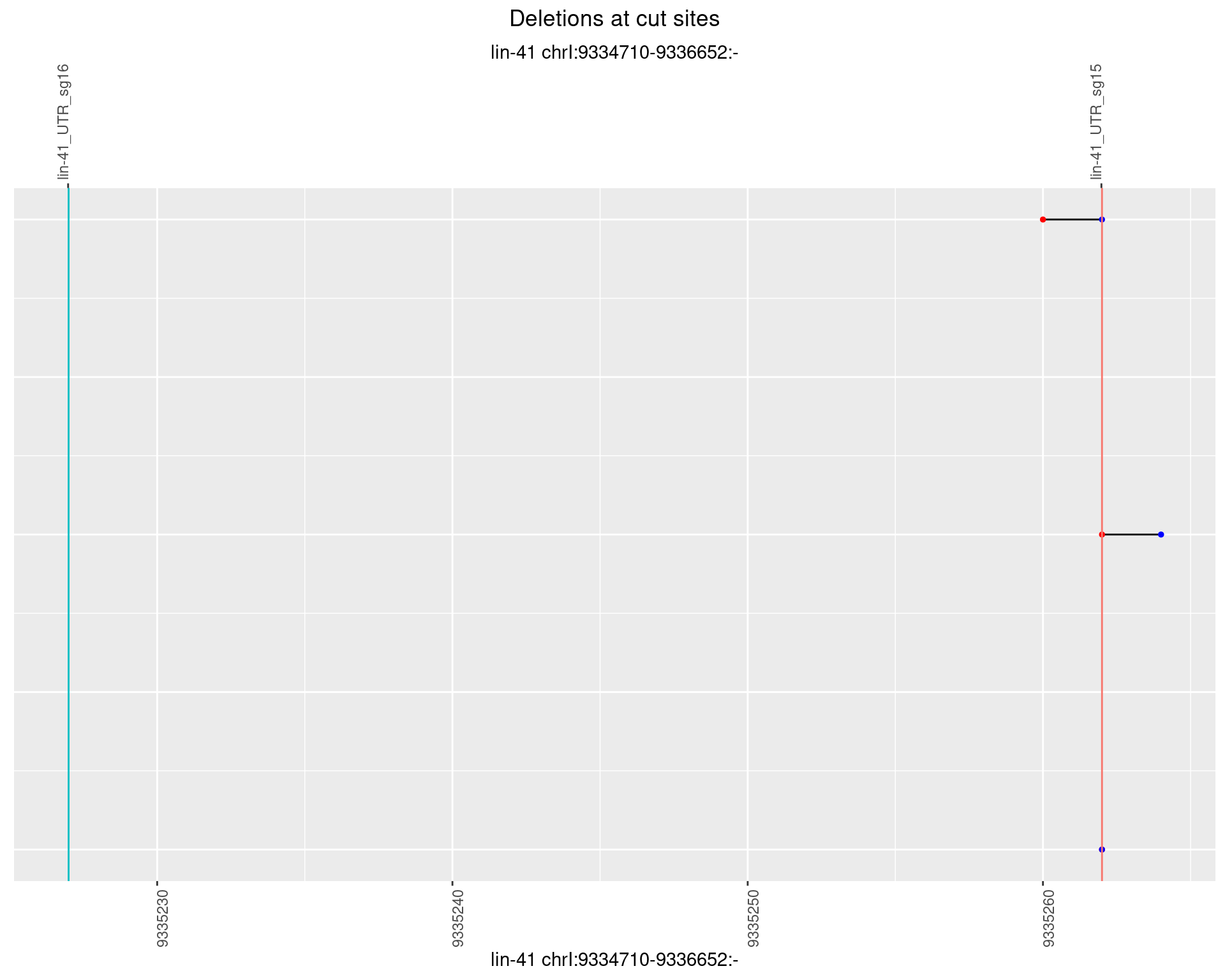
1.2.6 Freq: 0.1
No plot to show
1.3 gen_1624C_F1_lin-41_pool
1.3.1 Freq: 0
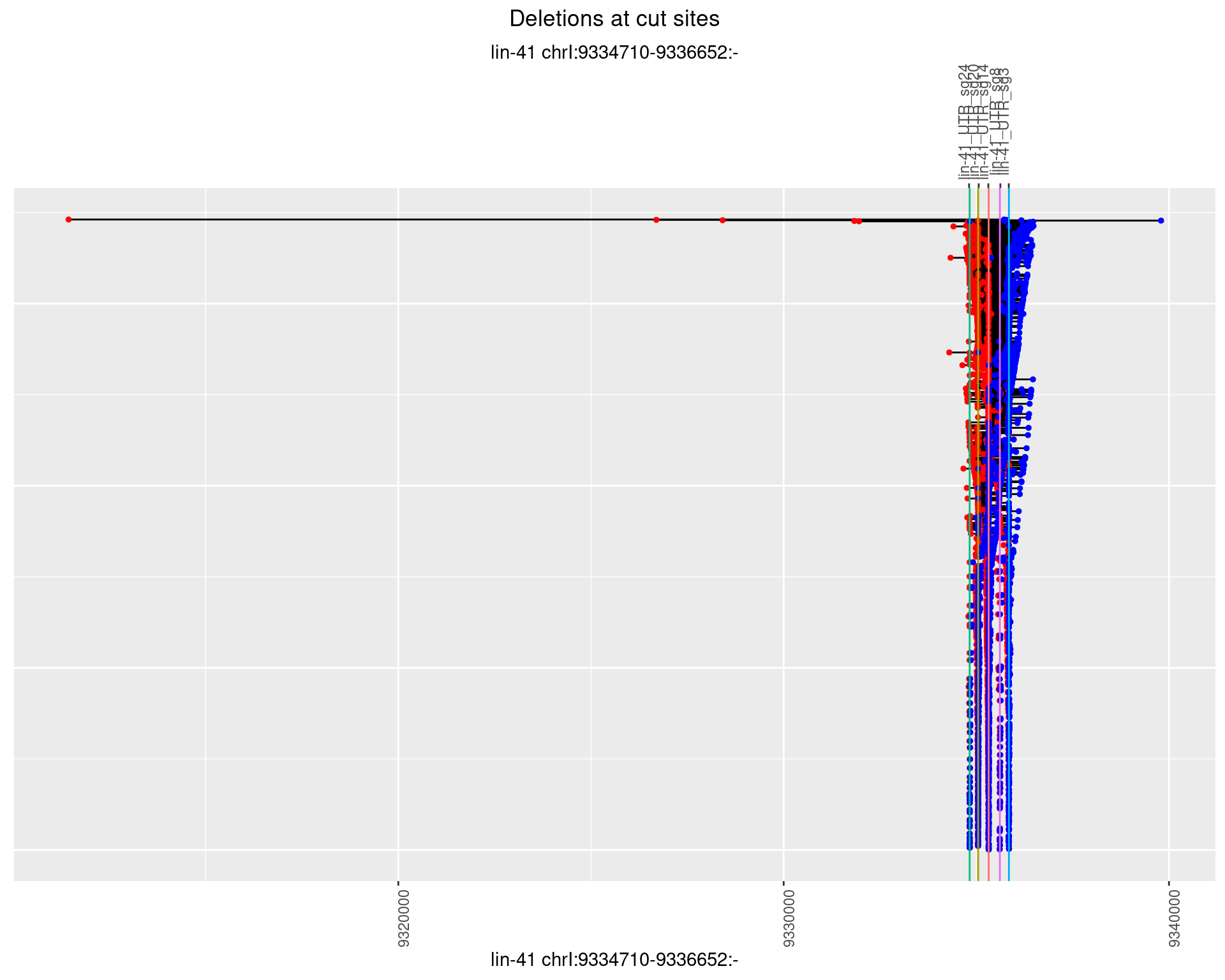
1.3.2 Freq: 1e-05
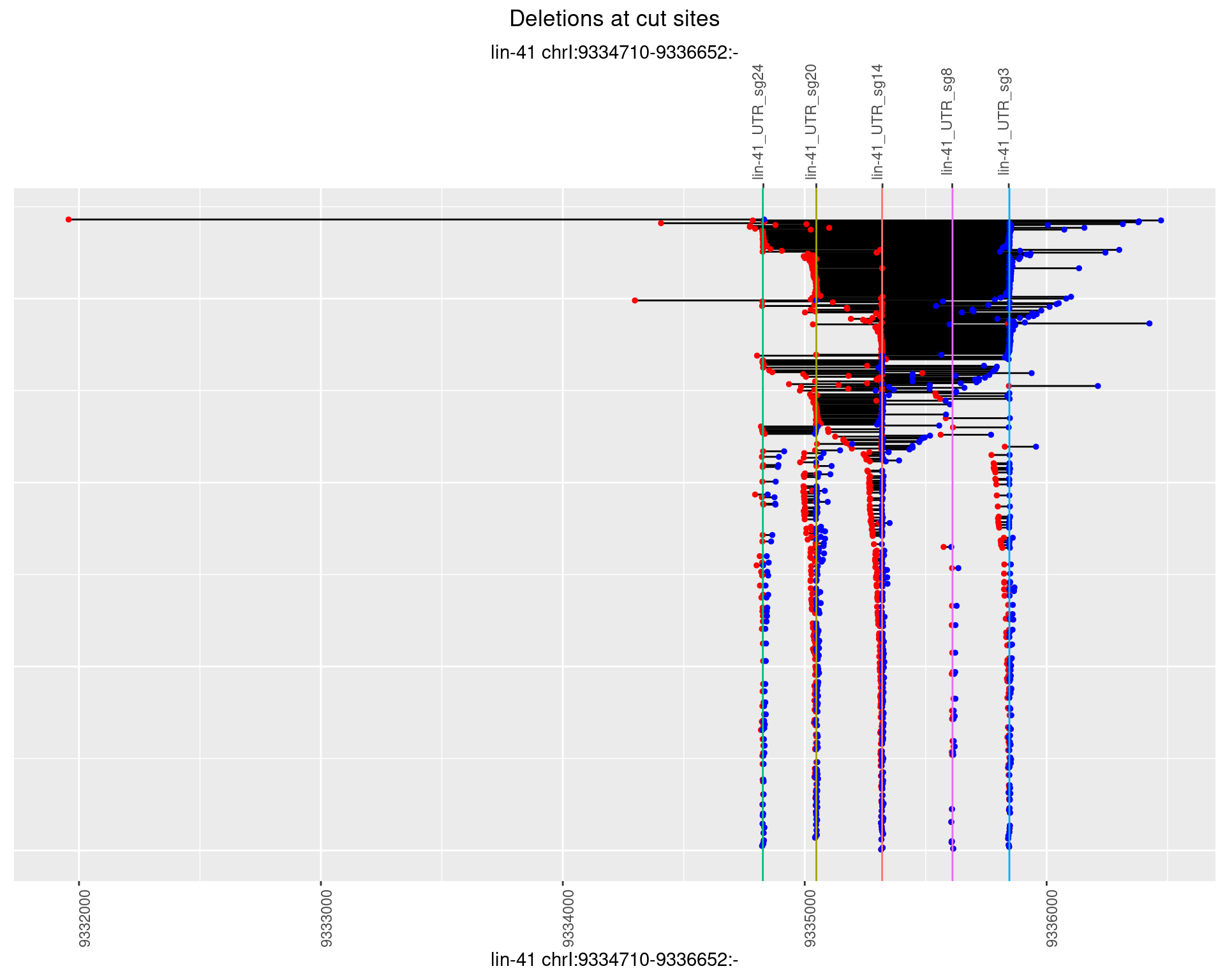
1.3.3 Freq: 1e-04
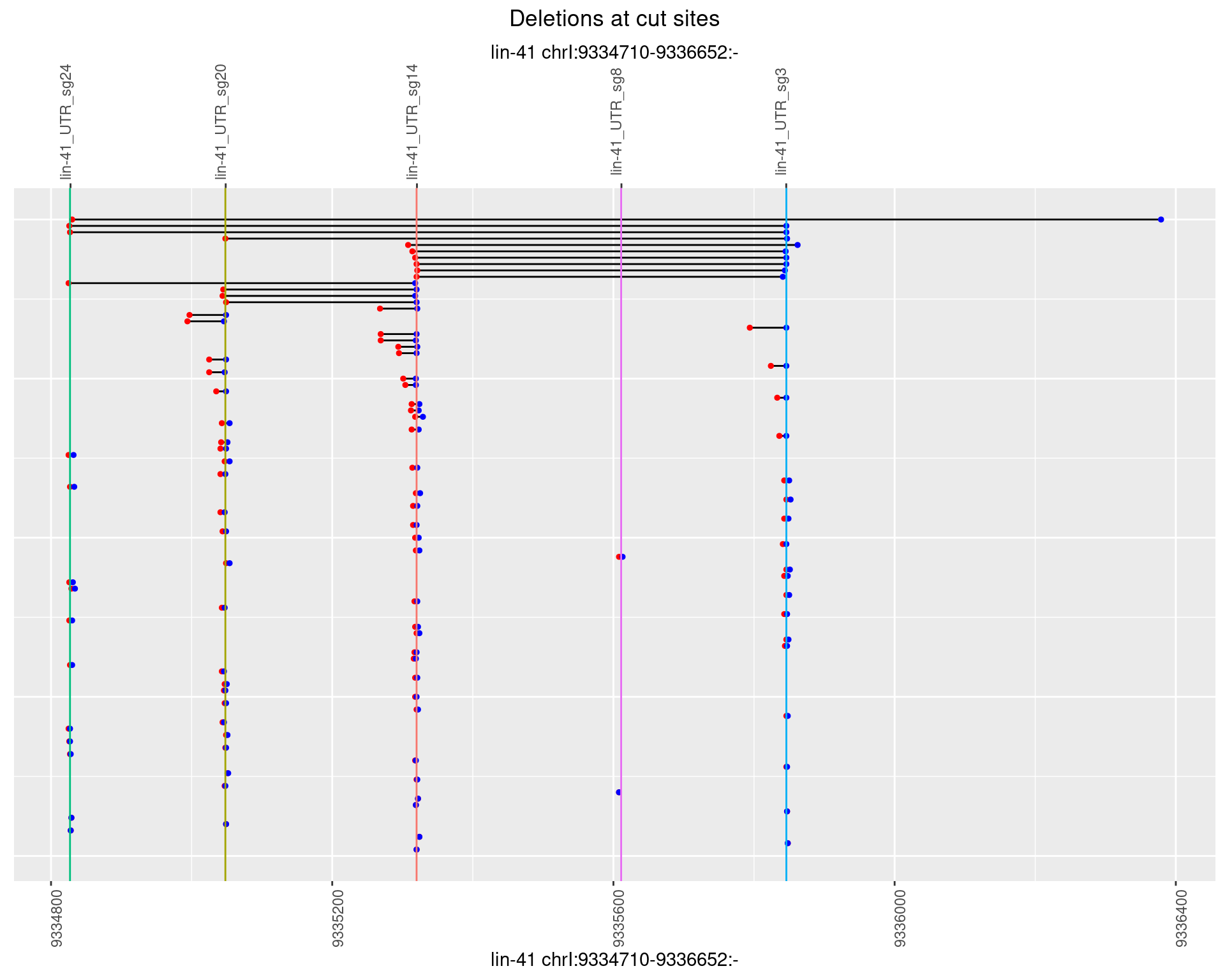
1.3.4 Freq: 0.001
1.3.5 Freq: 0.01
1.3.6 Freq: 0.1
No plot to show
1.4 gen_1624C_F1_lin-41_sg26sg27
1.4.1 Freq: 0
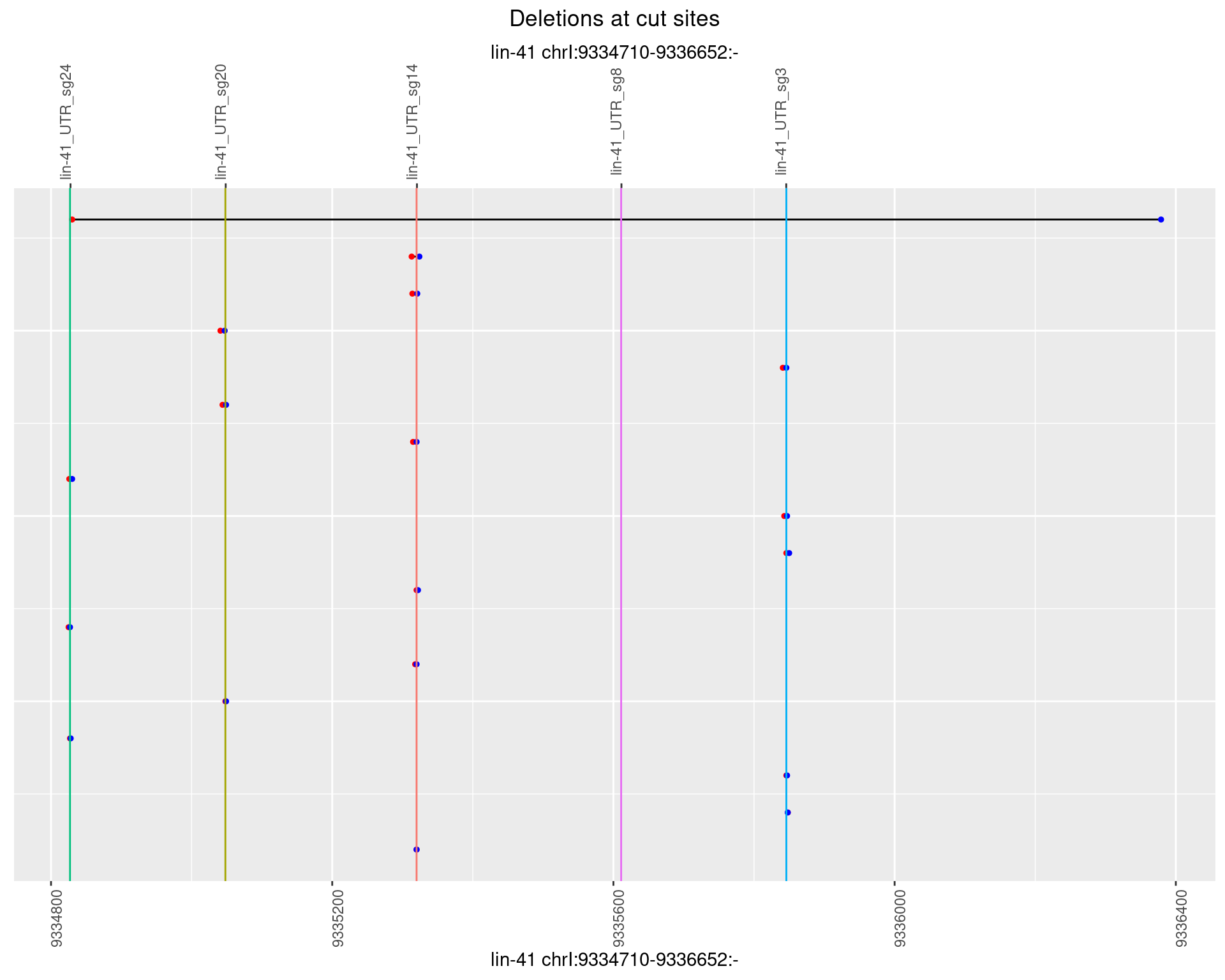
1.4.2 Freq: 1e-05
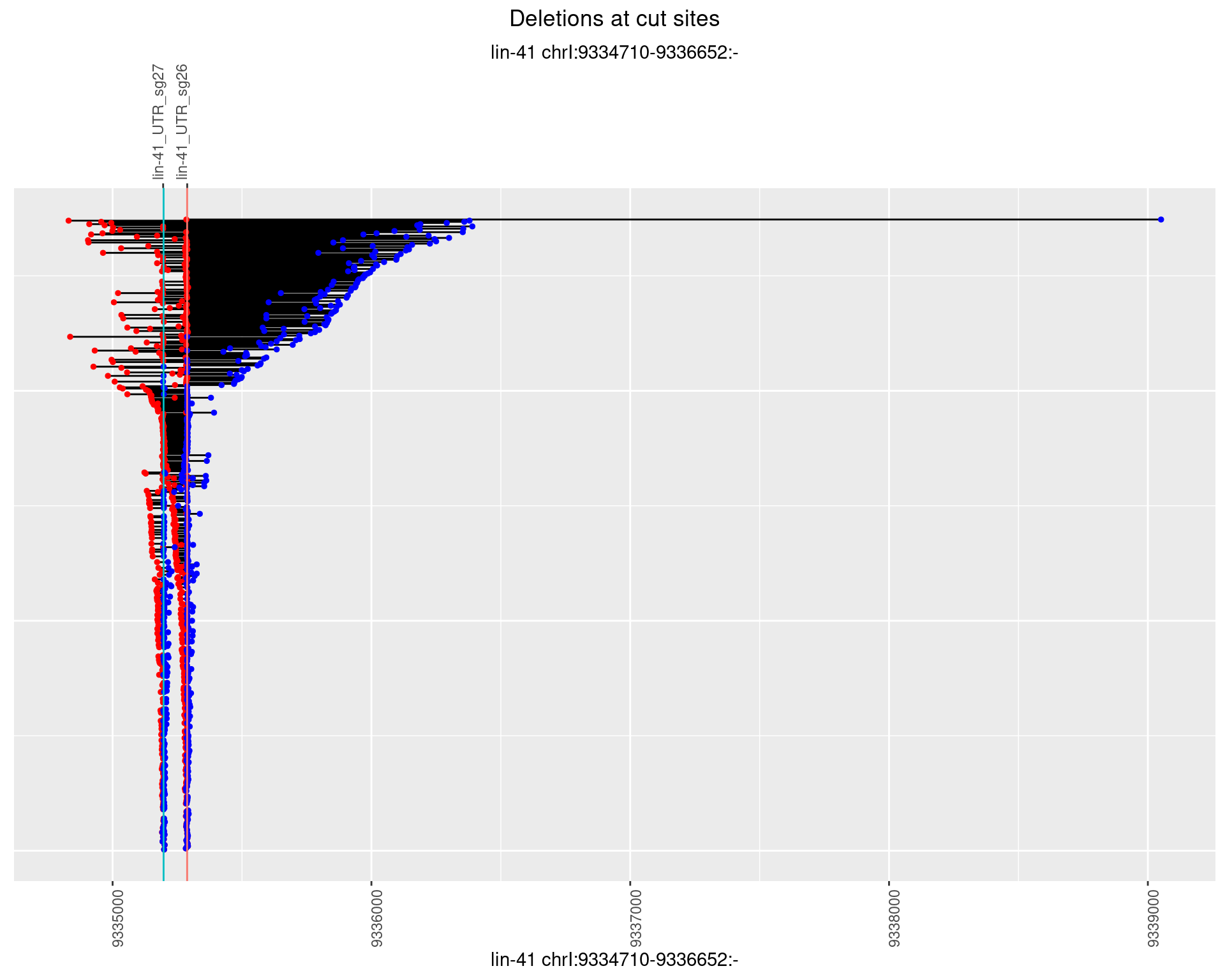
1.4.3 Freq: 1e-04
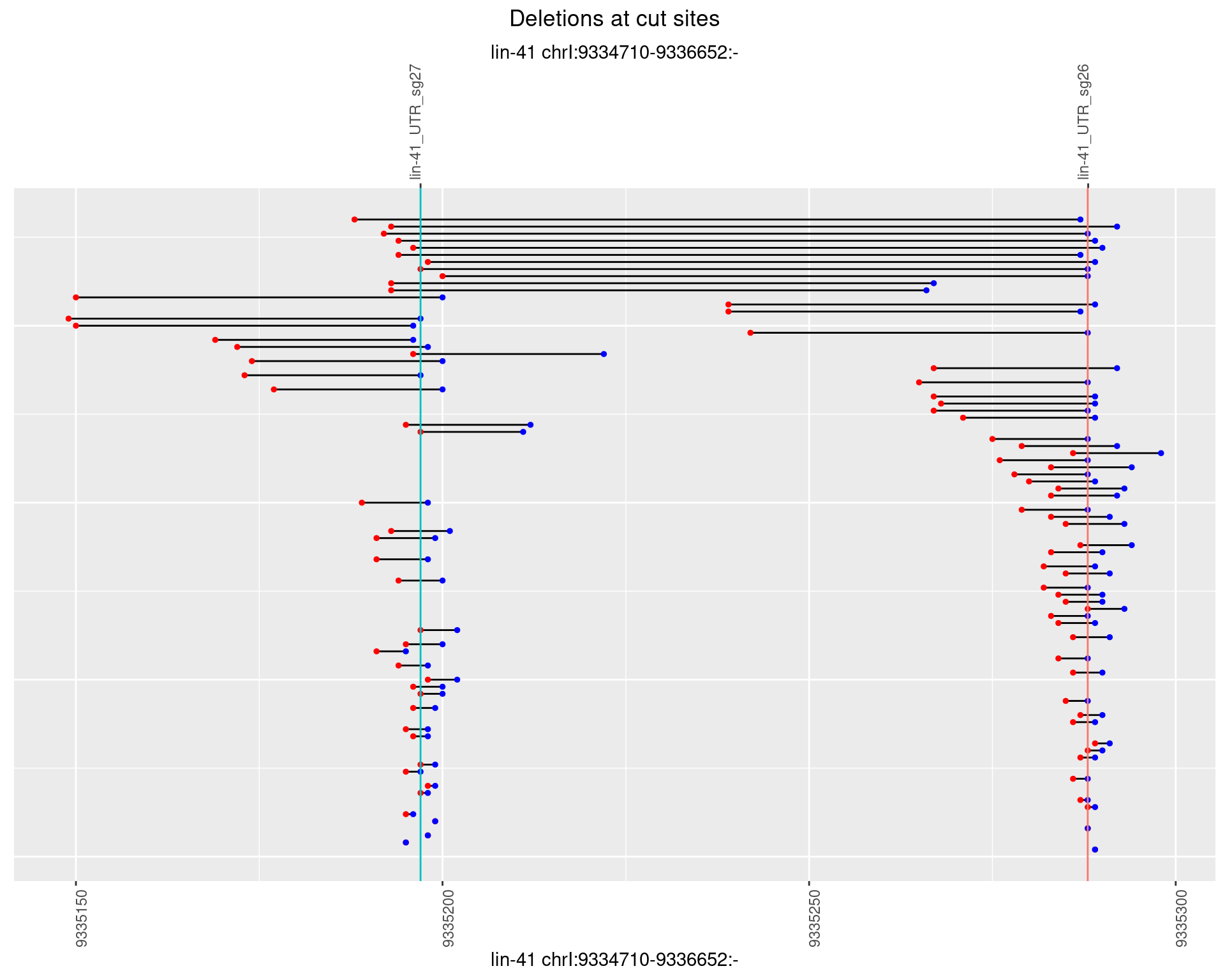
1.4.4 Freq: 0.001
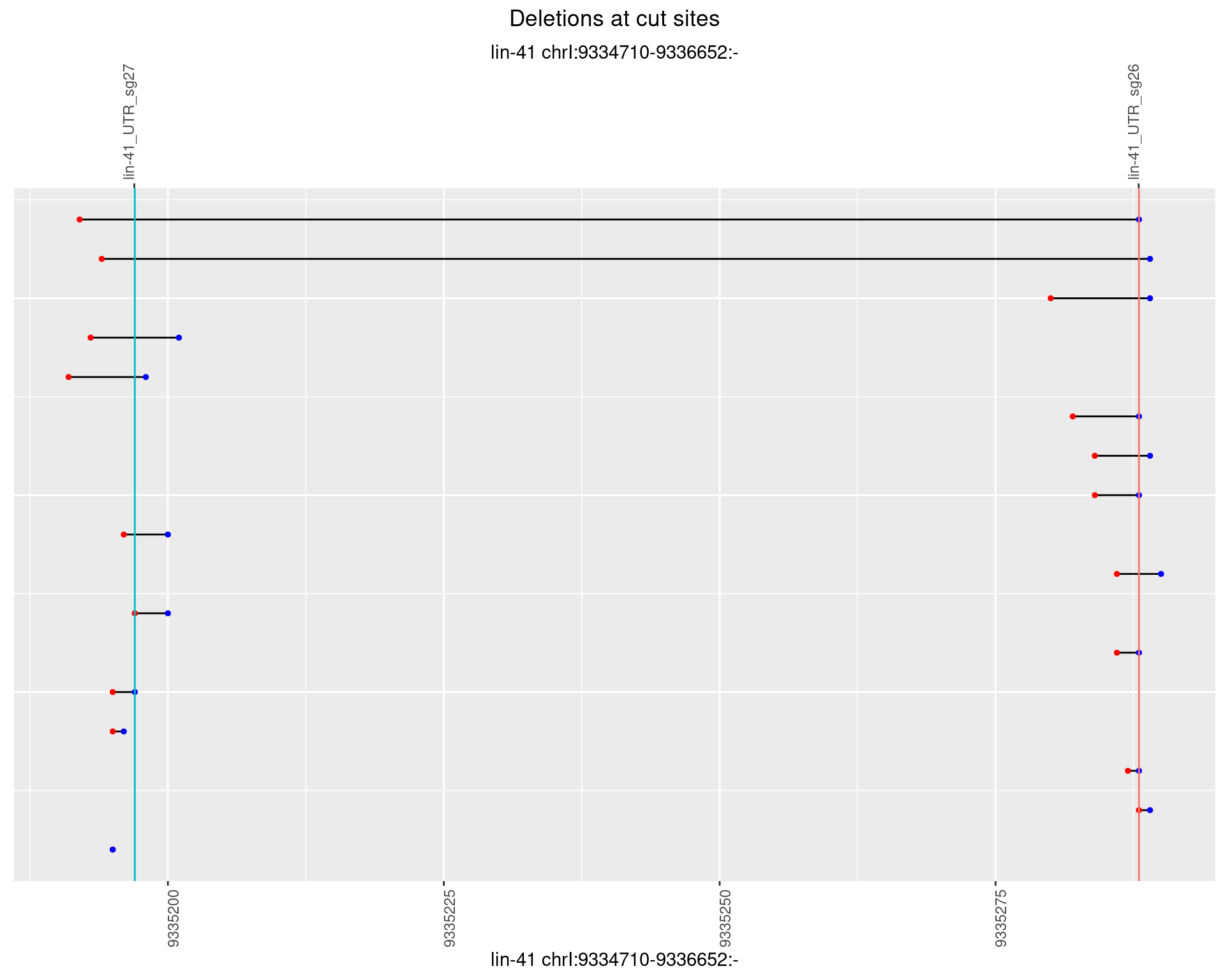
1.4.5 Freq: 0.01
No plot to show
1.4.6 Freq: 0.1
No plot to show
1.5 gen_24C_F2_lin-41_N2
1.5.1 Freq: 0
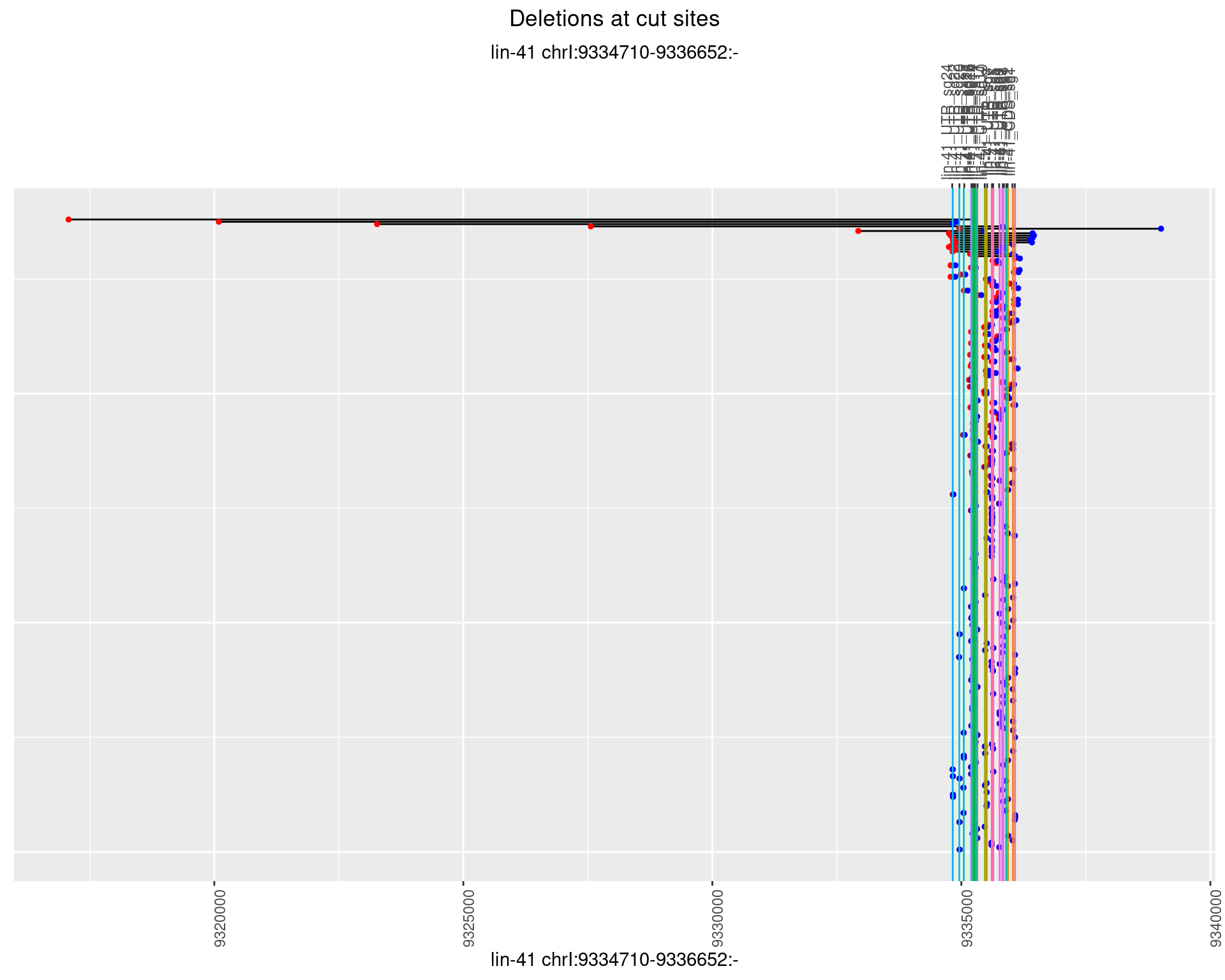
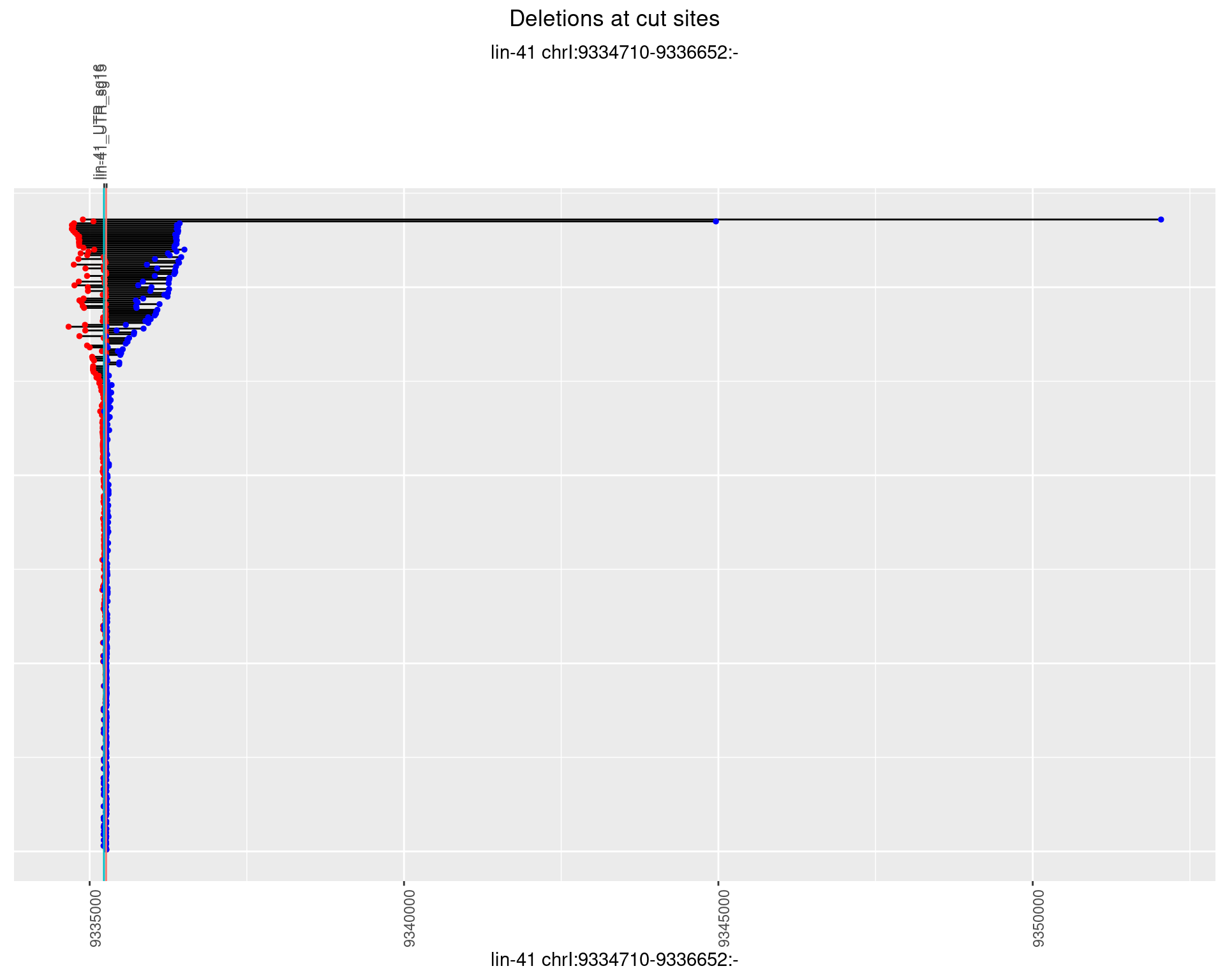
1.5.2 Freq: 1e-05
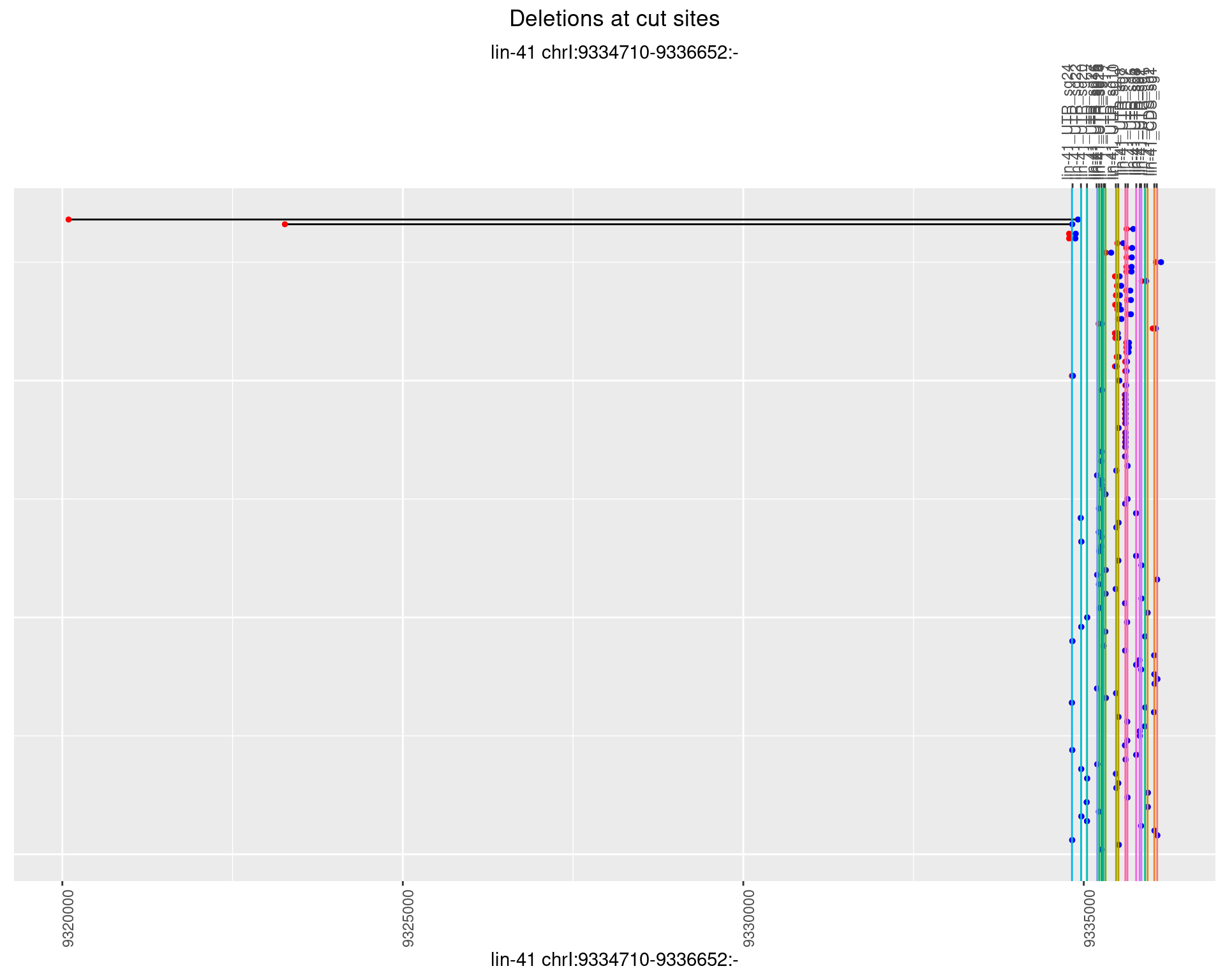
1.5.3 Freq: 1e-04
1.5.4 Freq: 0.001
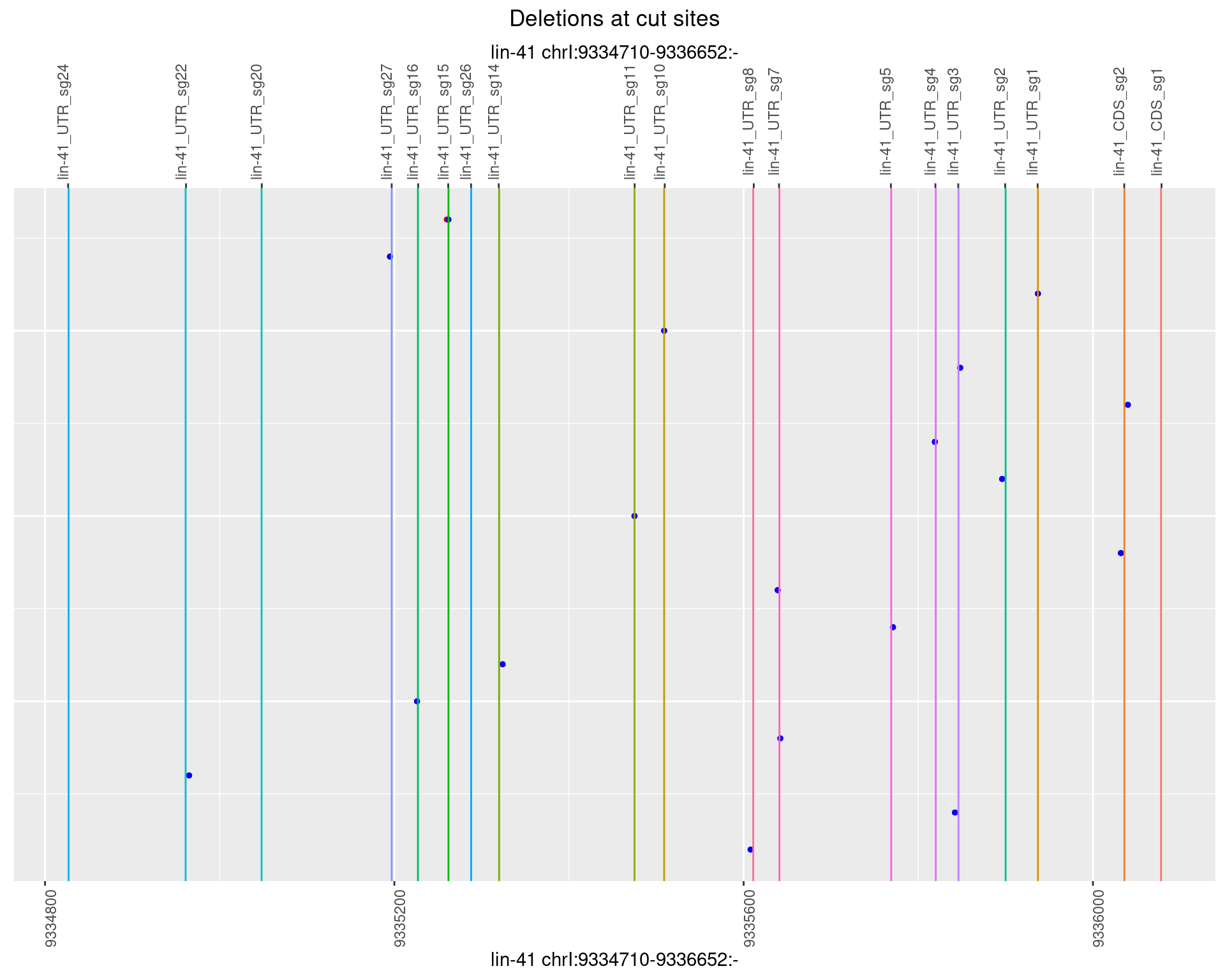
1.5.5 Freq: 0.01
No plot to show
1.5.6 Freq: 0.1
No plot to show
1.6 gen_24C_F2_lin-41_sg15sg16
1.6.1 Freq: 0
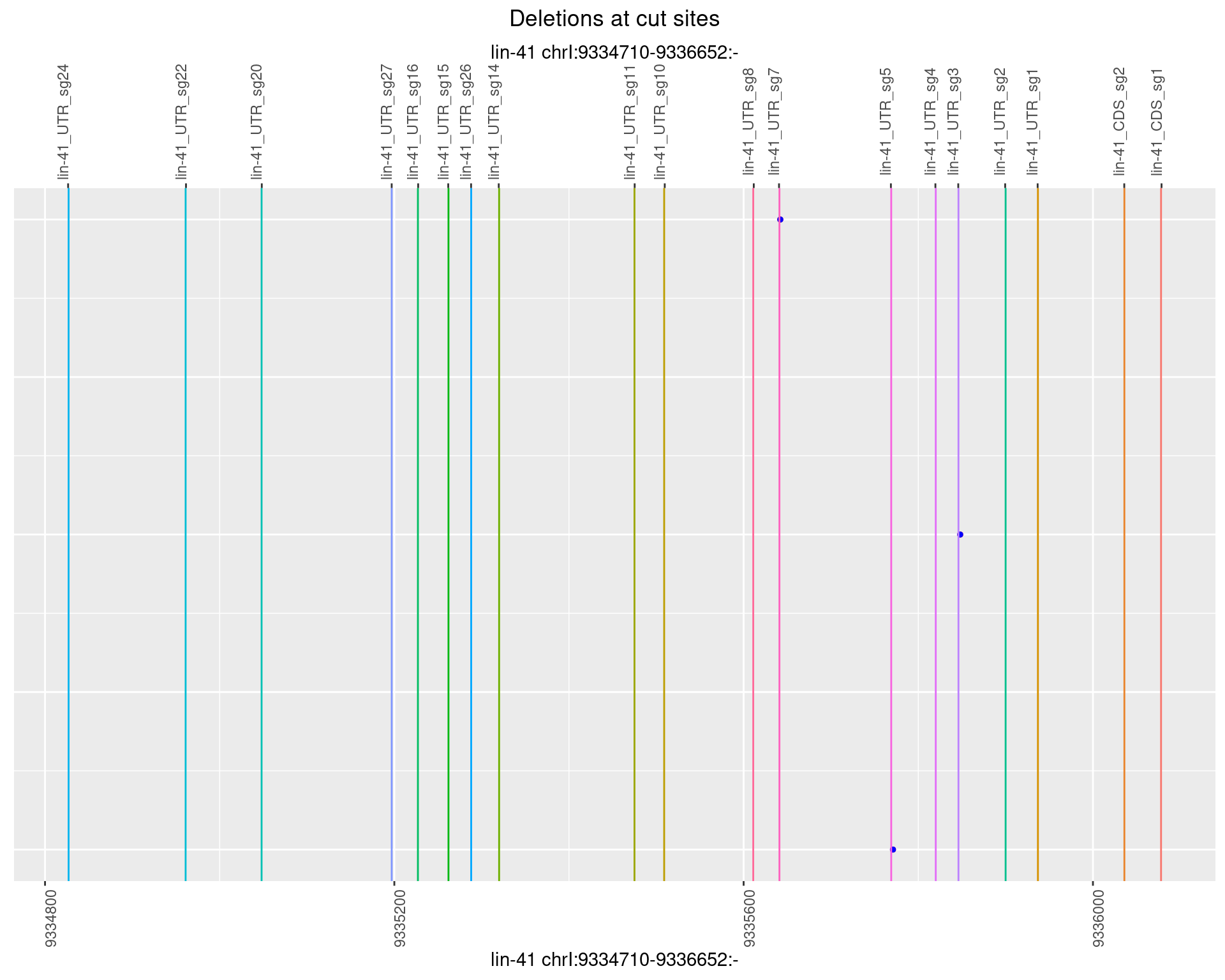
1.6.2 Freq: 1e-05
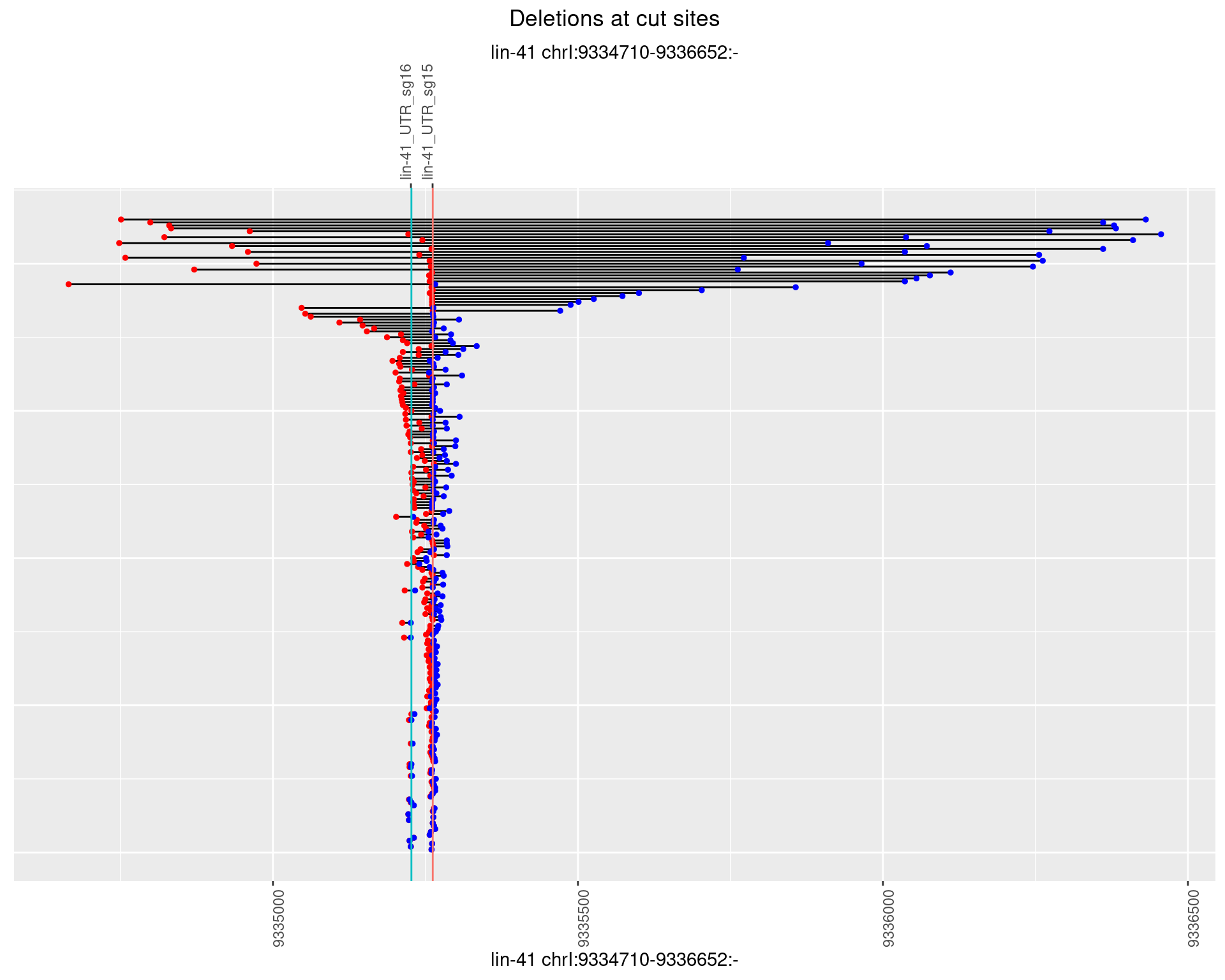
1.6.3 Freq: 1e-04
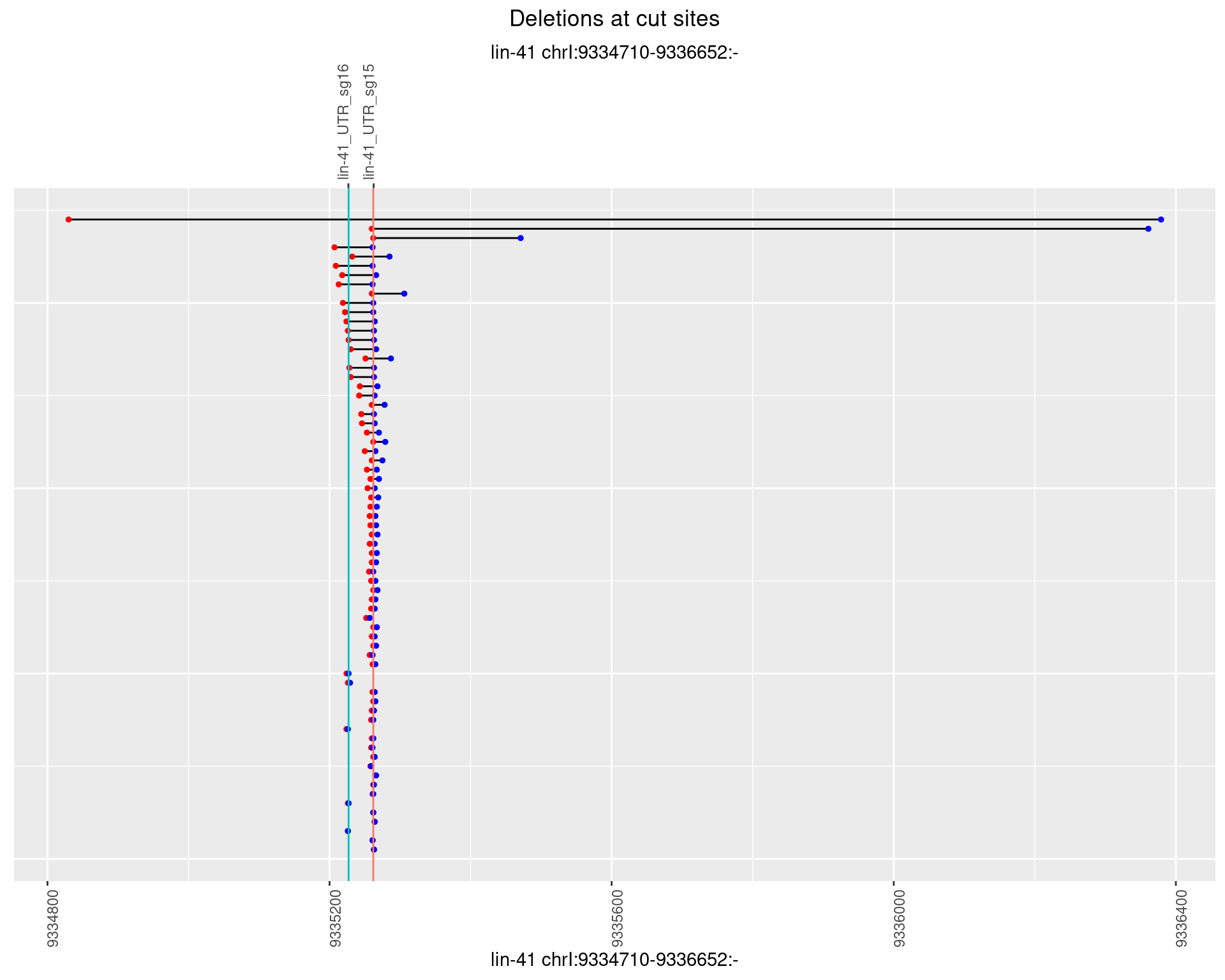
1.6.4 Freq: 0.001
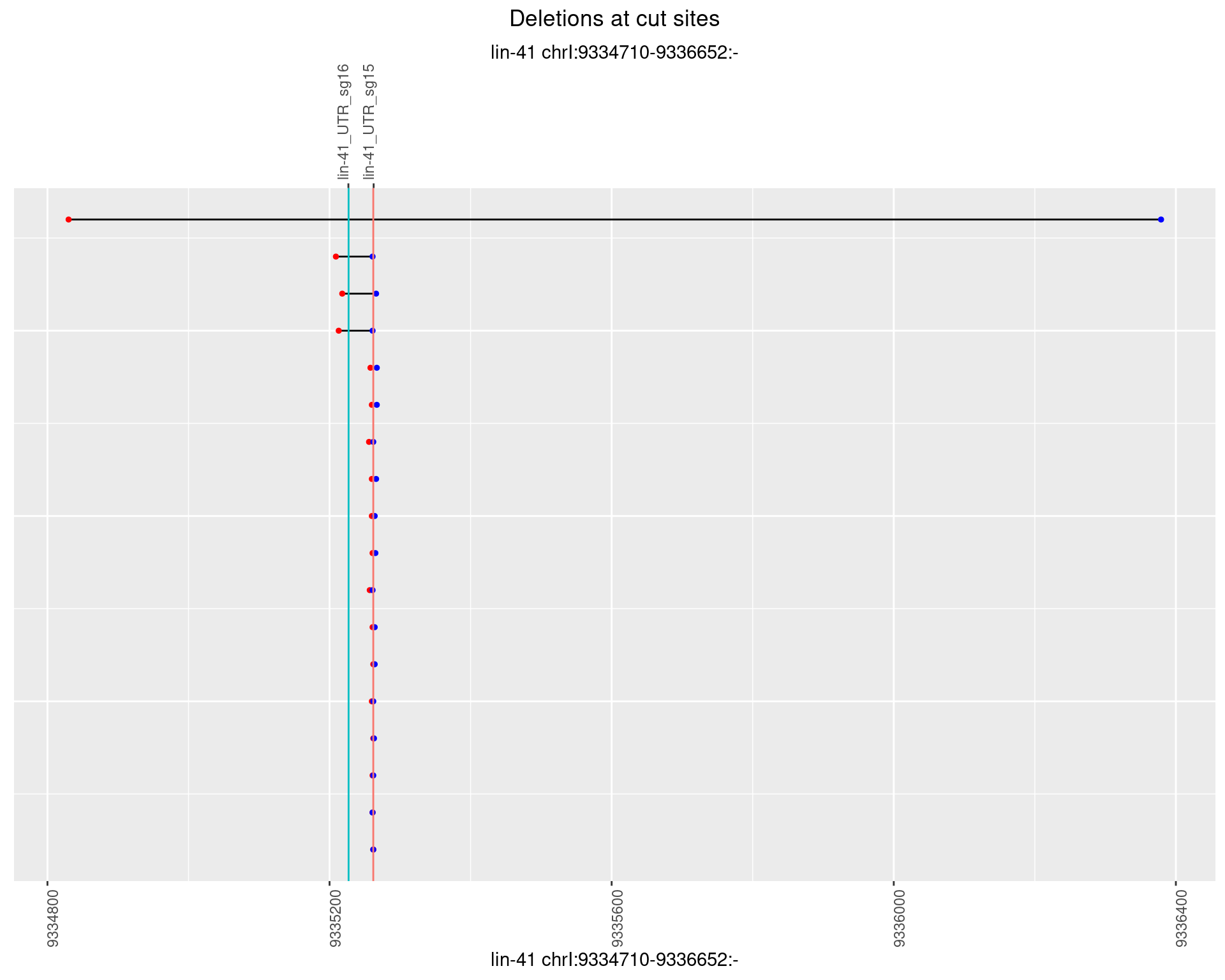
1.6.5 Freq: 0.01
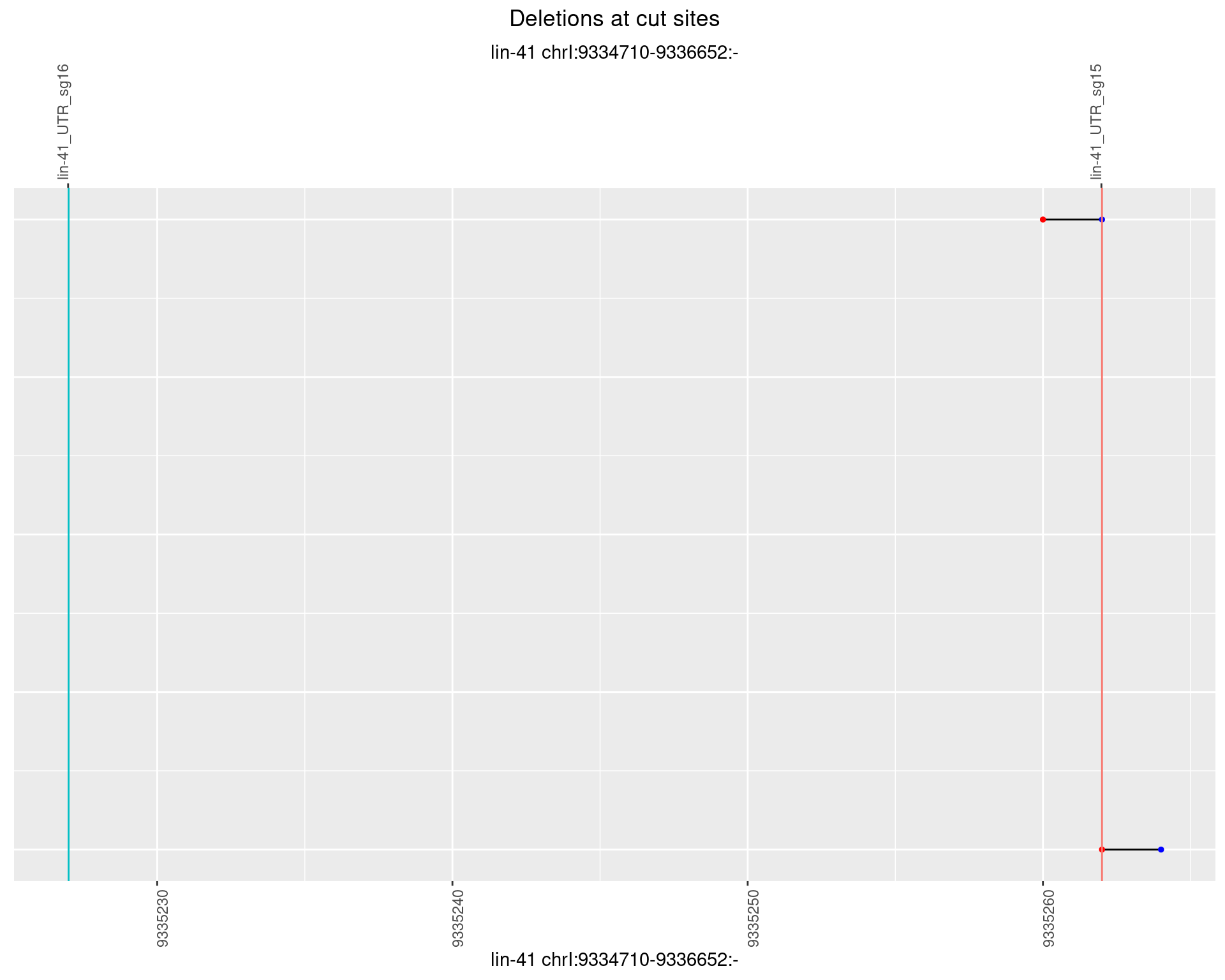
1.6.6 Freq: 0.1
No plot to show
1.7 gen_24C_F2_lin-41_pool
1.7.1 Freq: 0
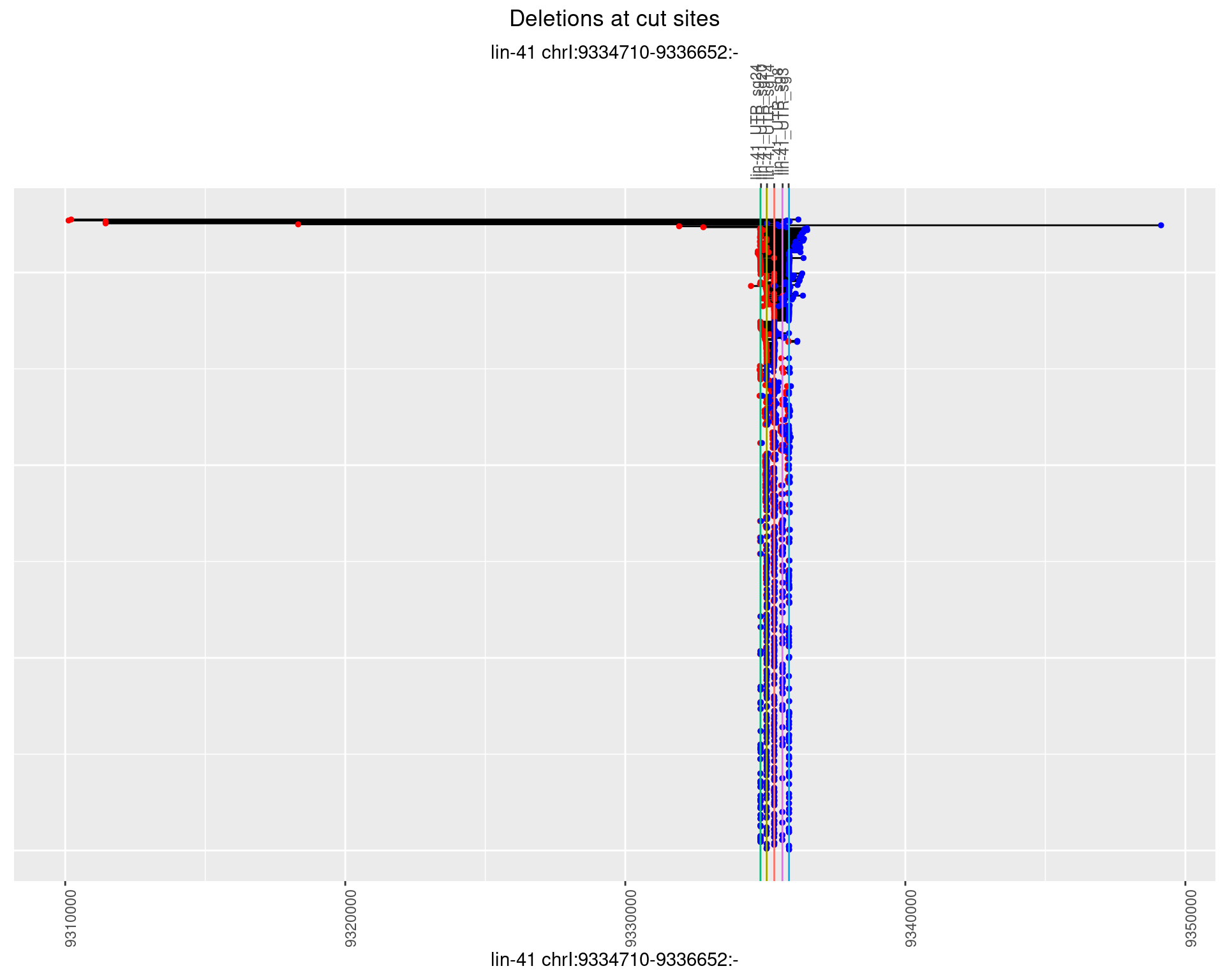
1.7.2 Freq: 1e-05
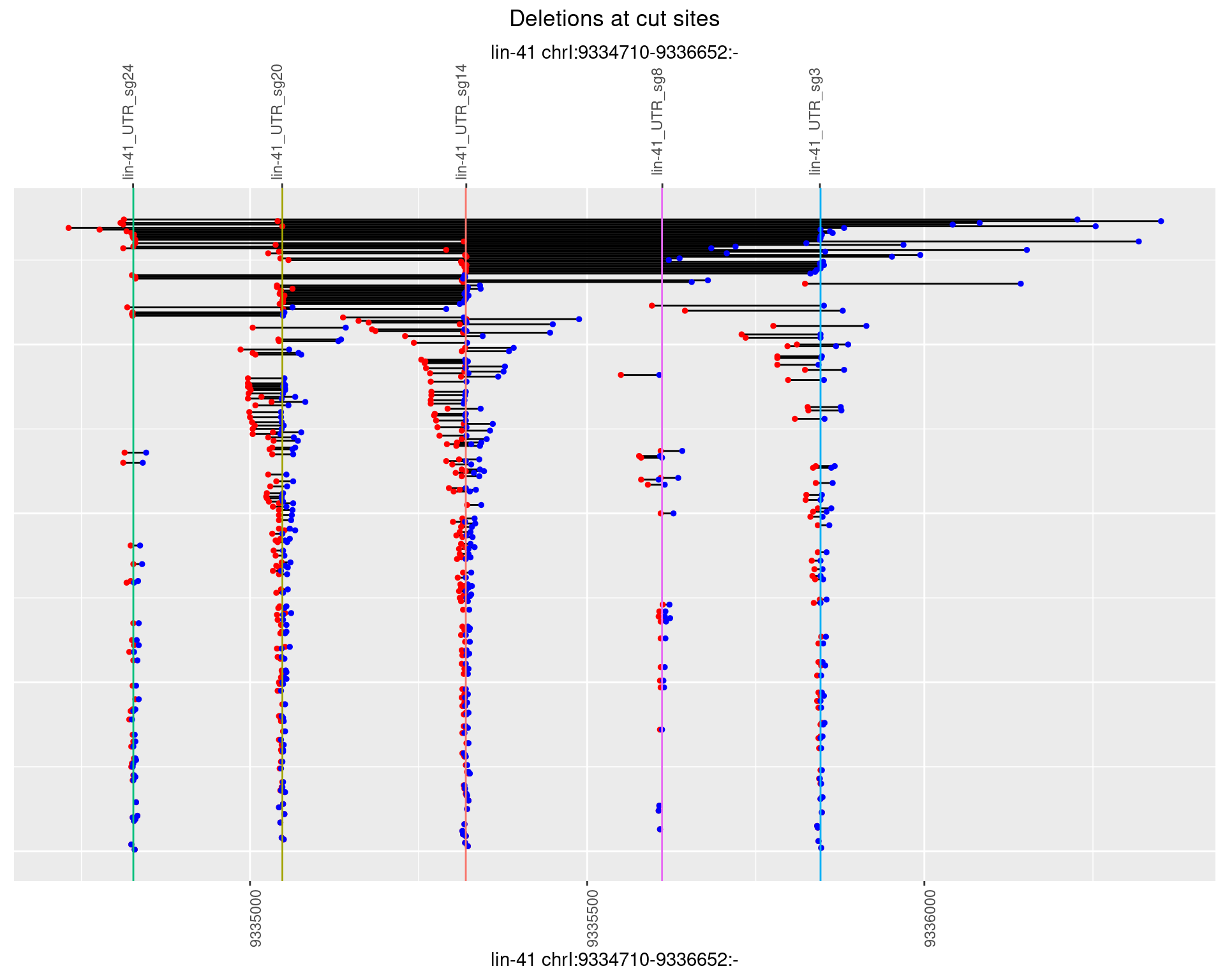
1.7.3 Freq: 1e-04
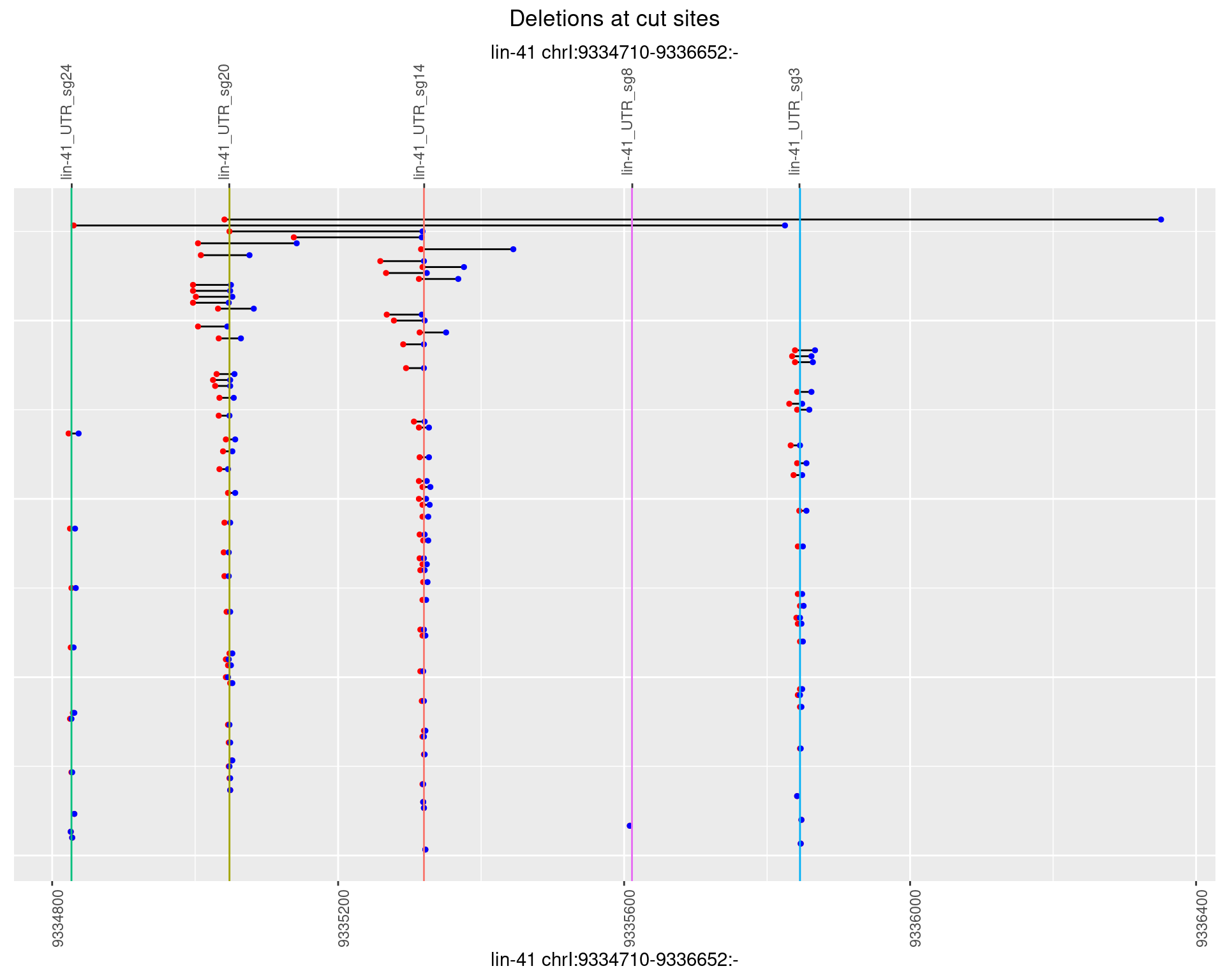
1.7.4 Freq: 0.001
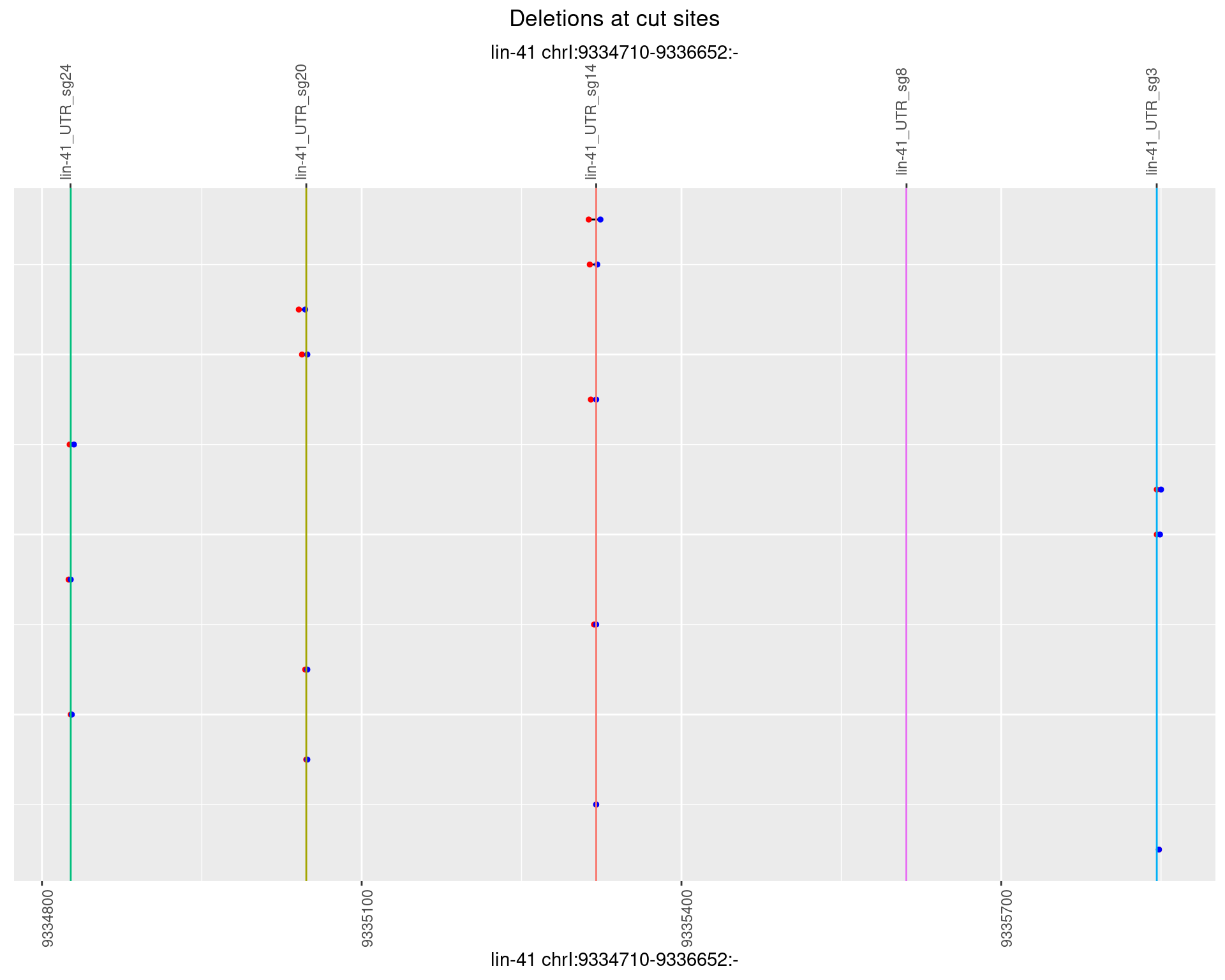
1.7.5 Freq: 0.01
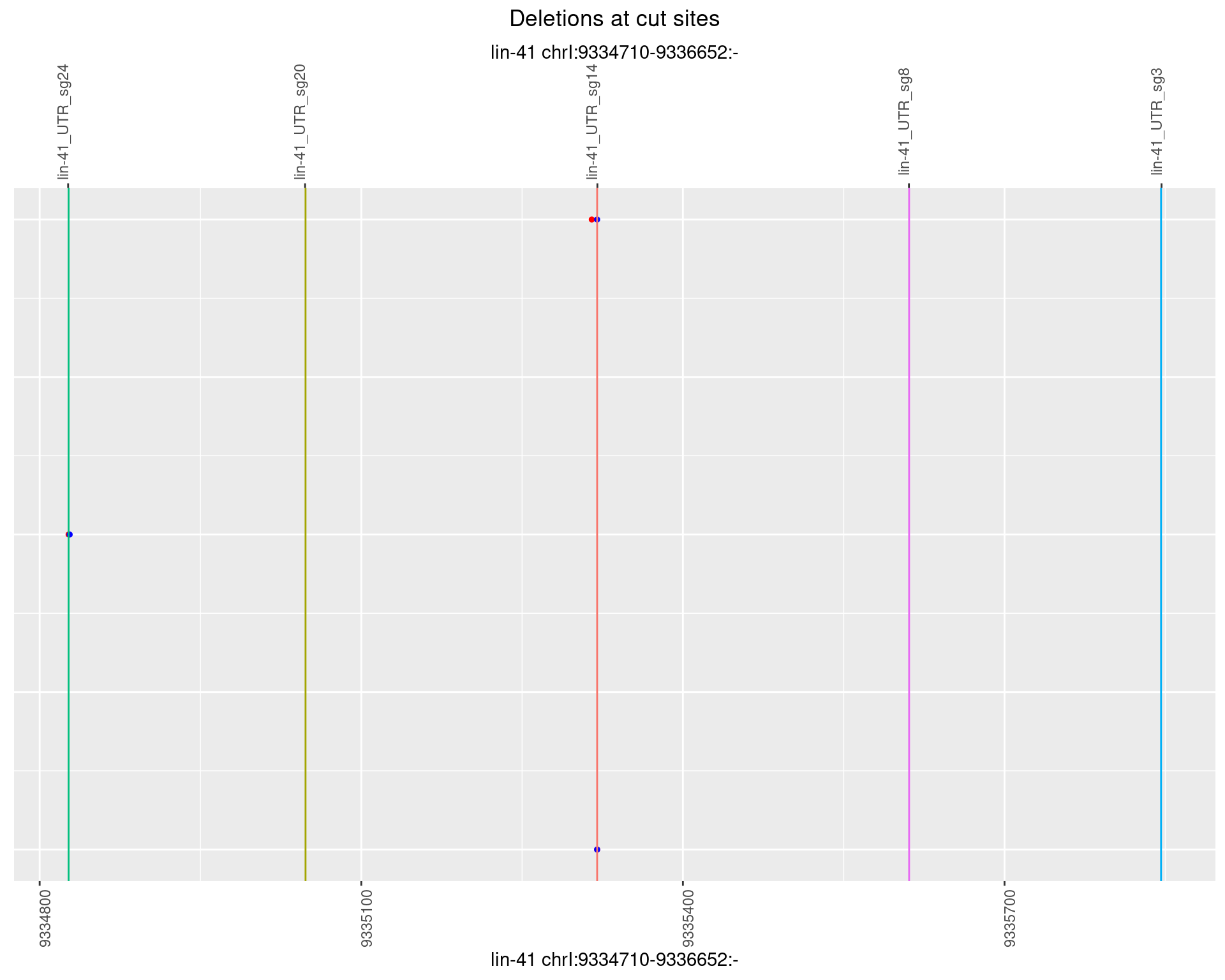
1.7.6 Freq: 0.1
No plot to show
1.8 gen_24C_F2_lin-41_sg26sg27
1.8.1 Freq: 0
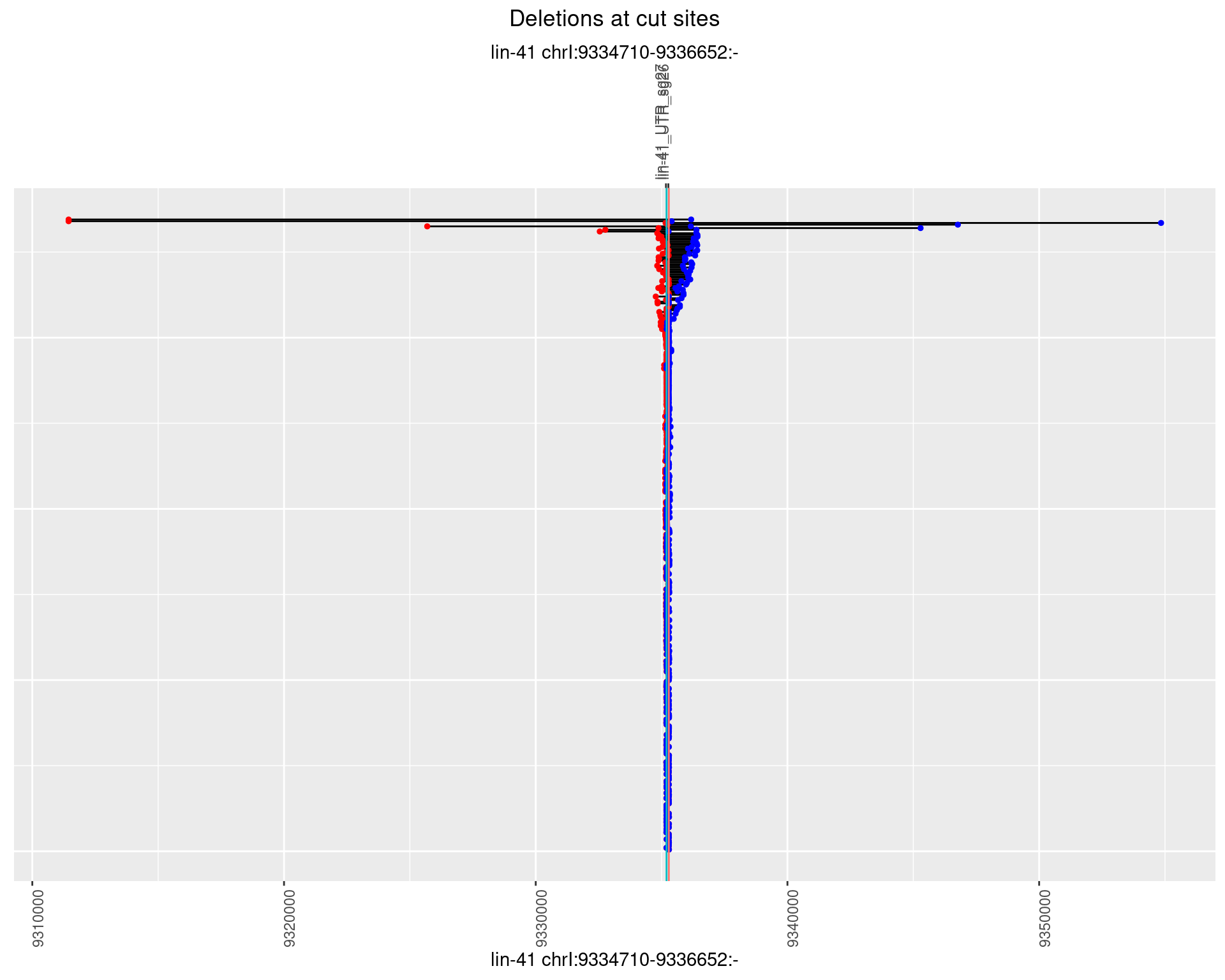
1.8.2 Freq: 1e-05
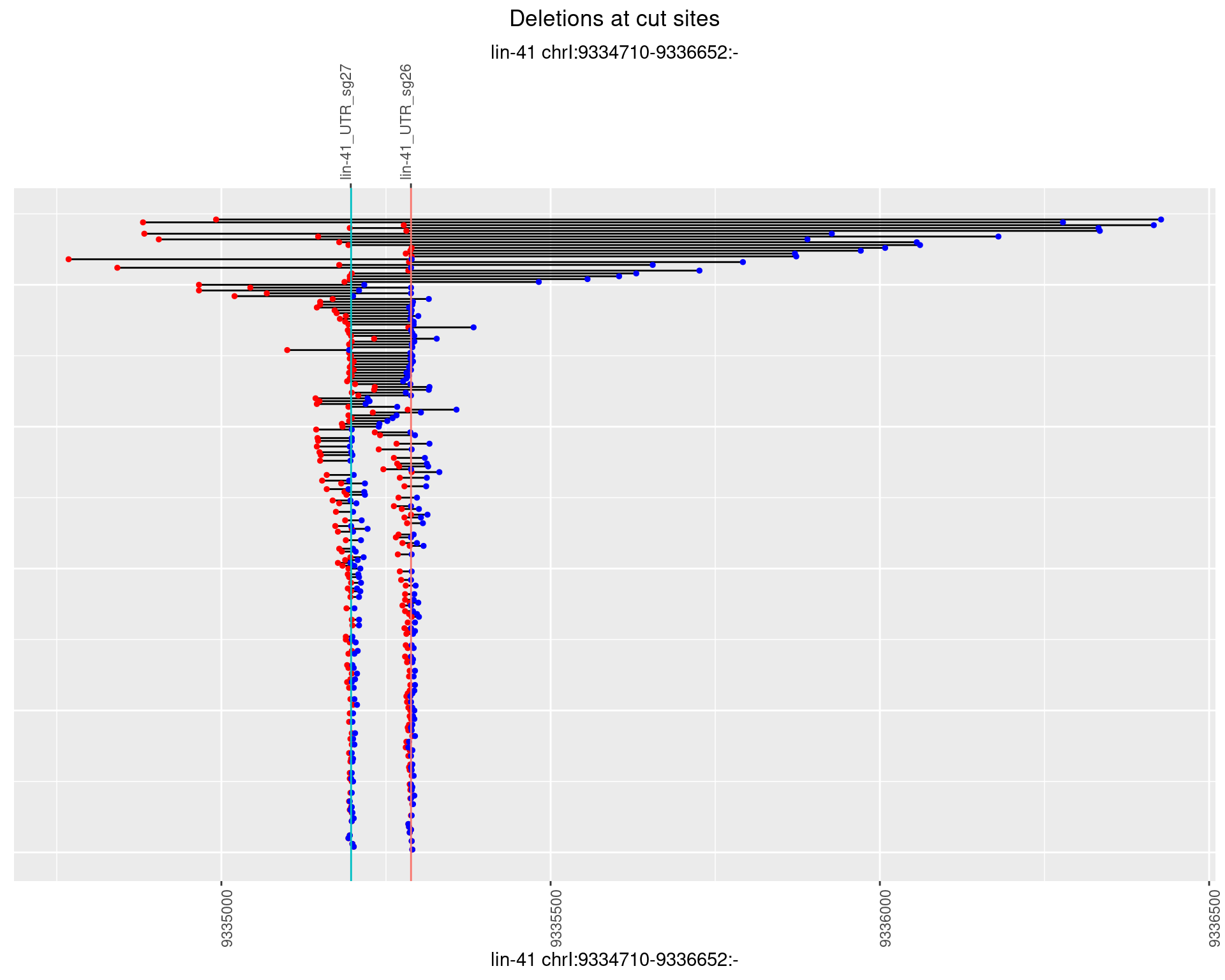
1.8.3 Freq: 1e-04
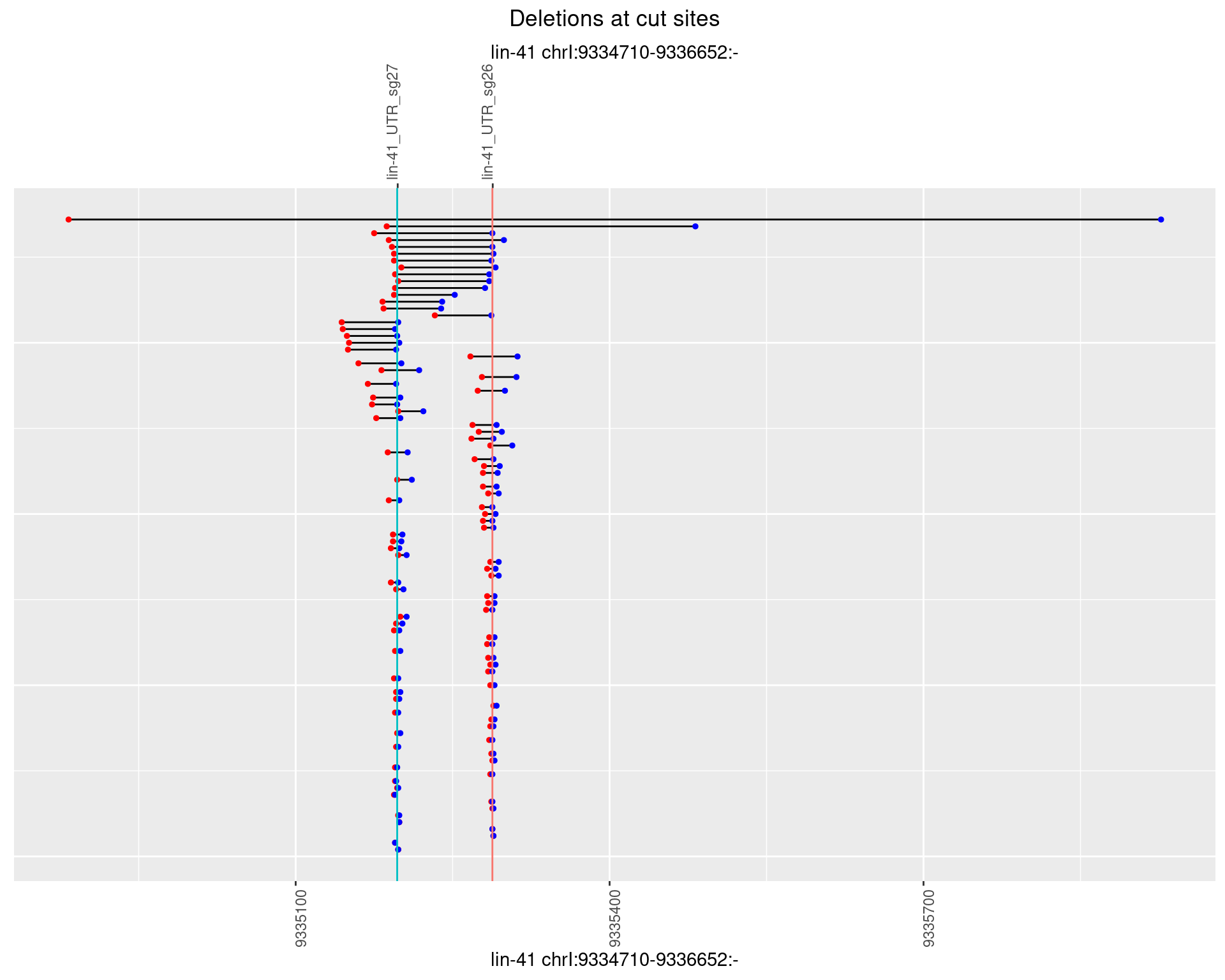
1.8.4 Freq: 0.001
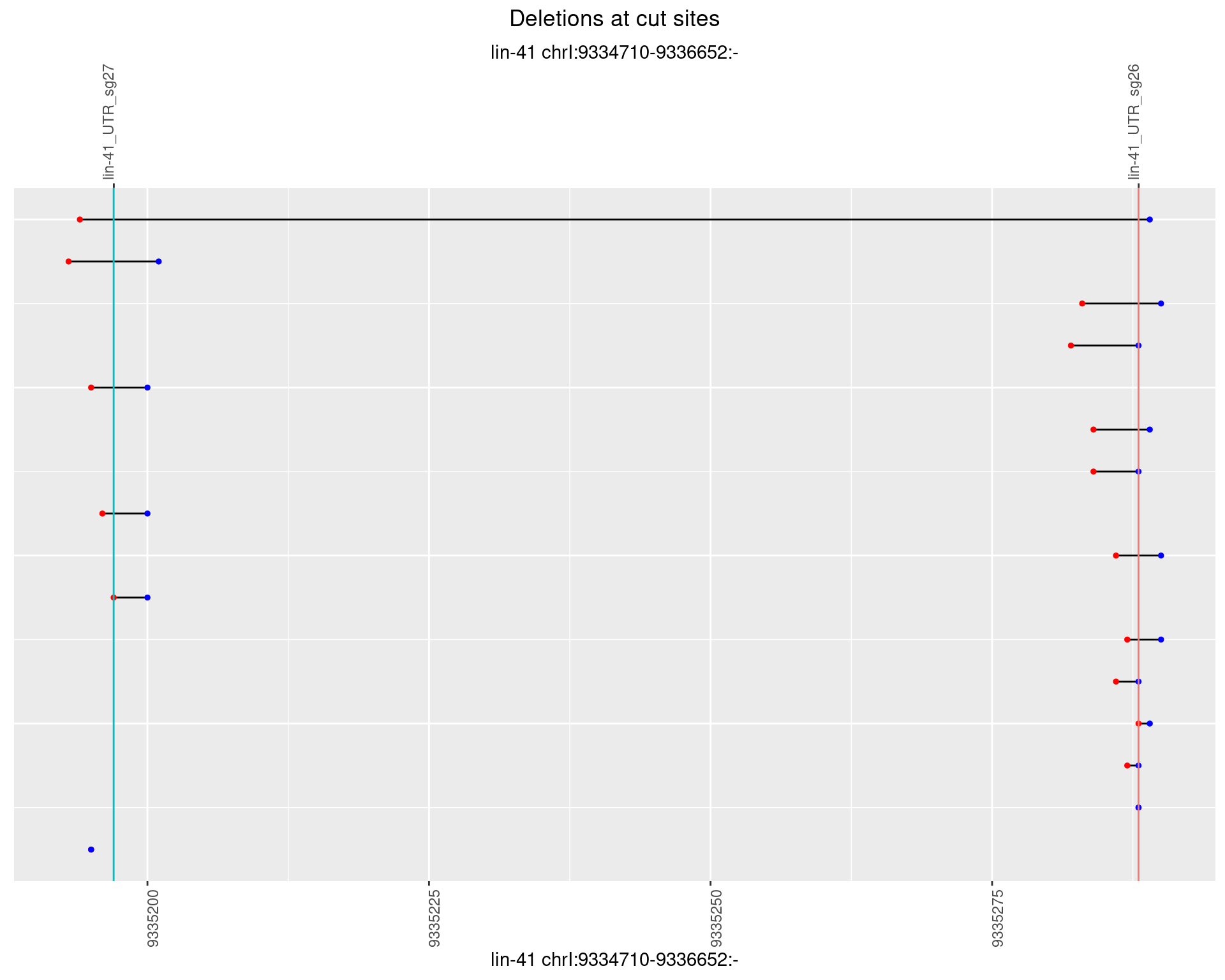
1.8.5 Freq: 0.01
1.8.6 Freq: 0.1
No plot to show
1.9 gen_24C_F3_lin-41_N2
1.9.1 Freq: 0
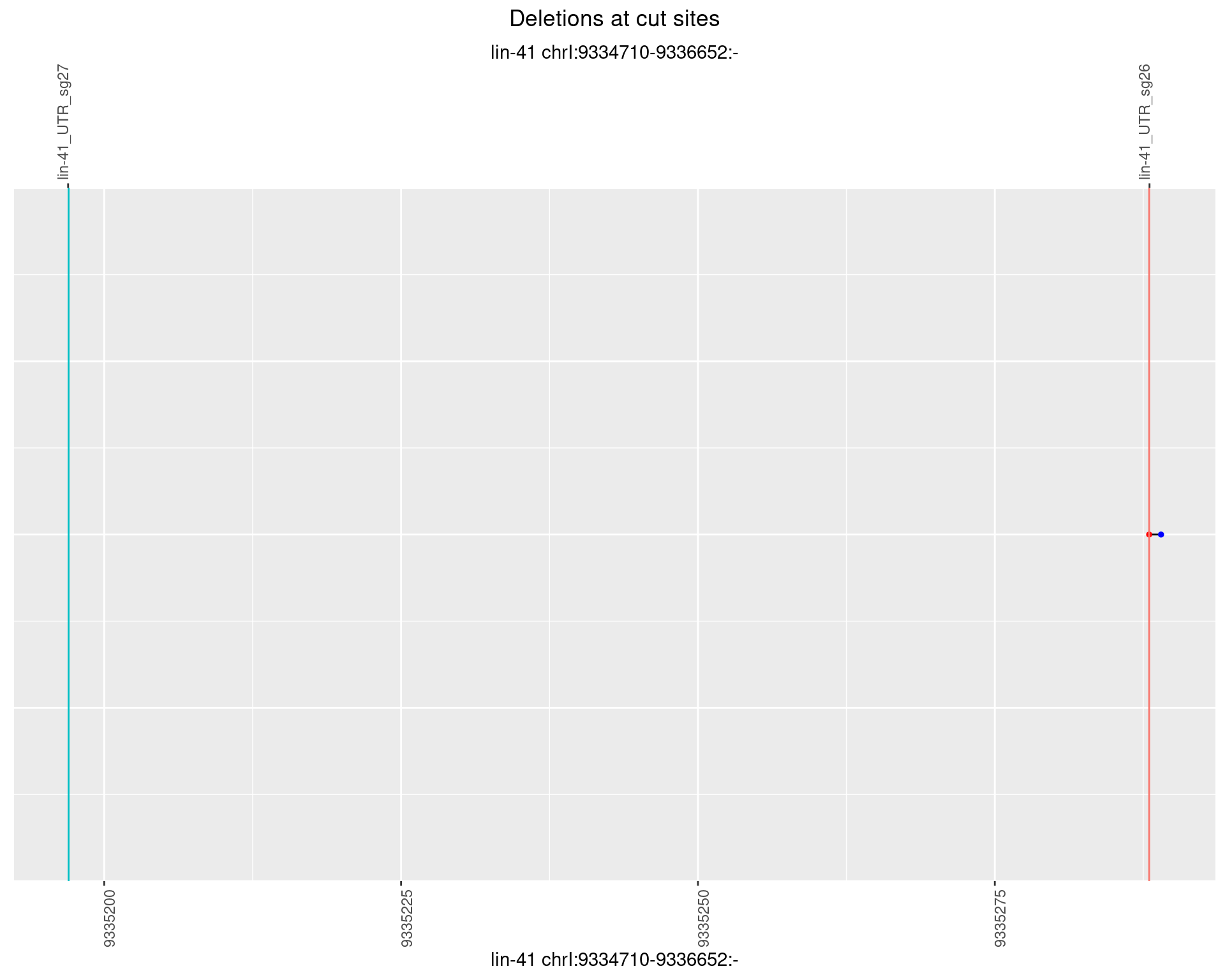
1.9.2 Freq: 1e-05
1.9.3 Freq: 1e-04
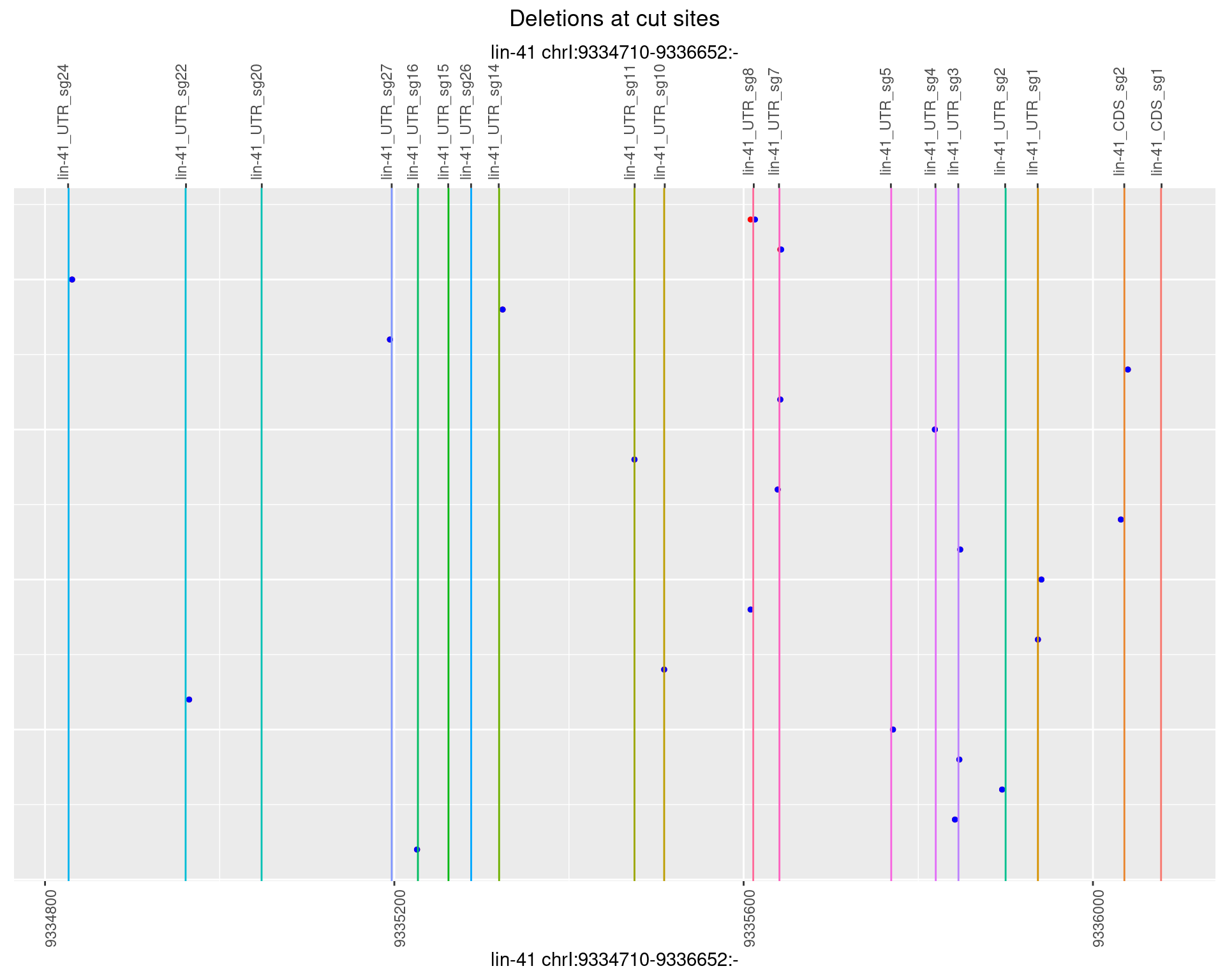
1.9.4 Freq: 0.001
1.9.5 Freq: 0.01
No plot to show
1.9.6 Freq: 0.1
No plot to show
1.10 gen_24C_F3_lin-41_sg15sg16
1.10.1 Freq: 0
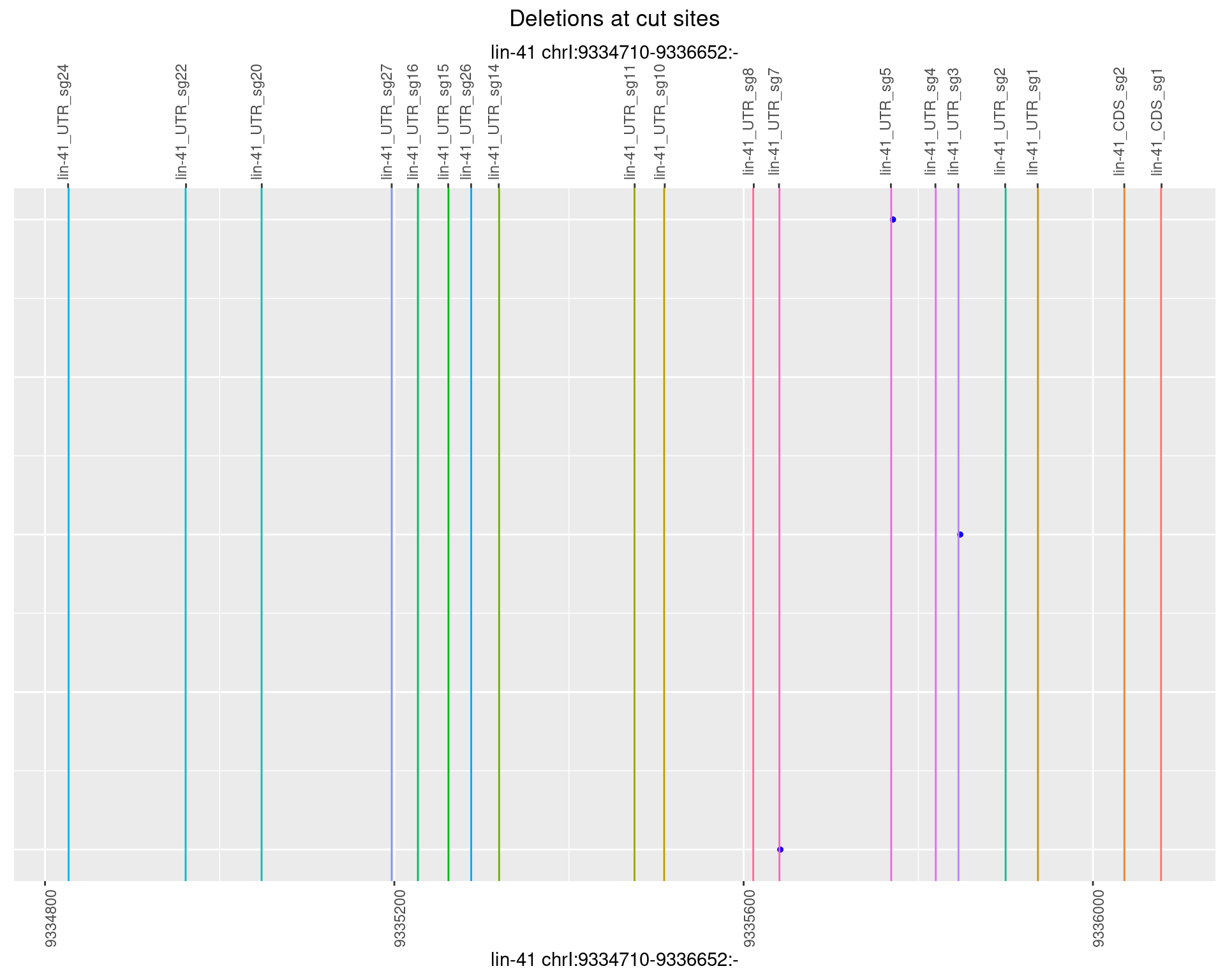
1.10.2 Freq: 1e-05
1.10.3 Freq: 1e-04
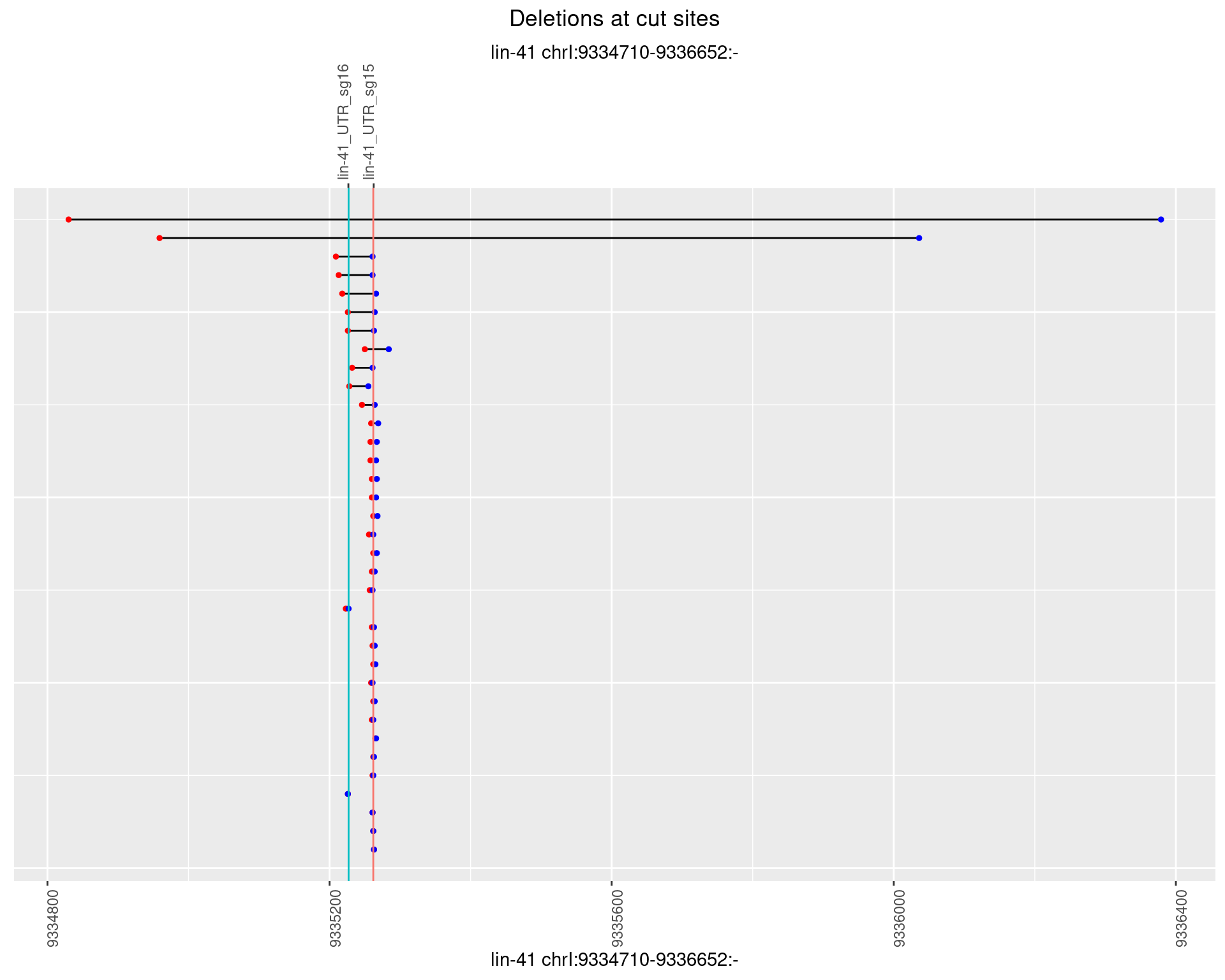
1.10.4 Freq: 0.001
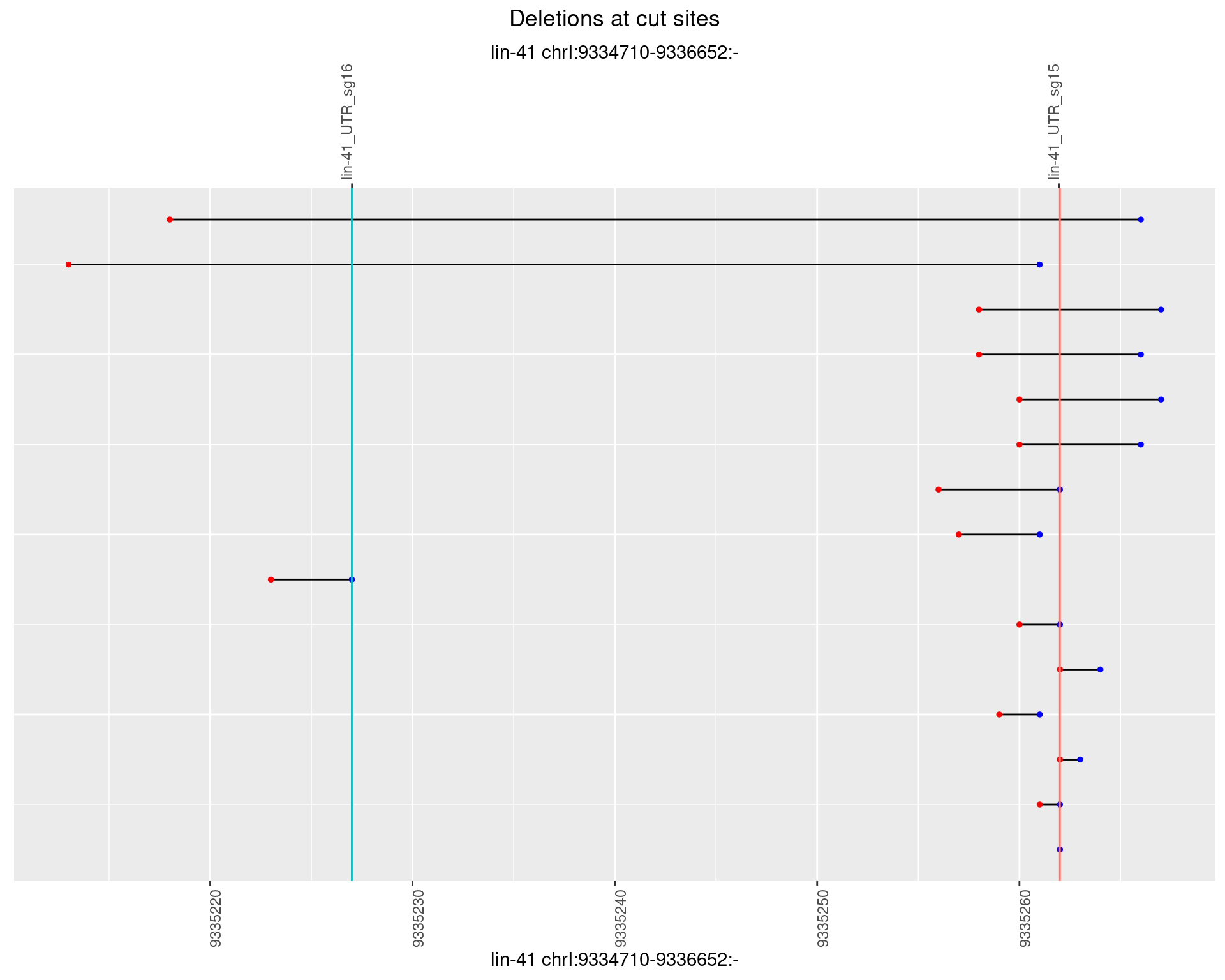
1.10.5 Freq: 0.01
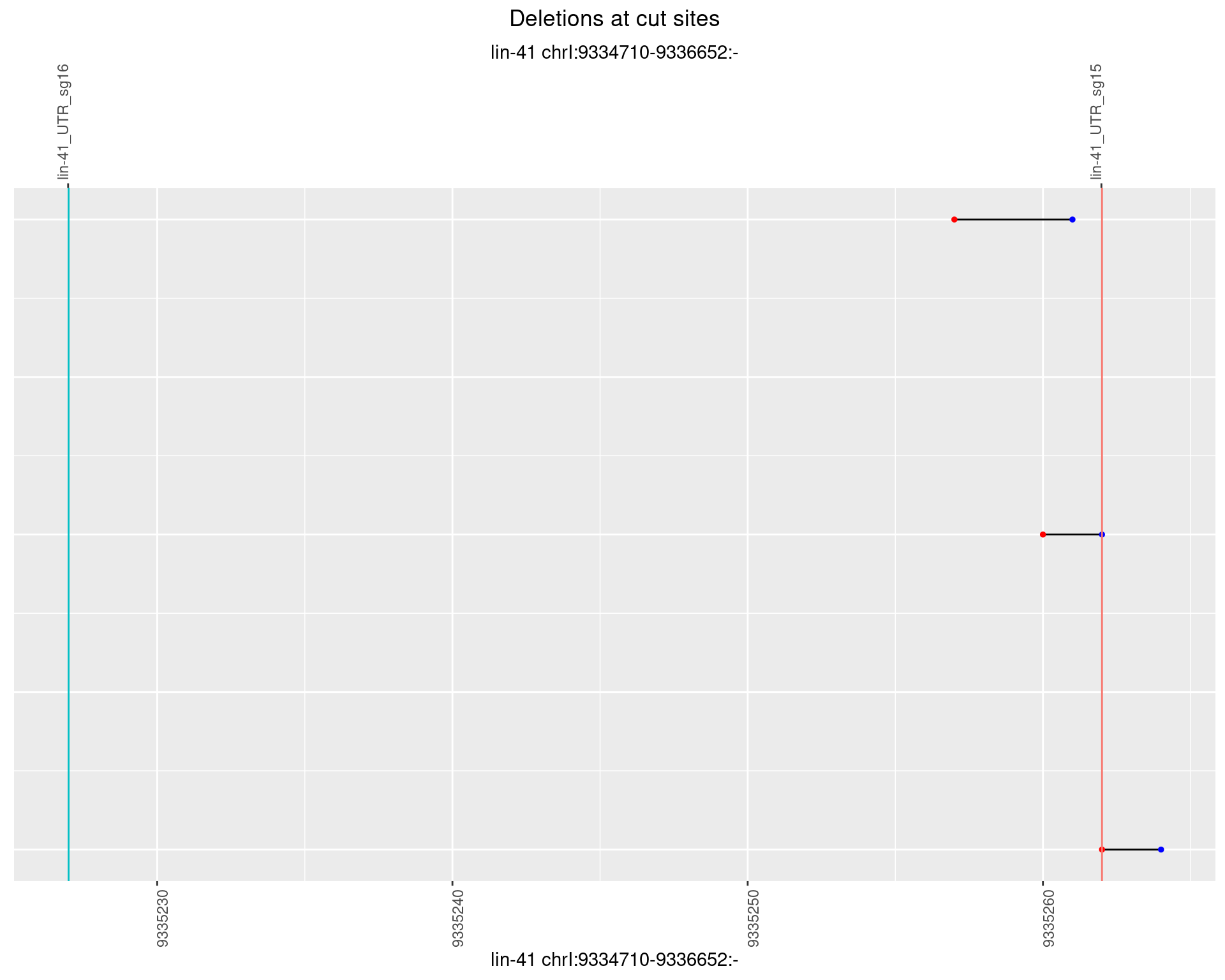
1.10.6 Freq: 0.1
No plot to show
1.11 gen_24C_F3_lin-41_pool
1.11.1 Freq: 0
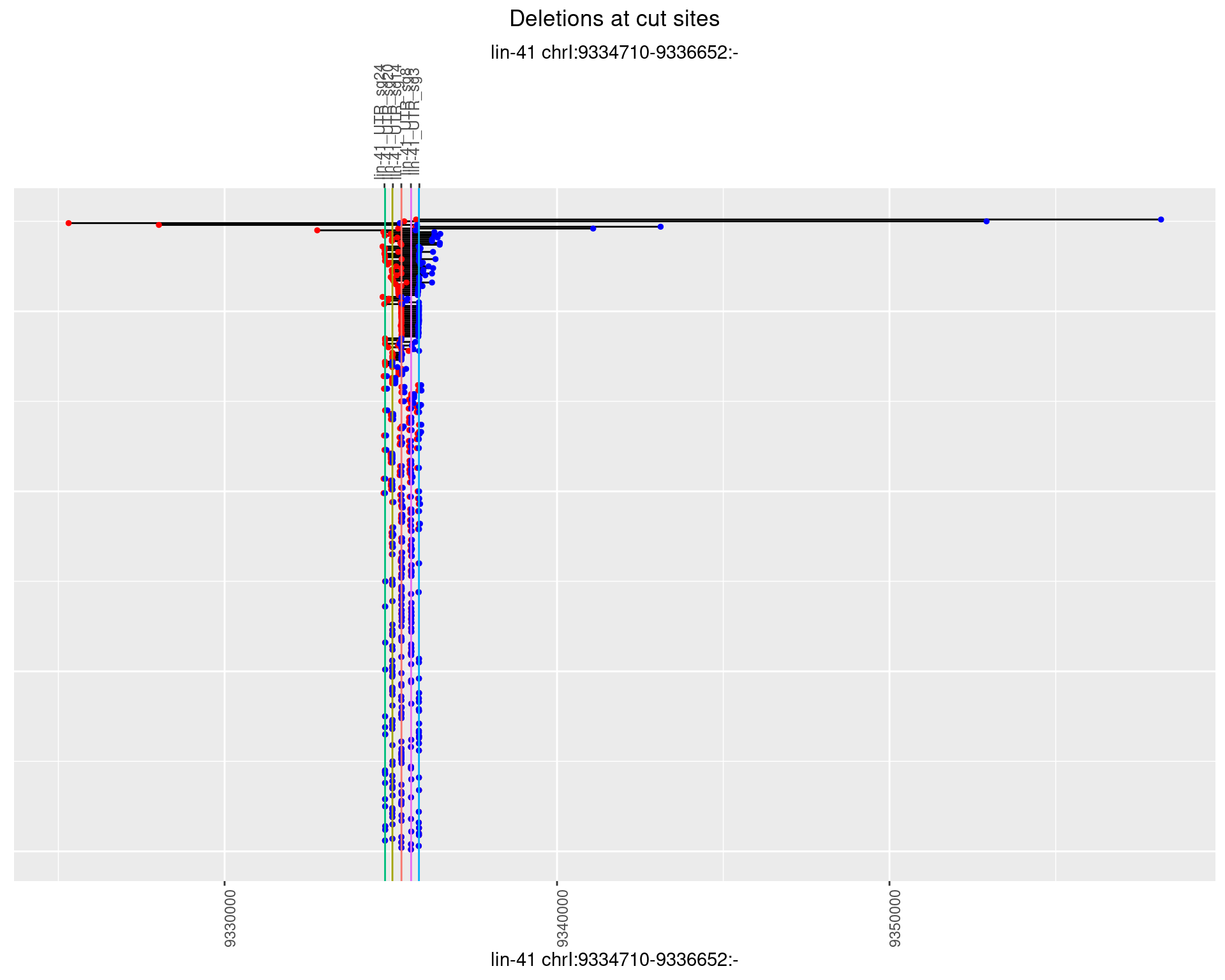
1.11.2 Freq: 1e-05
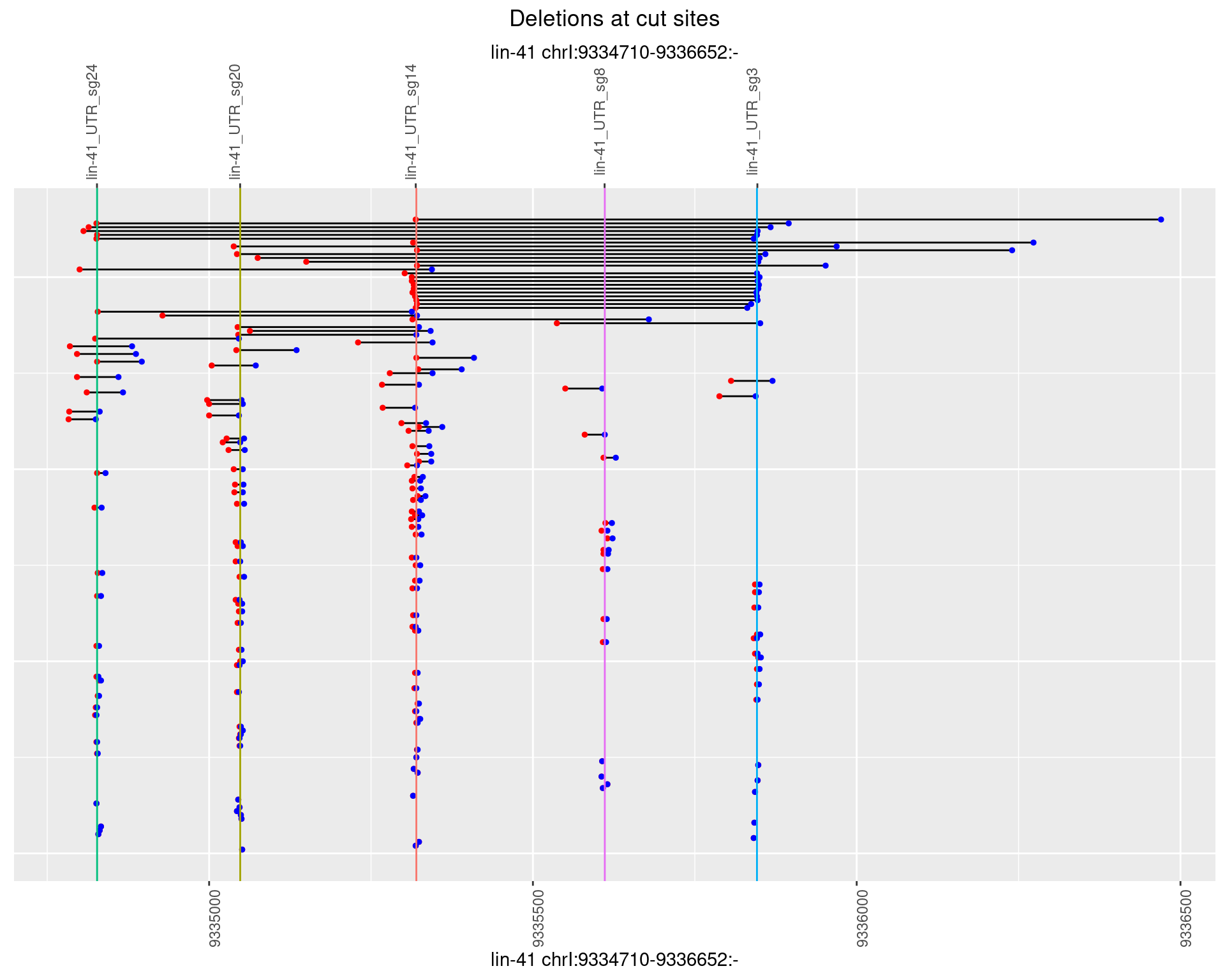
1.11.3 Freq: 1e-04
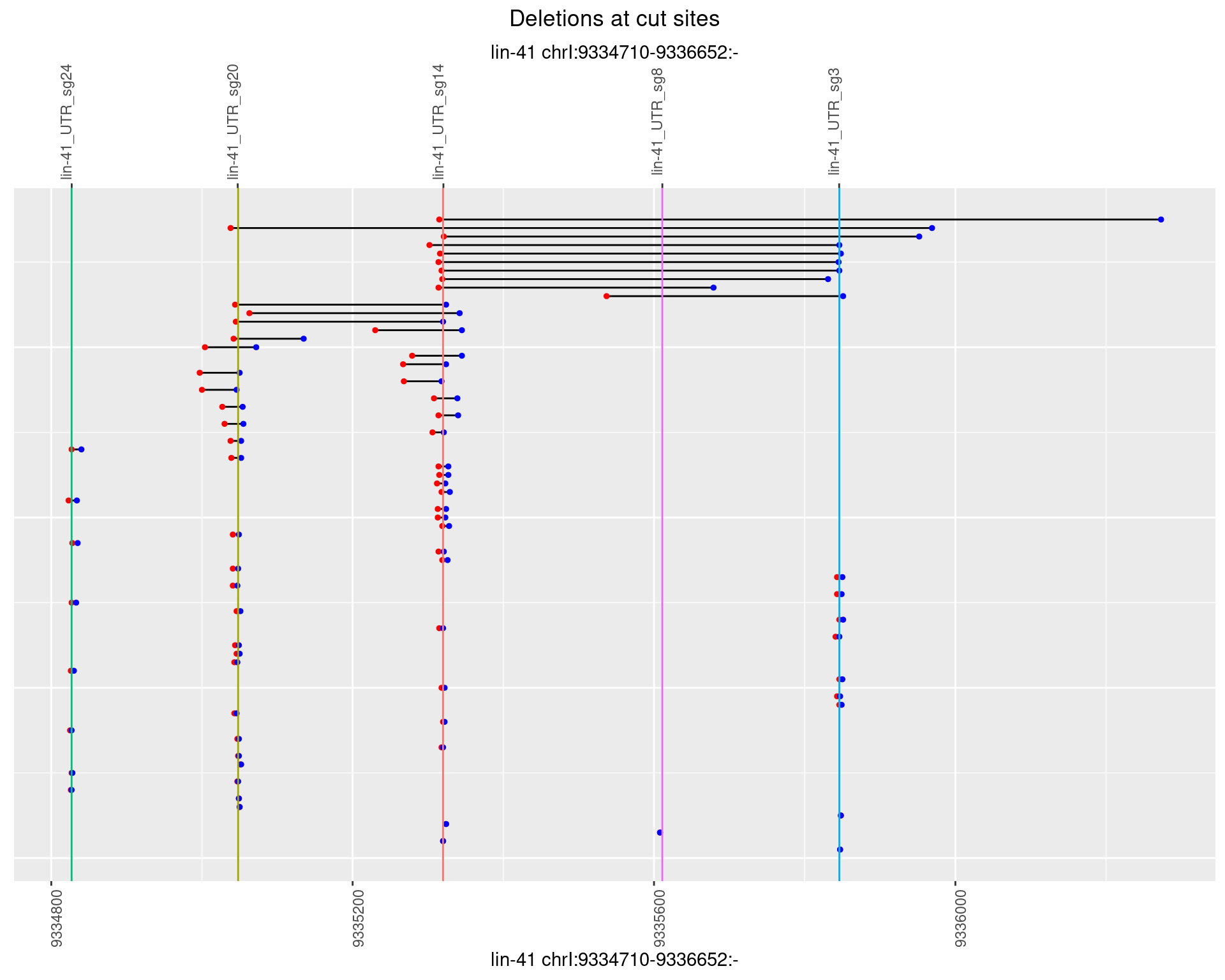
1.11.4 Freq: 0.001
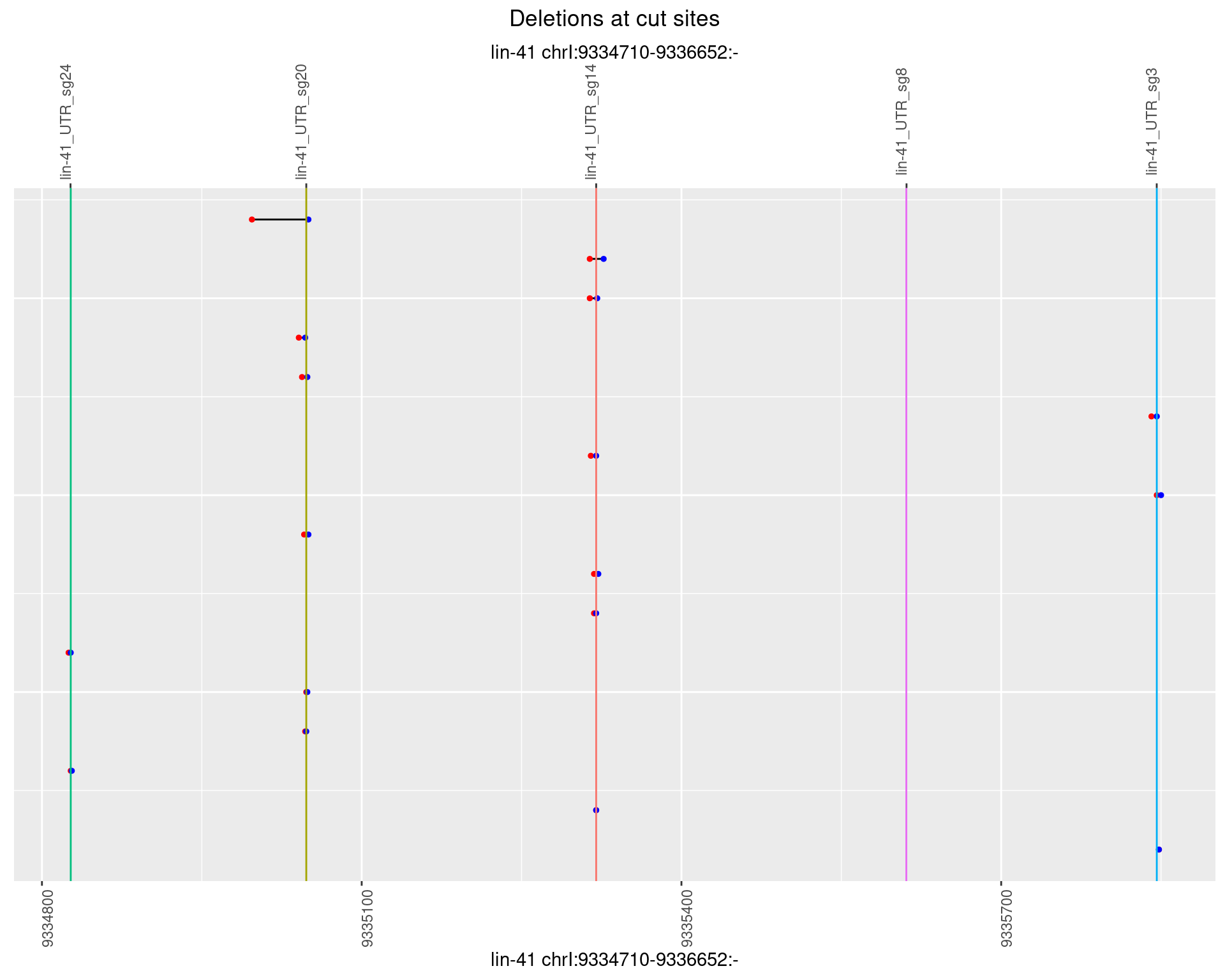
1.11.5 Freq: 0.01
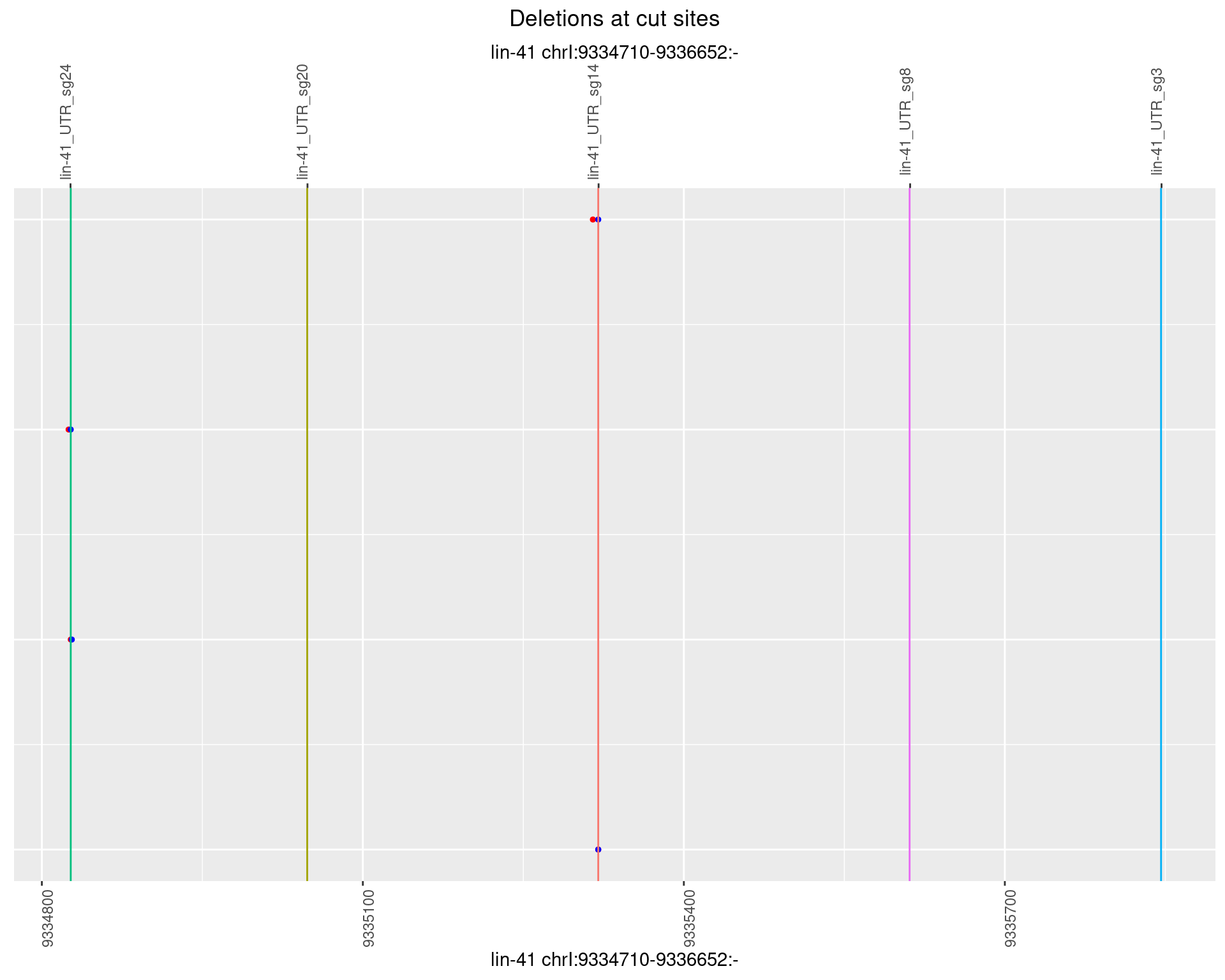
1.11.6 Freq: 0.1
No plot to show
1.12 gen_24C_F3_lin-41_sg26sg27
1.12.1 Freq: 0
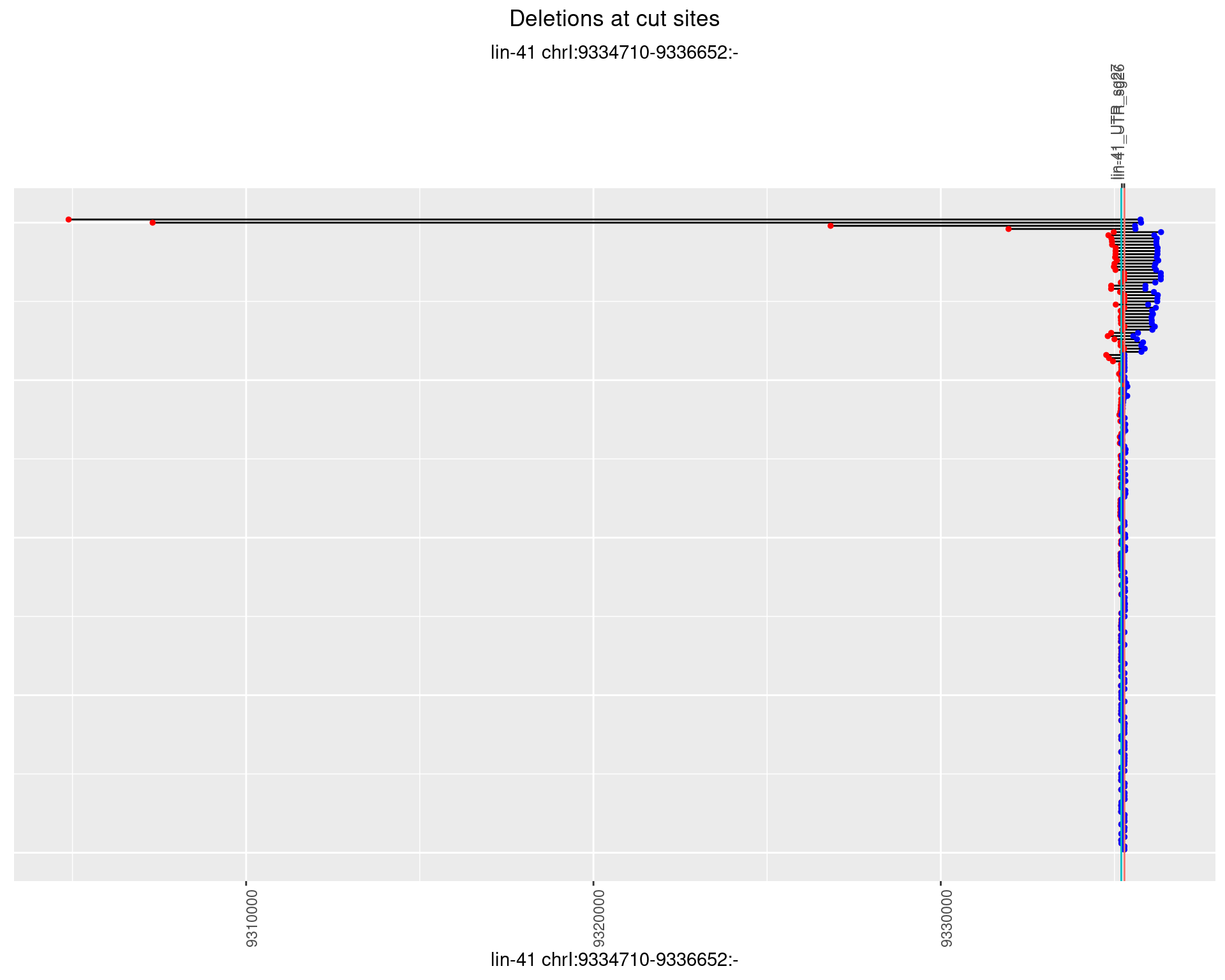
1.12.2 Freq: 1e-05
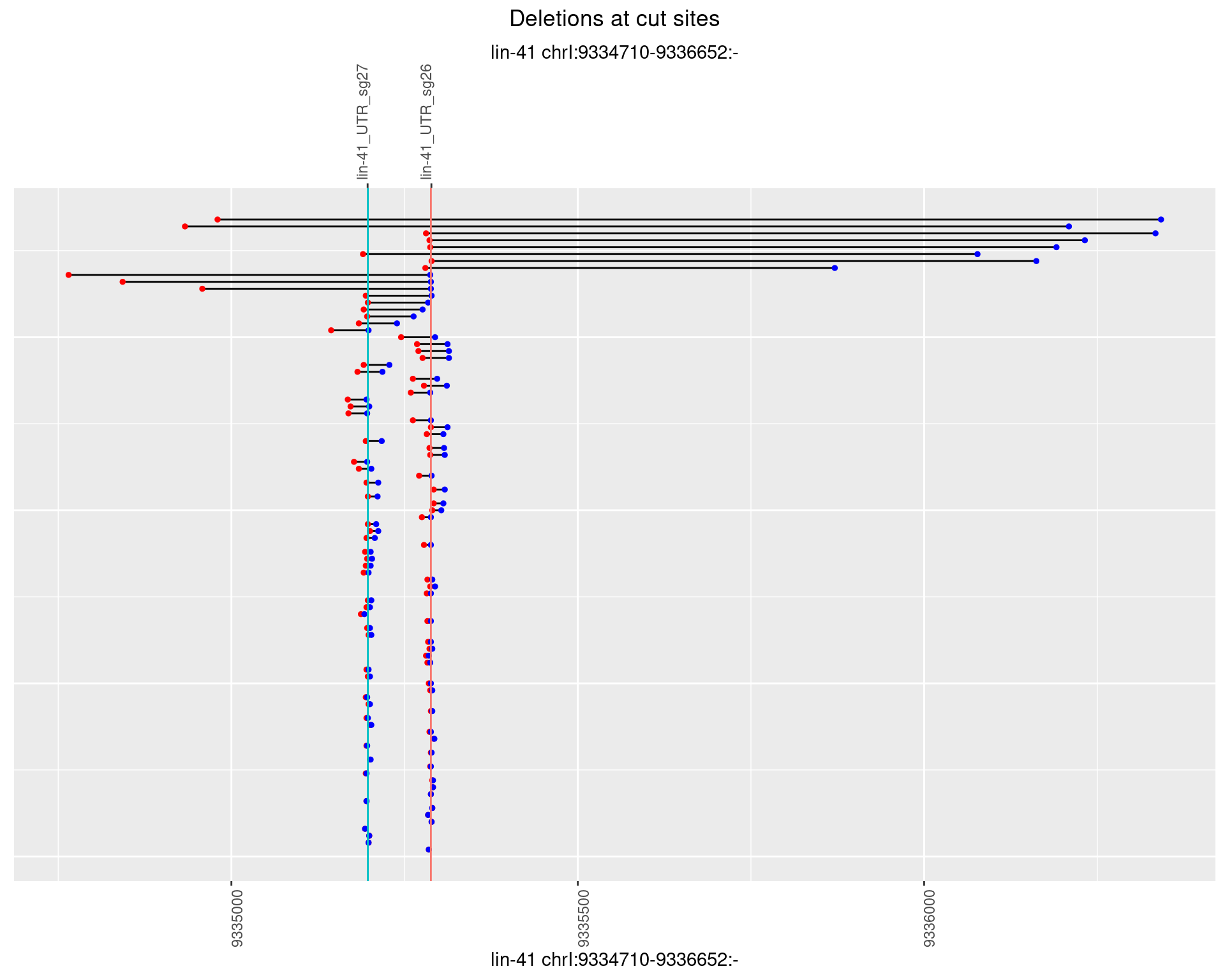
1.12.3 Freq: 1e-04
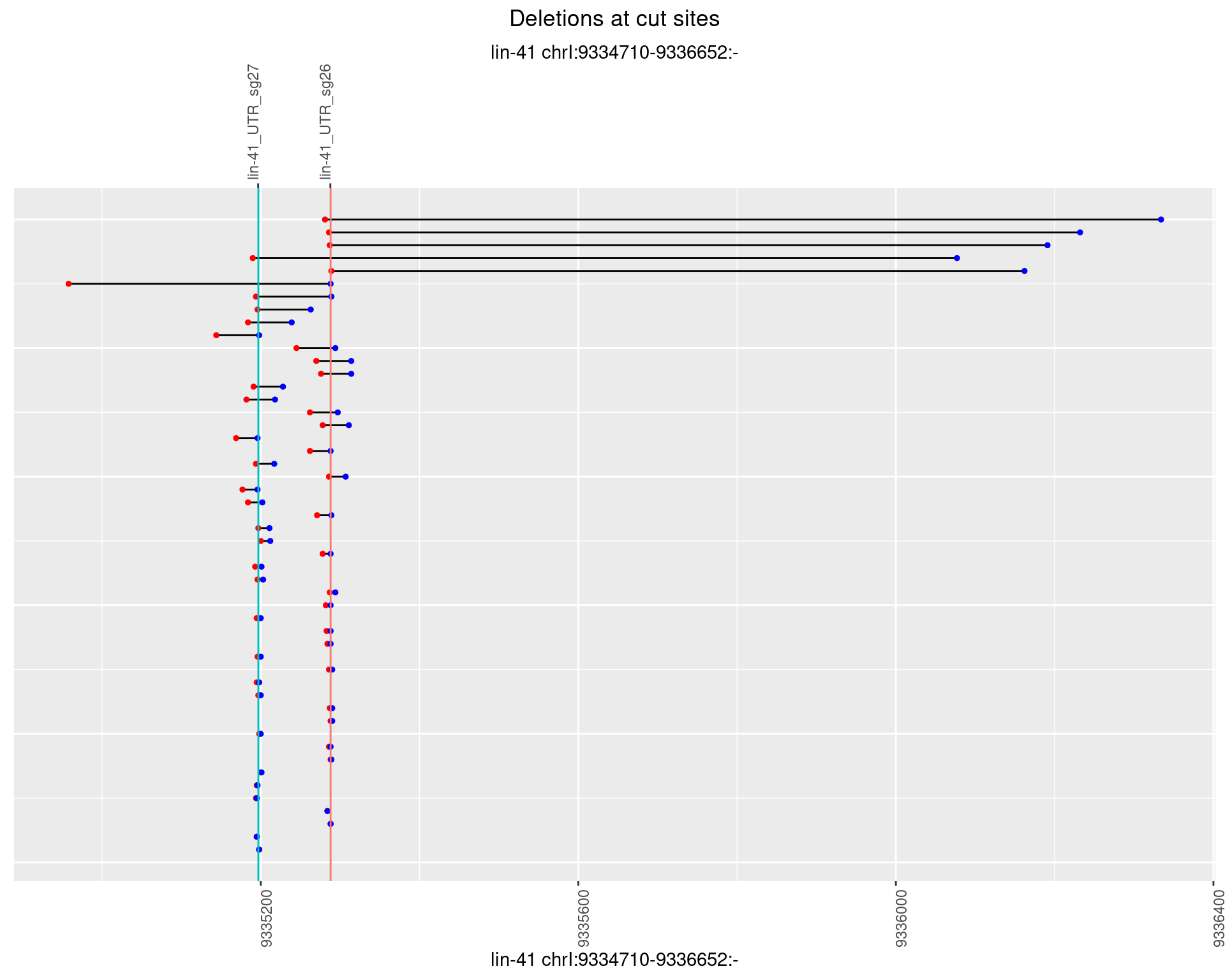
1.12.4 Freq: 0.001
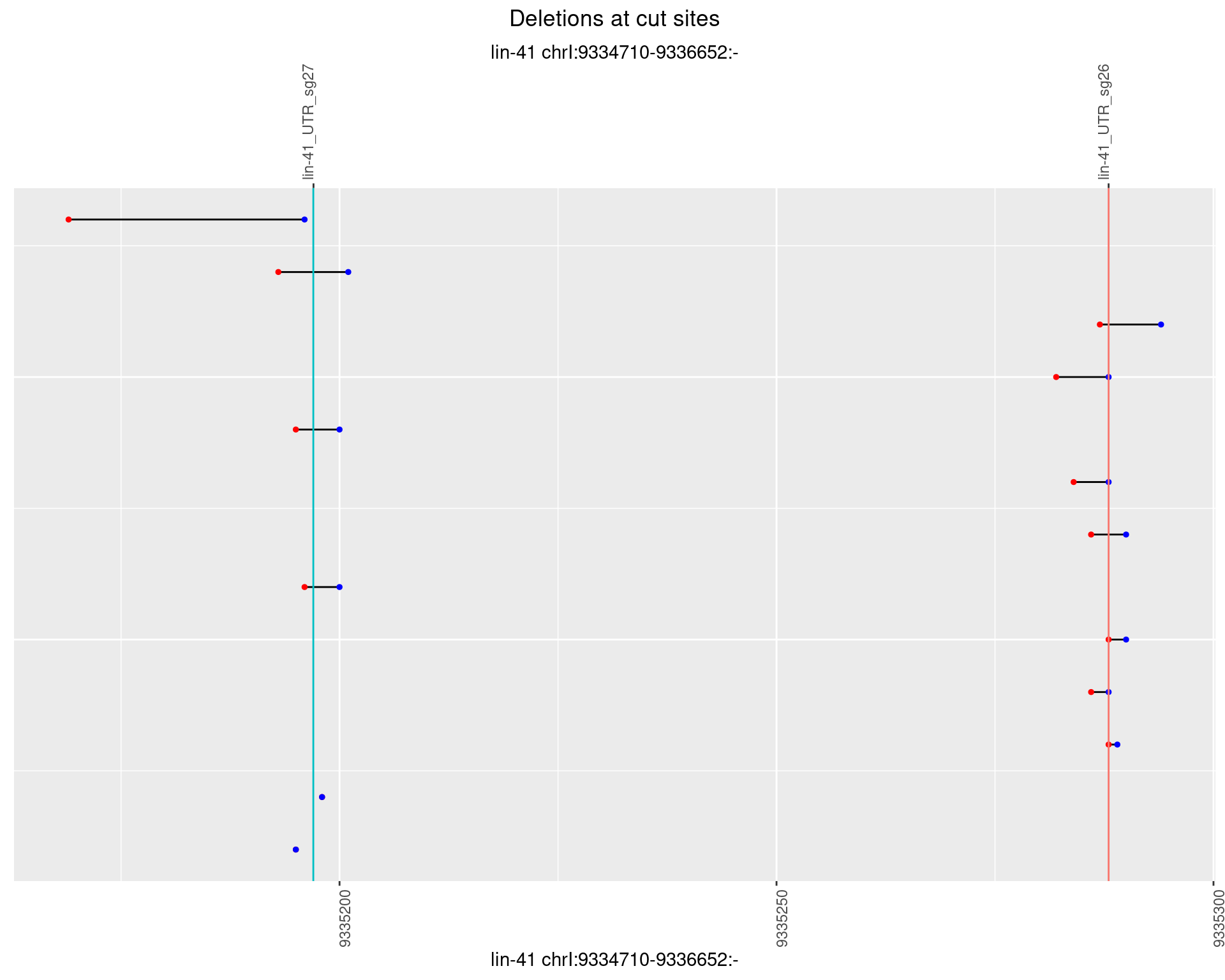
1.12.5 Freq: 0.01
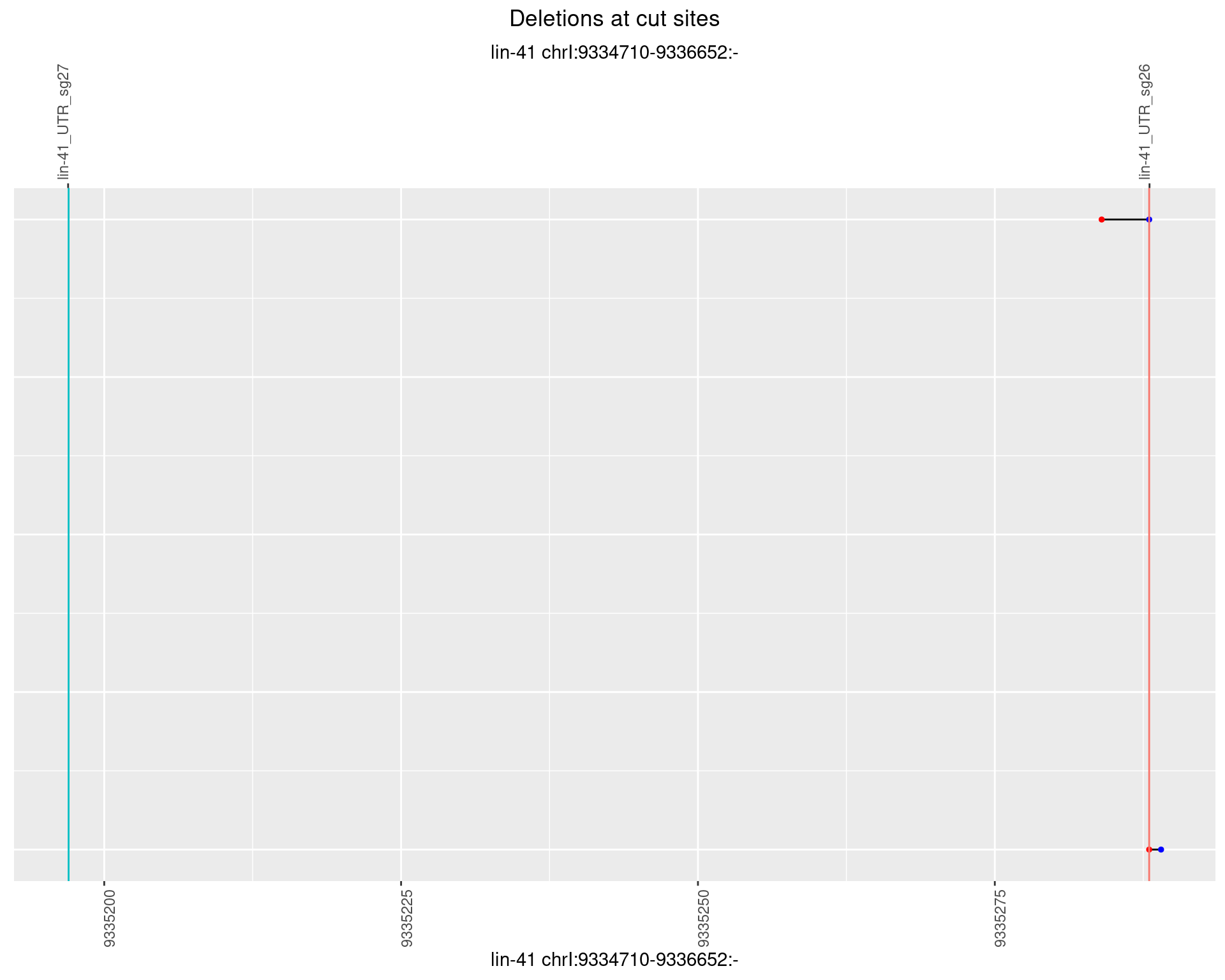
1.12.6 Freq: 0.1
No plot to show
1.13 gen_24C_F4_lin-41_N2
1.13.1 Freq: 0
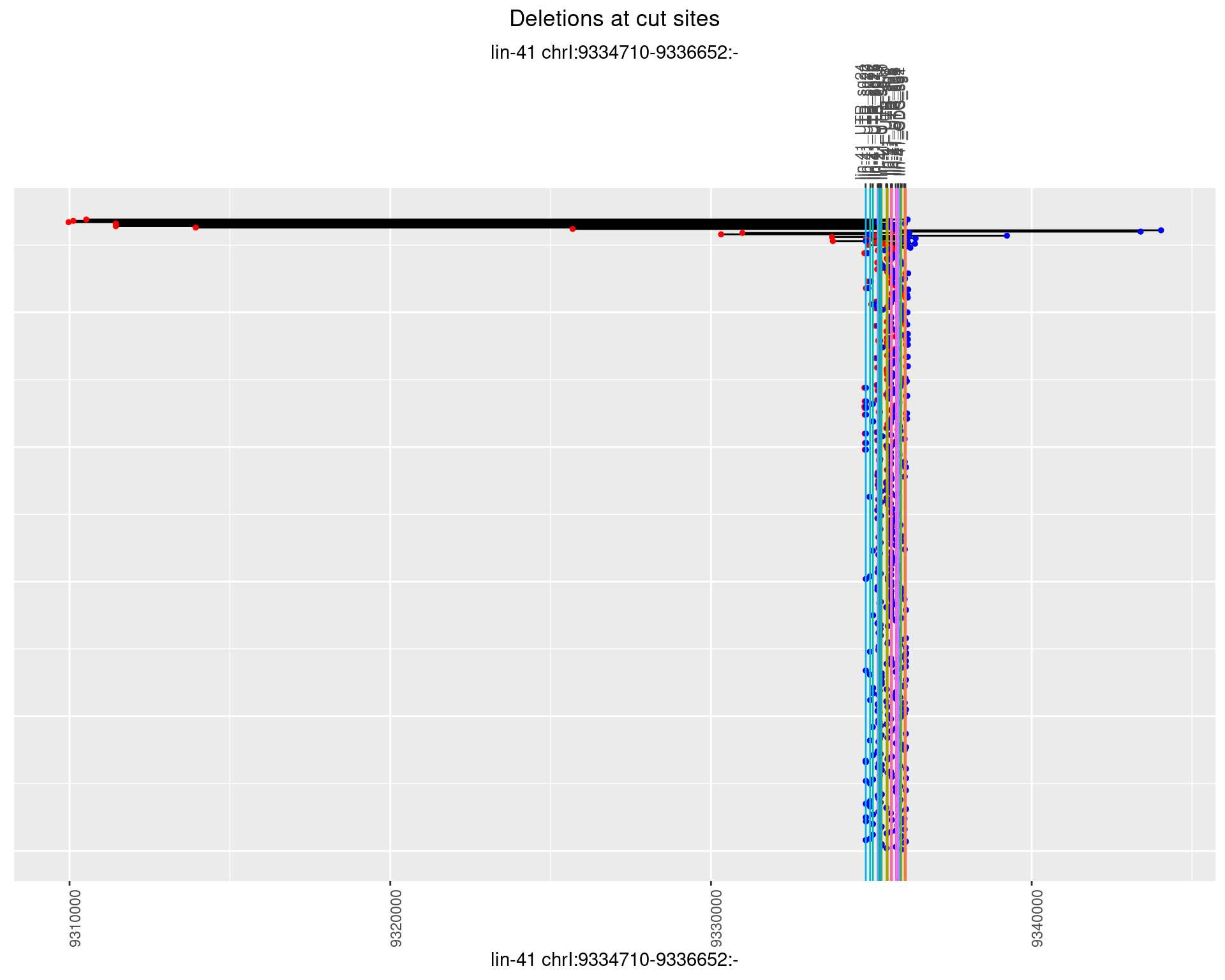
1.13.2 Freq: 1e-05
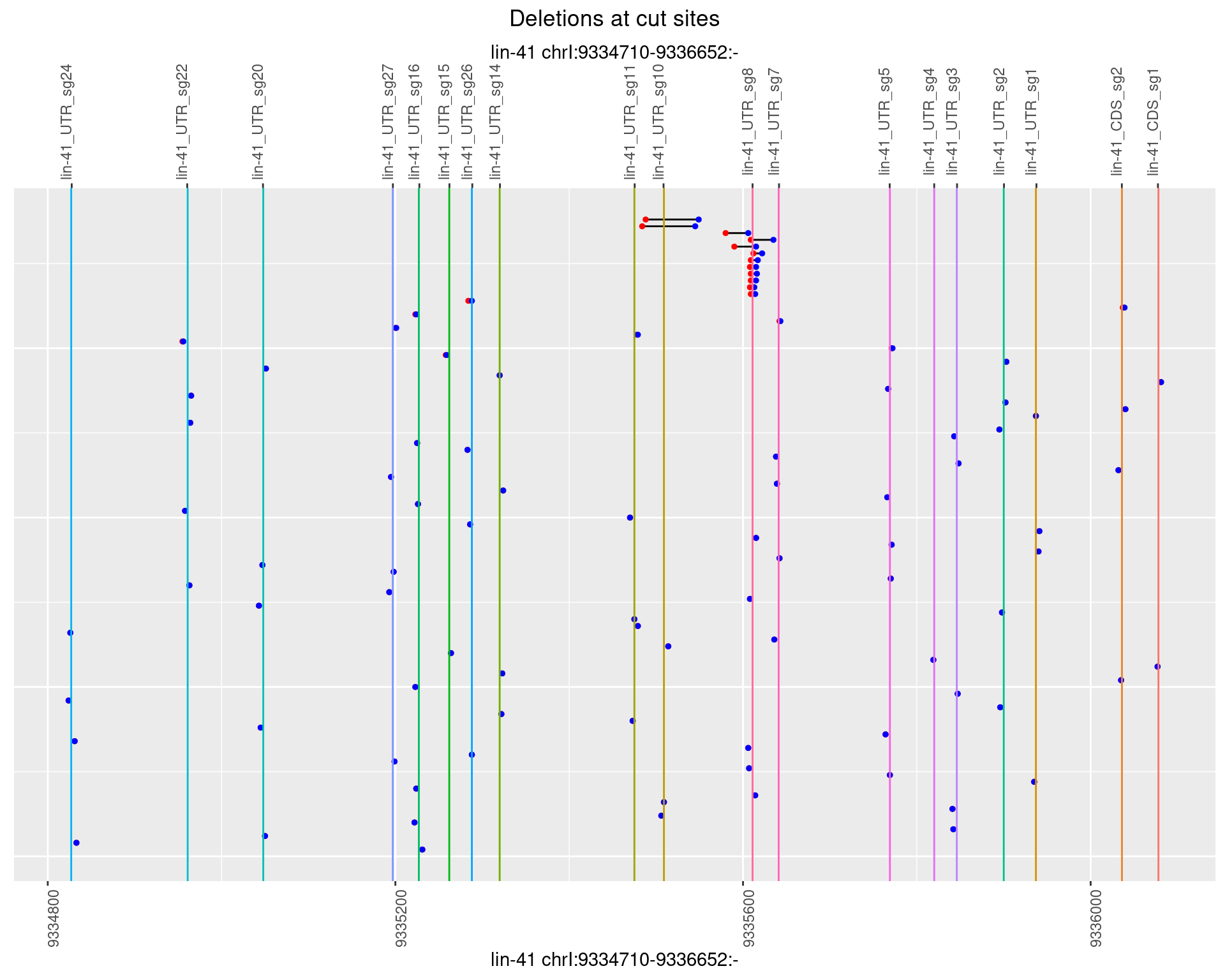
1.13.3 Freq: 1e-04
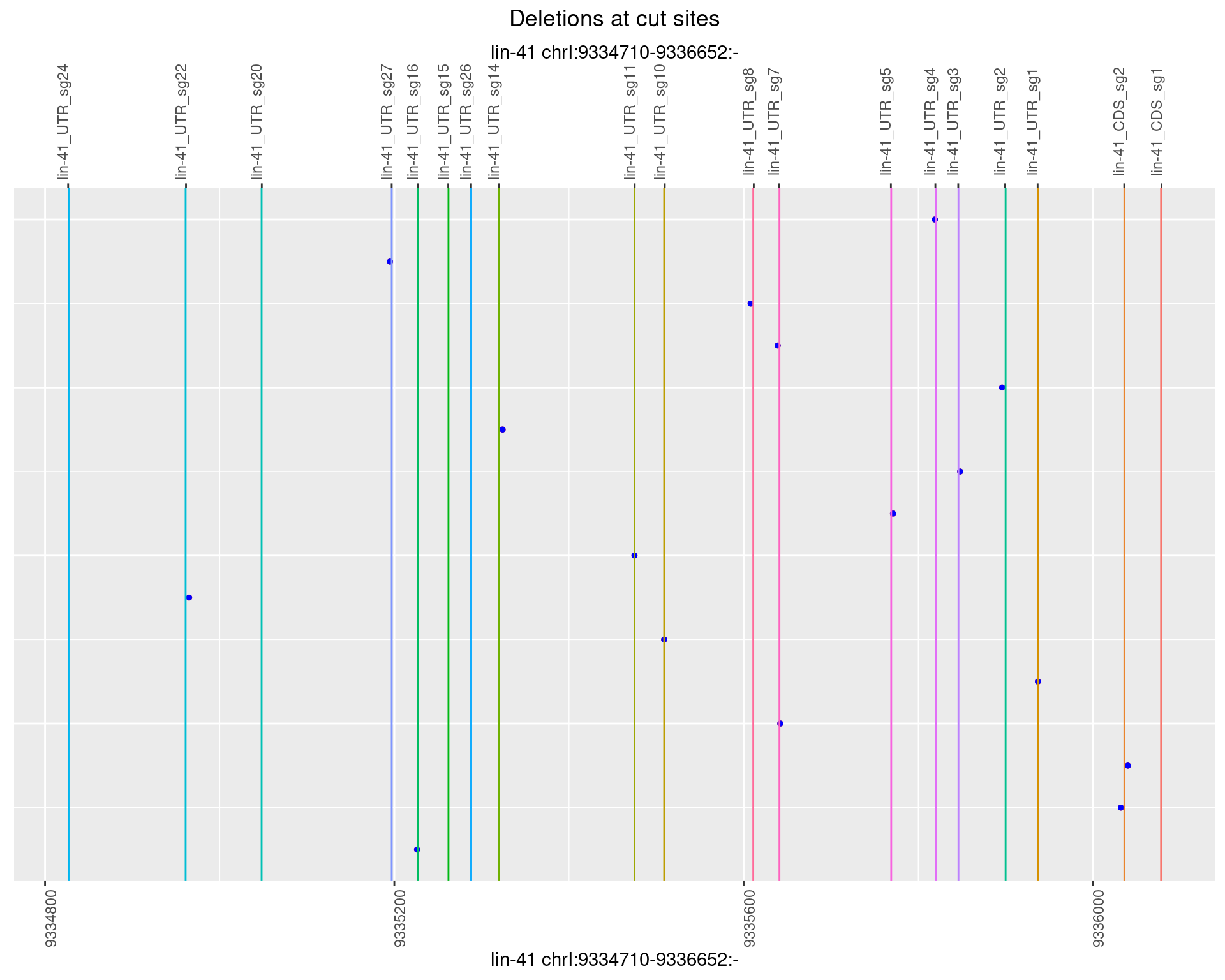
1.13.4 Freq: 0.001
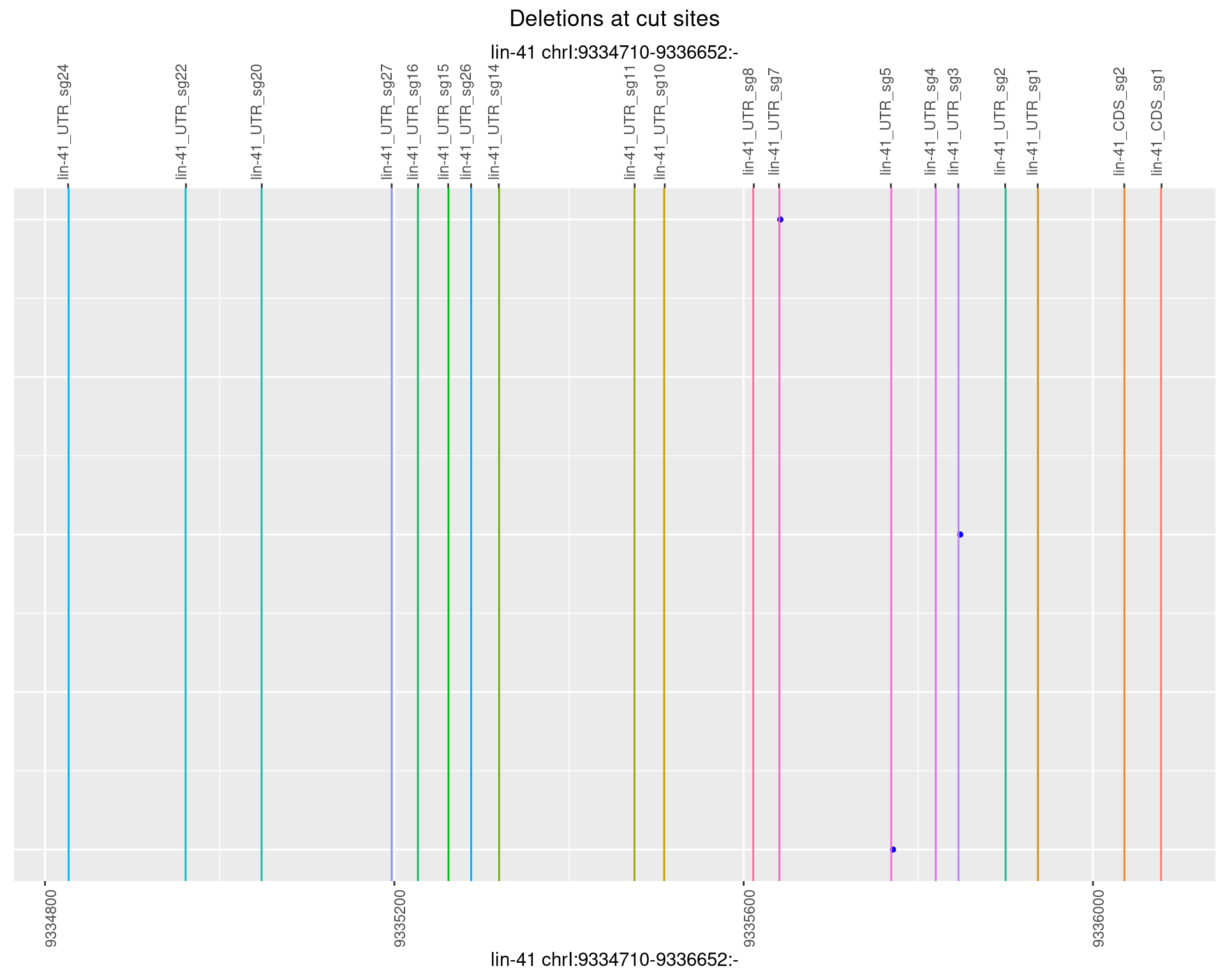
1.13.5 Freq: 0.01
No plot to show
1.13.6 Freq: 0.1
No plot to show
1.14 gen_24C_F4_lin-41_sg15sg16
1.14.1 Freq: 0
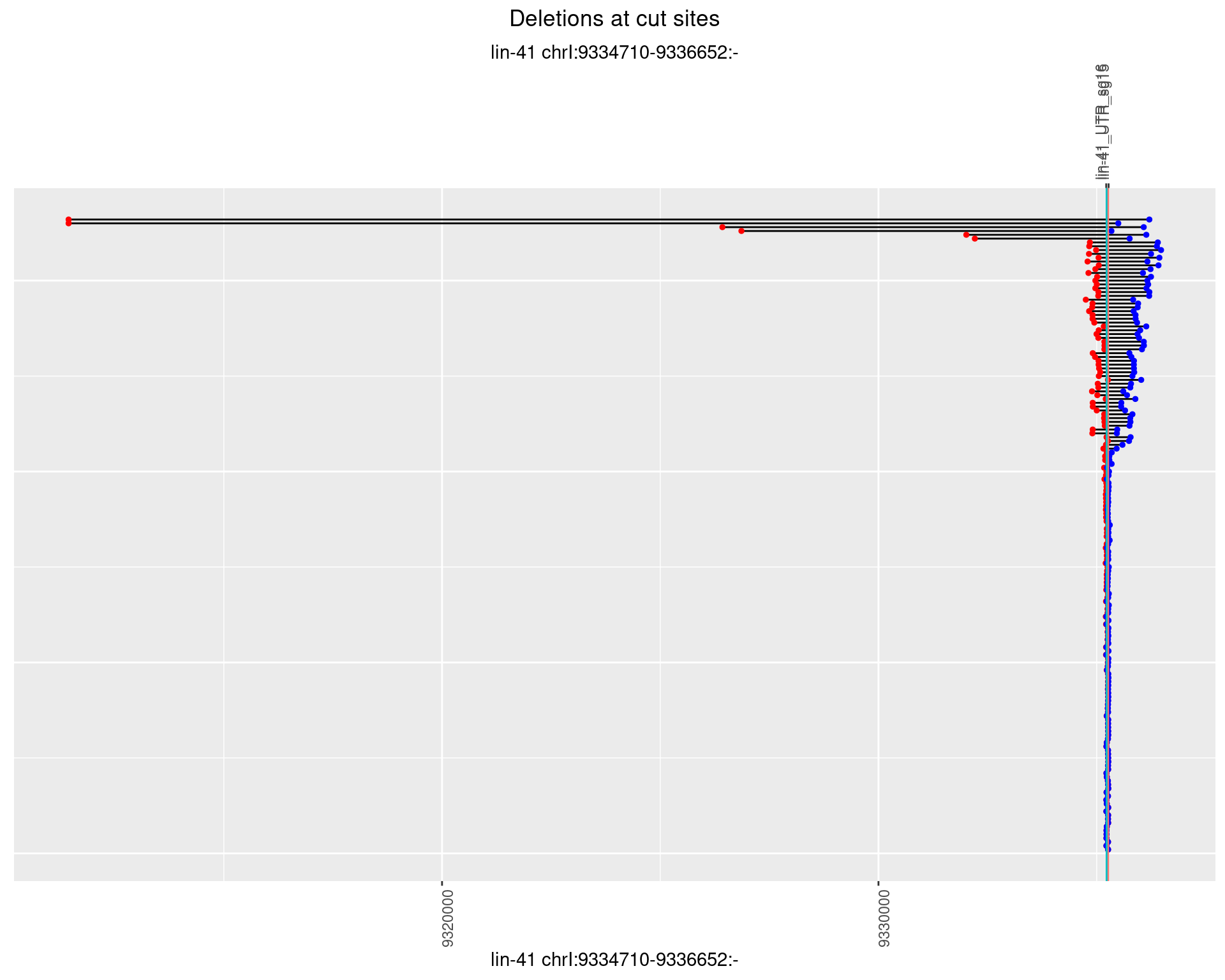
1.14.2 Freq: 1e-05
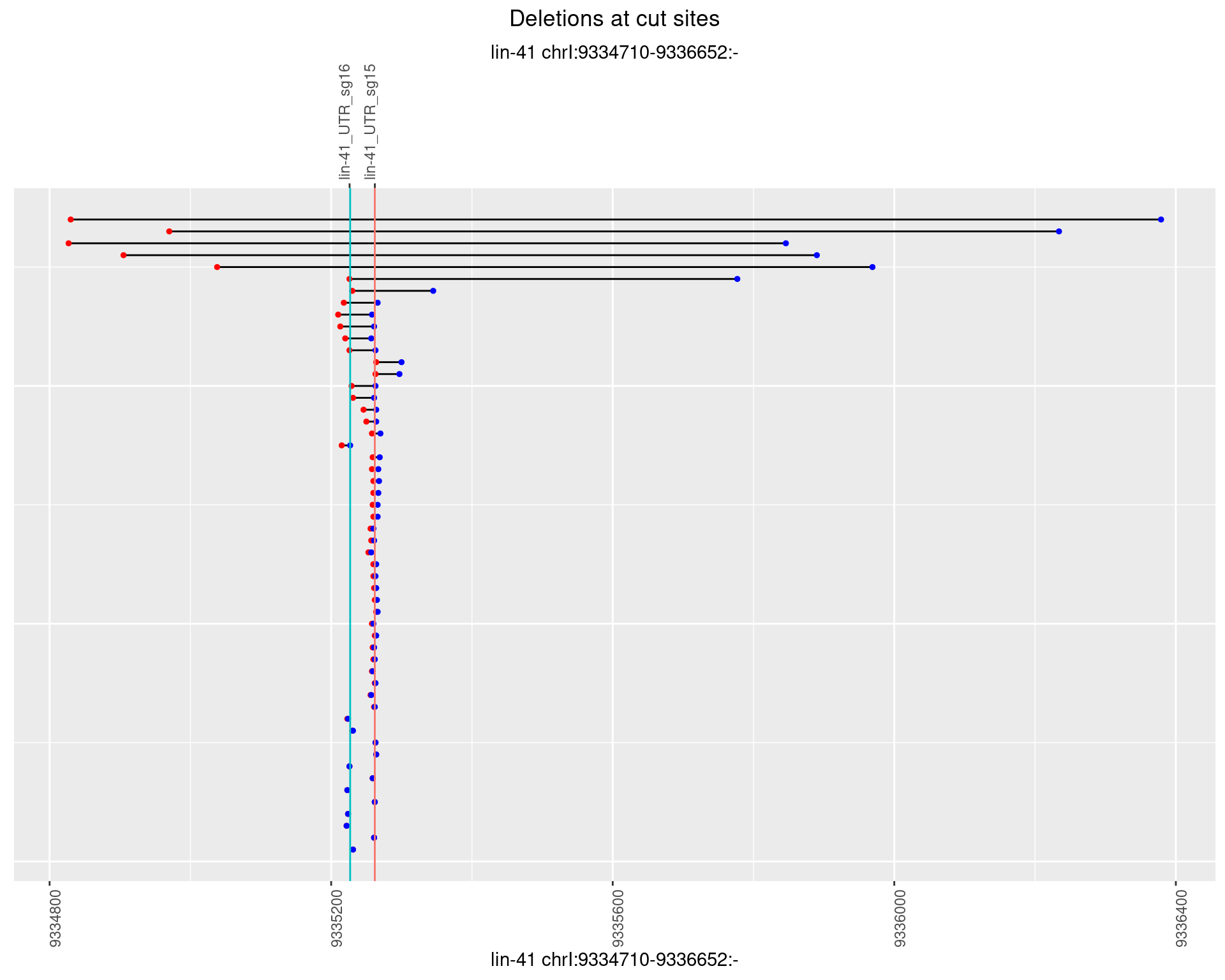
1.14.3 Freq: 1e-04
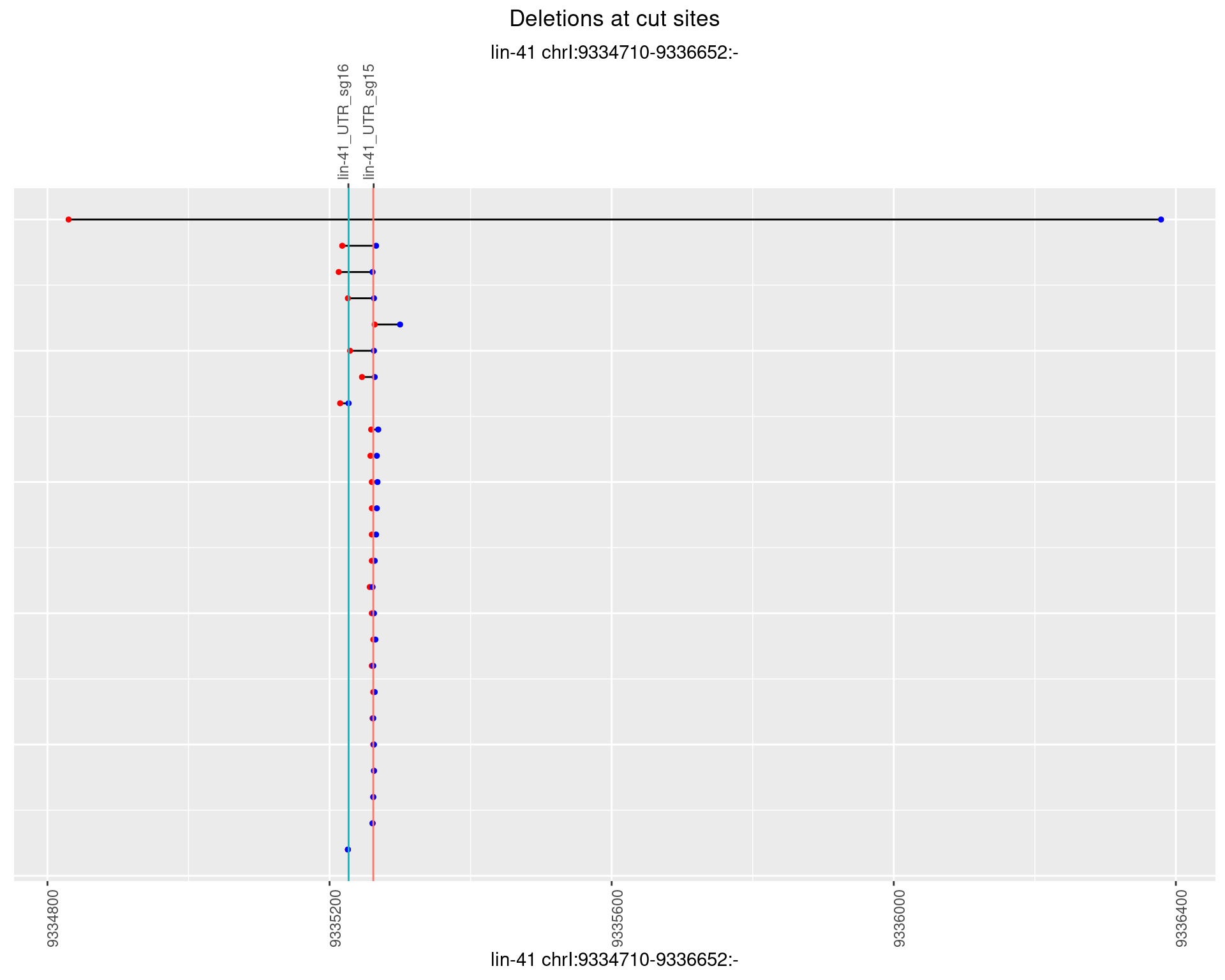
1.14.4 Freq: 0.001
1.14.5 Freq: 0.01
1.14.6 Freq: 0.1
No plot to show
1.15 gen_24C_F4_lin-41_pool
1.15.1 Freq: 0
1.15.2 Freq: 1e-05
1.15.3 Freq: 1e-04
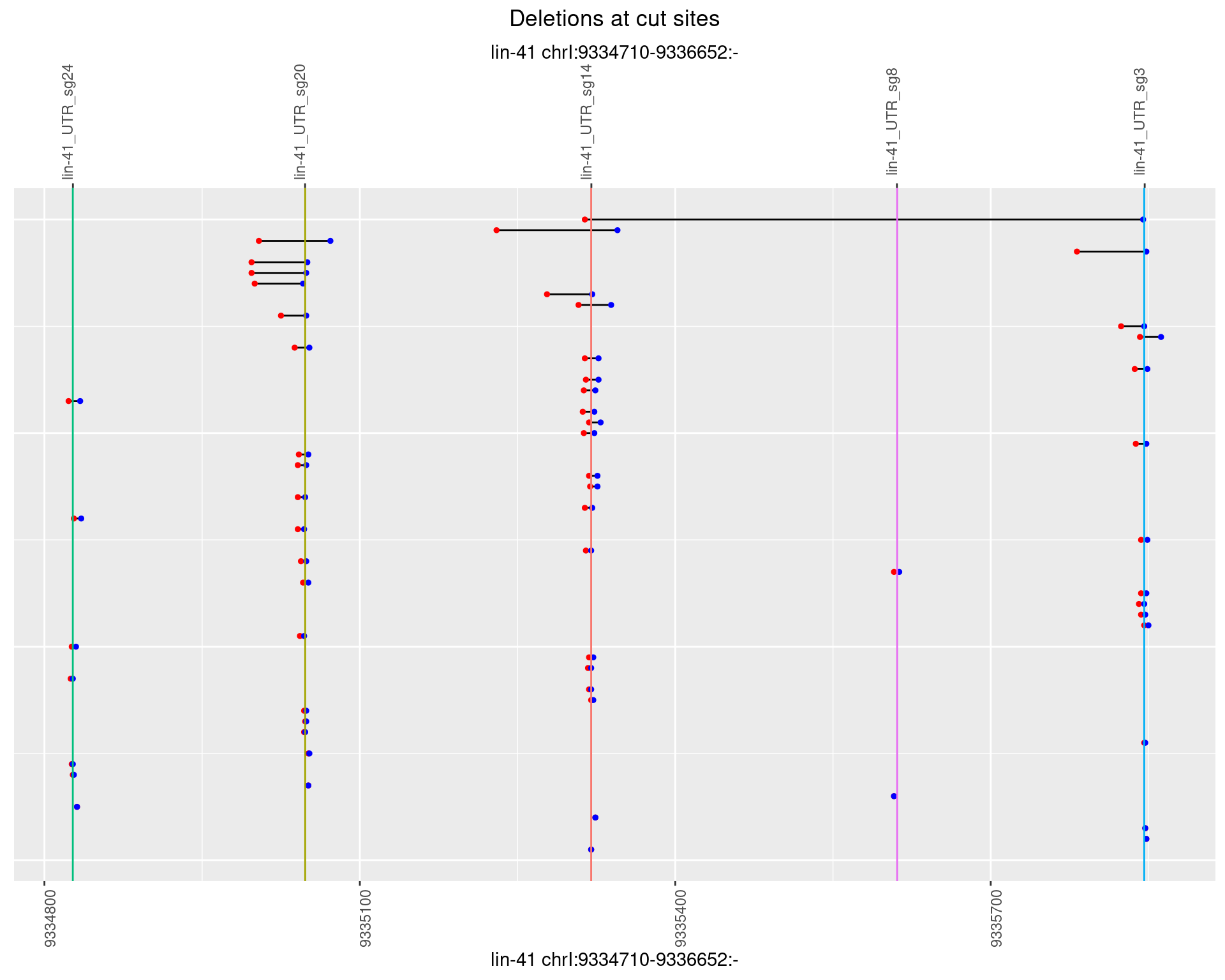
1.15.4 Freq: 0.001
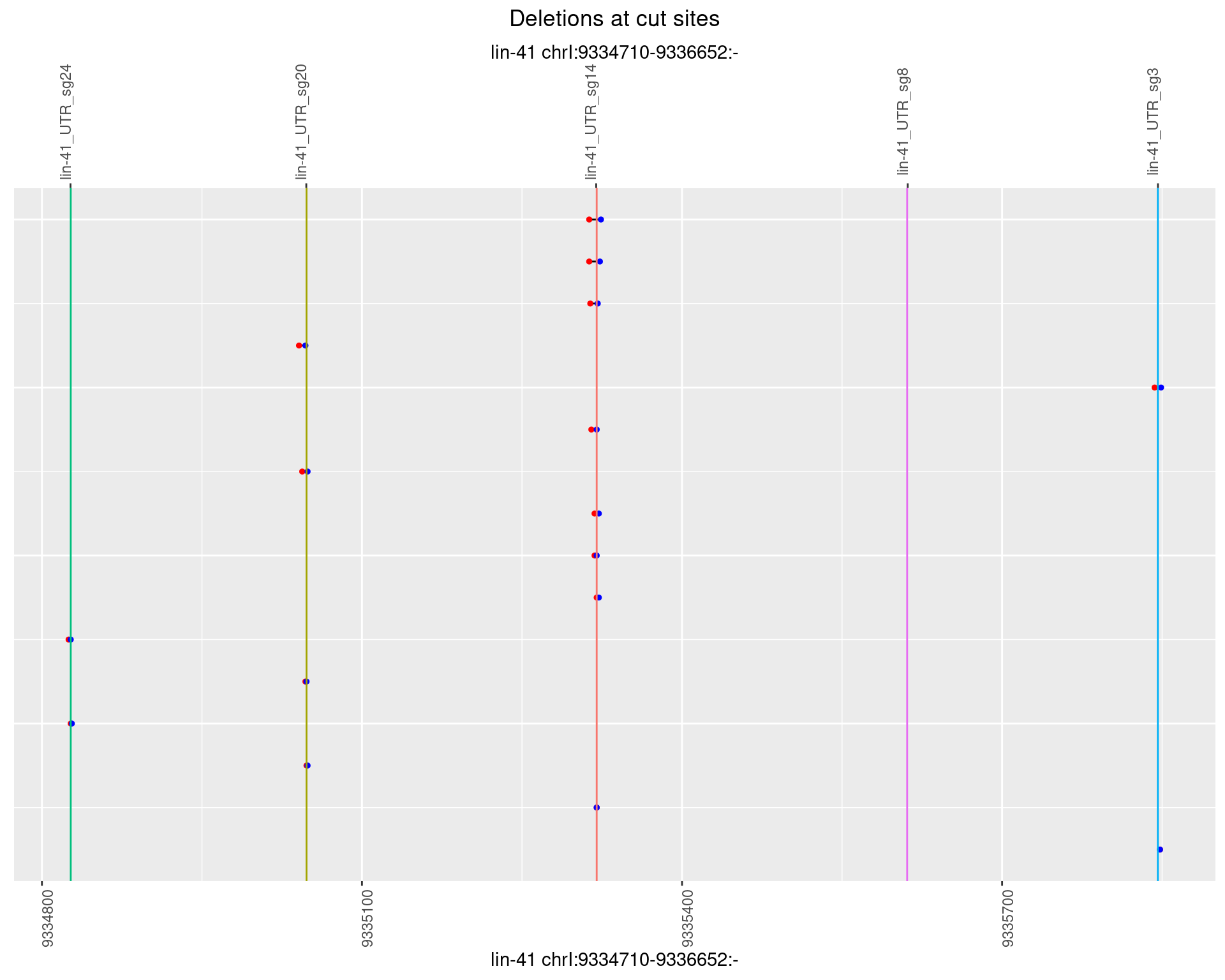
1.15.5 Freq: 0.01
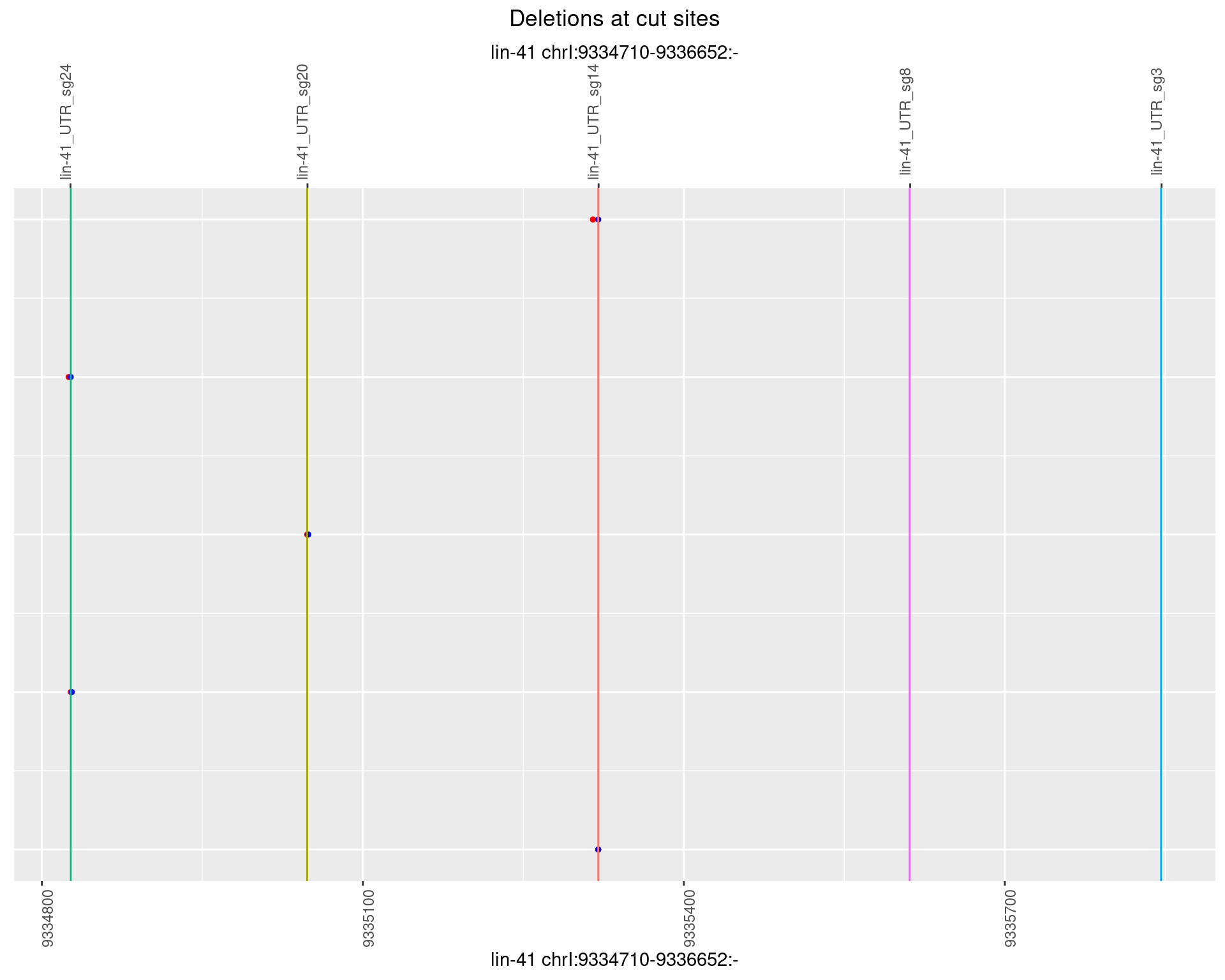
1.15.6 Freq: 0.1
No plot to show
1.16 gen_24C_F4_lin-41_sg26sg27
1.16.1 Freq: 0
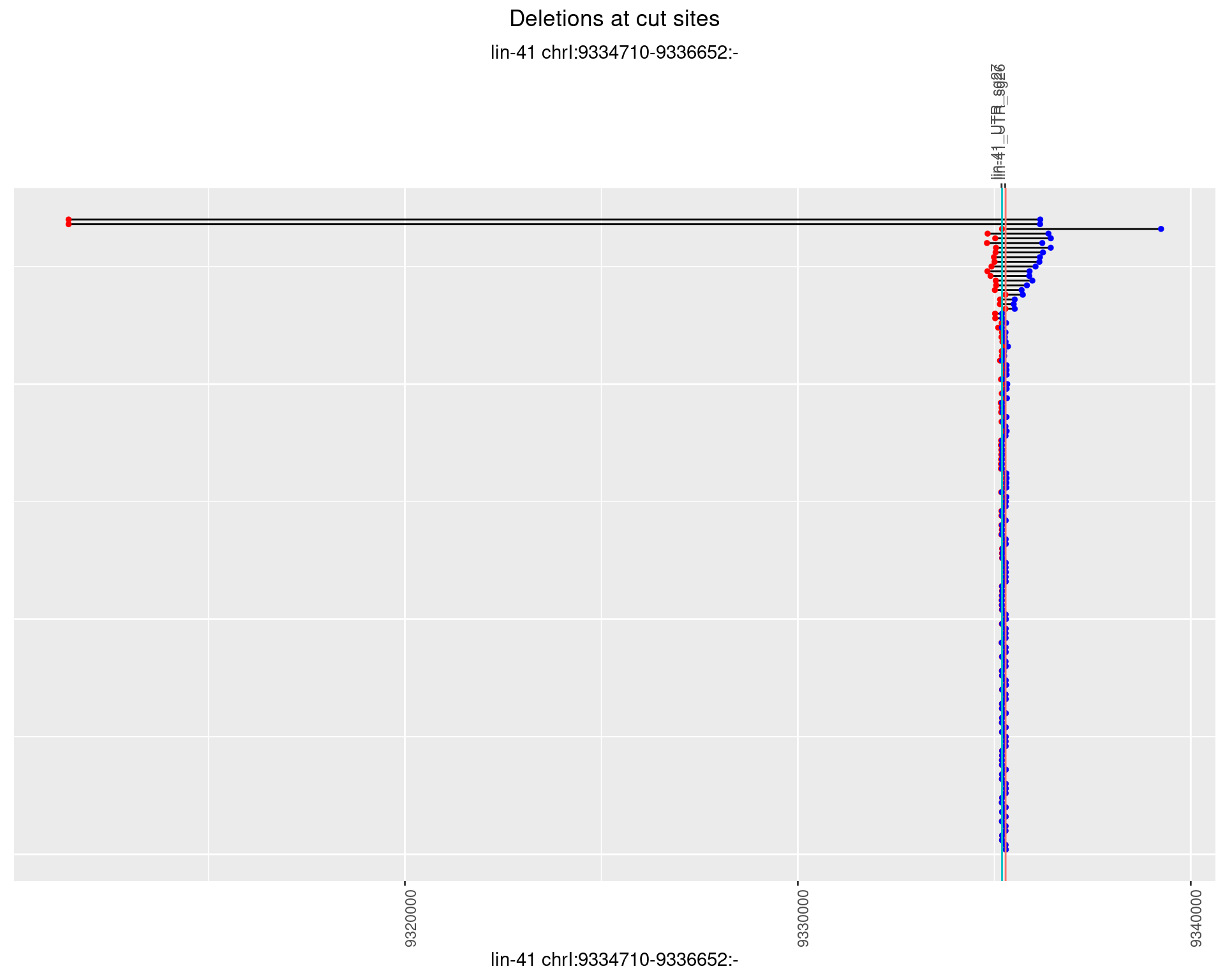
1.16.2 Freq: 1e-05
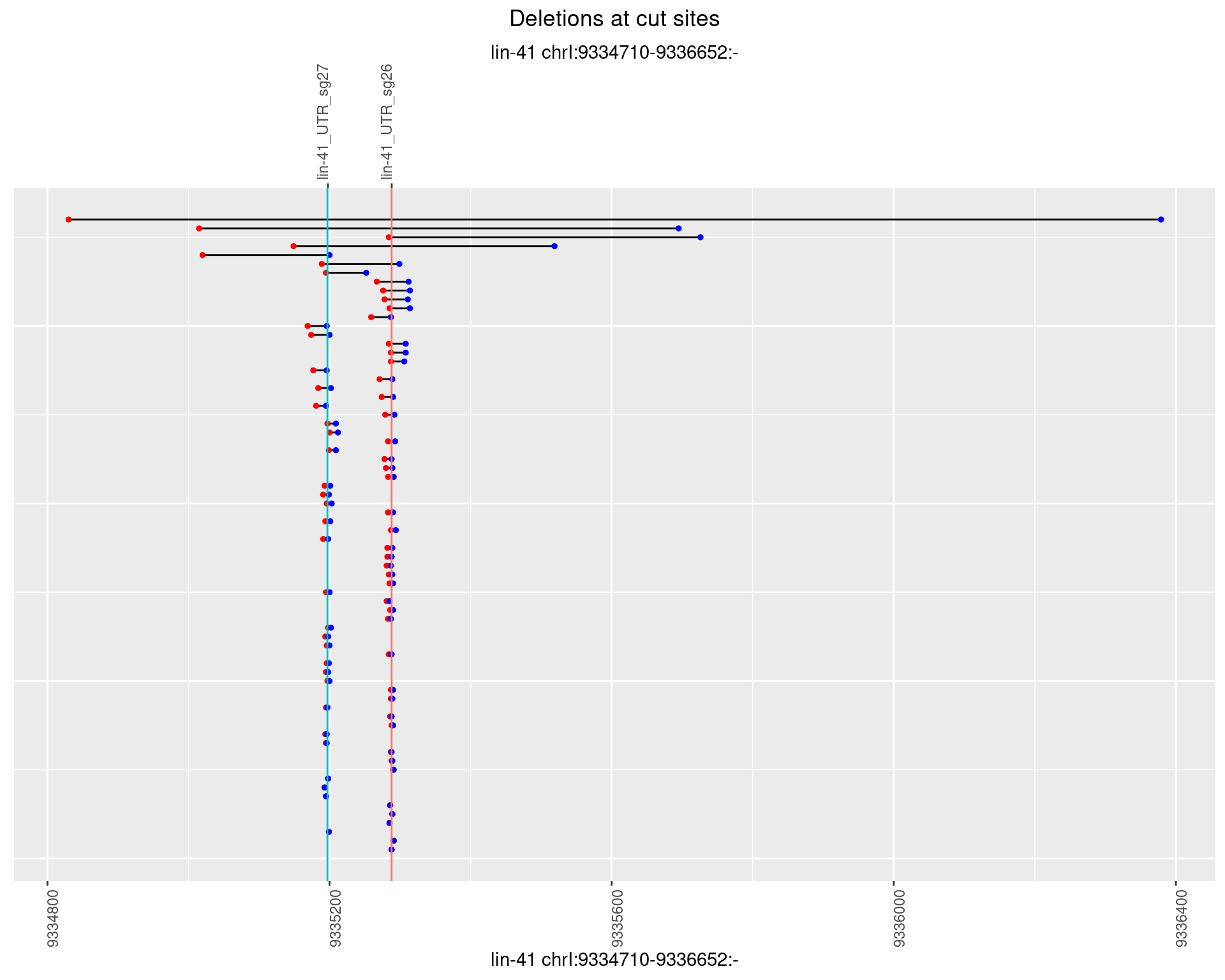
1.16.3 Freq: 1e-04
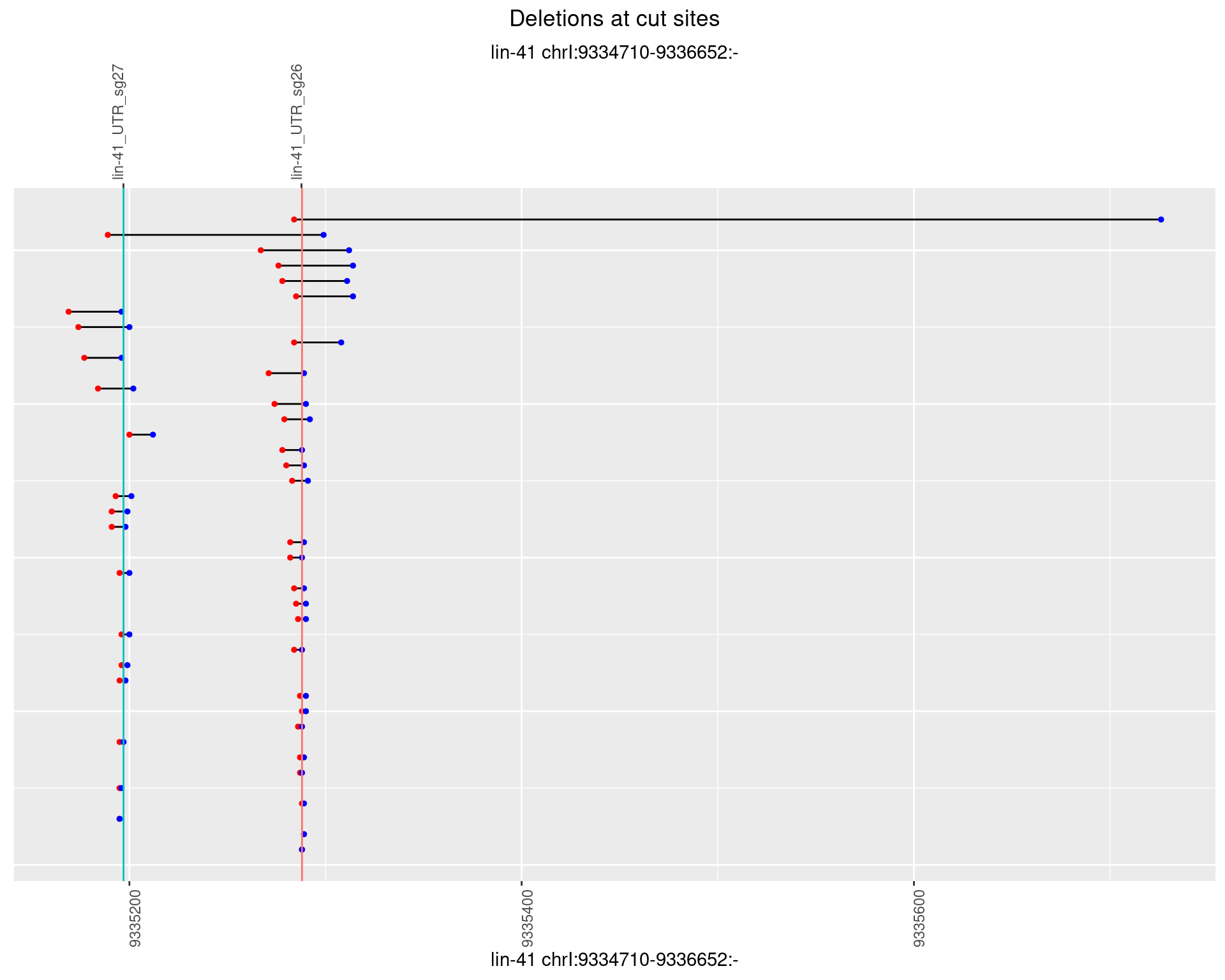
1.16.4 Freq: 0.001
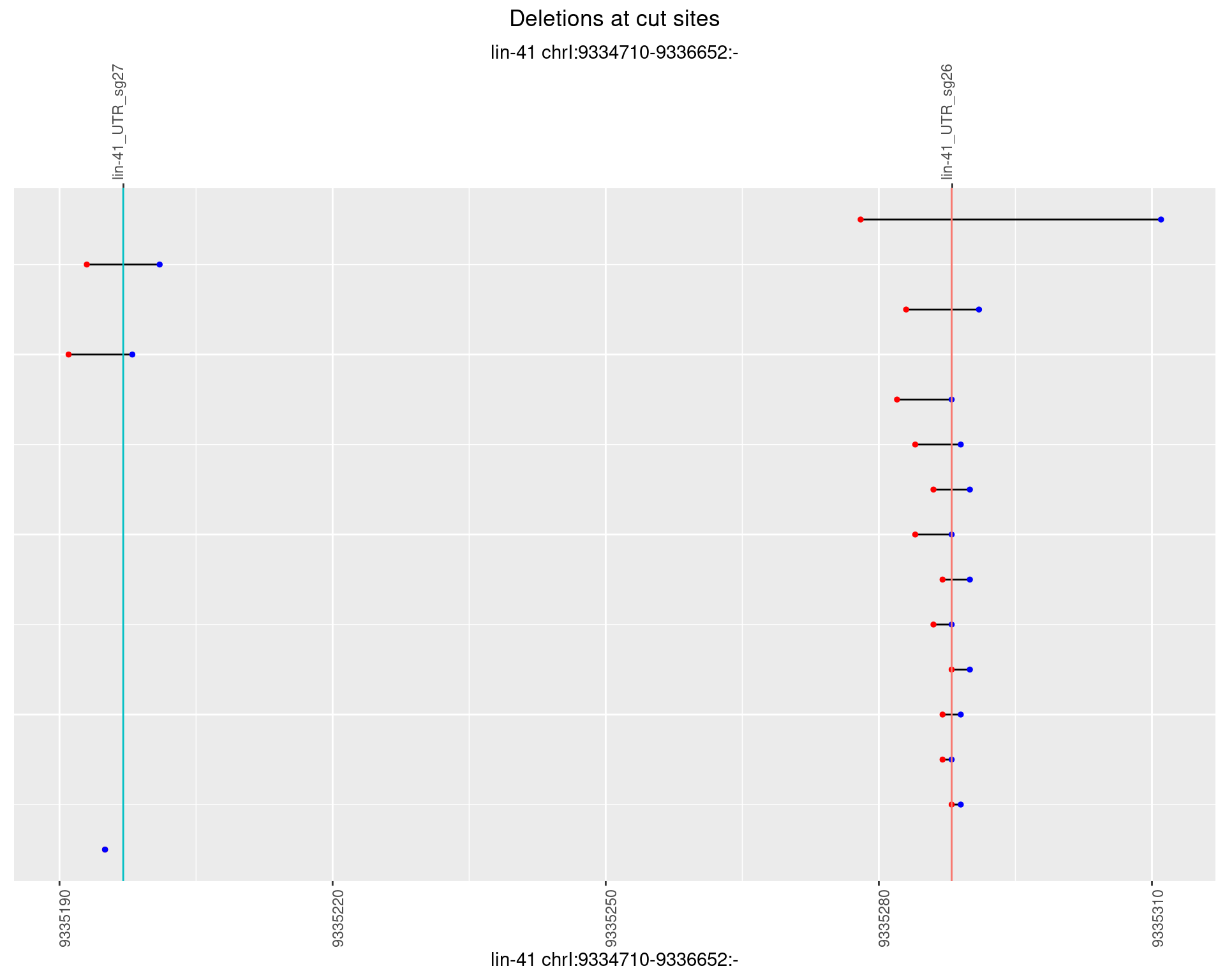
1.16.5 Freq: 0.01
1.16.6 Freq: 0.1
No plot to show
1.17 gen_24C_F5_lin-41_N2
1.17.1 Freq: 0
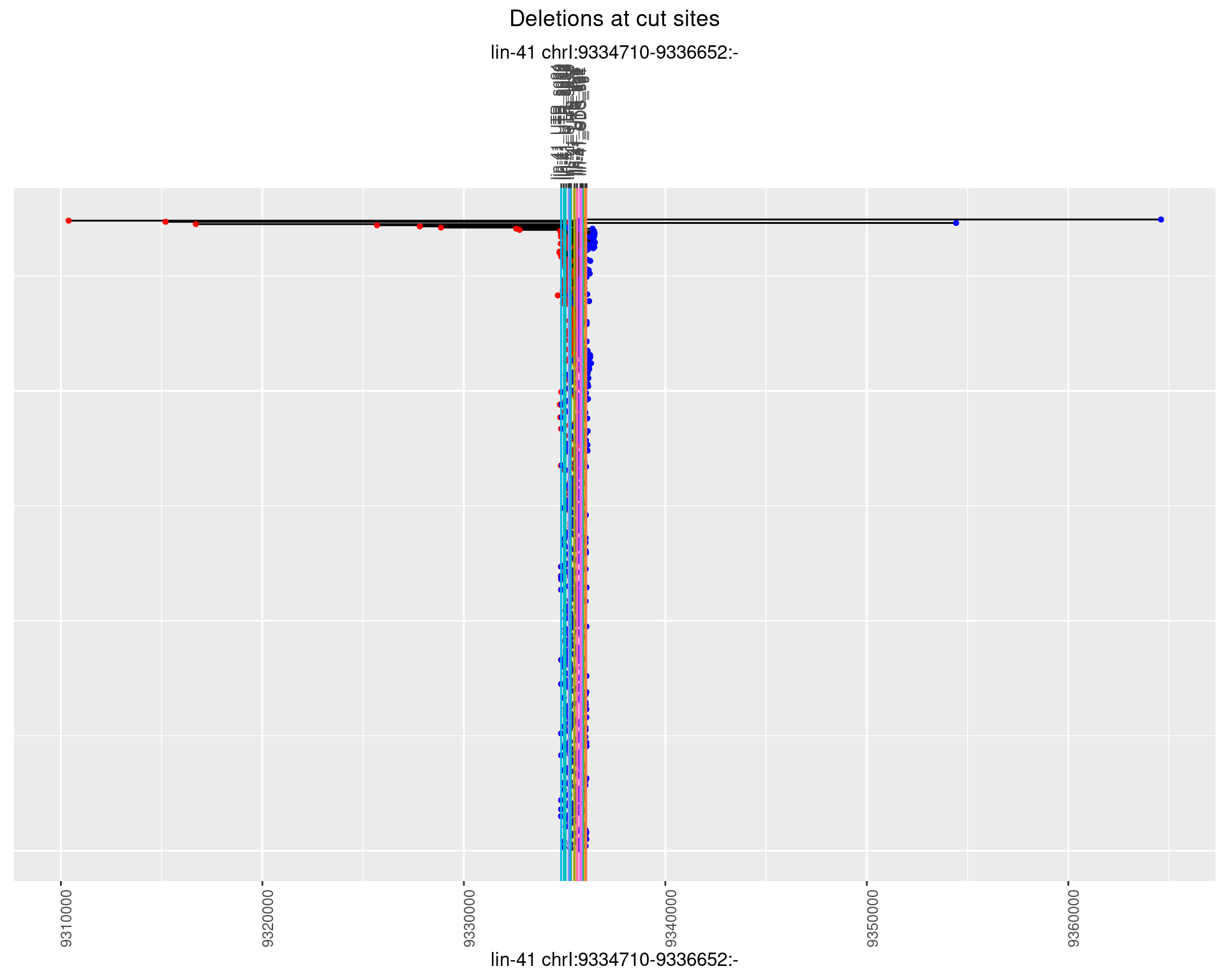
1.17.2 Freq: 1e-05
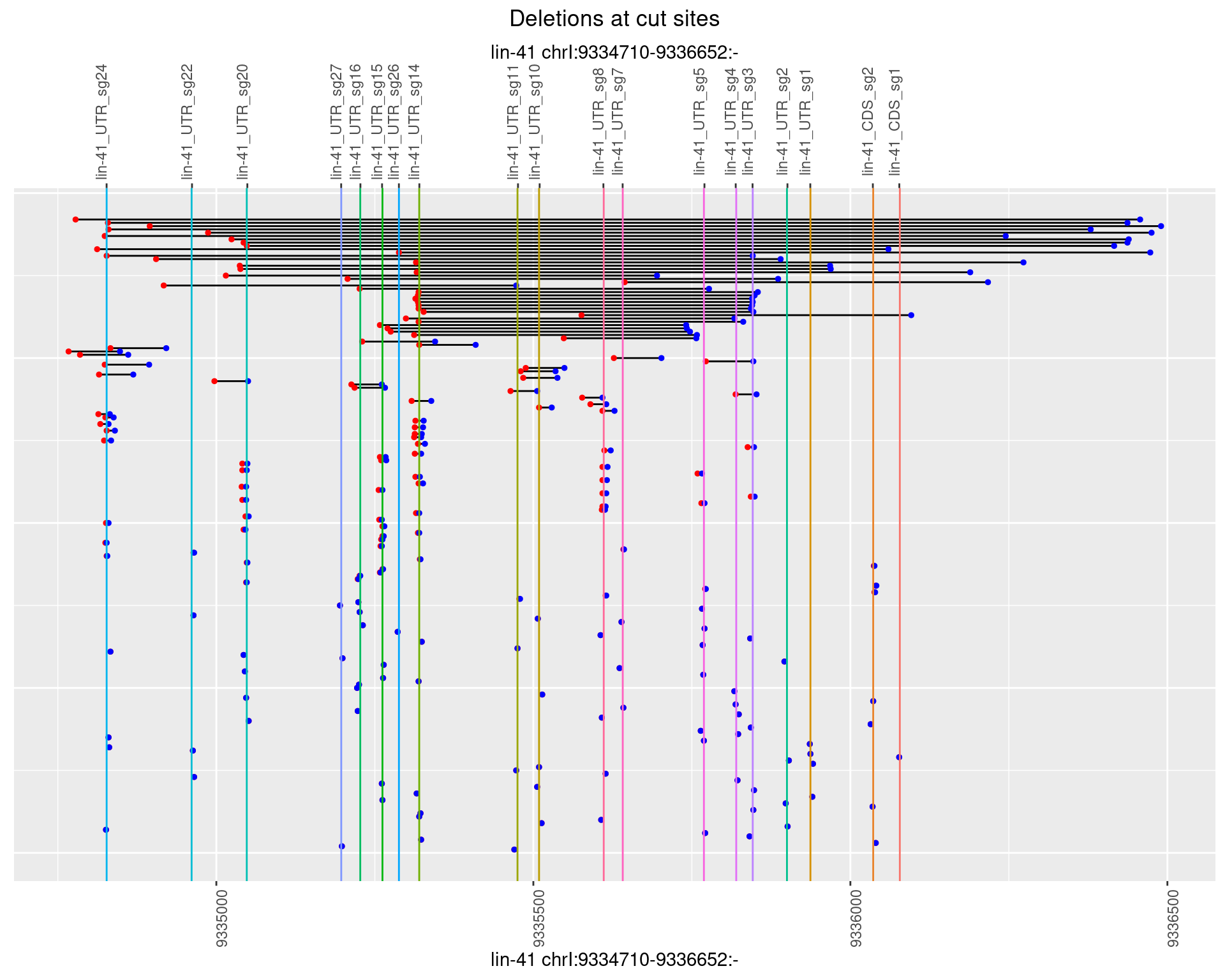
1.17.3 Freq: 1e-04
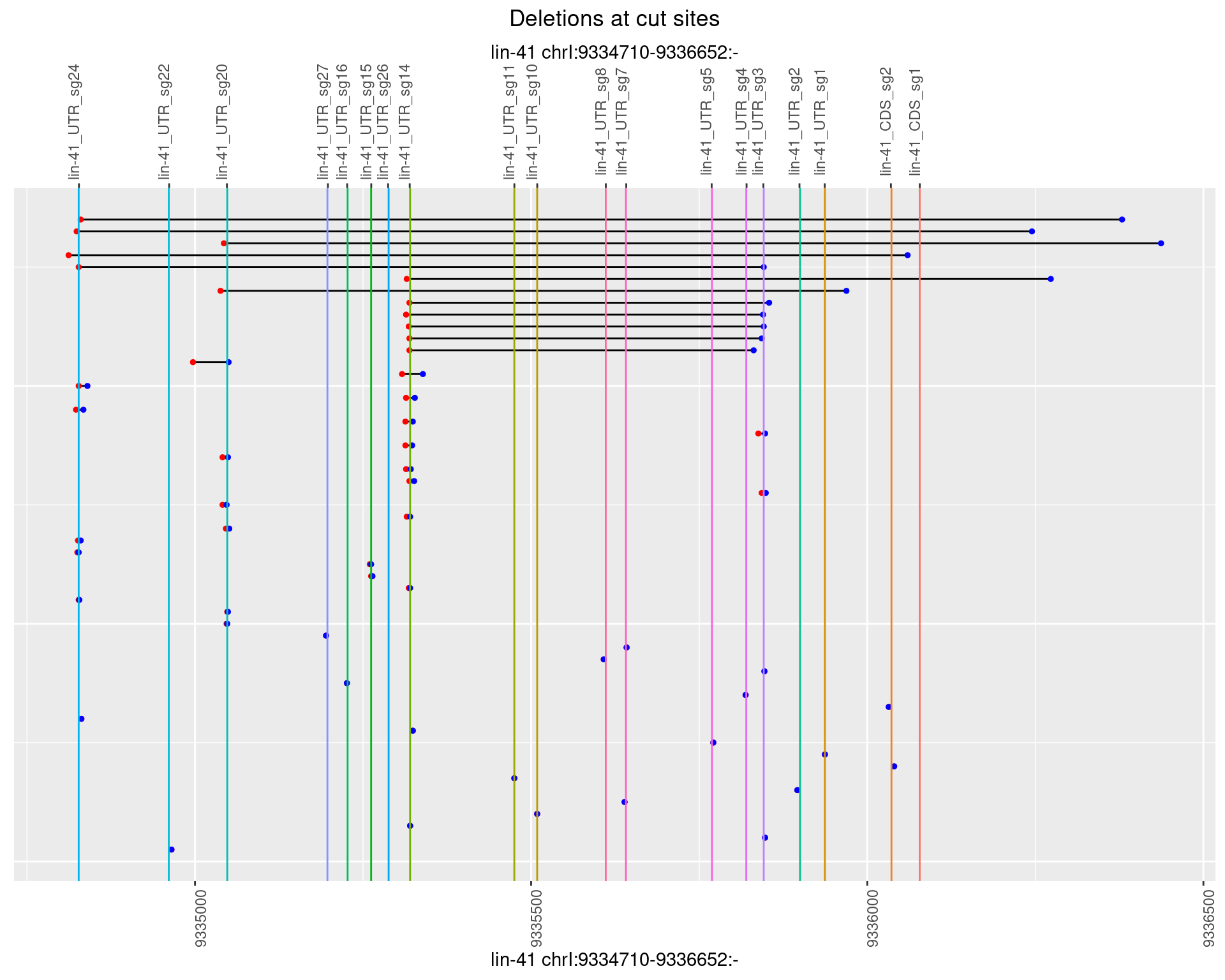
1.17.4 Freq: 0.001
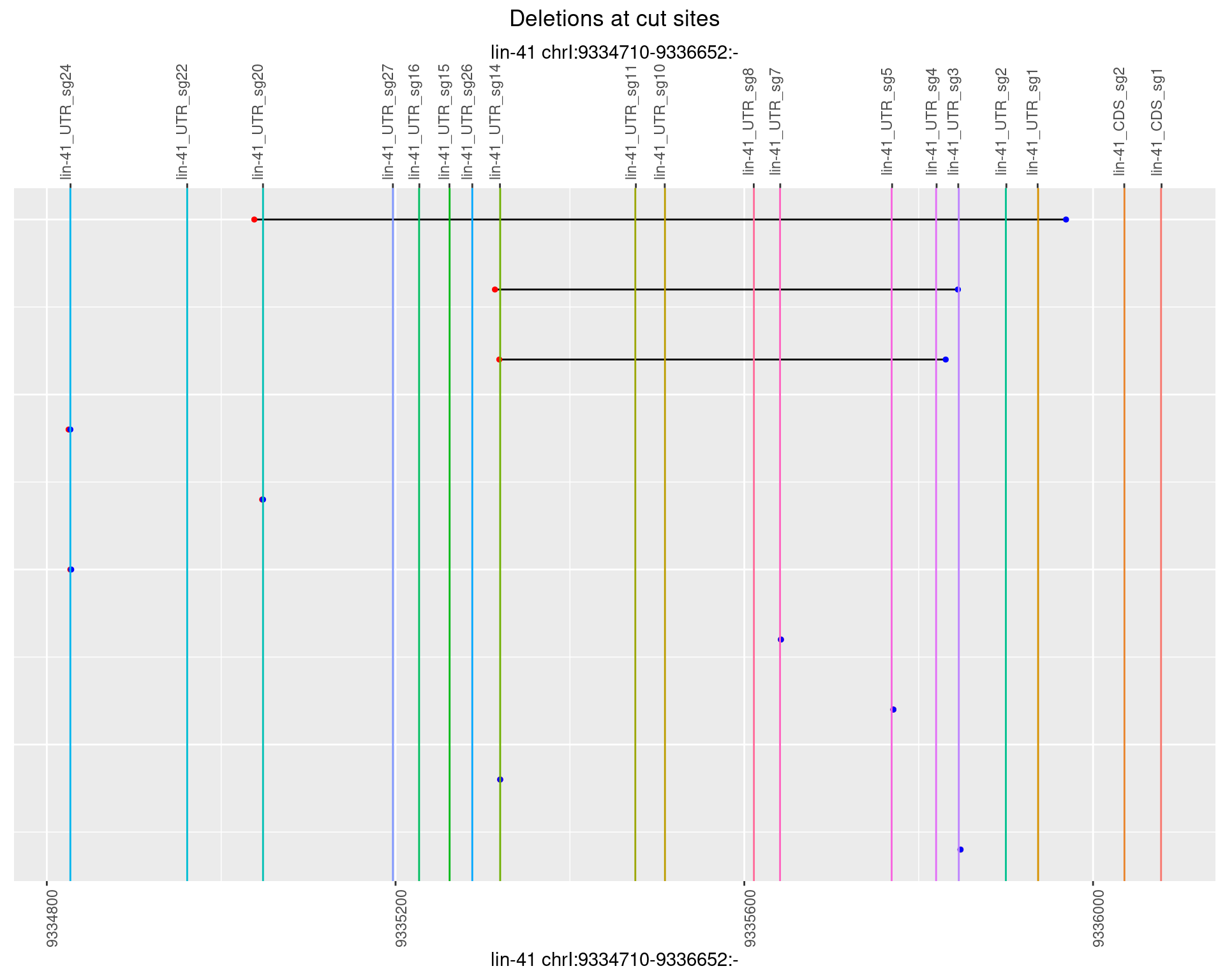
1.17.5 Freq: 0.01
No plot to show
1.17.6 Freq: 0.1
No plot to show
1.18 gen_24C_F5_lin-41_sg15sg16
1.18.1 Freq: 0
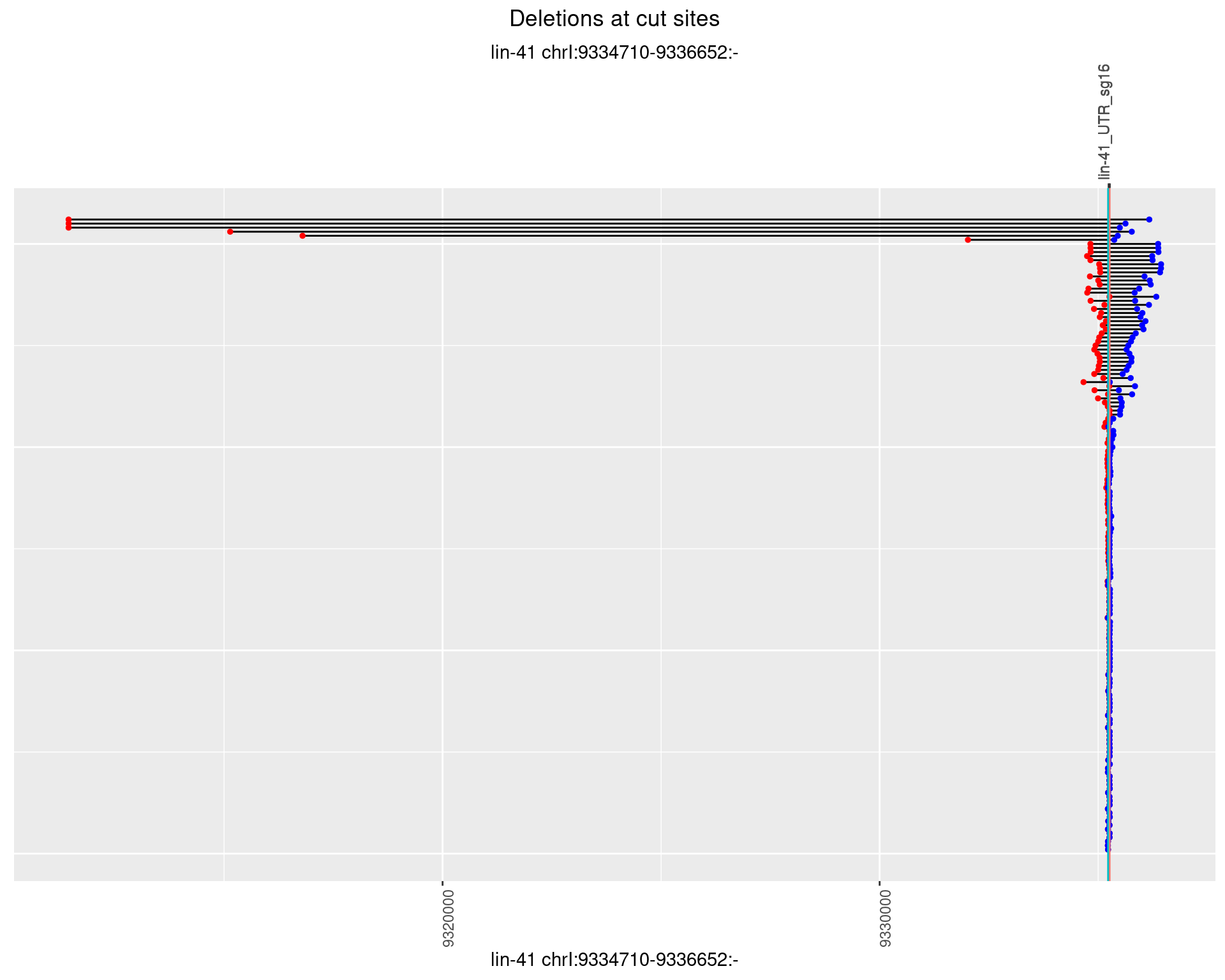
1.18.2 Freq: 1e-05
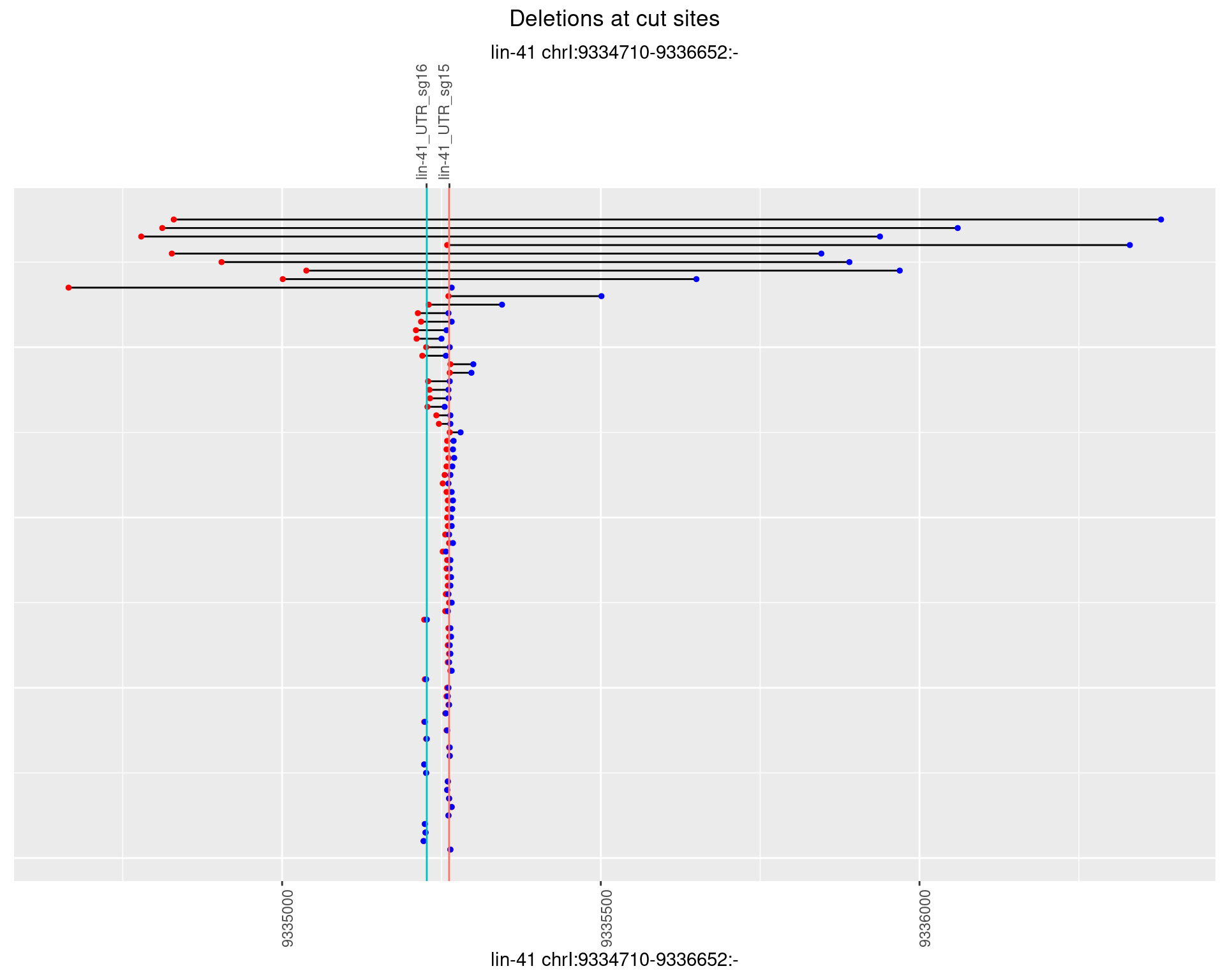
1.18.3 Freq: 1e-04
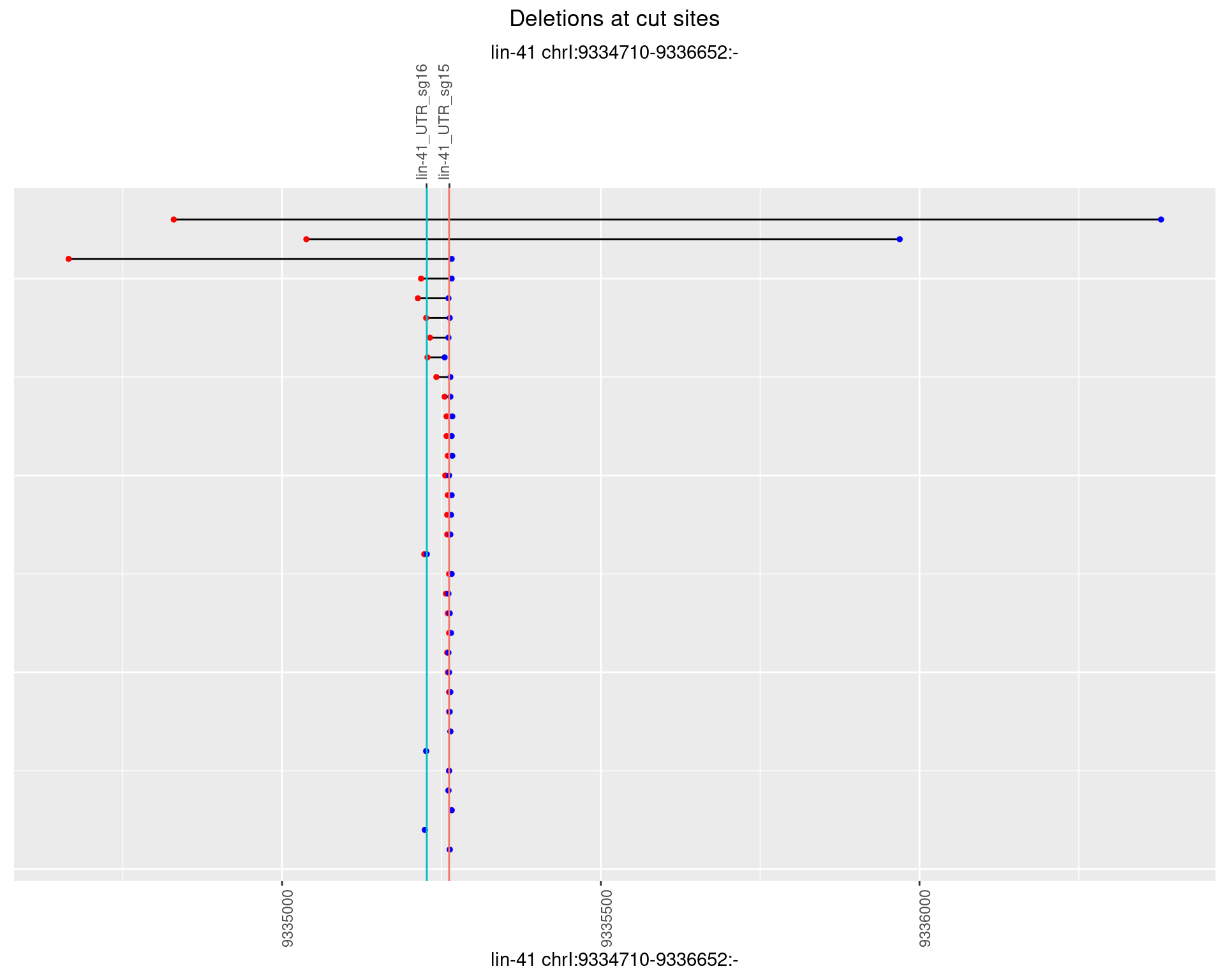
1.18.4 Freq: 0.001
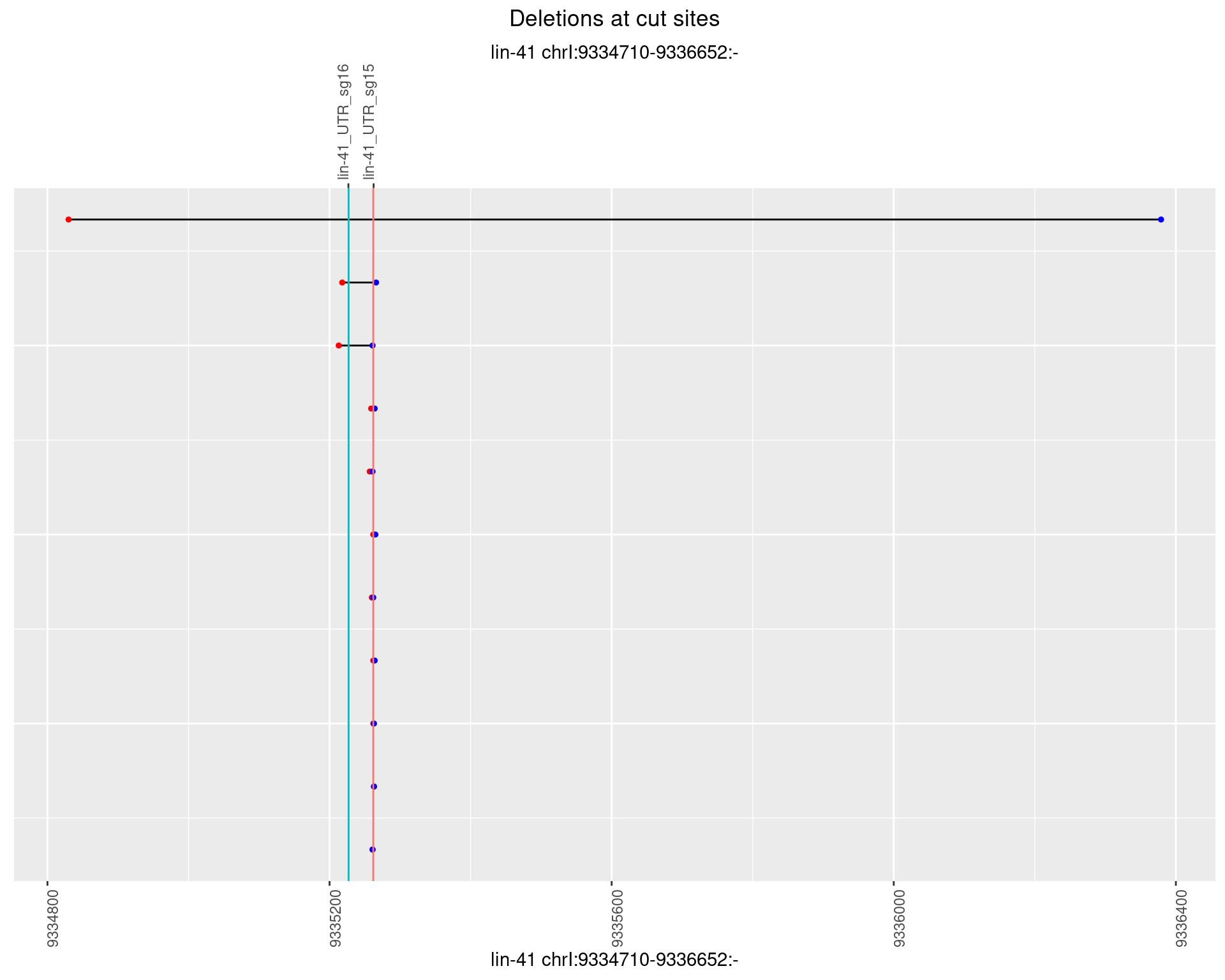
1.18.5 Freq: 0.01
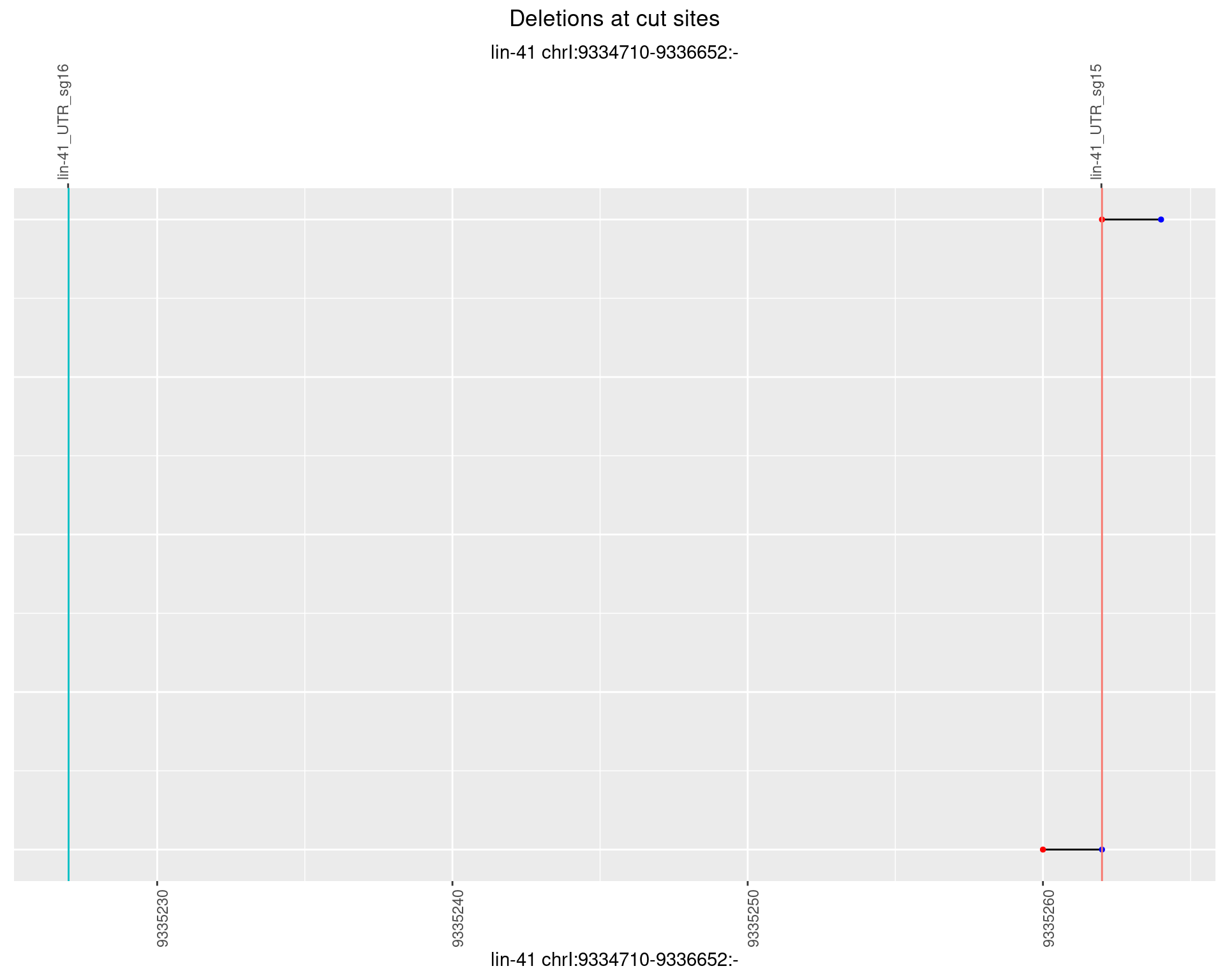
1.18.6 Freq: 0.1
No plot to show
1.19 gen_24C_F5_lin-41_pool
1.19.1 Freq: 0
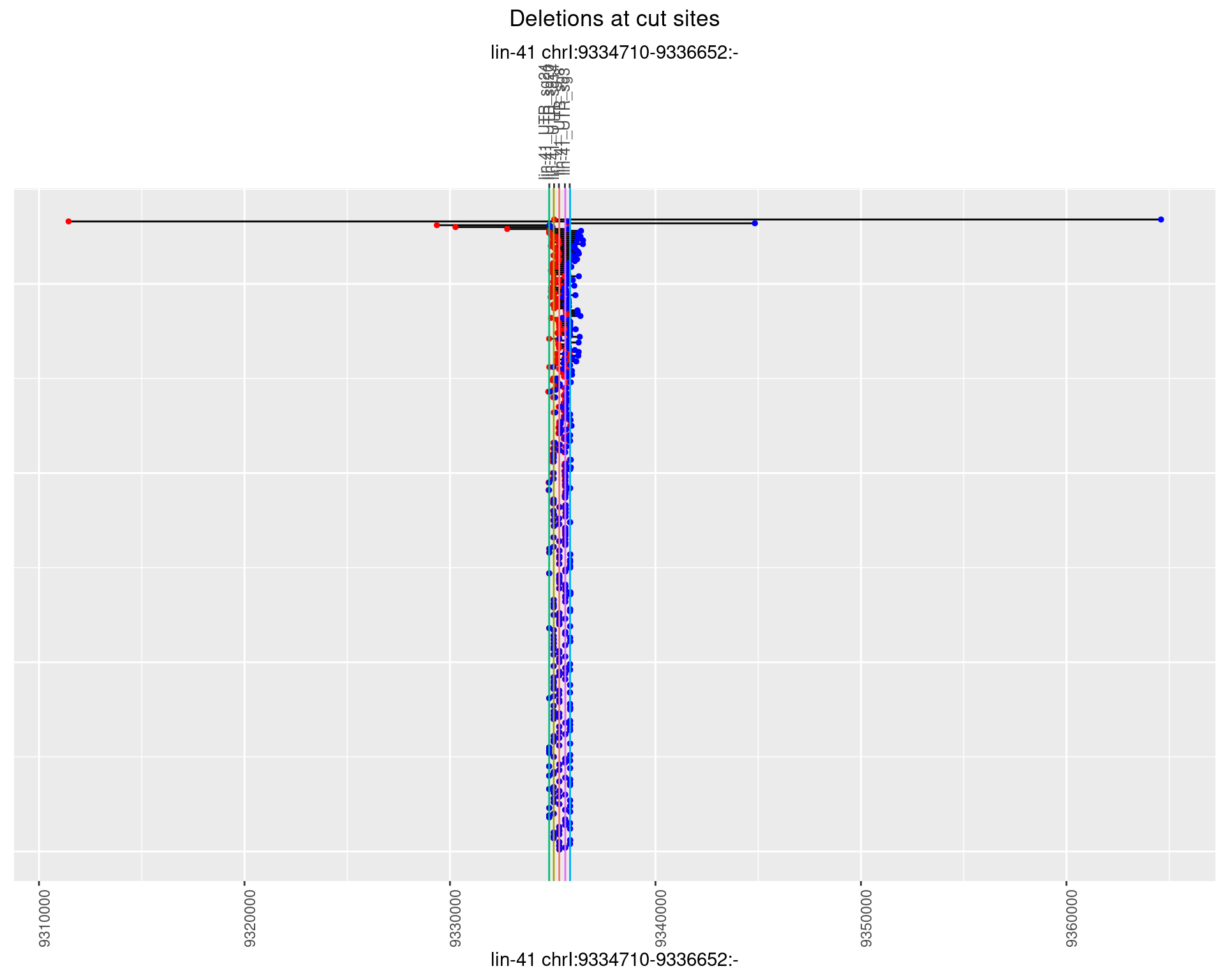
1.19.2 Freq: 1e-05
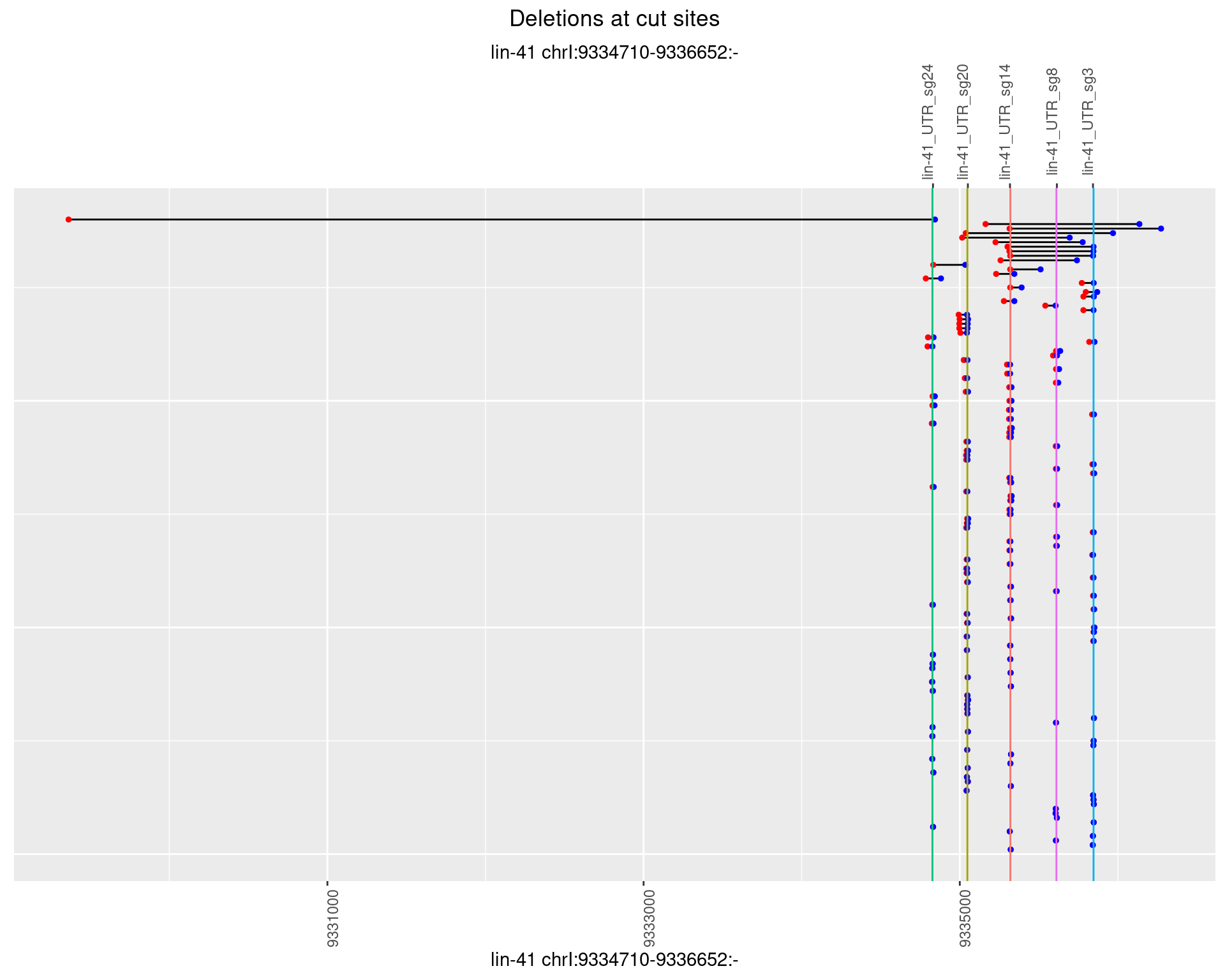
1.19.3 Freq: 1e-04
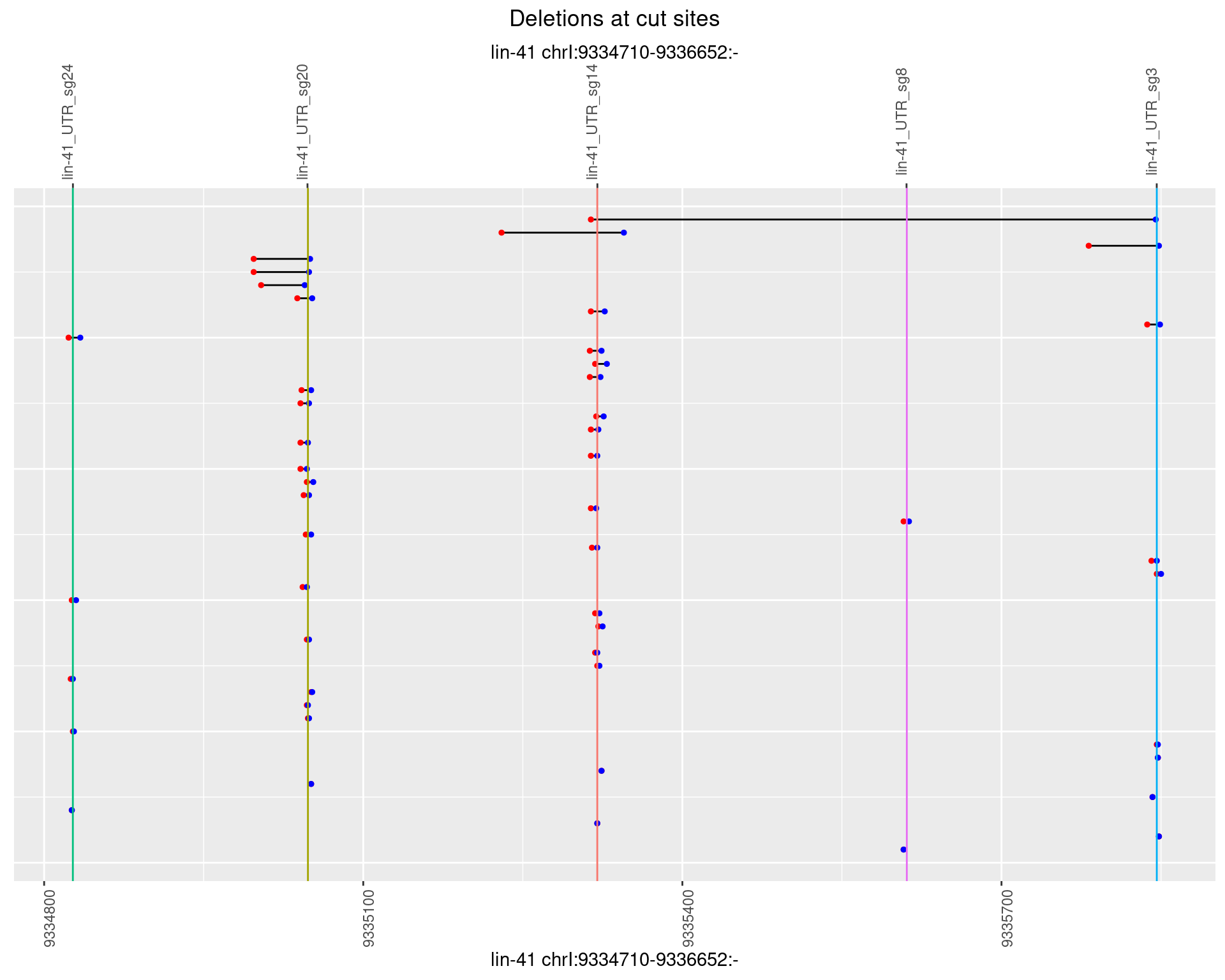
1.19.4 Freq: 0.001
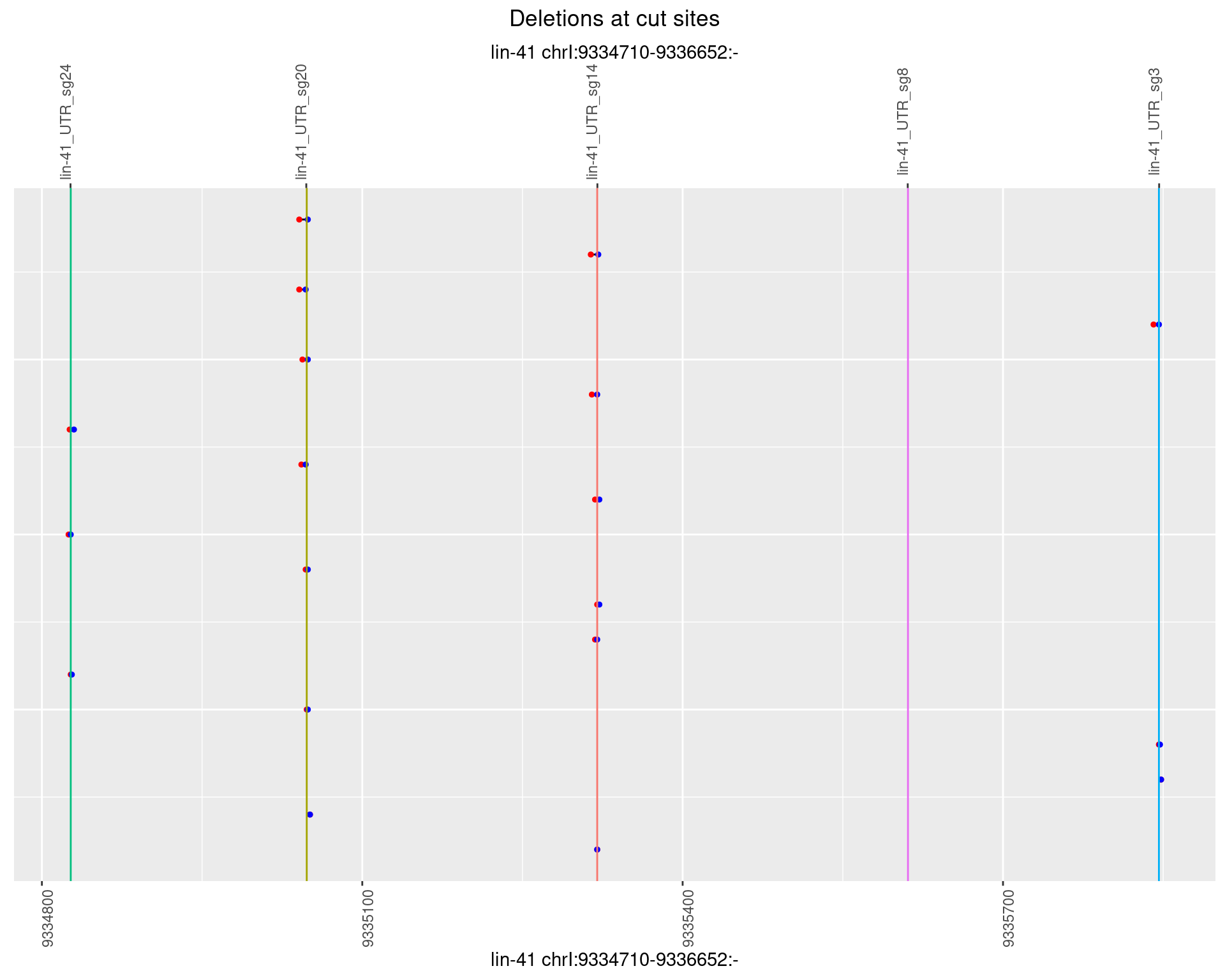
1.19.5 Freq: 0.01
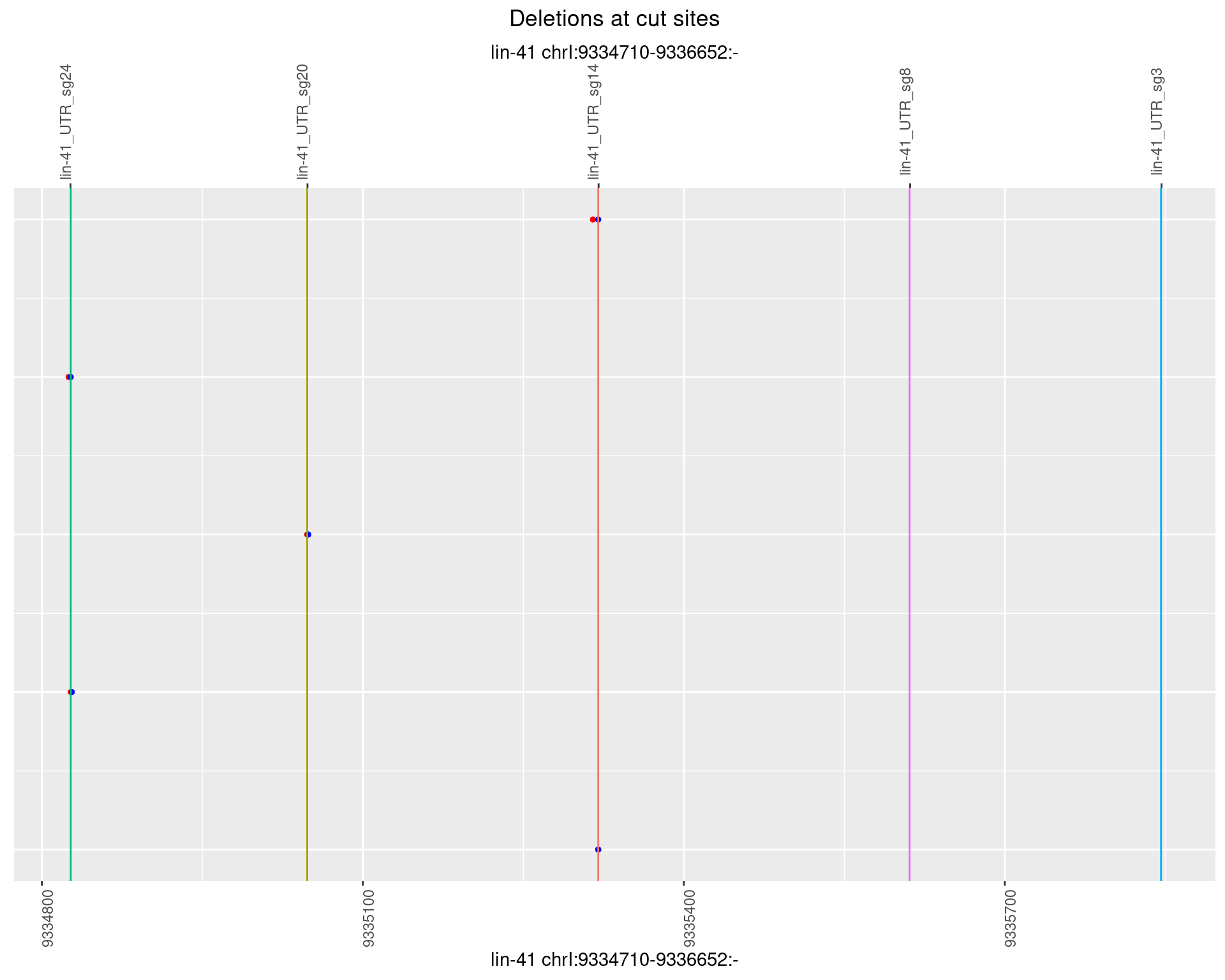
1.19.6 Freq: 0.1
No plot to show
1.20 gen_24C_F5_lin-41_sg26sg27
1.20.1 Freq: 0
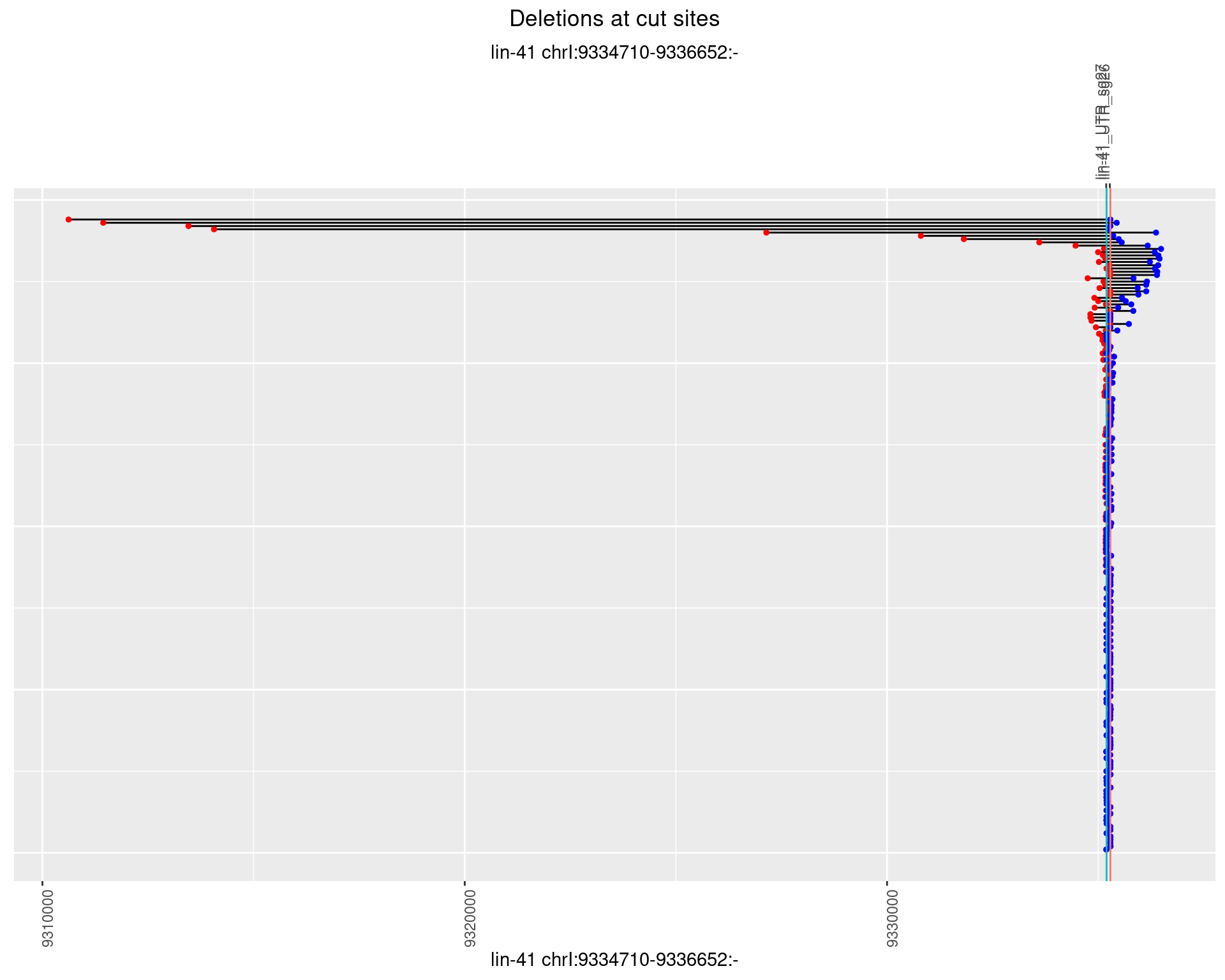
1.20.2 Freq: 1e-05
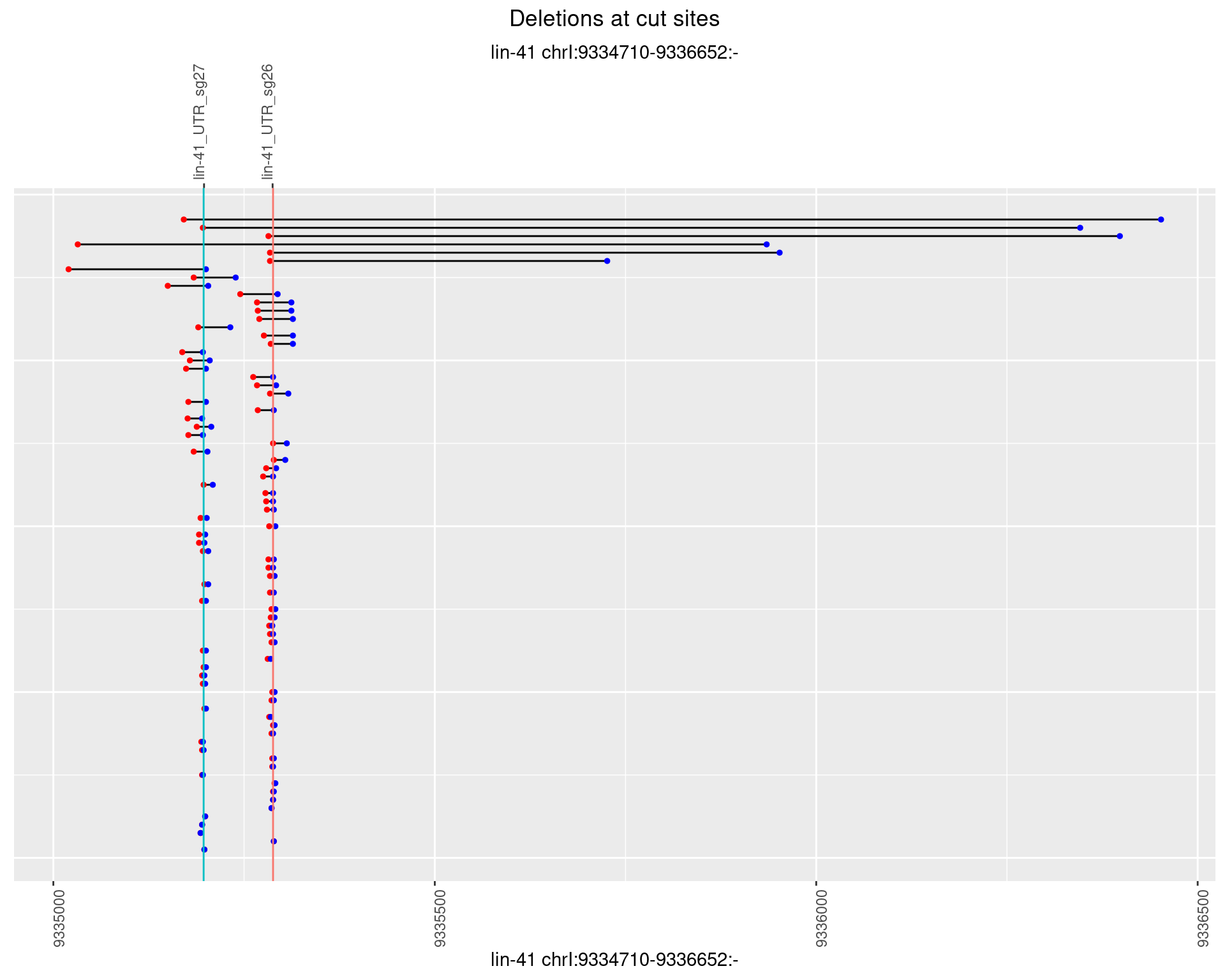
1.20.3 Freq: 1e-04
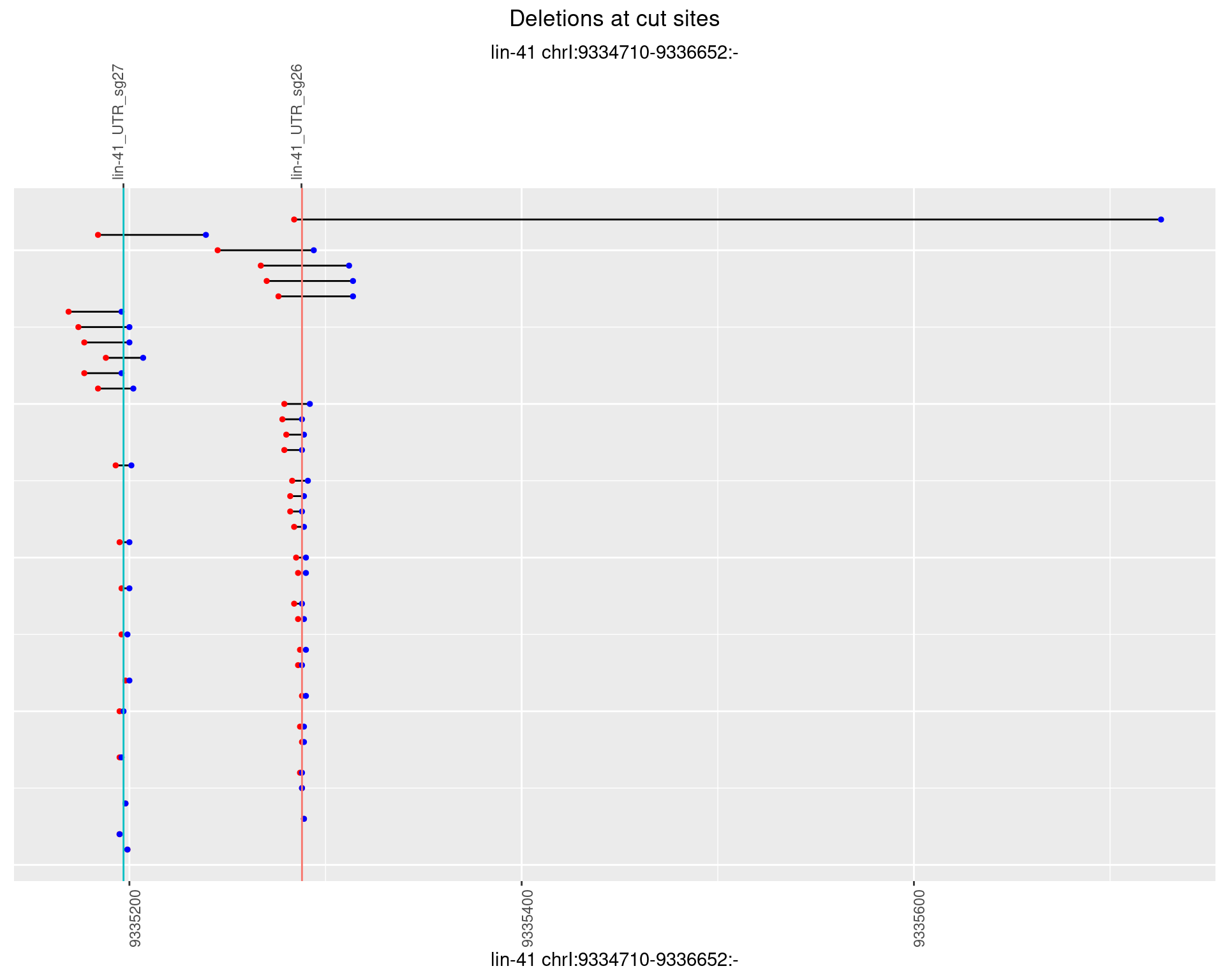
1.20.4 Freq: 0.001
1.20.5 Freq: 0.01
No plot to show
1.20.6 Freq: 0.1
No plot to show
1.21 gen_16C_F2_lin-41_N2
1.21.1 Freq: 0
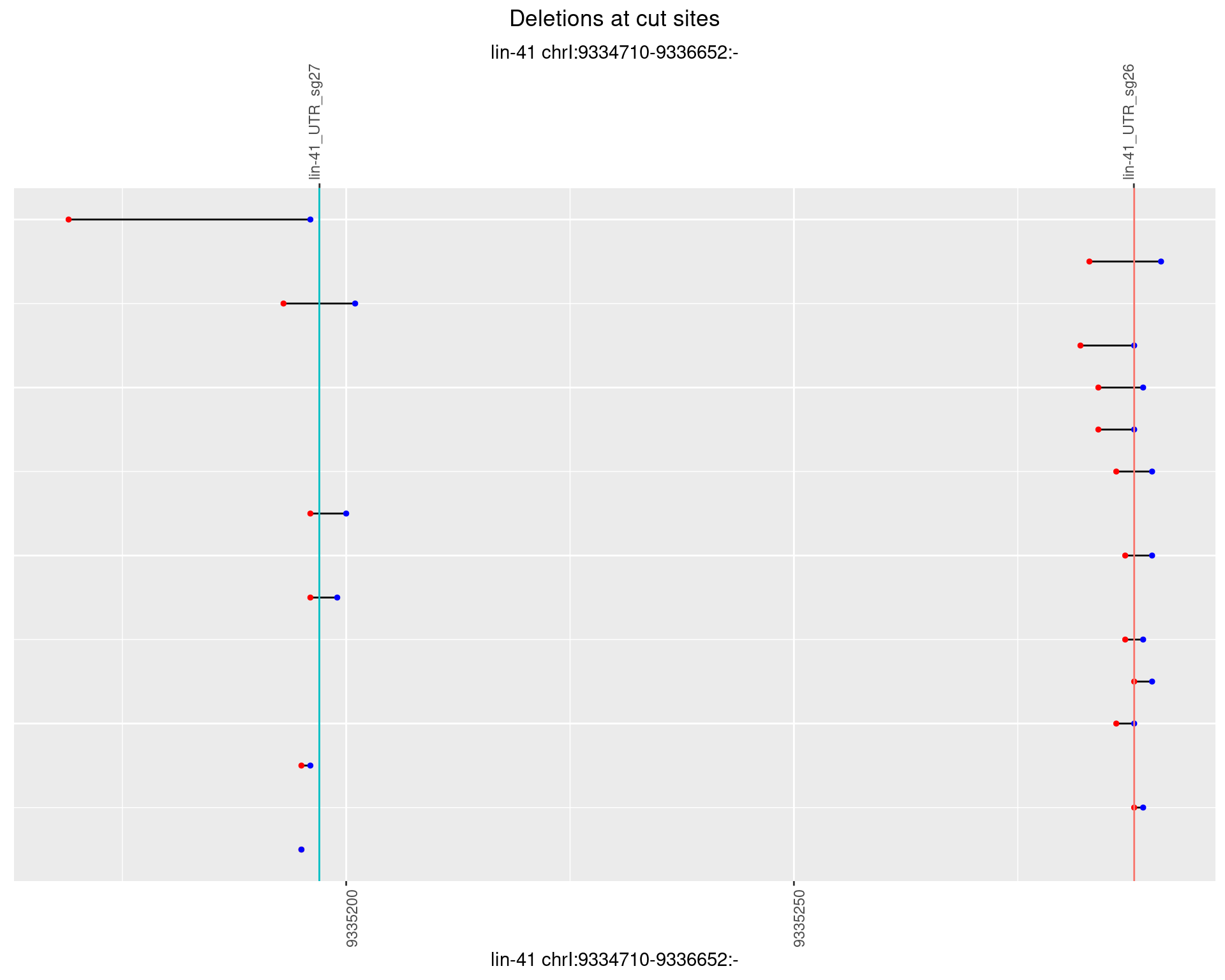
1.21.2 Freq: 1e-05
1.21.3 Freq: 1e-04
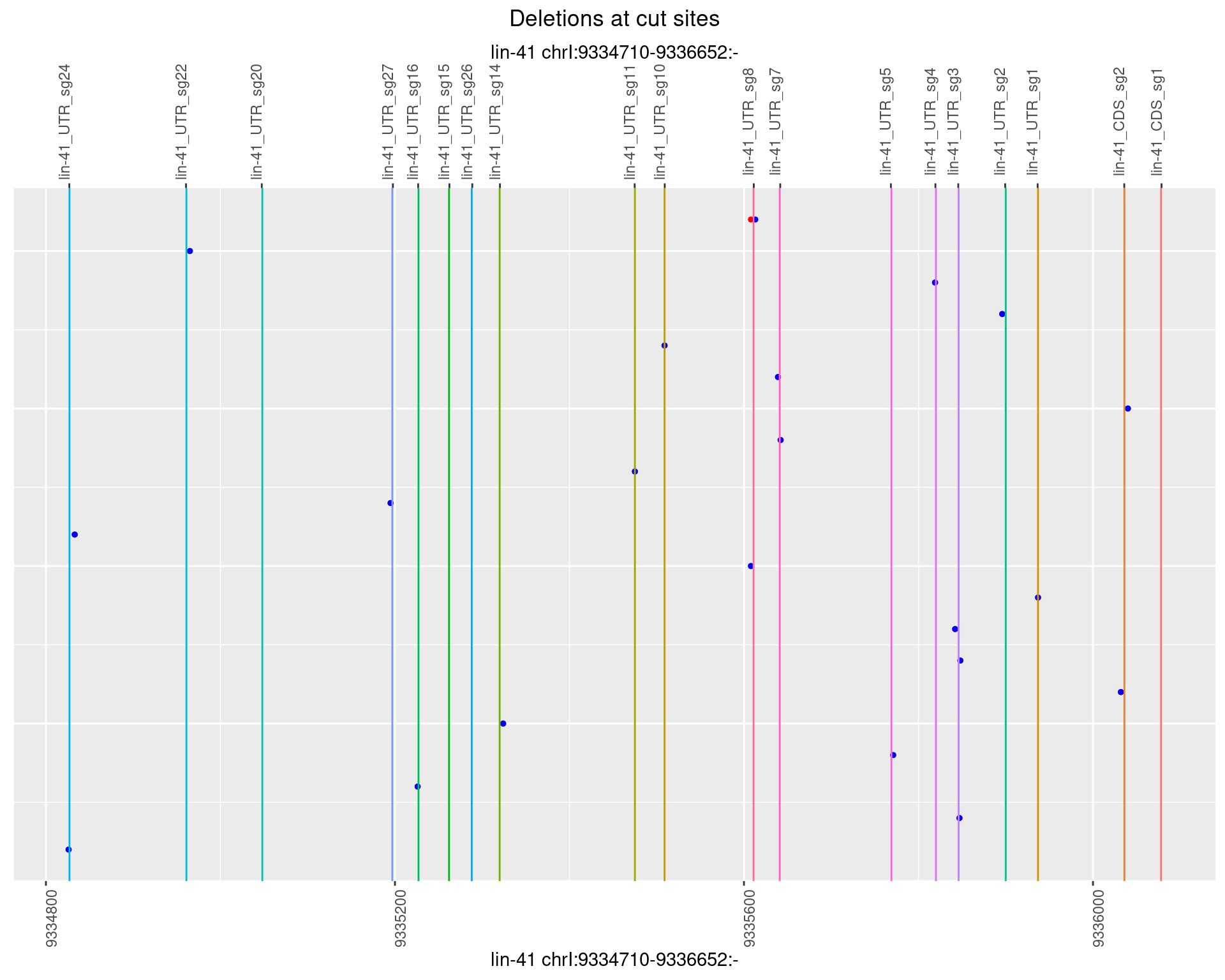
1.21.4 Freq: 0.001
1.21.5 Freq: 0.01
No plot to show
1.21.6 Freq: 0.1
No plot to show
1.22 gen_16C_F2_lin-41_sg15sg16
1.22.1 Freq: 0
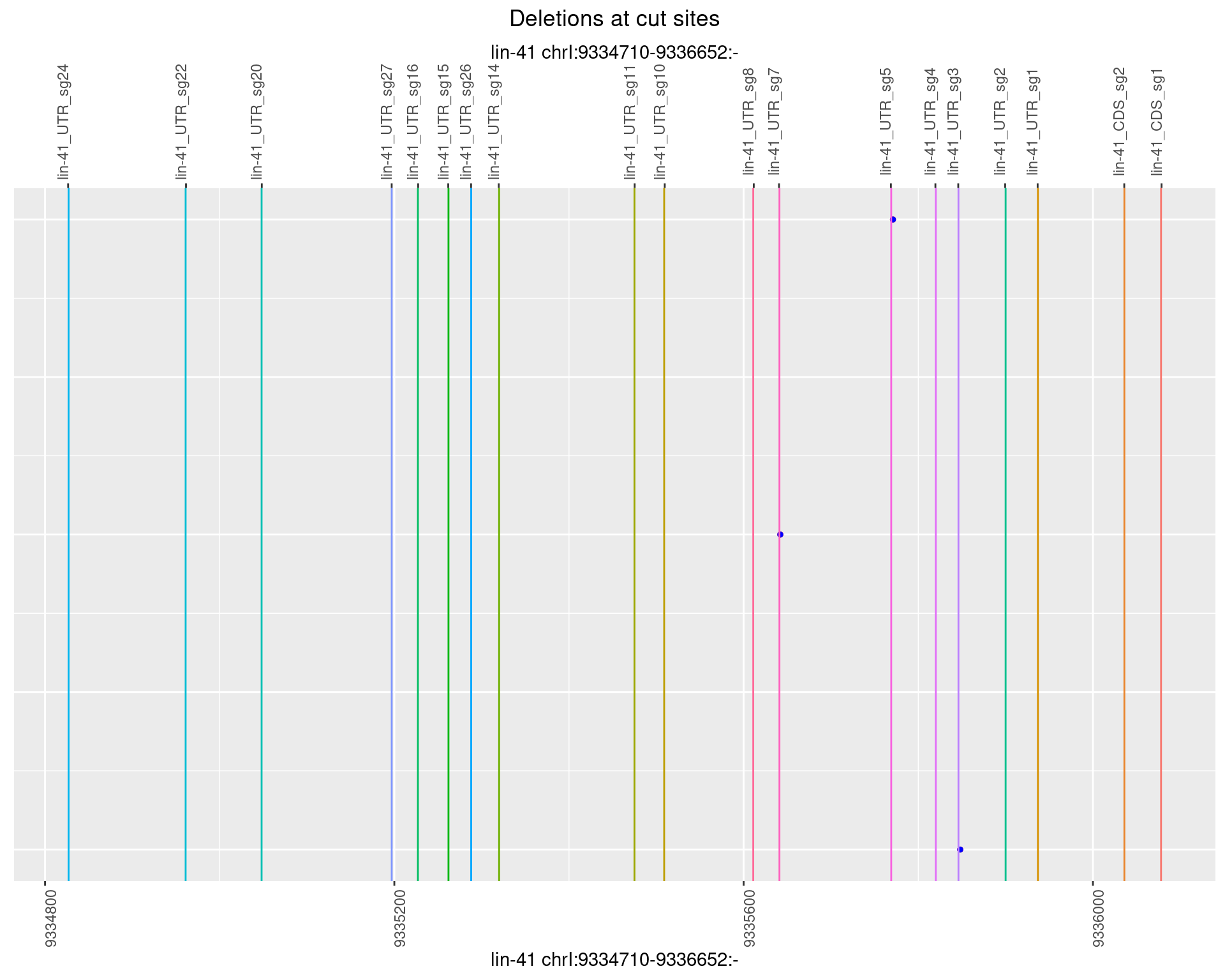
1.22.2 Freq: 1e-05
1.22.3 Freq: 1e-04
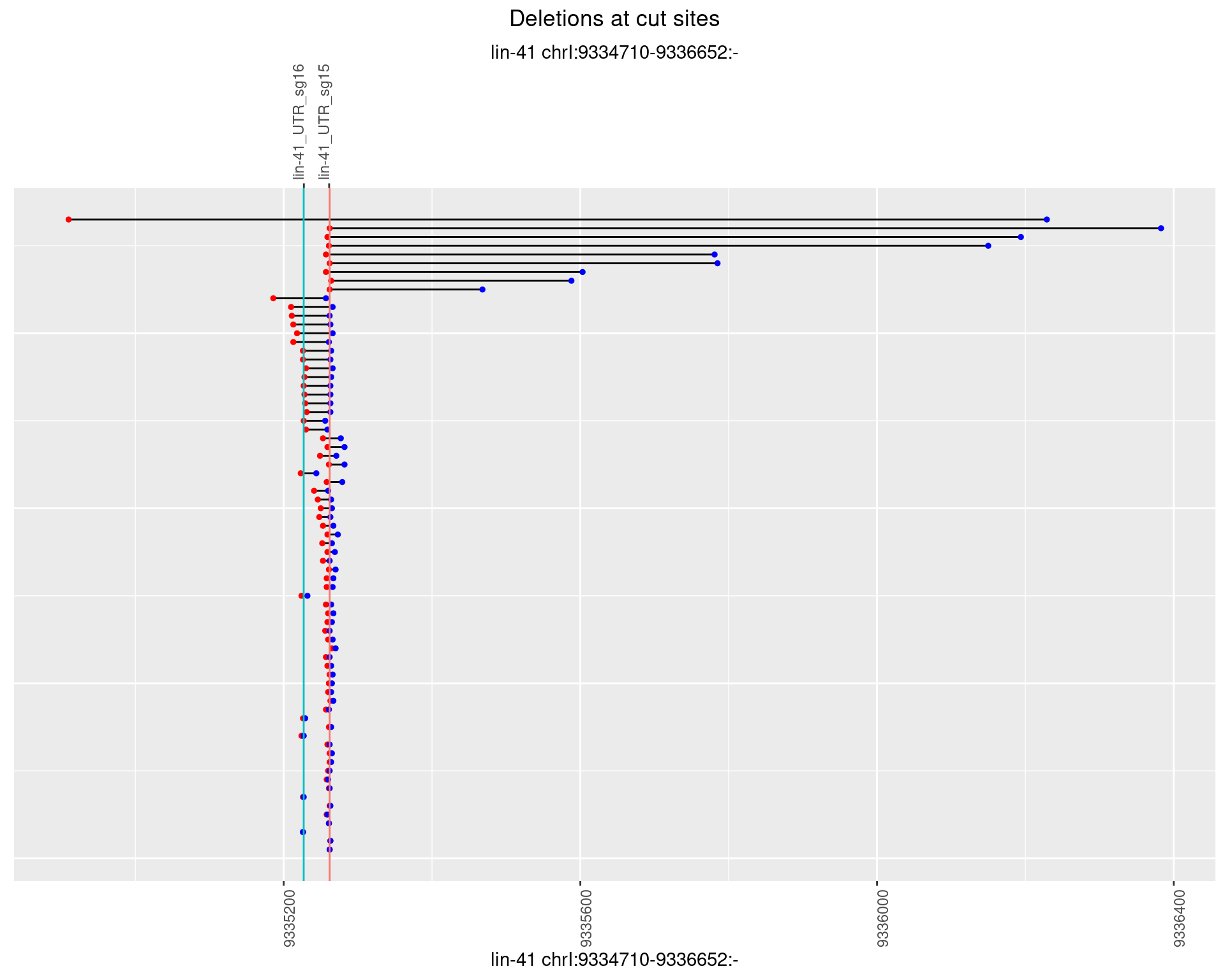
1.22.4 Freq: 0.001
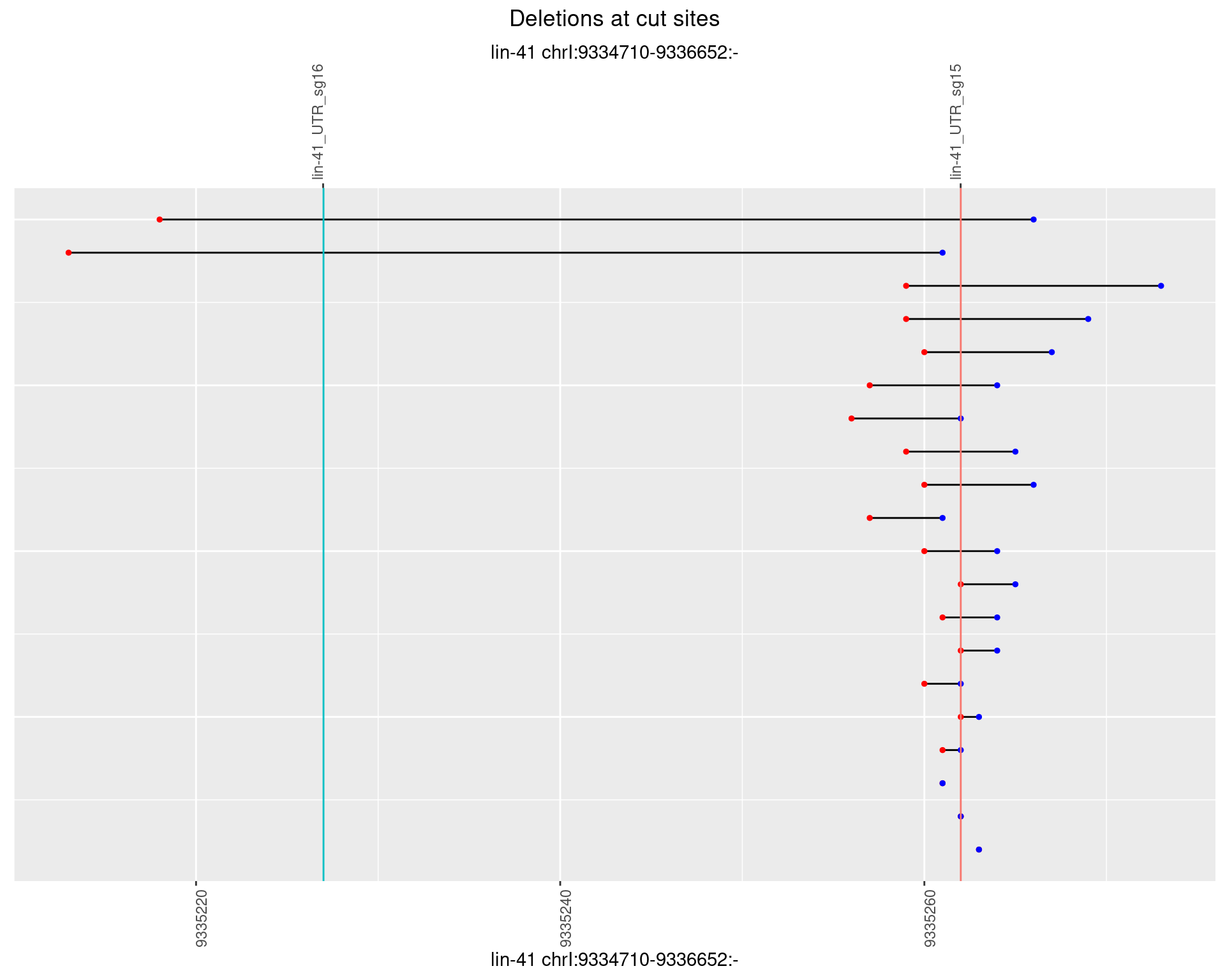
1.22.5 Freq: 0.01
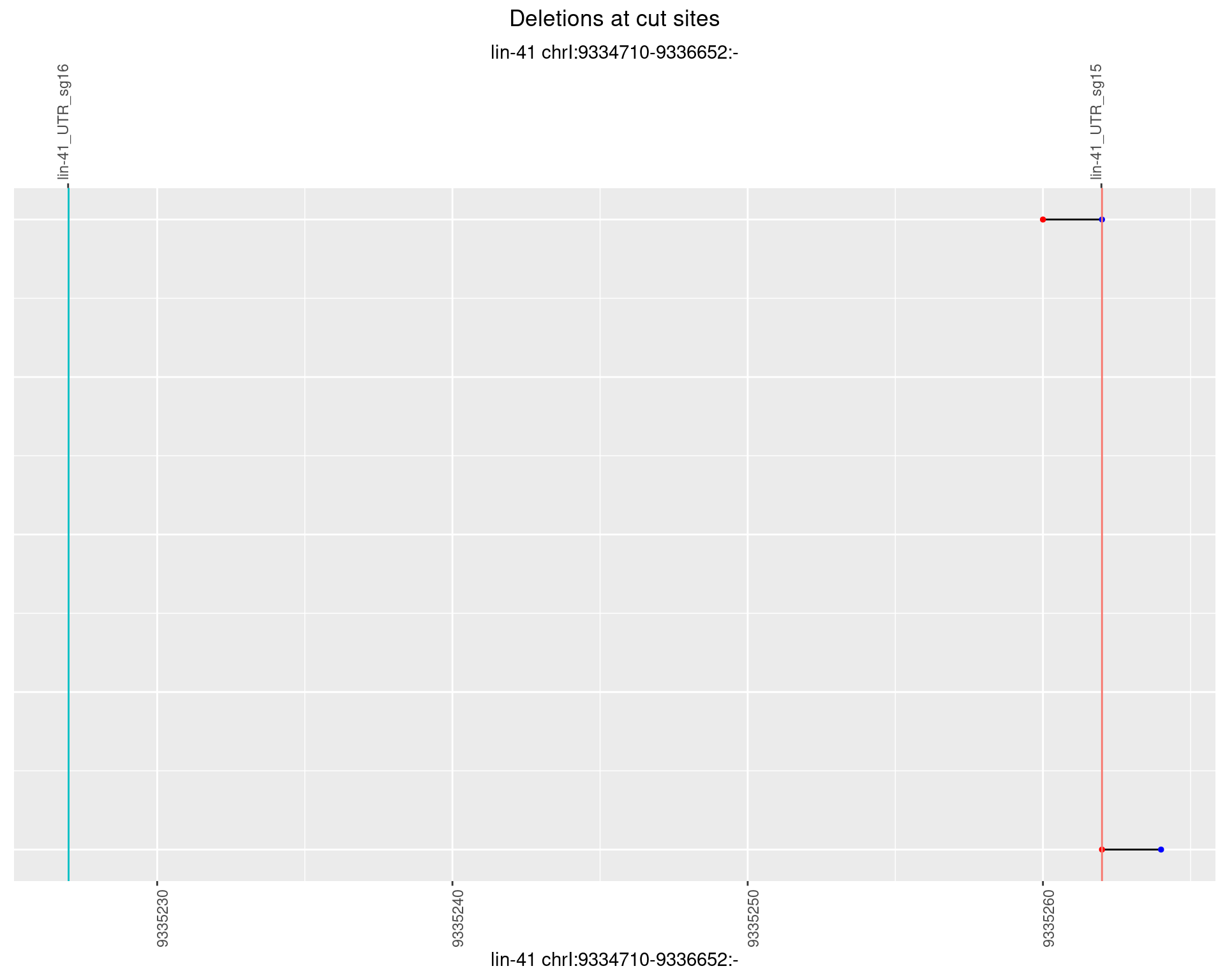
1.22.6 Freq: 0.1
No plot to show
1.23 gen_16C_F2_lin-41_pool
1.23.1 Freq: 0
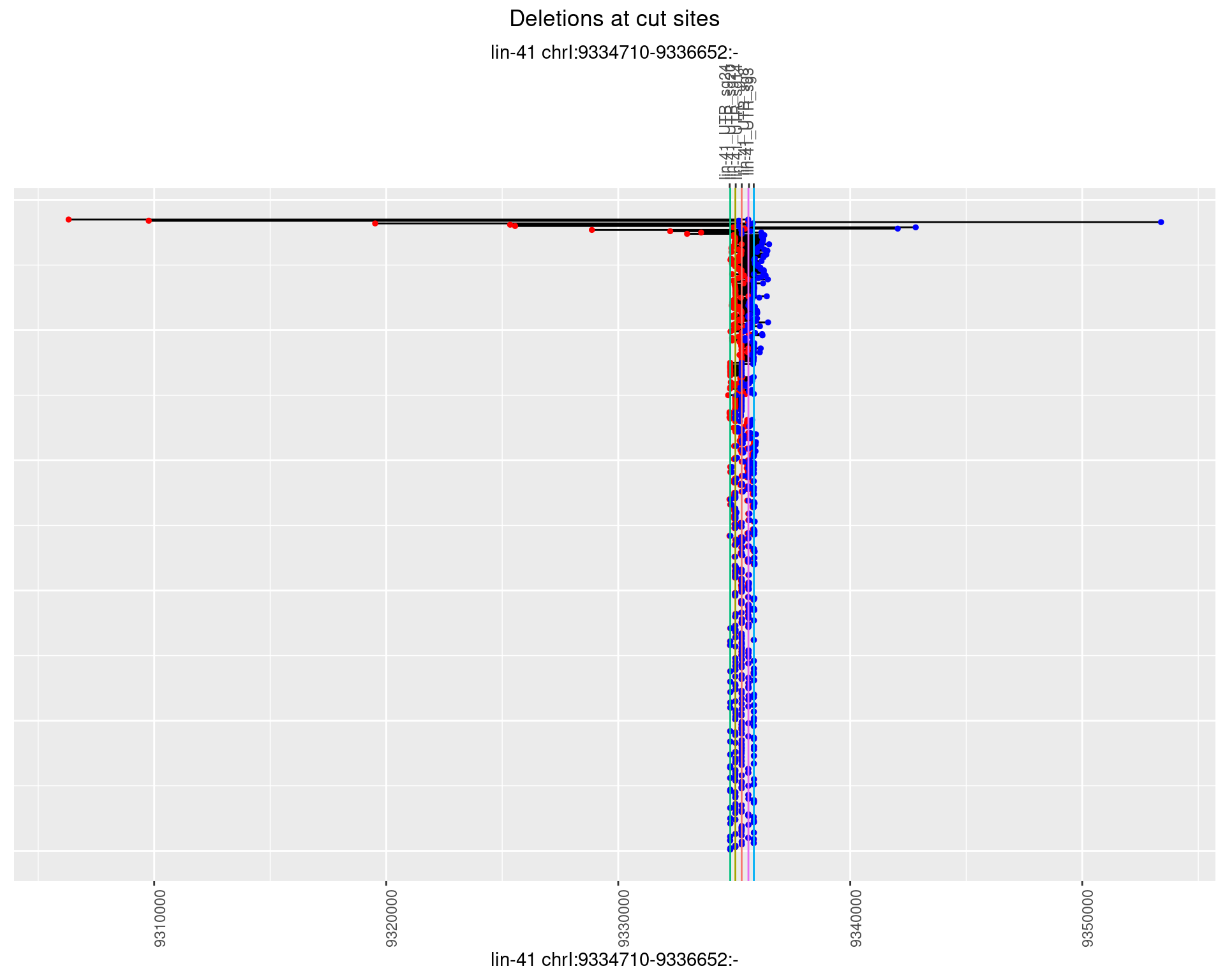
1.23.2 Freq: 1e-05
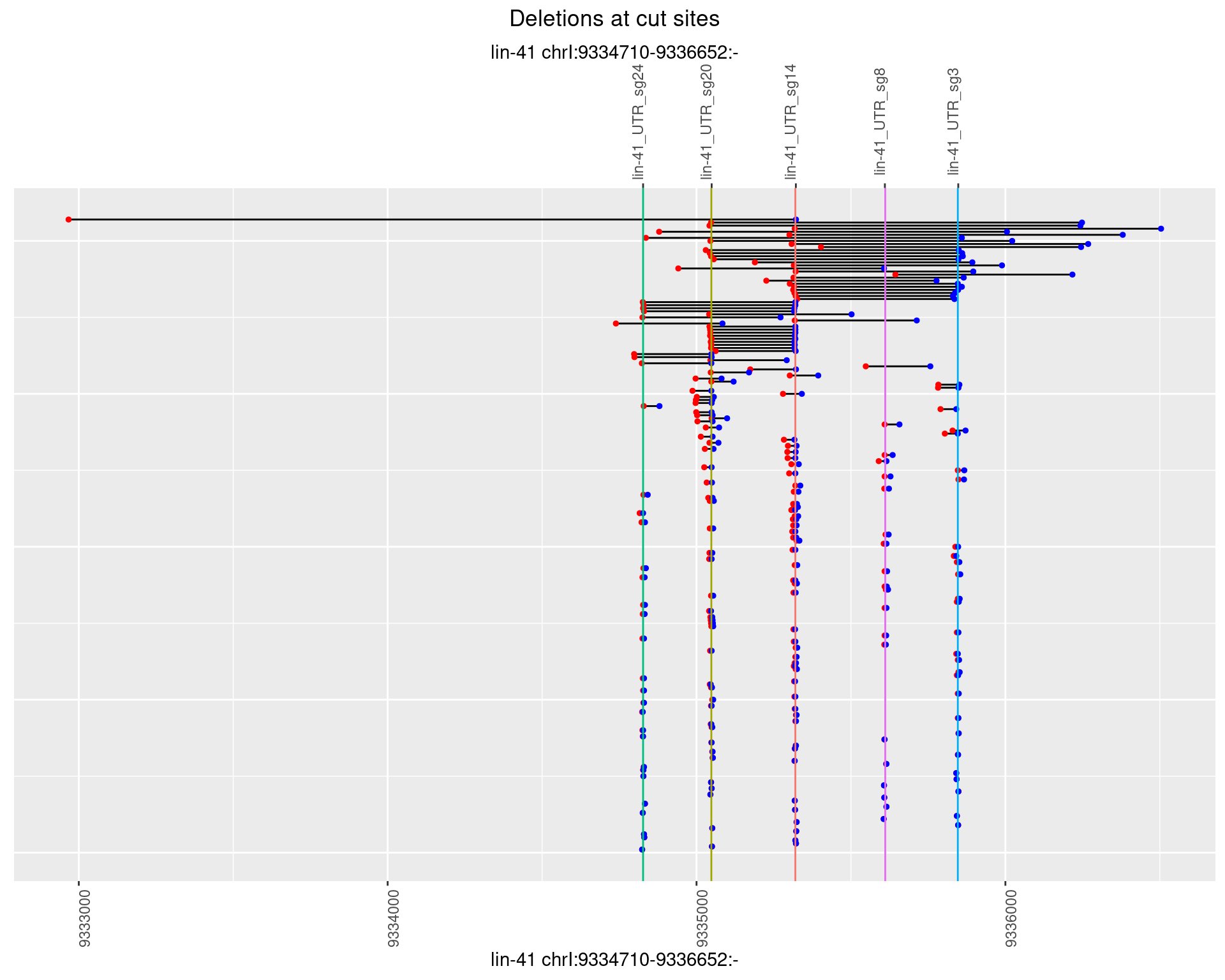
1.23.3 Freq: 1e-04
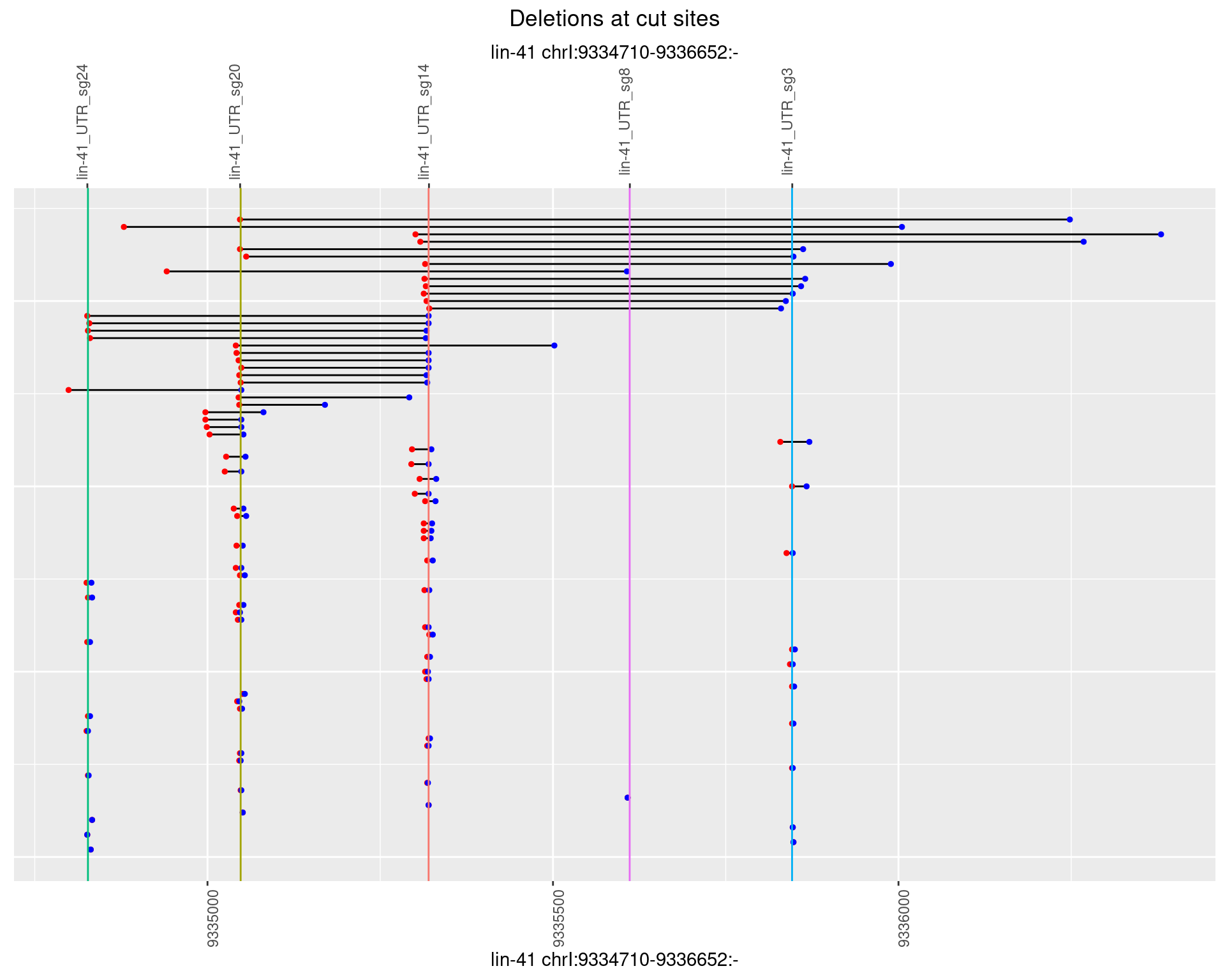
1.23.4 Freq: 0.001
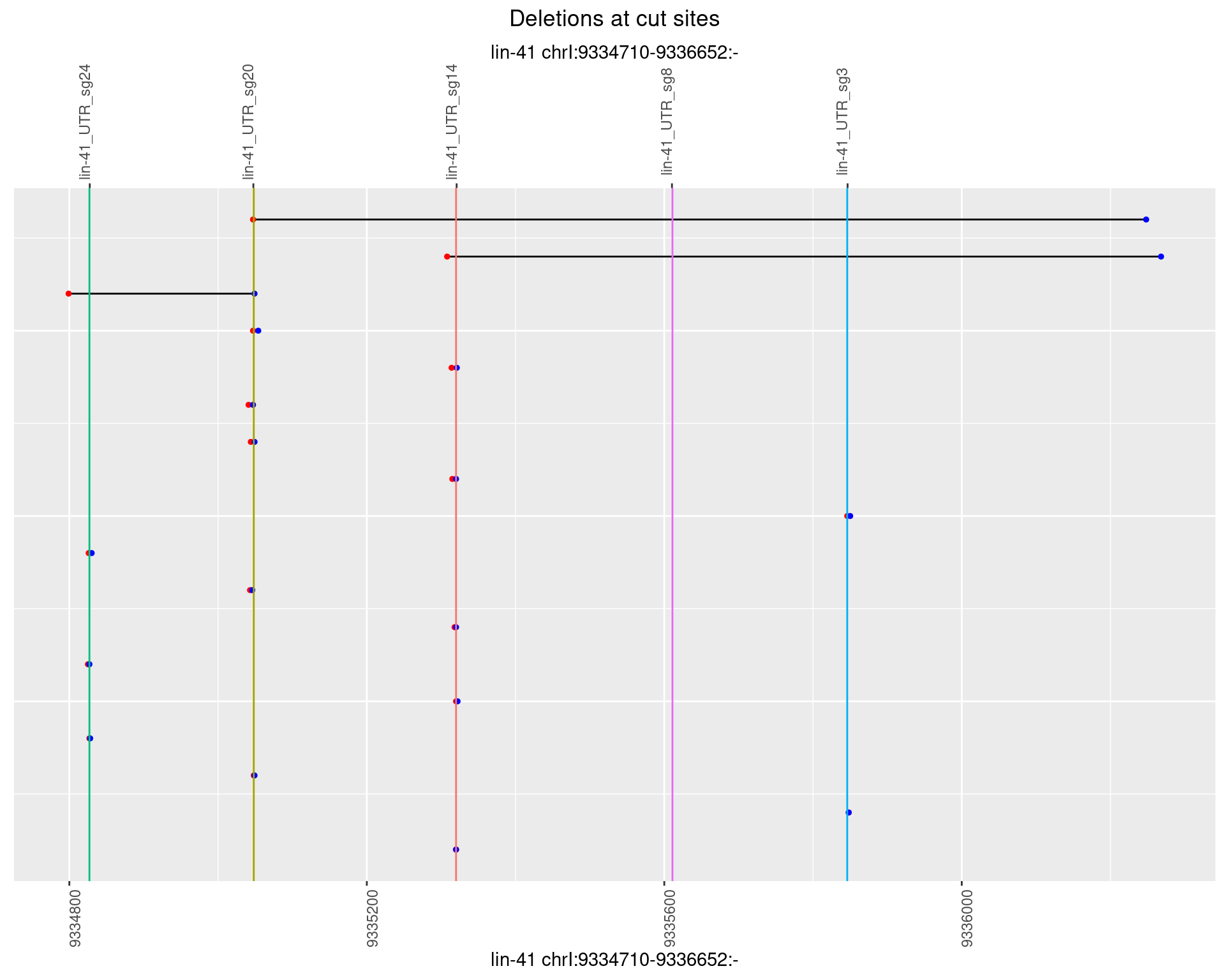
1.23.5 Freq: 0.01
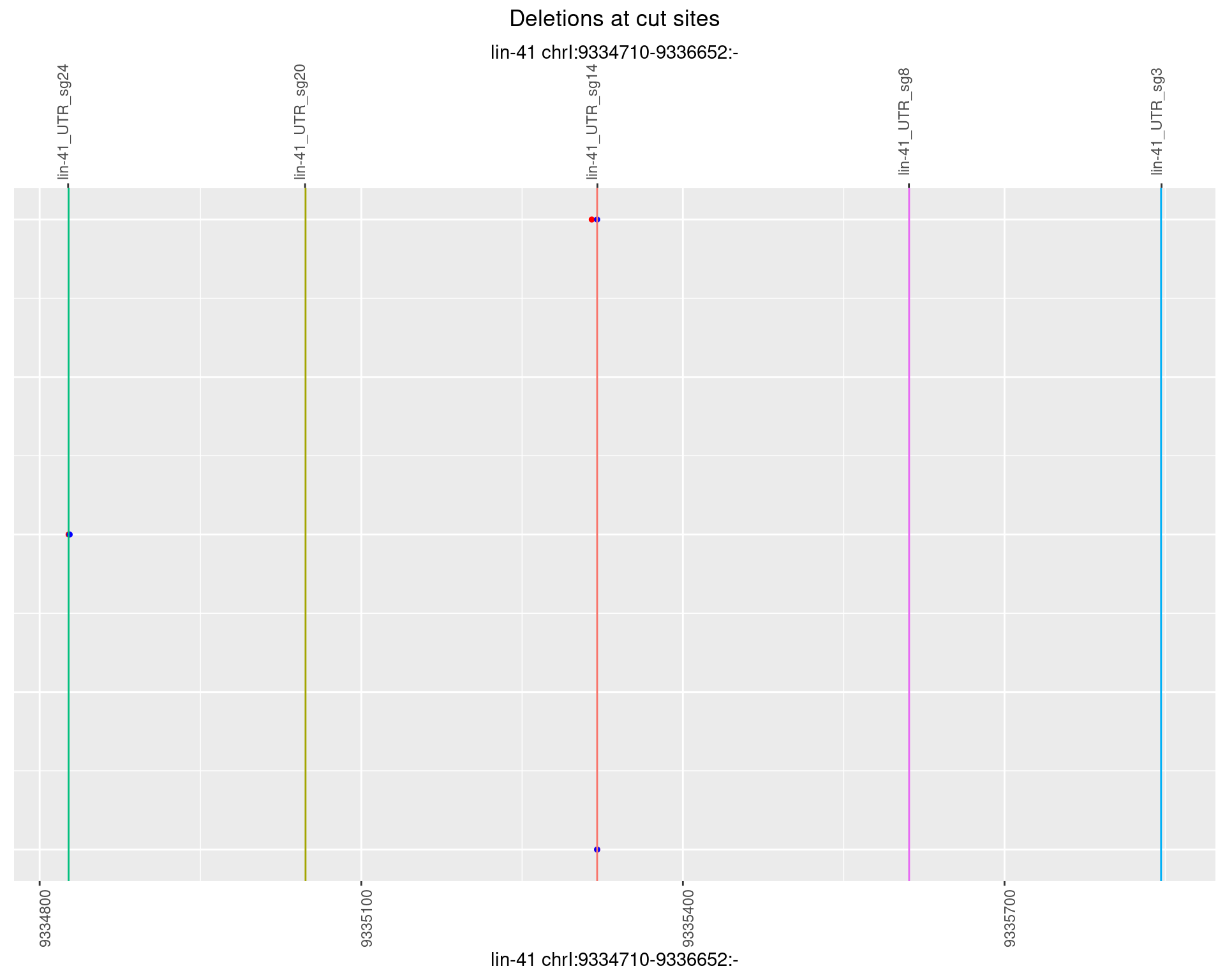
1.23.6 Freq: 0.1
No plot to show
1.24 gen_16C_F2_lin-41_sg26sg27
1.24.1 Freq: 0
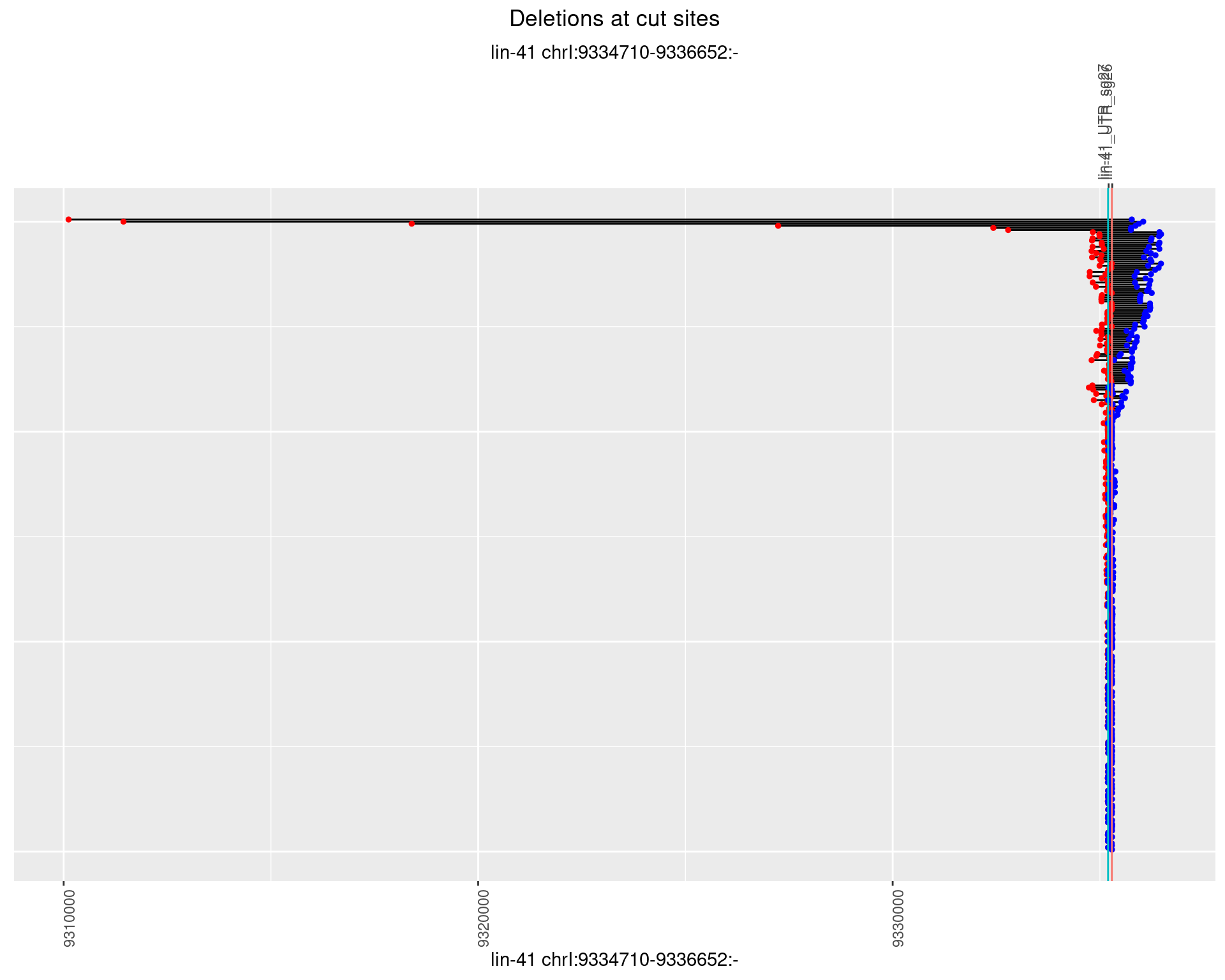
1.24.2 Freq: 1e-05
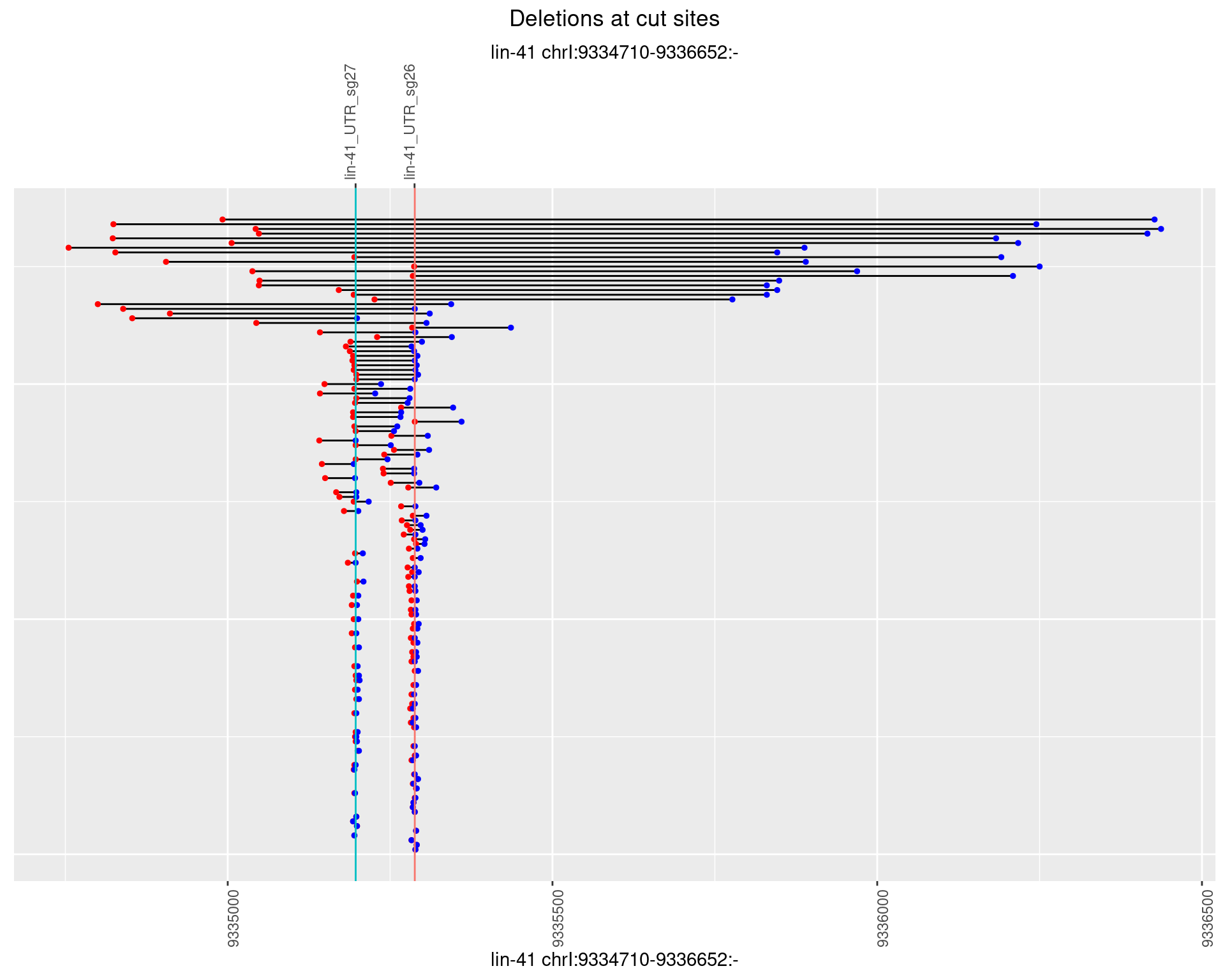
1.24.3 Freq: 1e-04
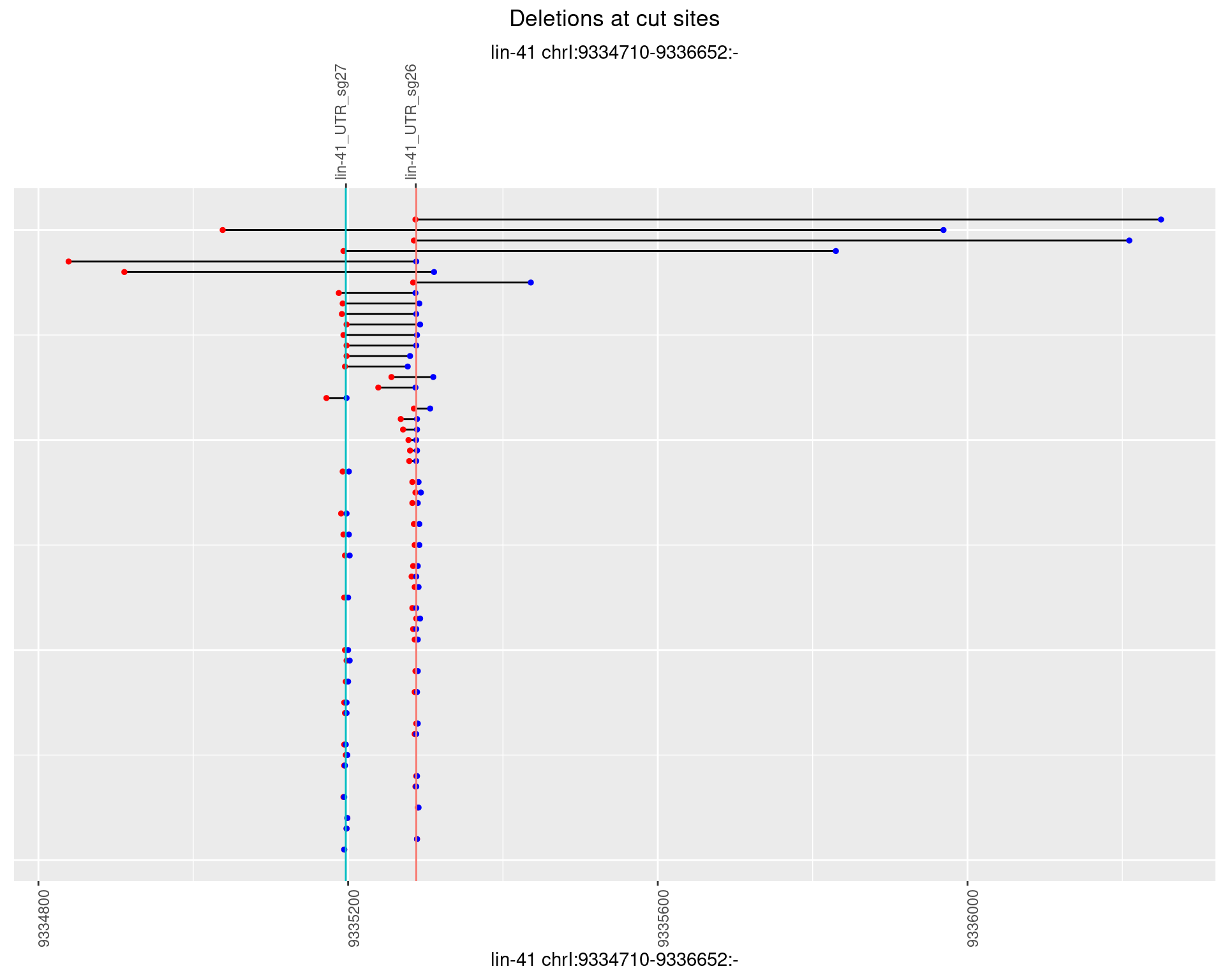
1.24.4 Freq: 0.001
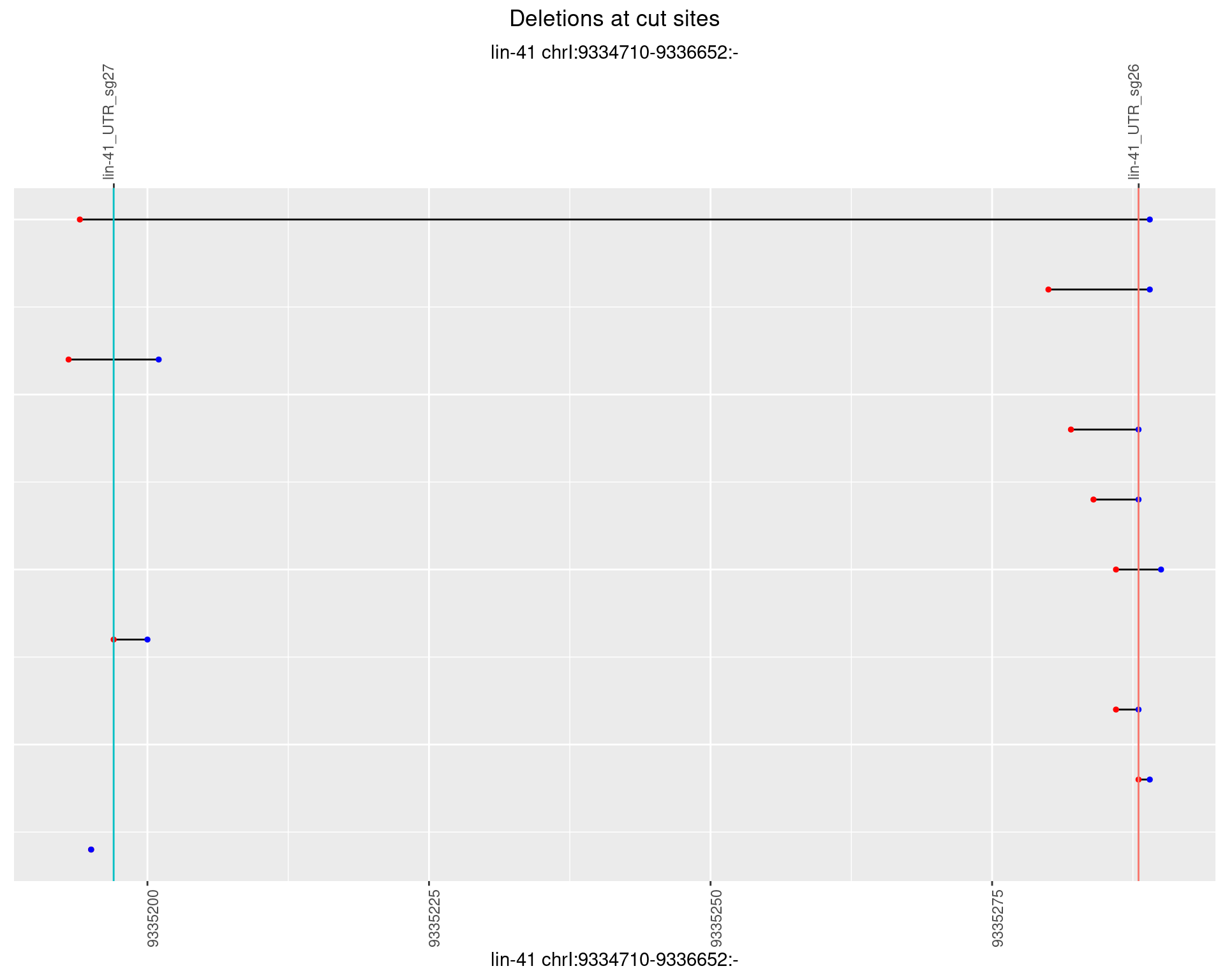
1.24.5 Freq: 0.01
No plot to show
1.24.6 Freq: 0.1
No plot to show
1.25 gen_16C_F3_lin-41_N2
1.25.1 Freq: 0
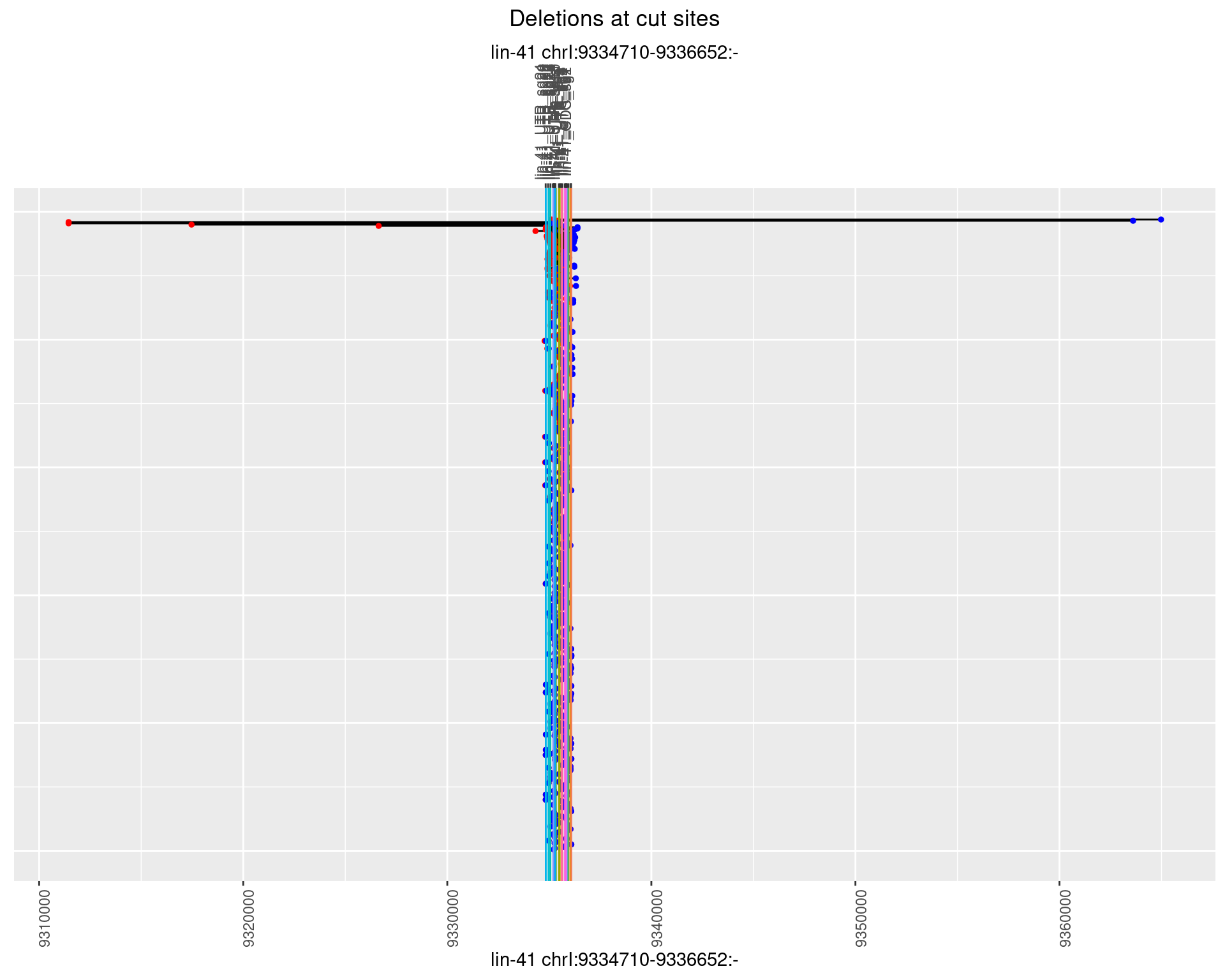
1.25.2 Freq: 1e-05
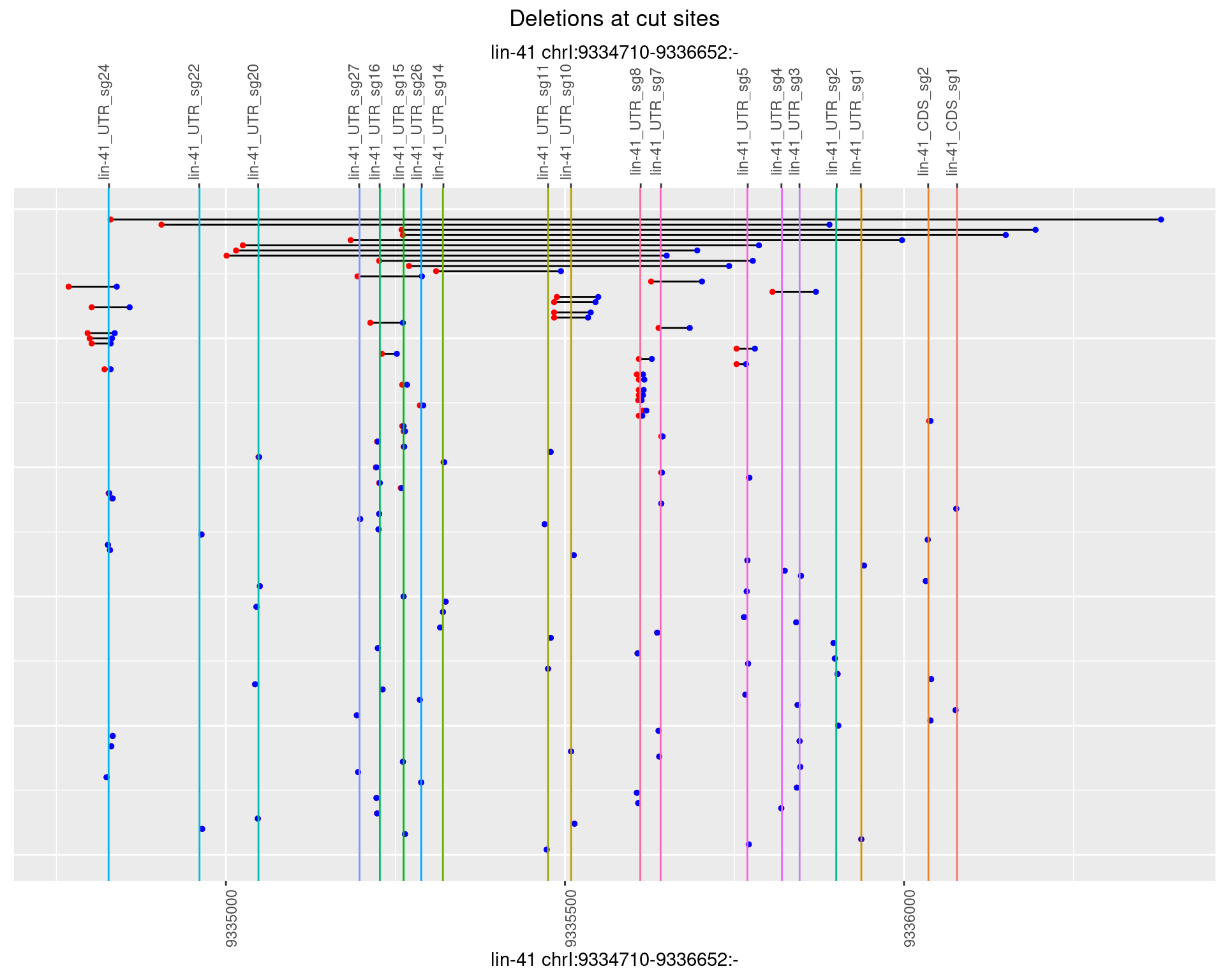
1.25.3 Freq: 1e-04
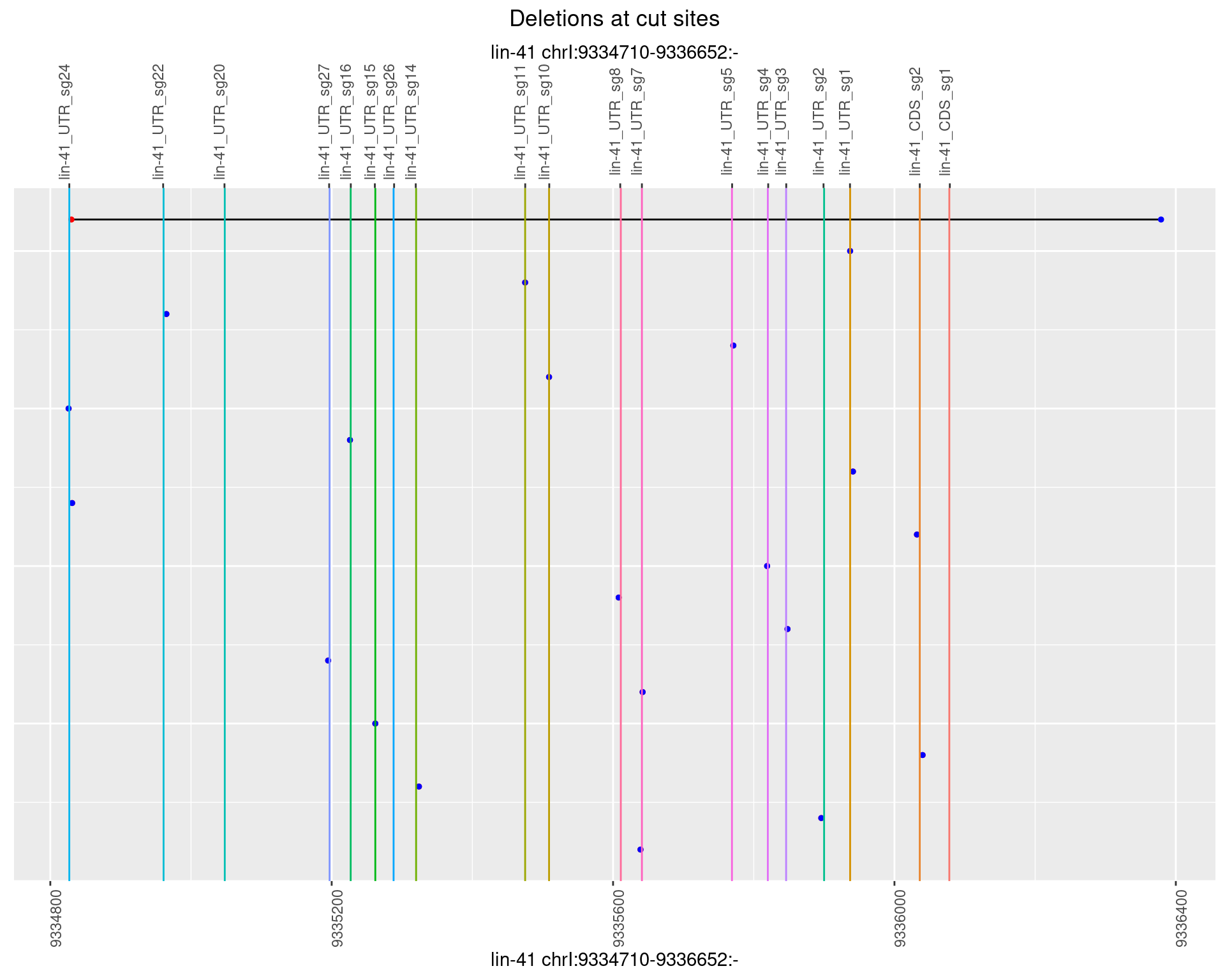
1.25.4 Freq: 0.001
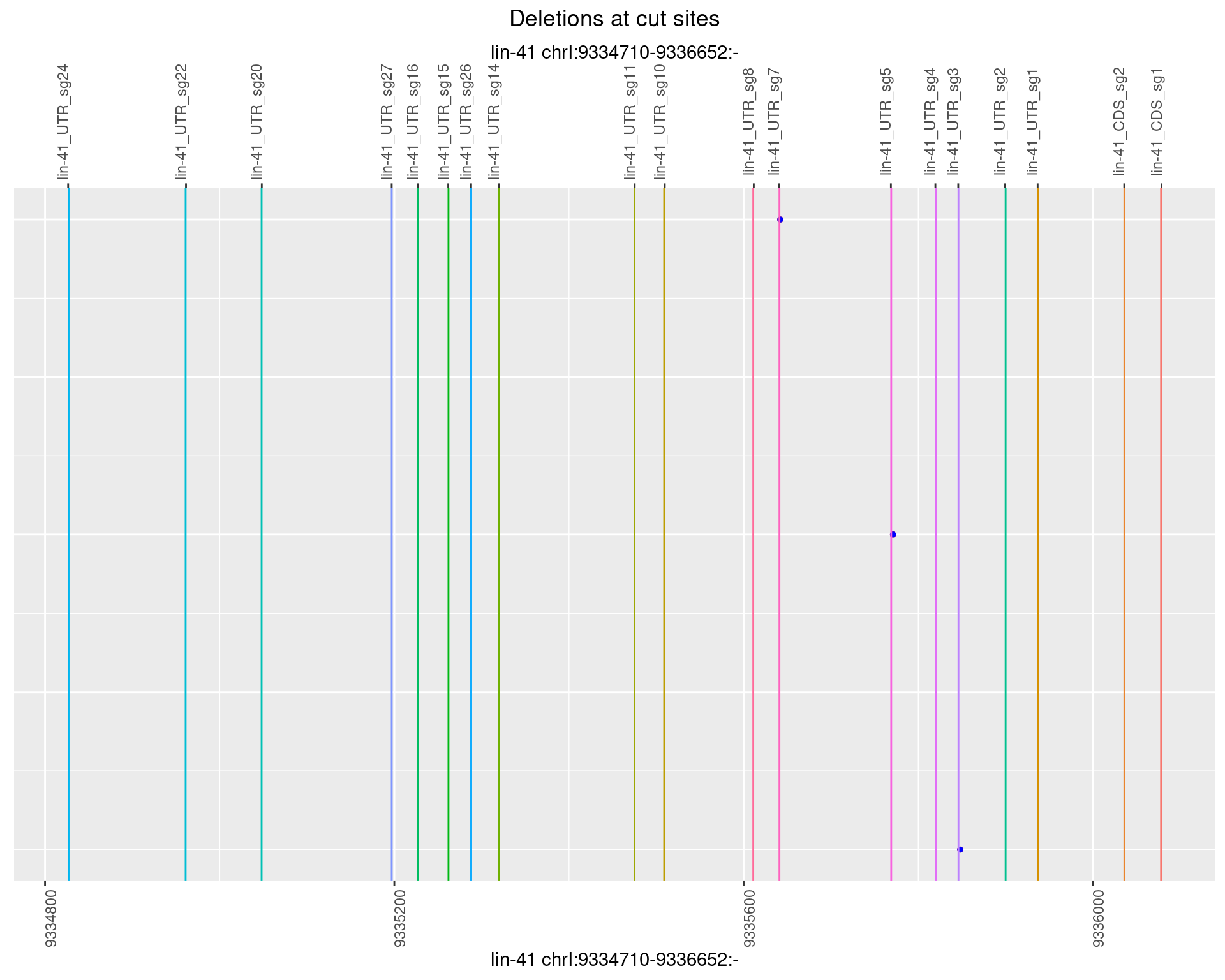
1.25.5 Freq: 0.01
No plot to show
1.25.6 Freq: 0.1
No plot to show
1.26 gen_16C_F3_lin-41_sg15sg16
1.26.1 Freq: 0
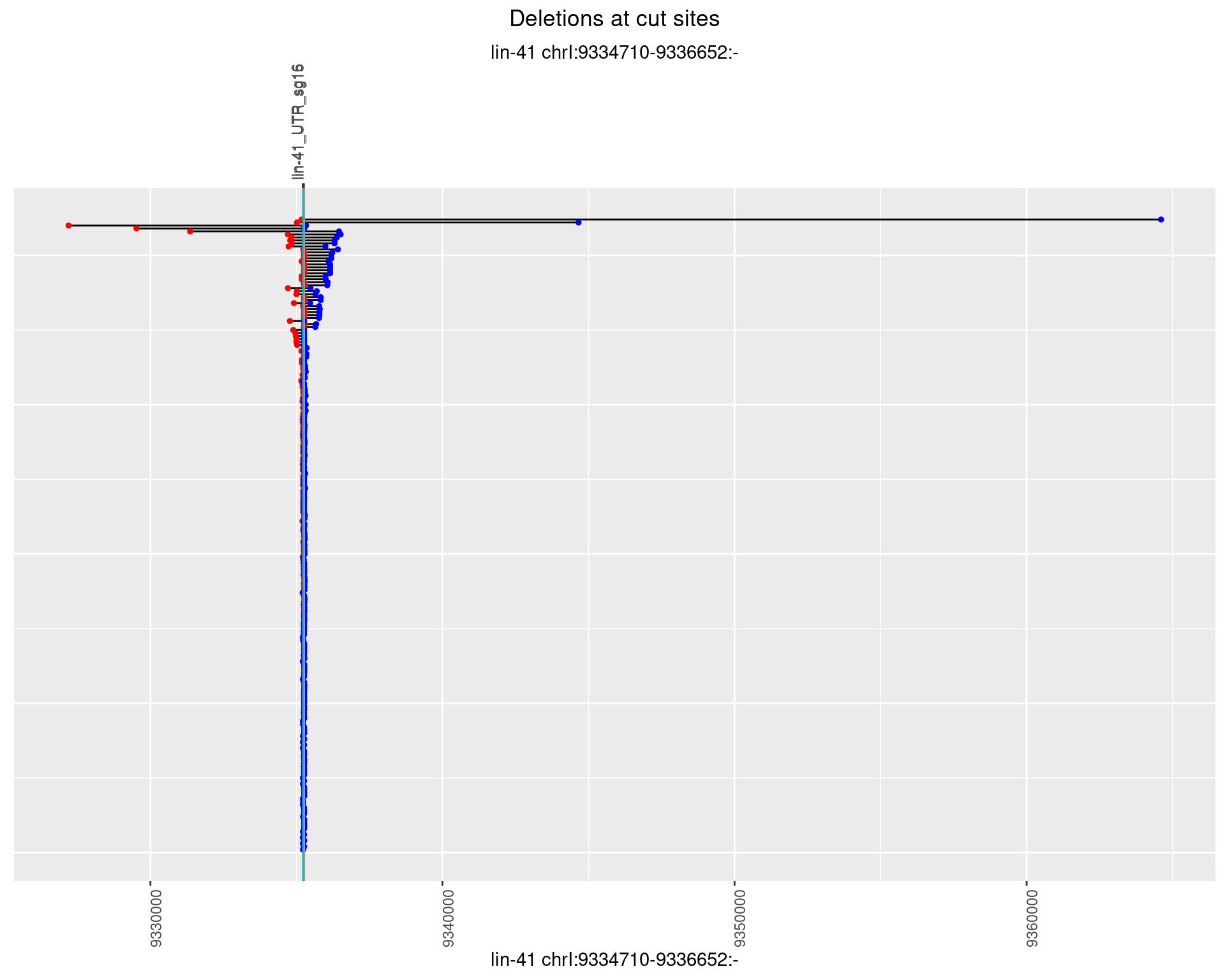
1.26.2 Freq: 1e-05
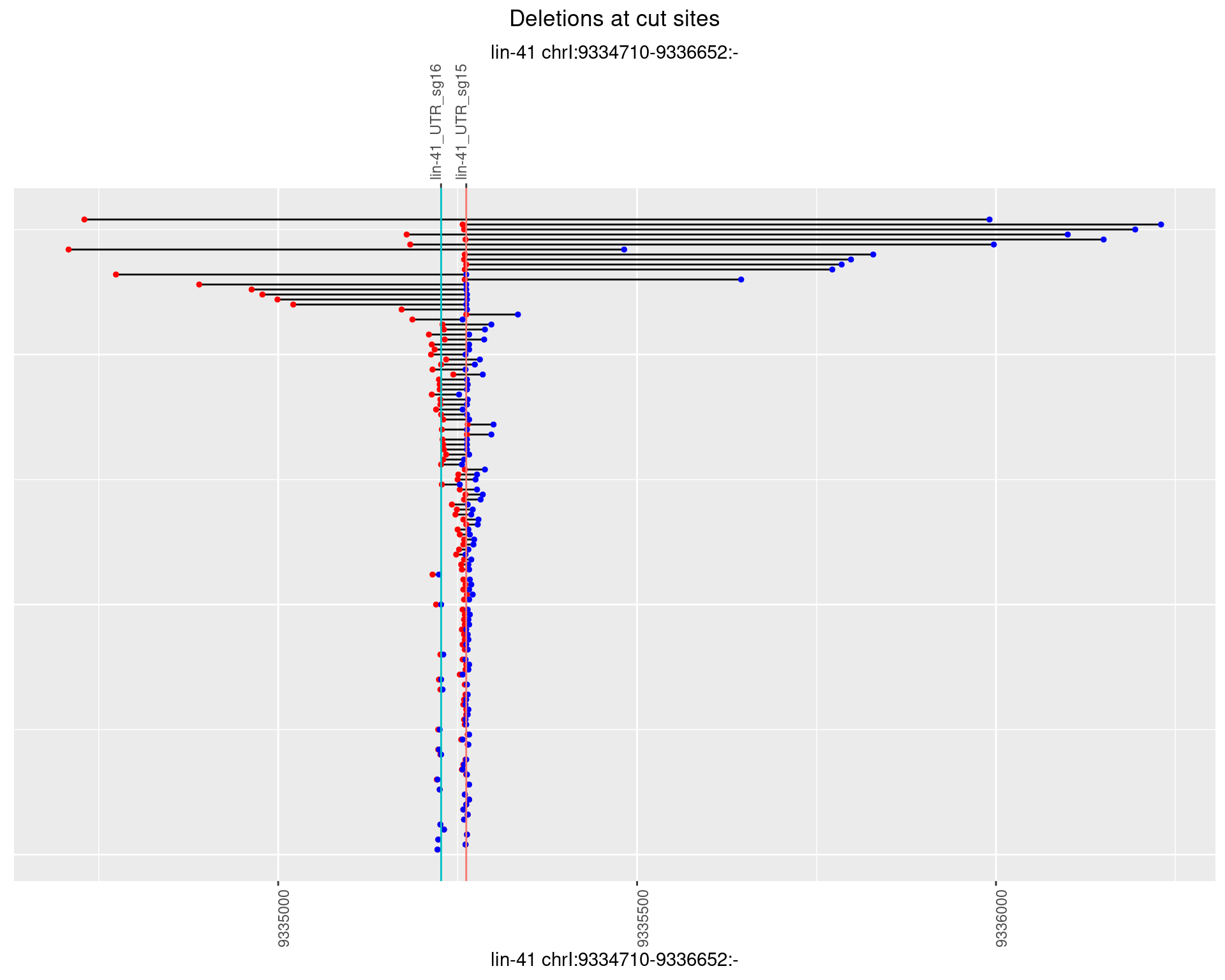
1.26.3 Freq: 1e-04
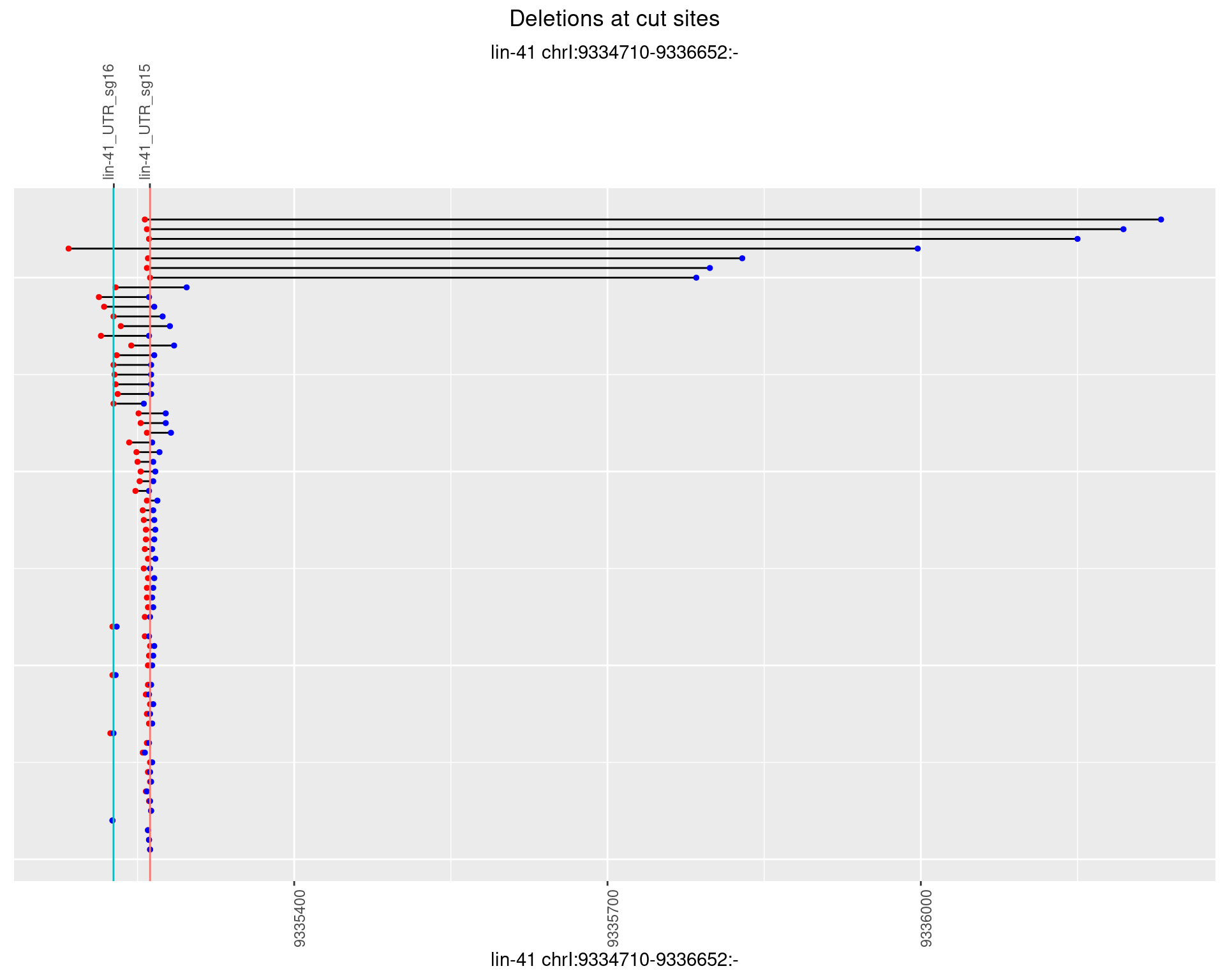
1.26.4 Freq: 0.001
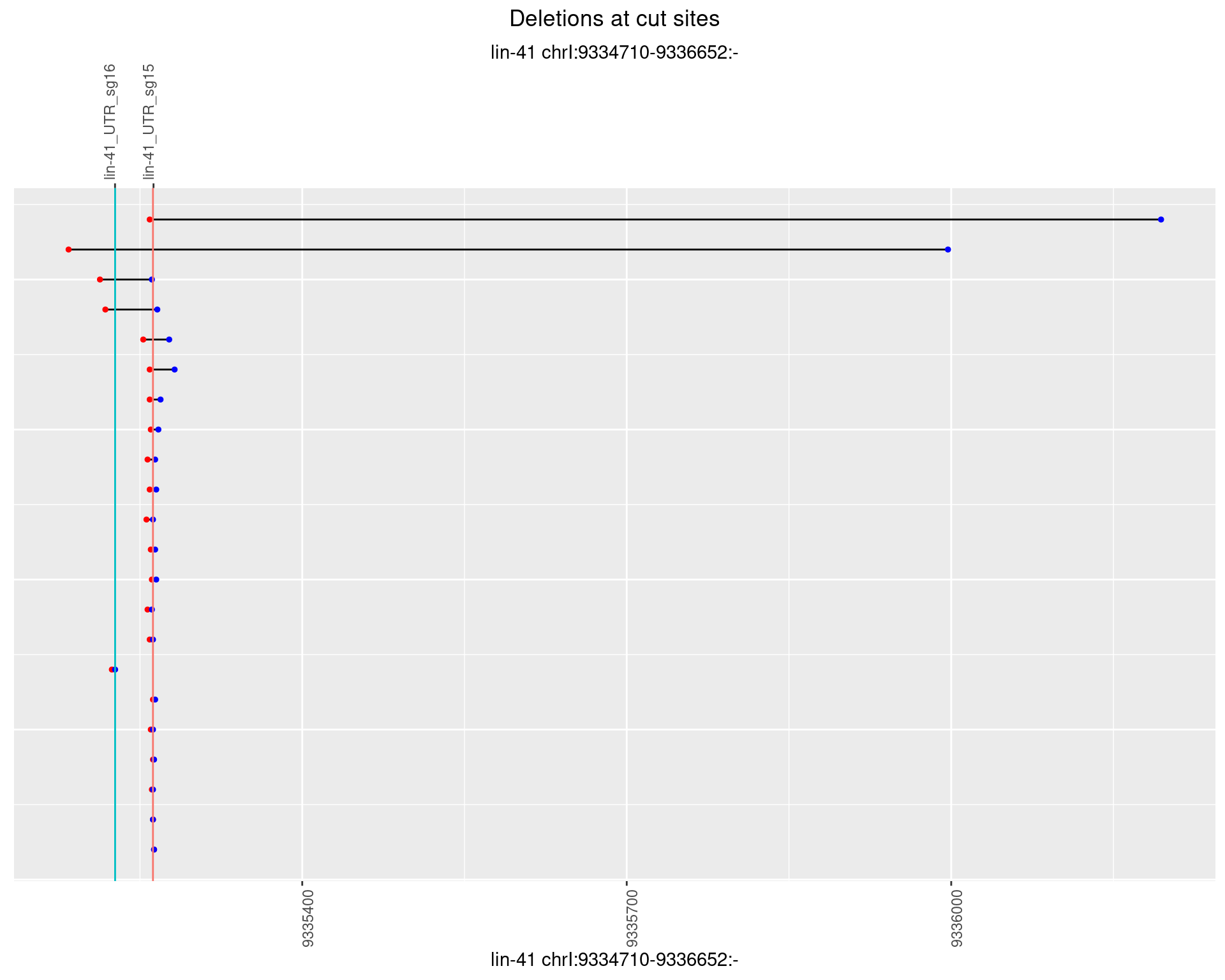
1.26.5 Freq: 0.01
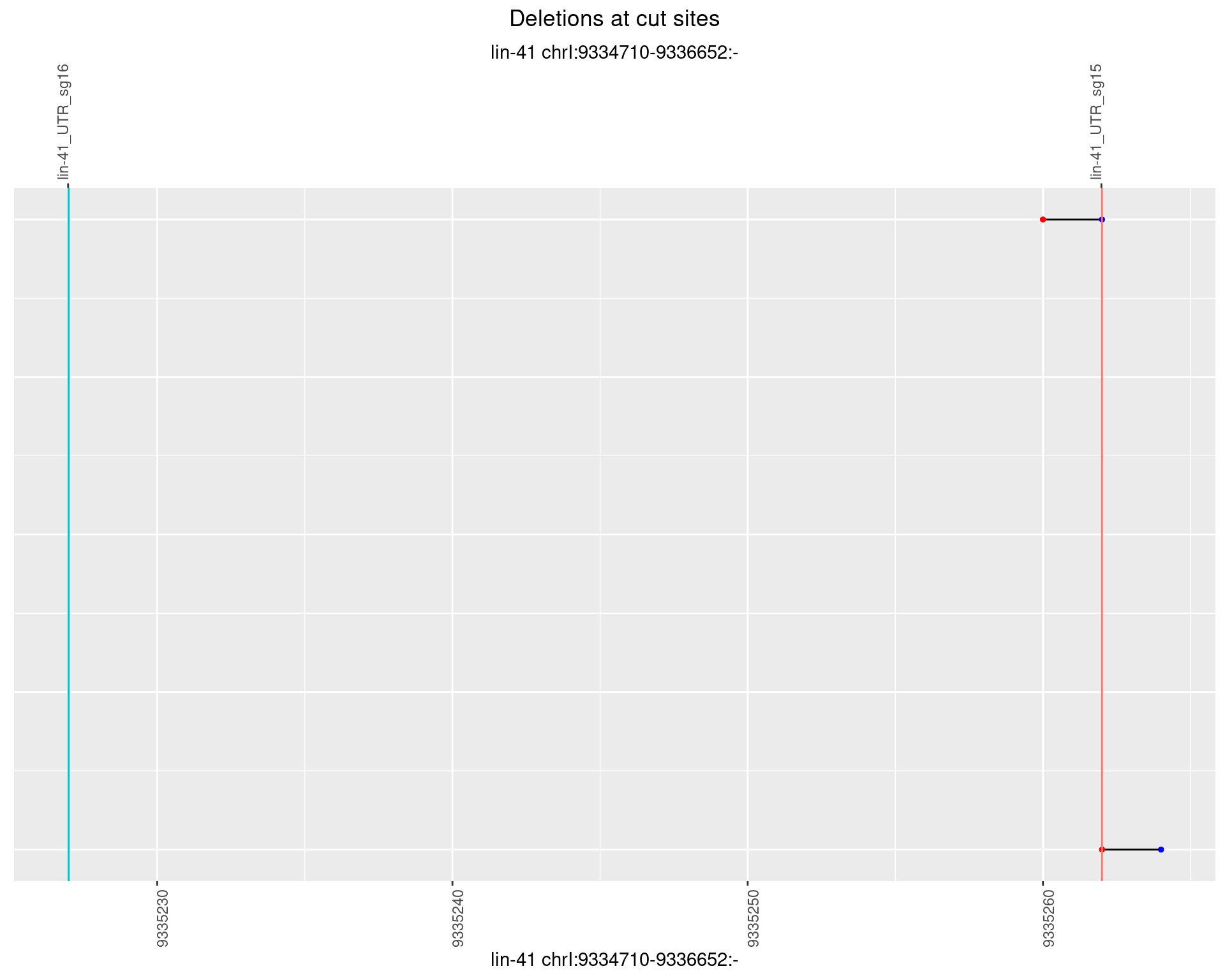
1.26.6 Freq: 0.1
No plot to show
1.27 gen_16C_F3_lin-41_pool
1.27.1 Freq: 0
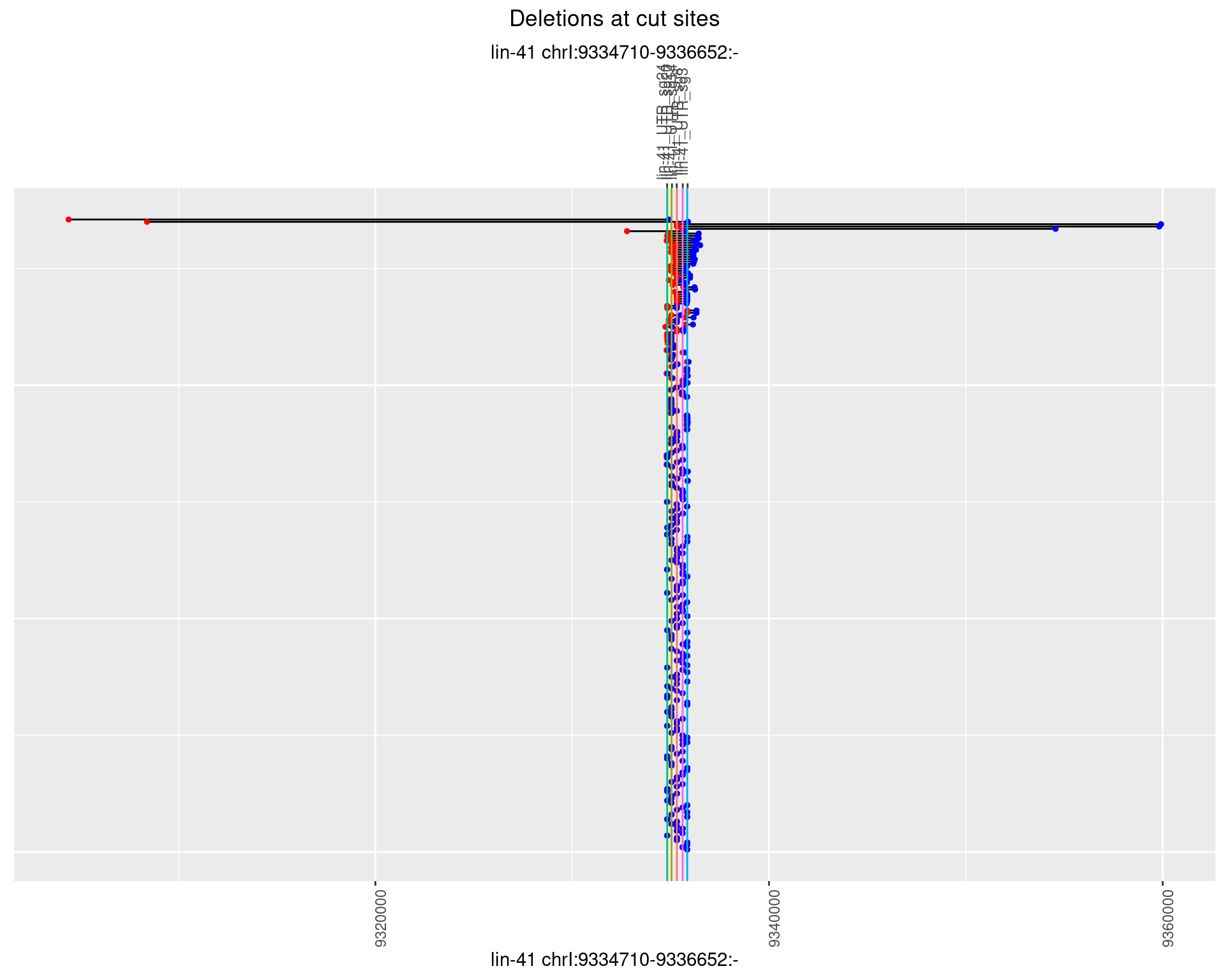
1.27.2 Freq: 1e-05
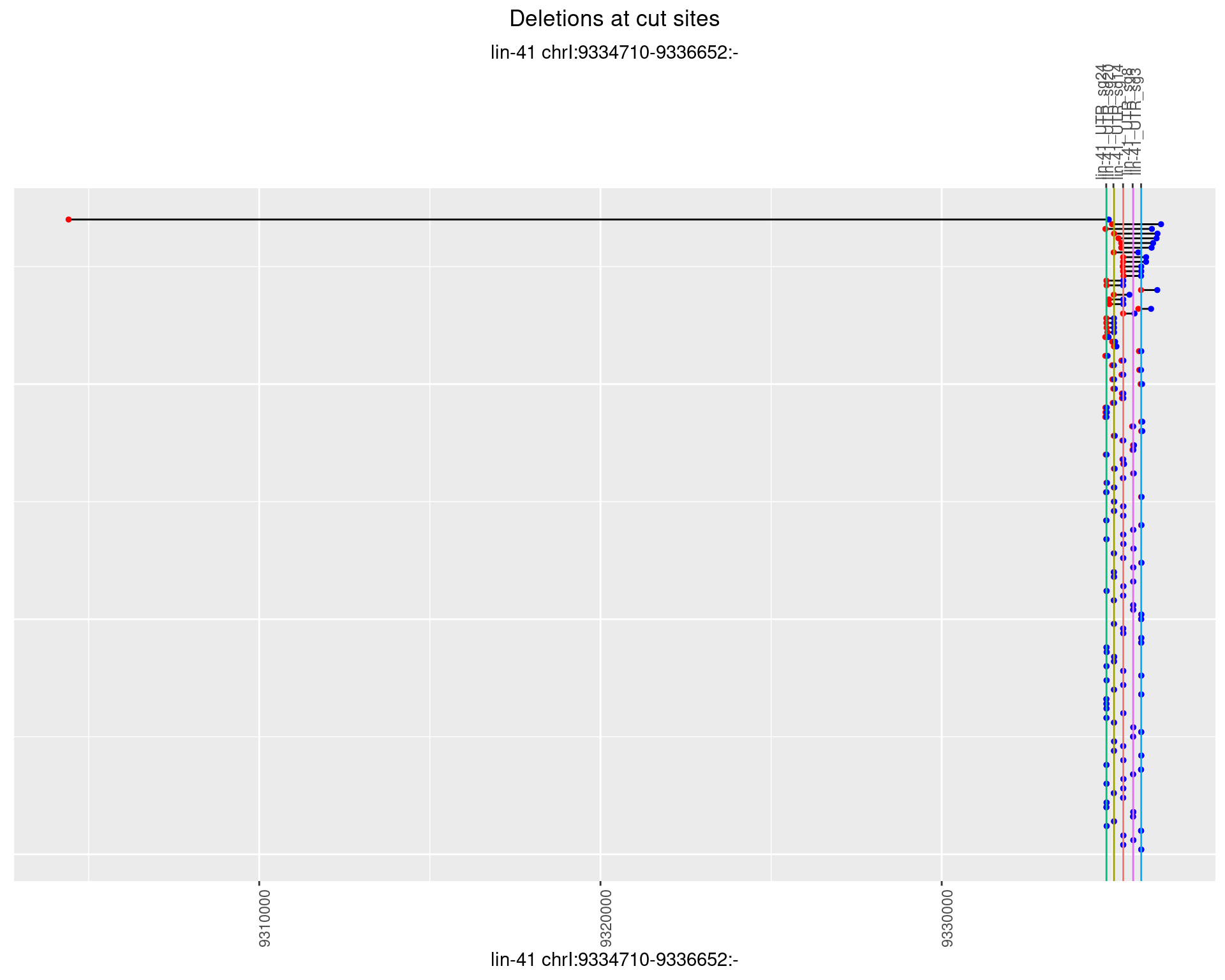
1.27.3 Freq: 1e-04
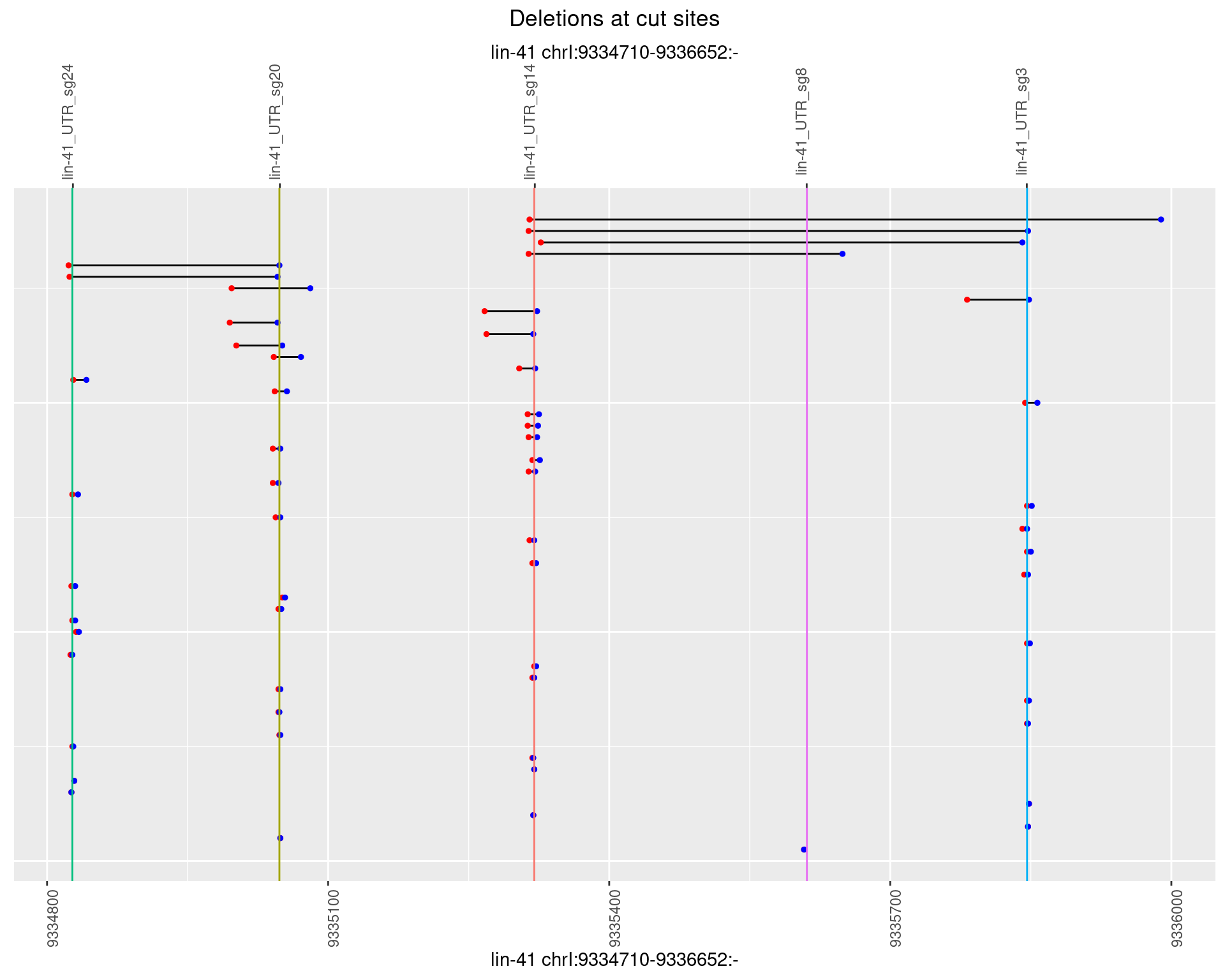
1.27.4 Freq: 0.001
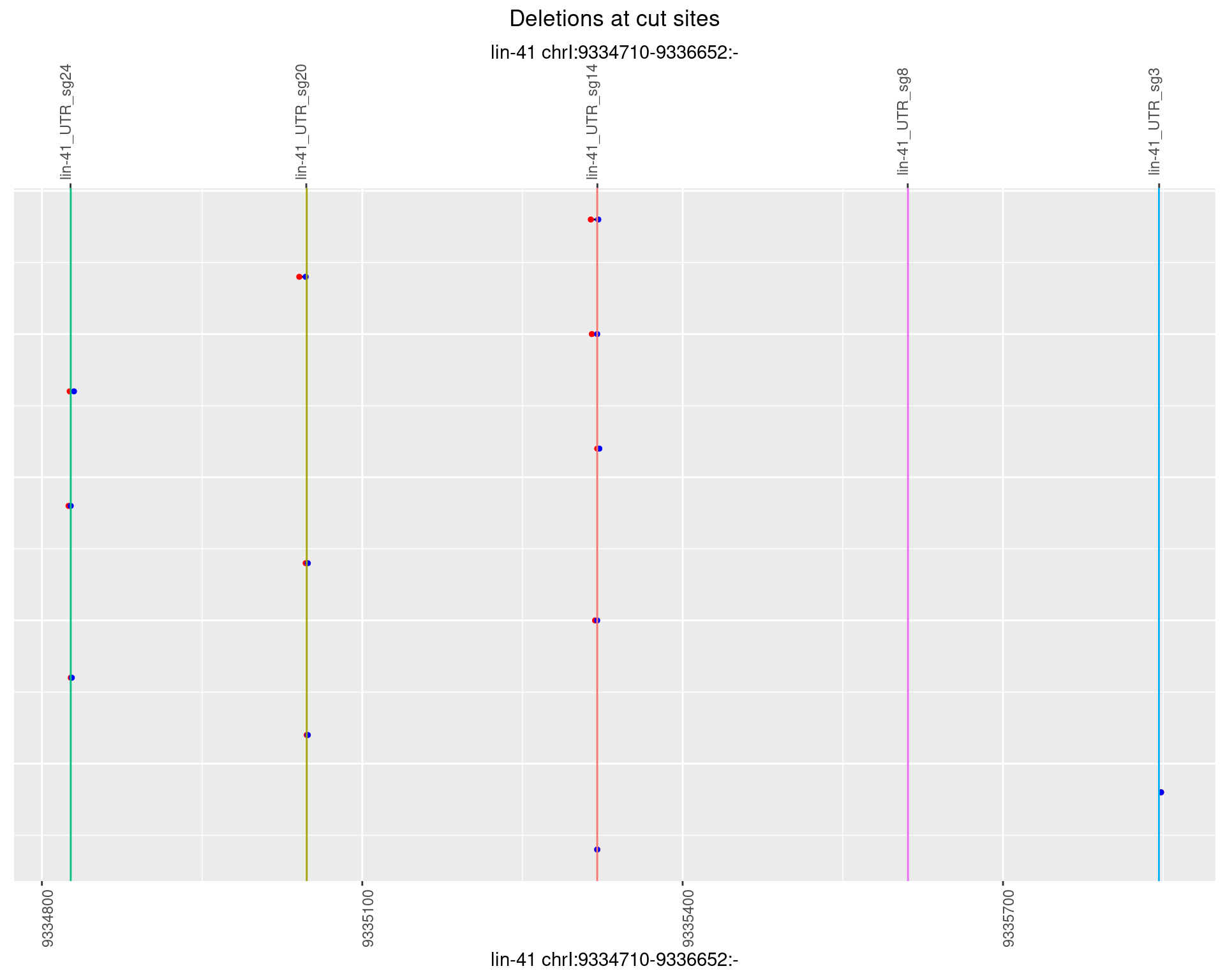
1.27.5 Freq: 0.01
1.27.6 Freq: 0.1
No plot to show
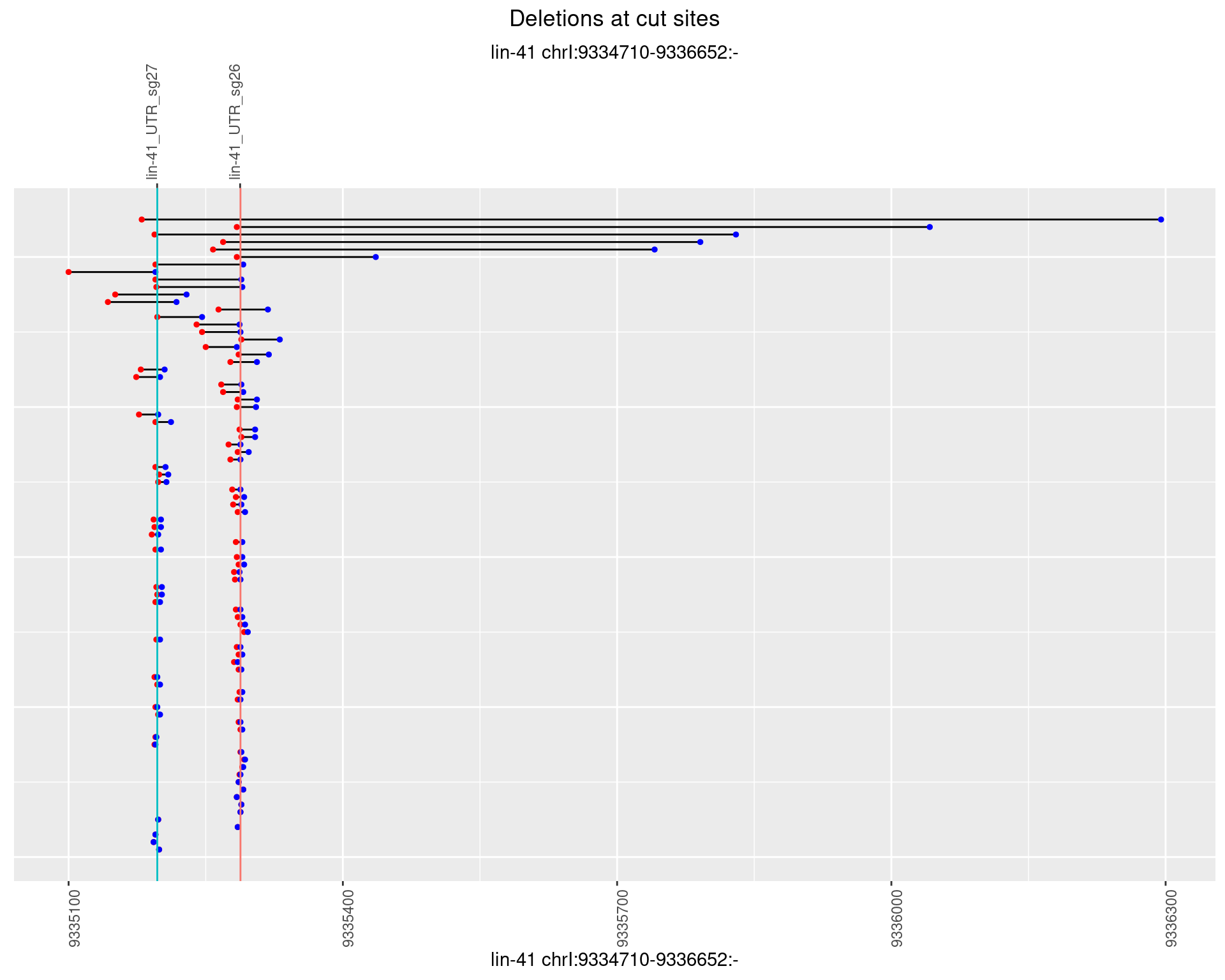
1.28 gen_16C_F3_lin-41_sg26sg27
1.28.1 Freq: 0
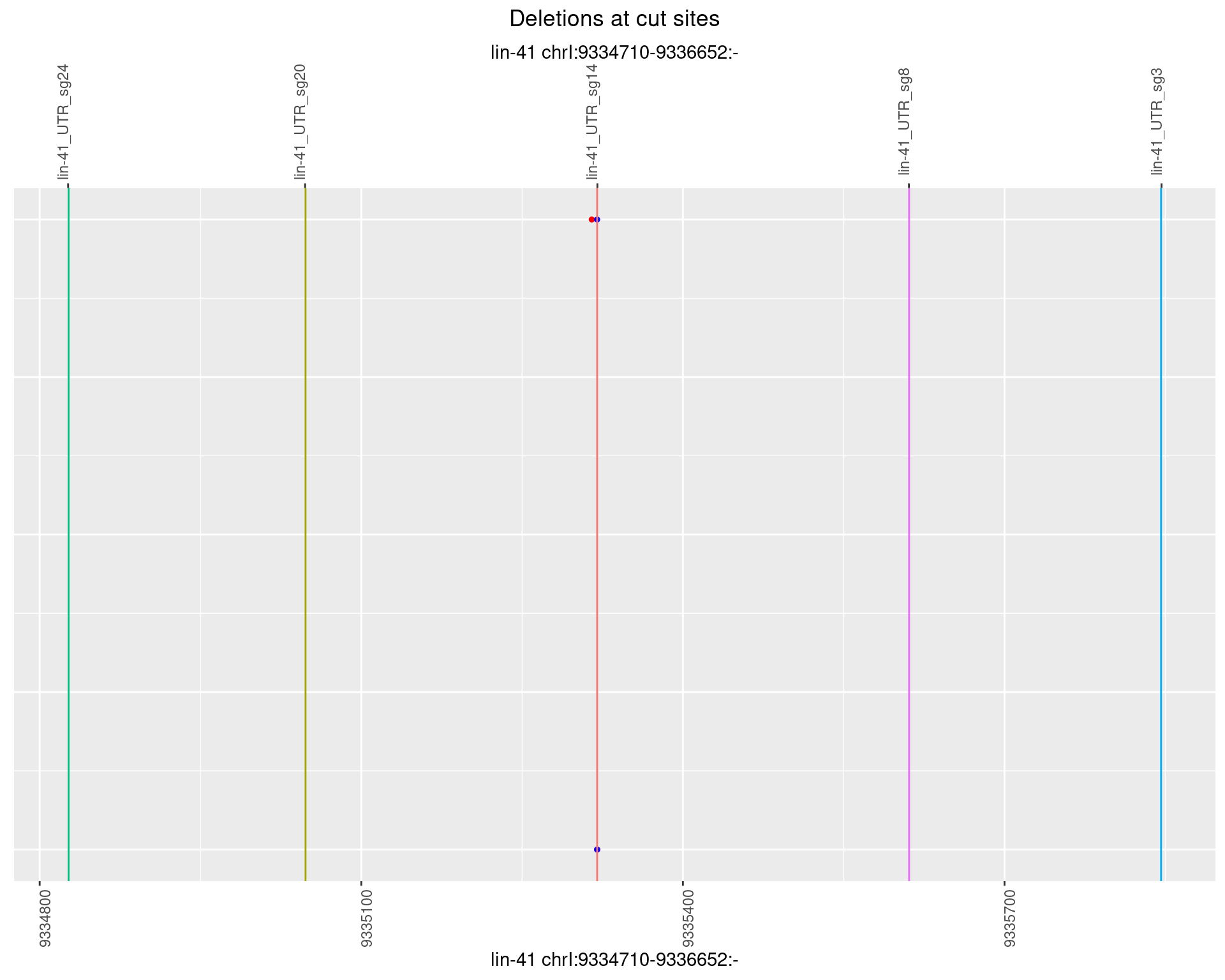
1.28.2 Freq: 1e-05
1.28.3 Freq: 1e-04
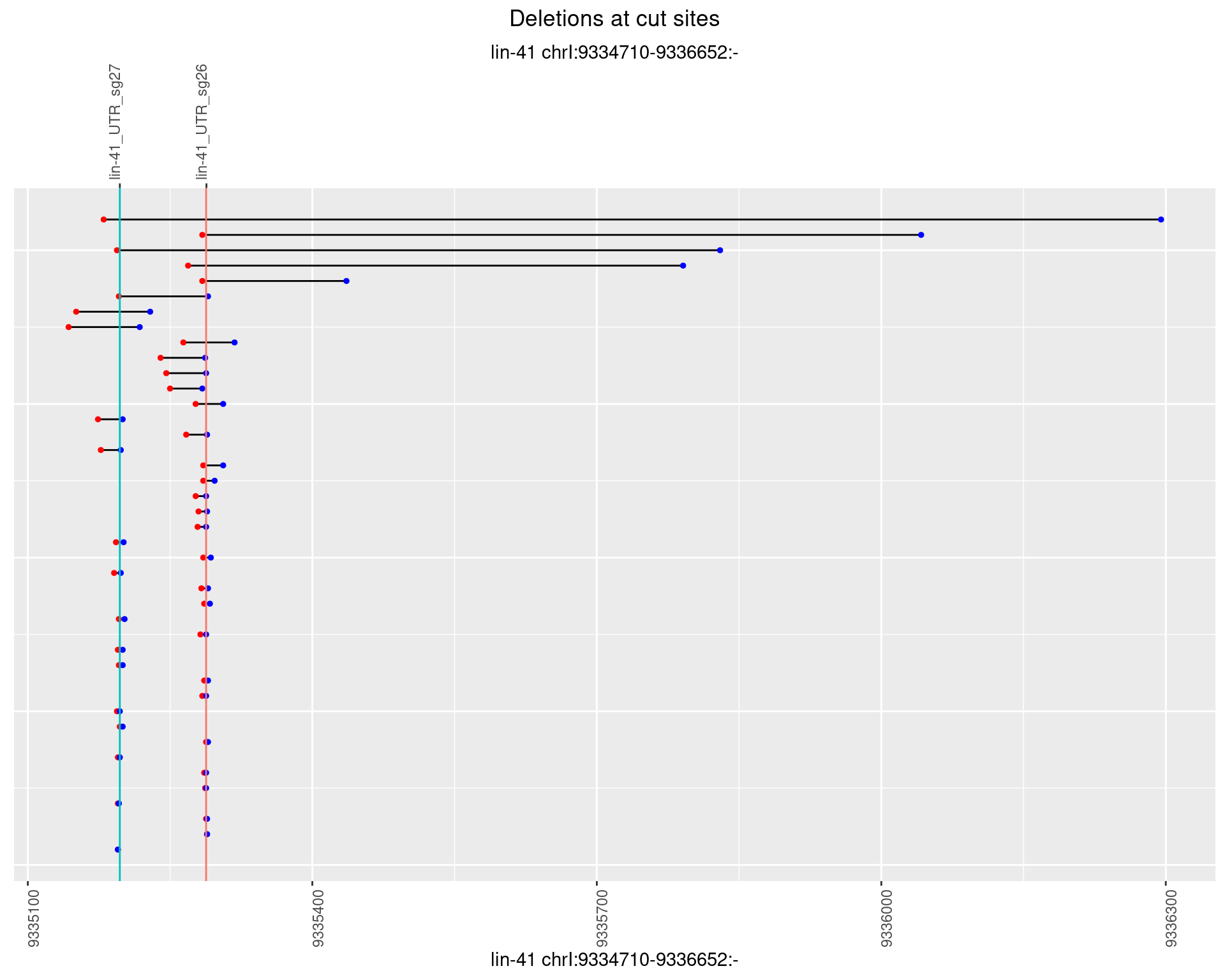
1.28.4 Freq: 0.001
1.28.5 Freq: 0.01
1.28.6 Freq: 0.1
No plot to show
1.29 gen_16C_F4_lin-41_N2
1.29.1 Freq: 0
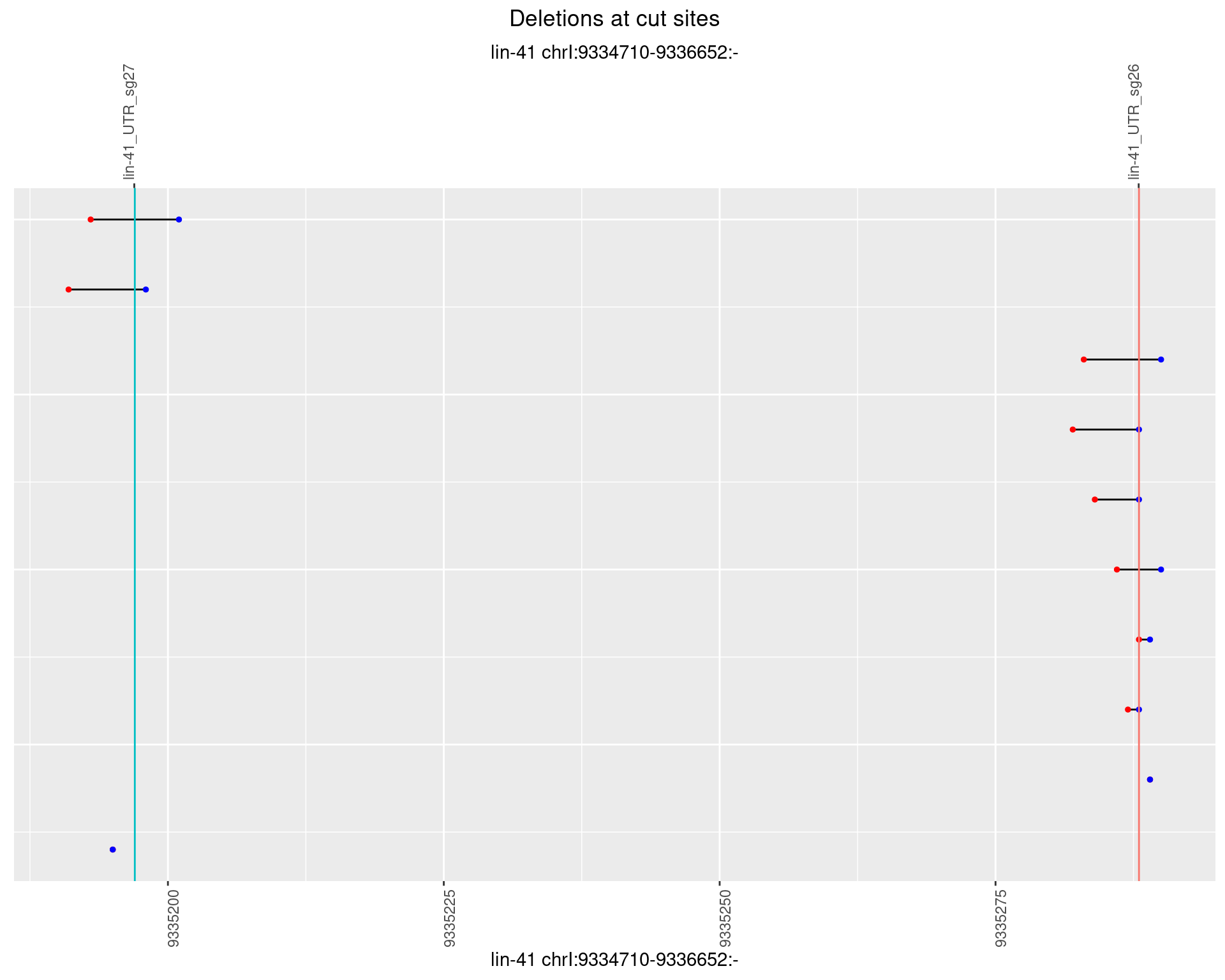
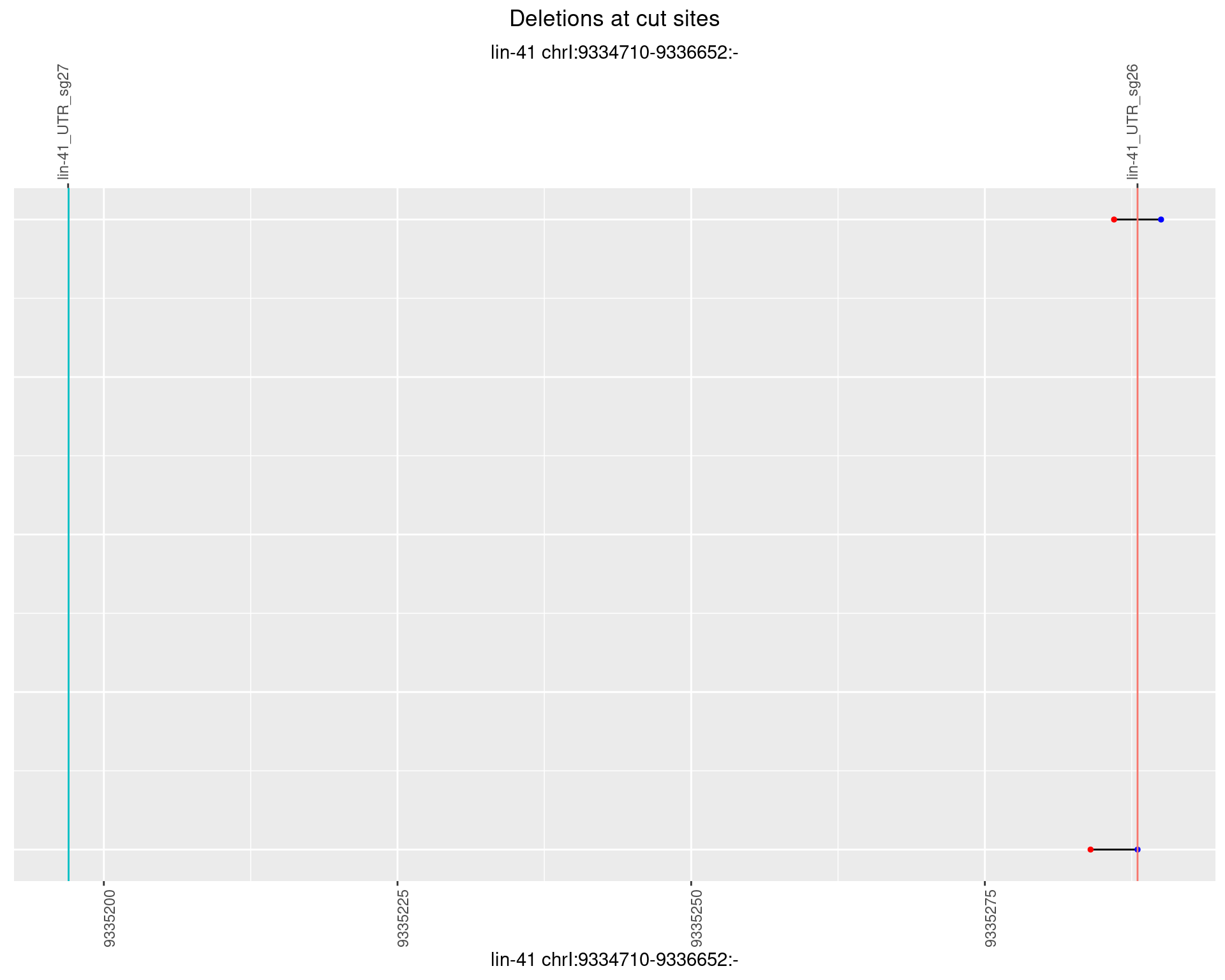
1.29.2 Freq: 1e-05
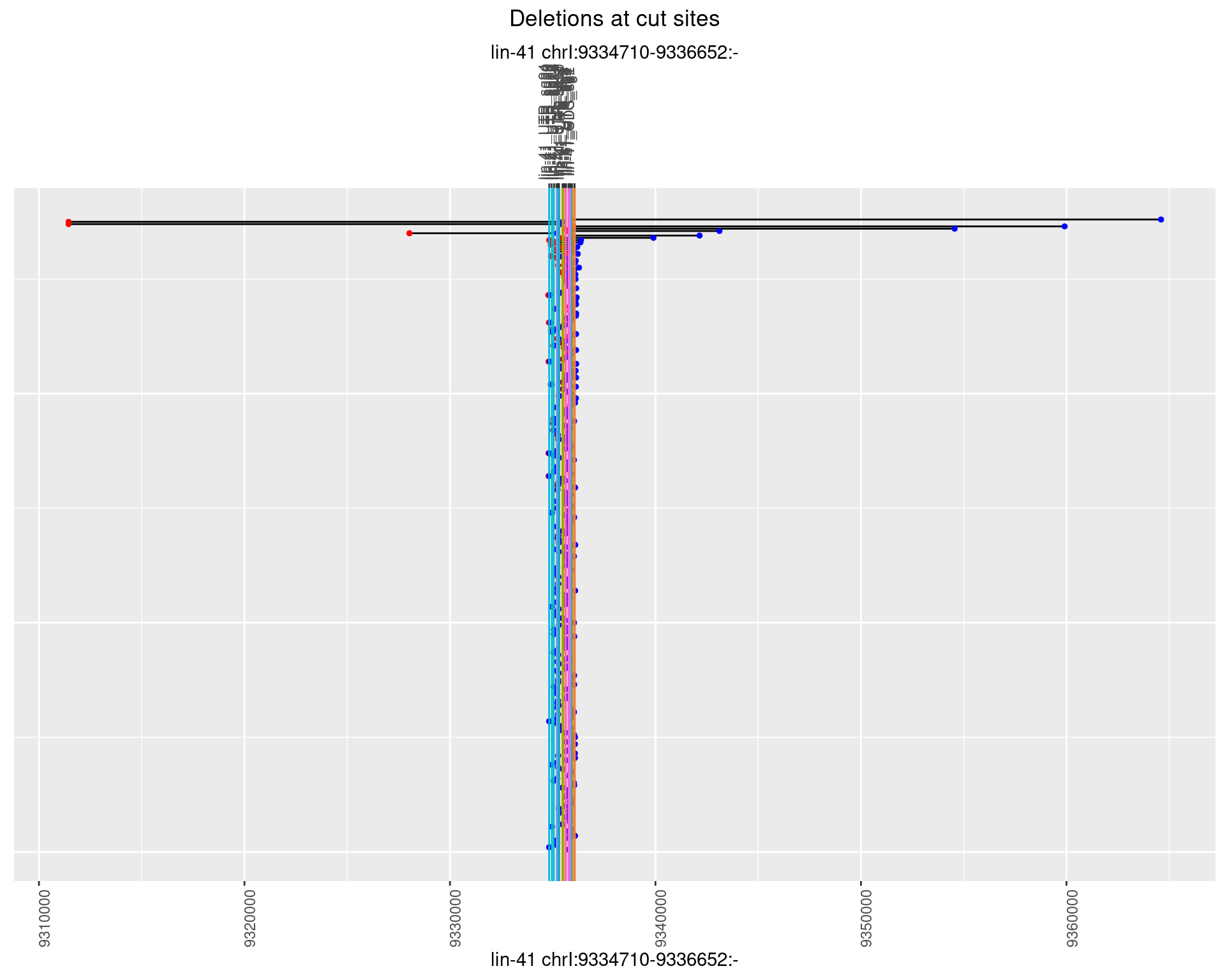
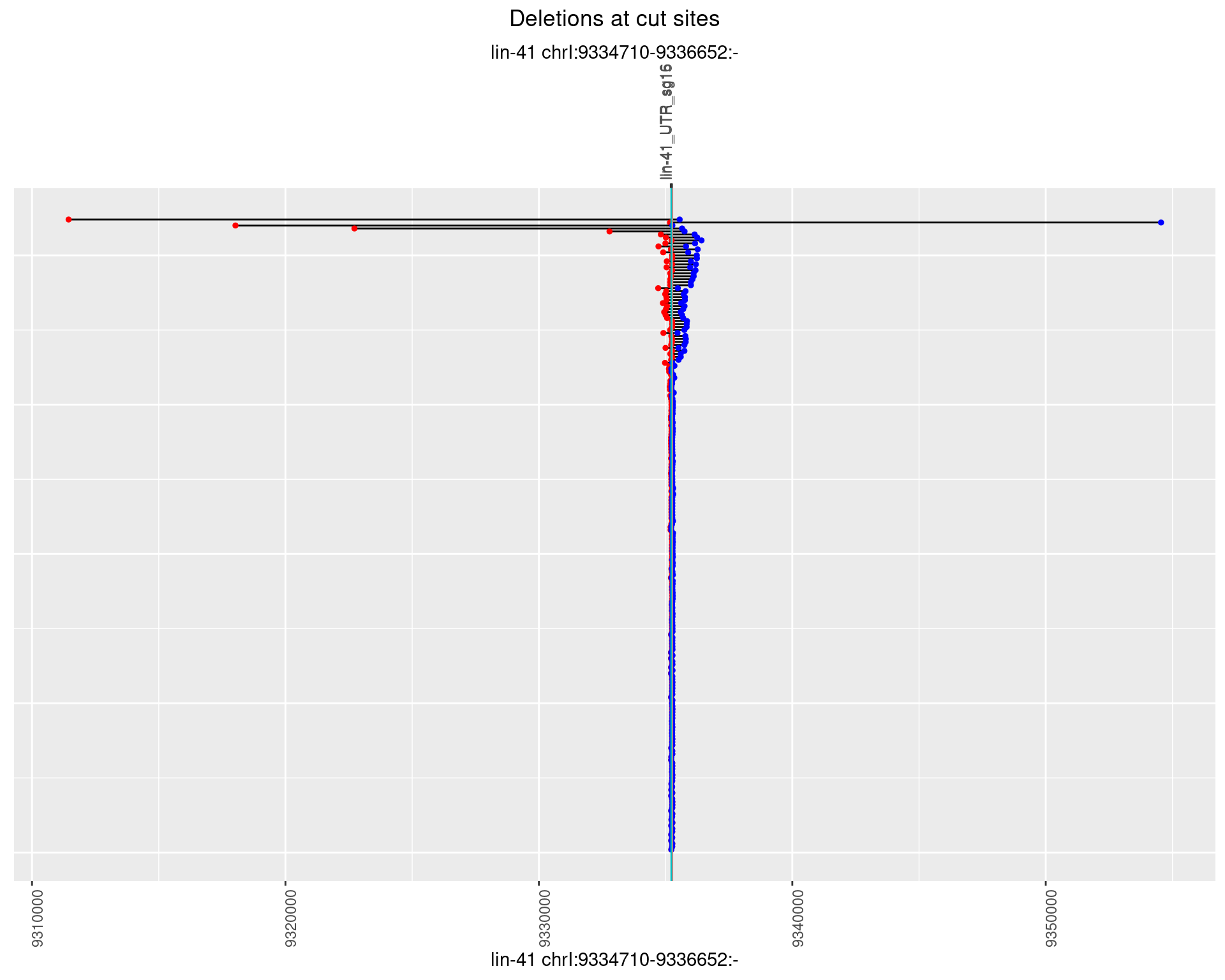
1.29.3 Freq: 1e-04
1.29.4 Freq: 0.001
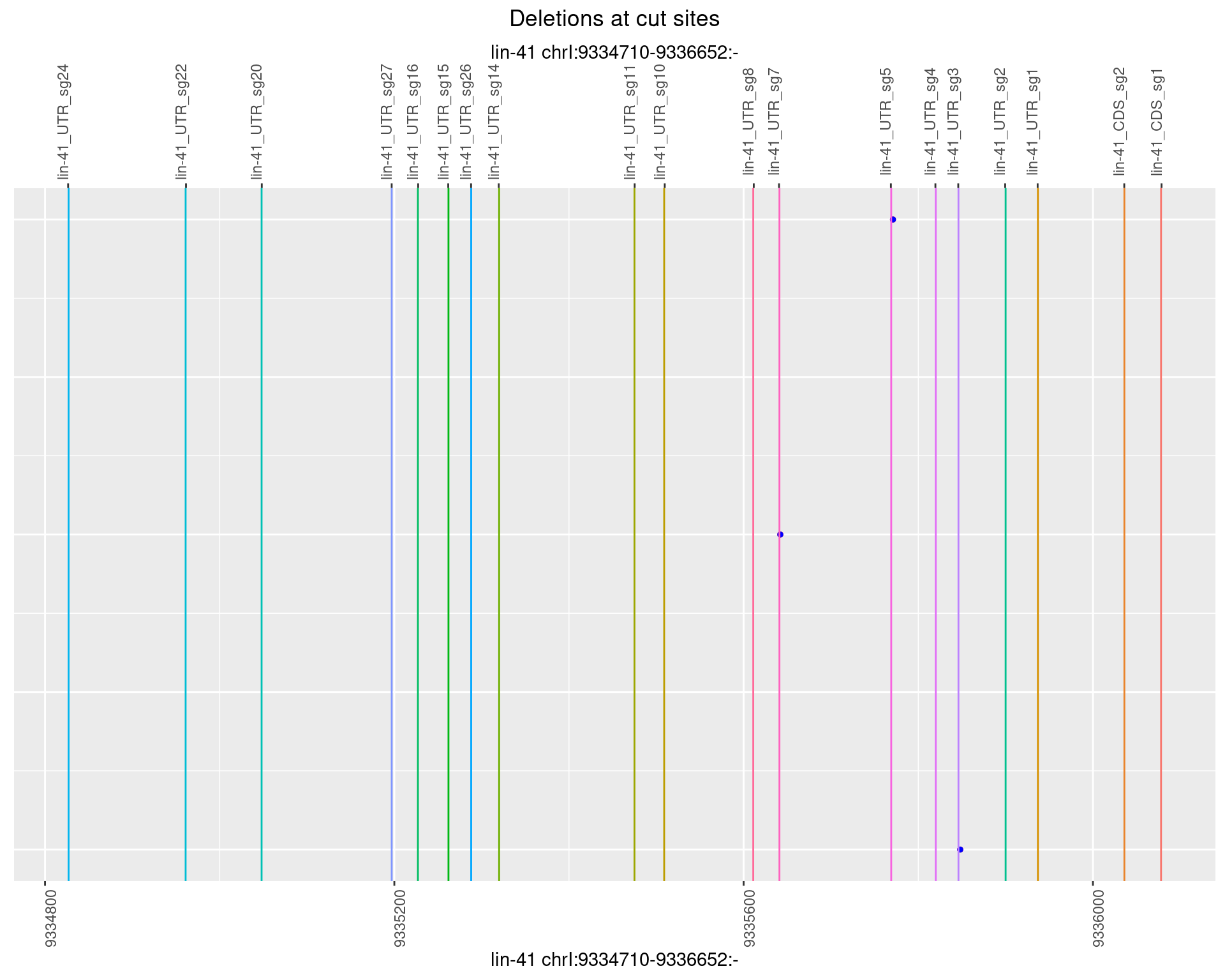
1.29.5 Freq: 0.01
No plot to show
1.29.6 Freq: 0.1
No plot to show
1.30 gen_16C_F4_lin-41_sg15sg16
1.30.1 Freq: 0
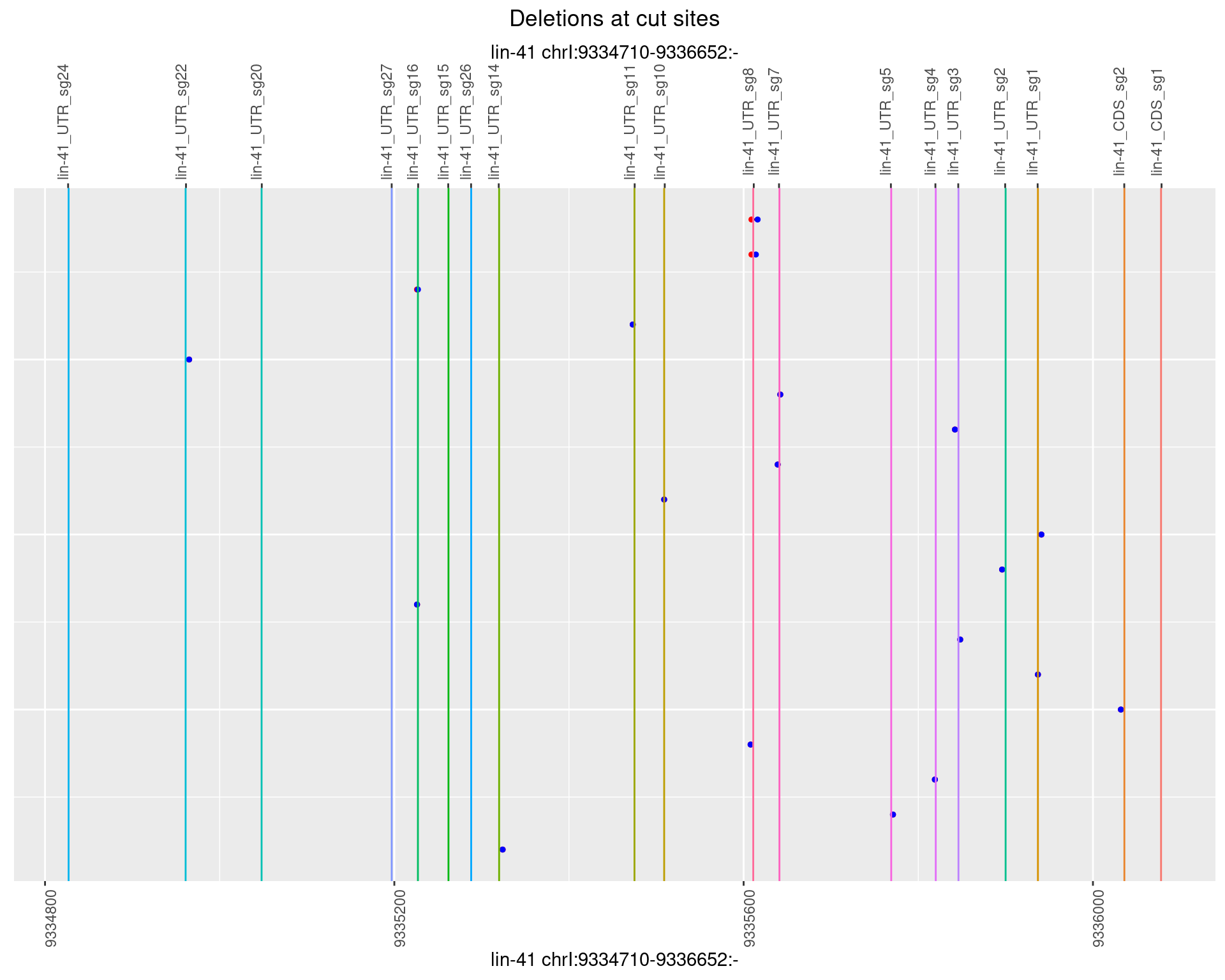
1.30.2 Freq: 1e-05
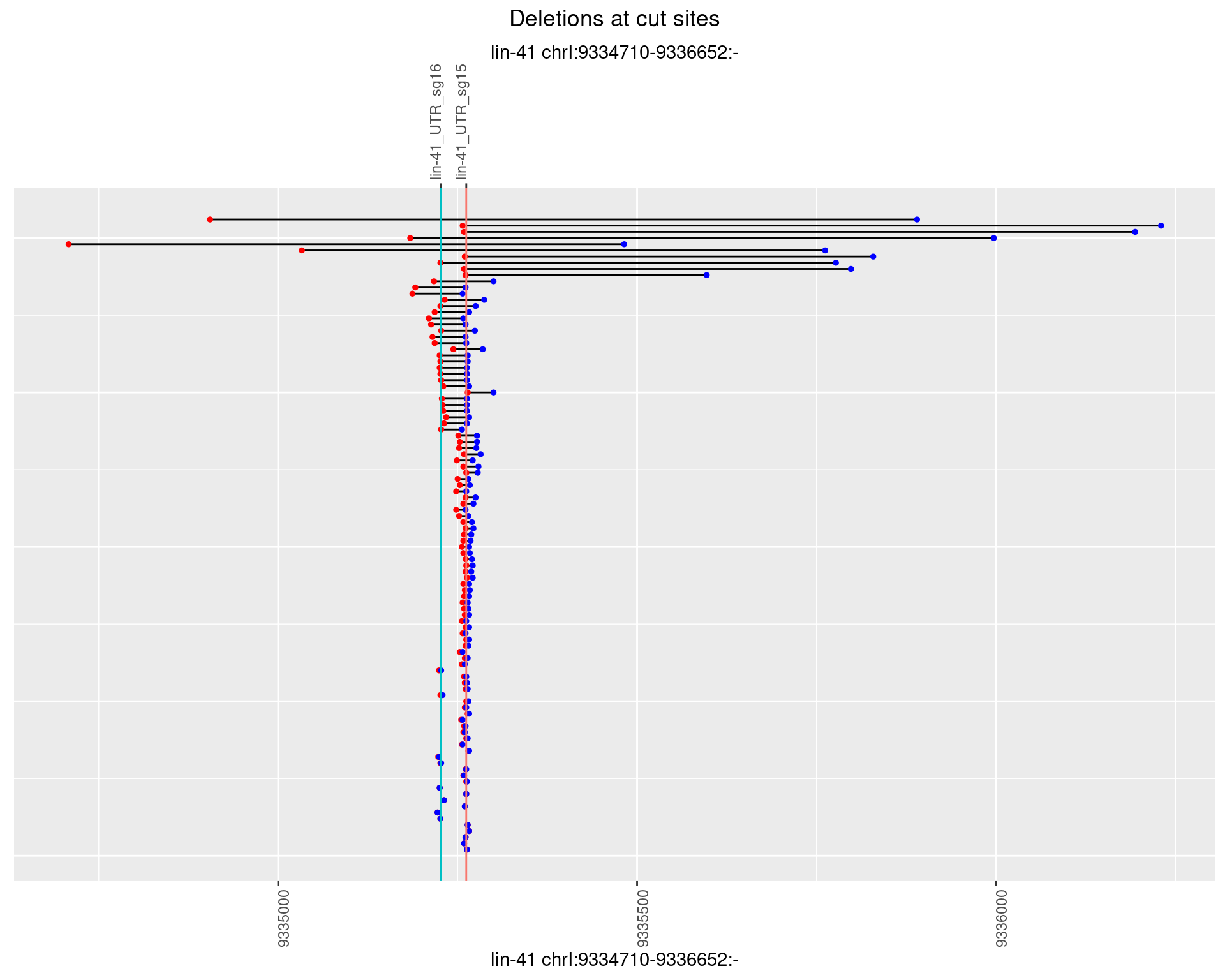
1.30.3 Freq: 1e-04
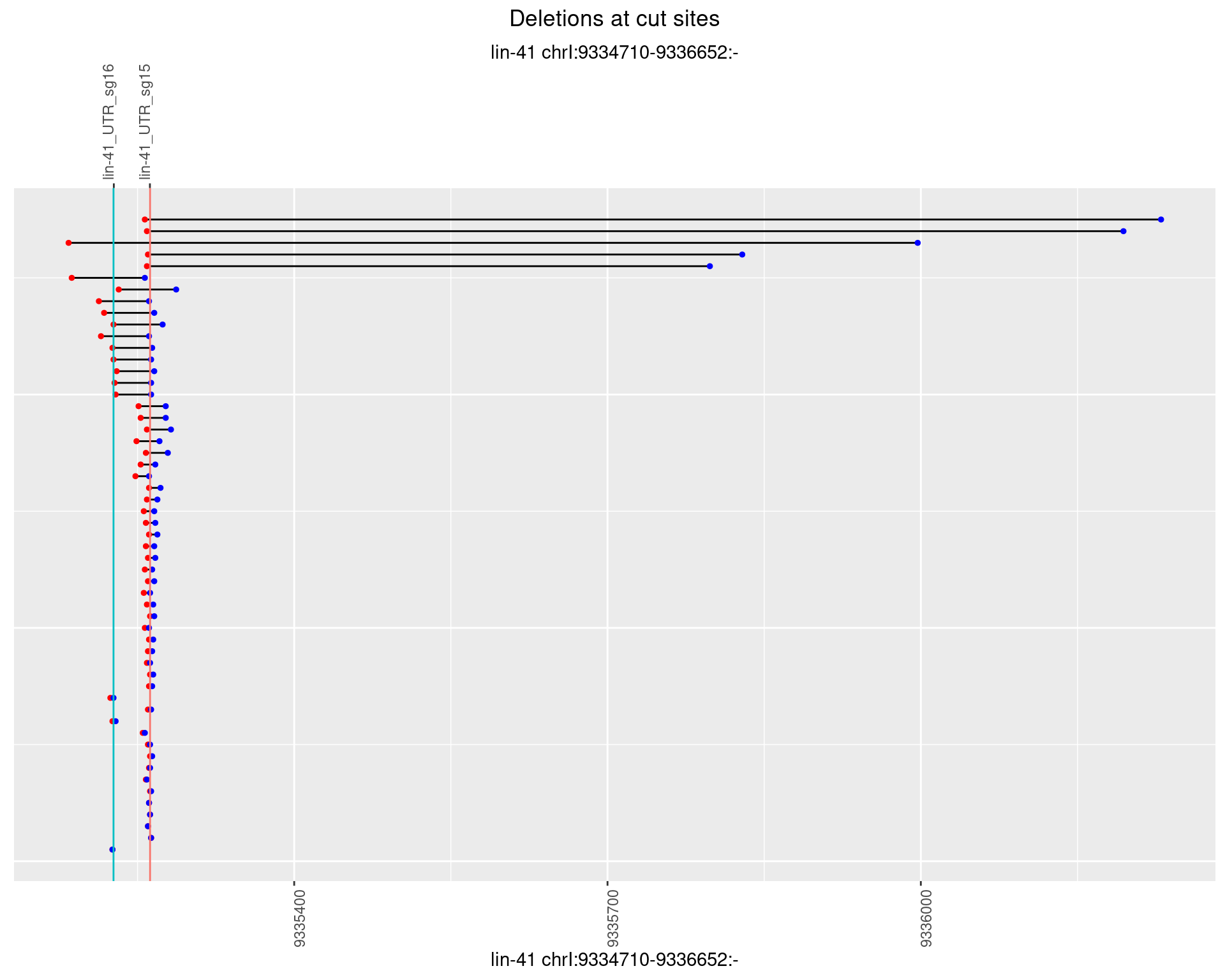
1.30.4 Freq: 0.001
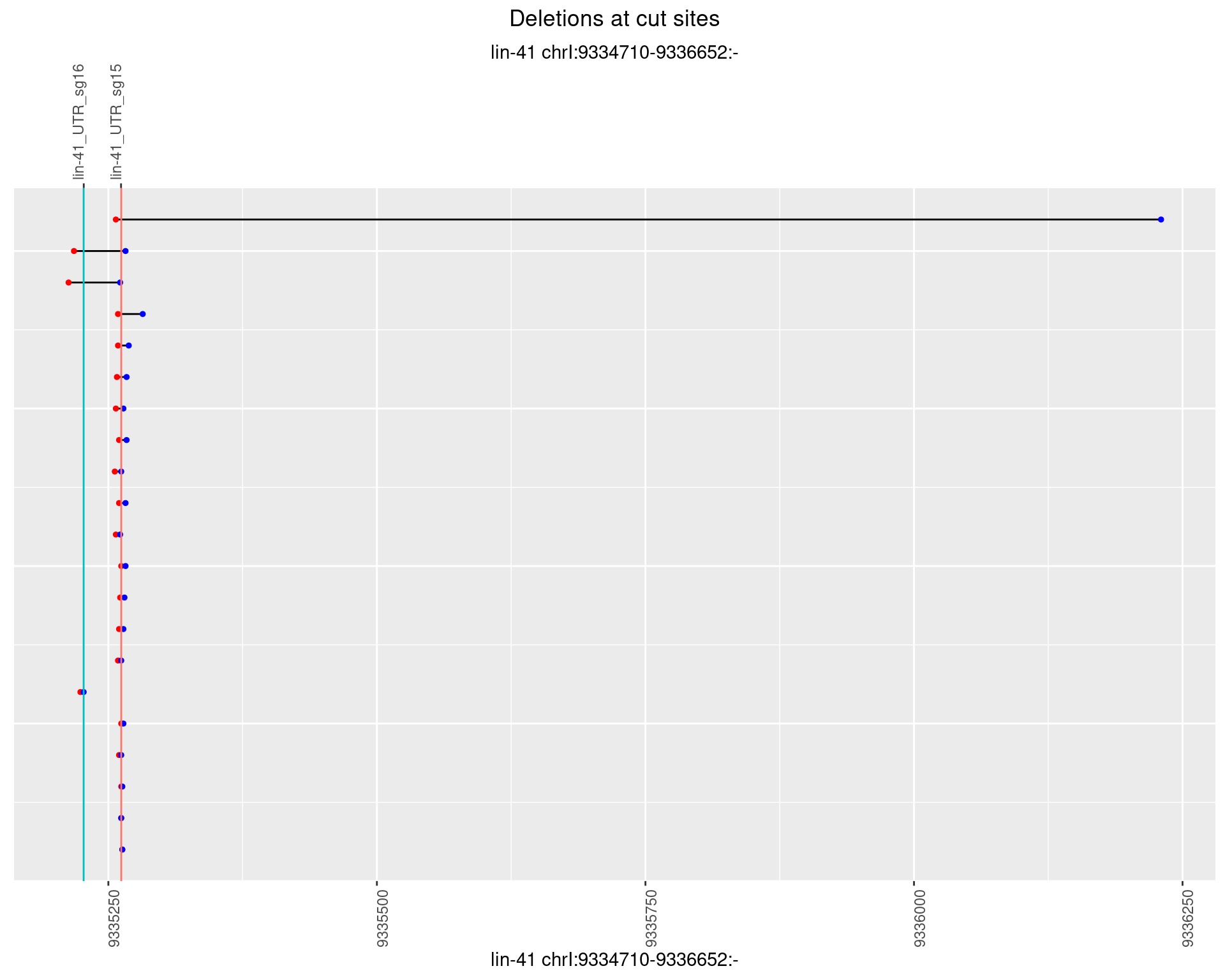
1.30.5 Freq: 0.01
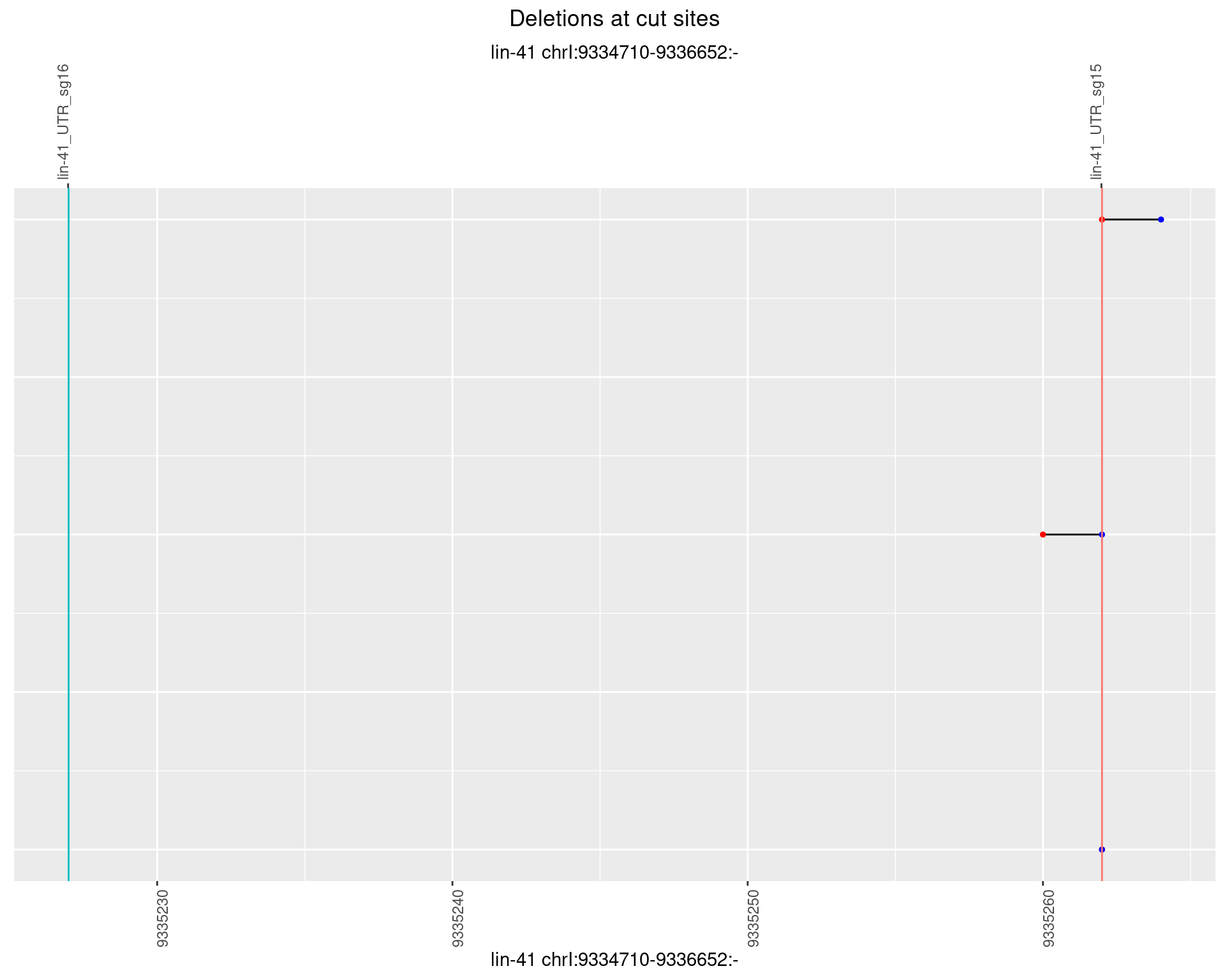
1.30.6 Freq: 0.1
No plot to show
1.31 gen_16C_F4_lin-41_pool
1.31.1 Freq: 0
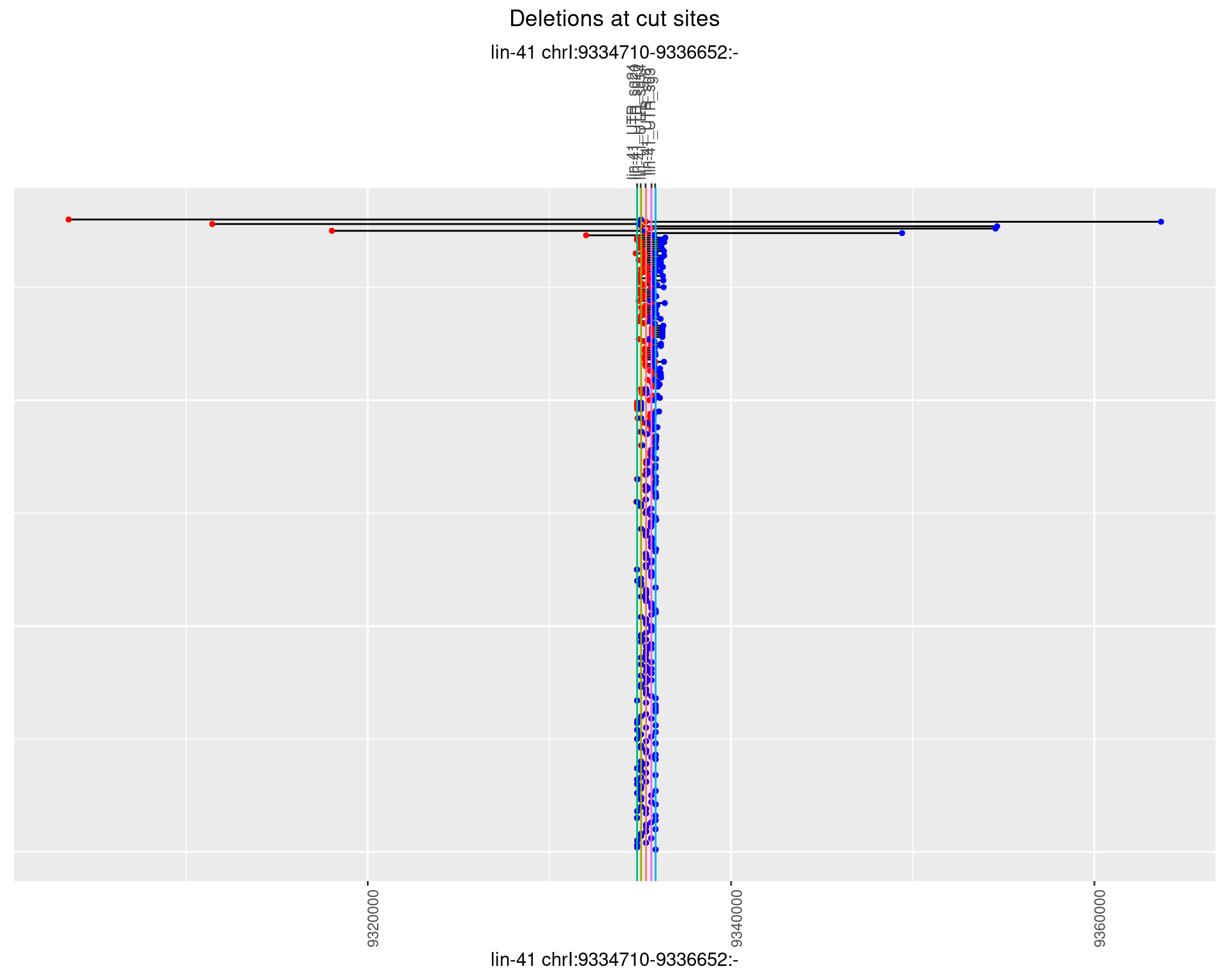
1.31.2 Freq: 1e-05
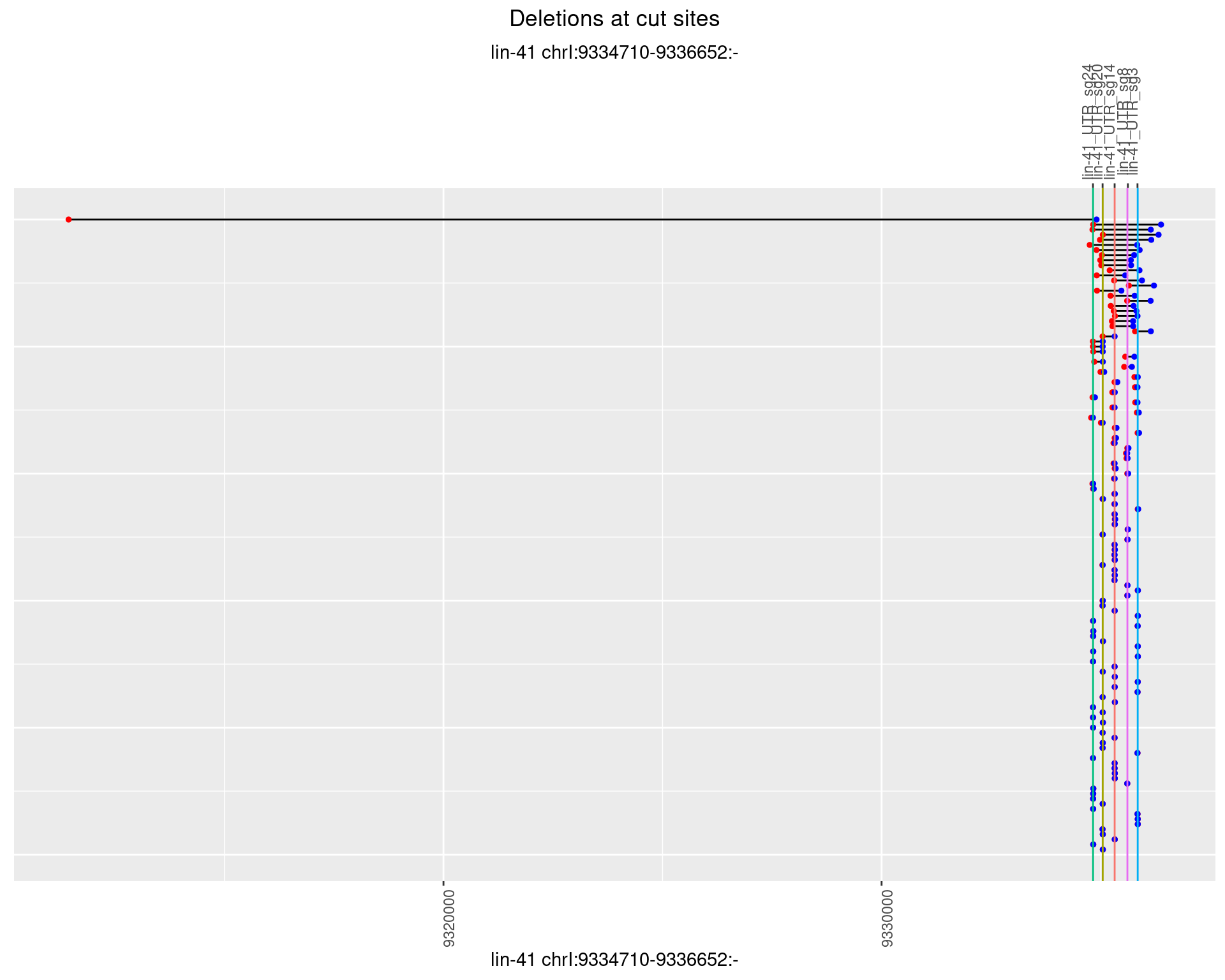
1.31.3 Freq: 1e-04
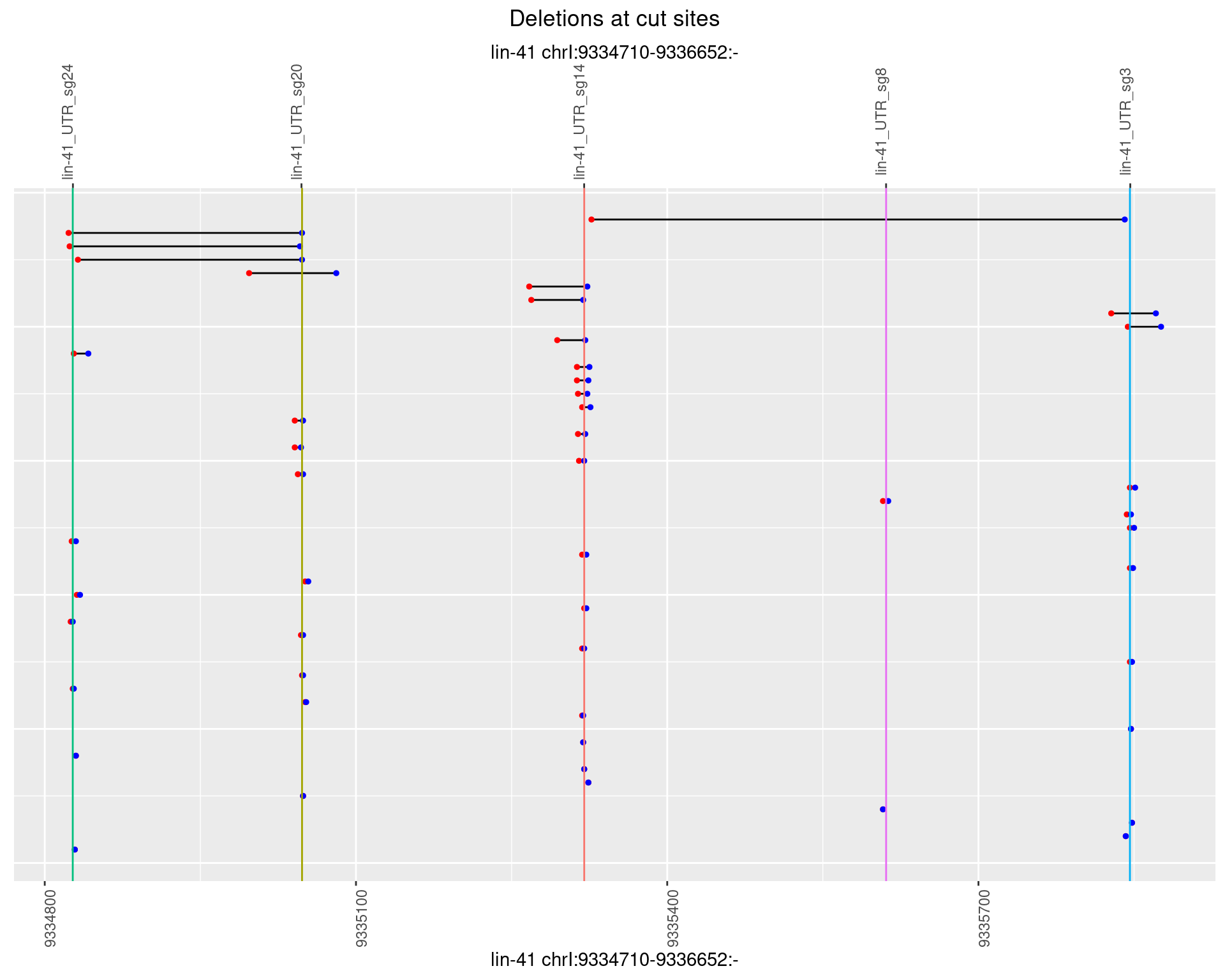
1.31.4 Freq: 0.001
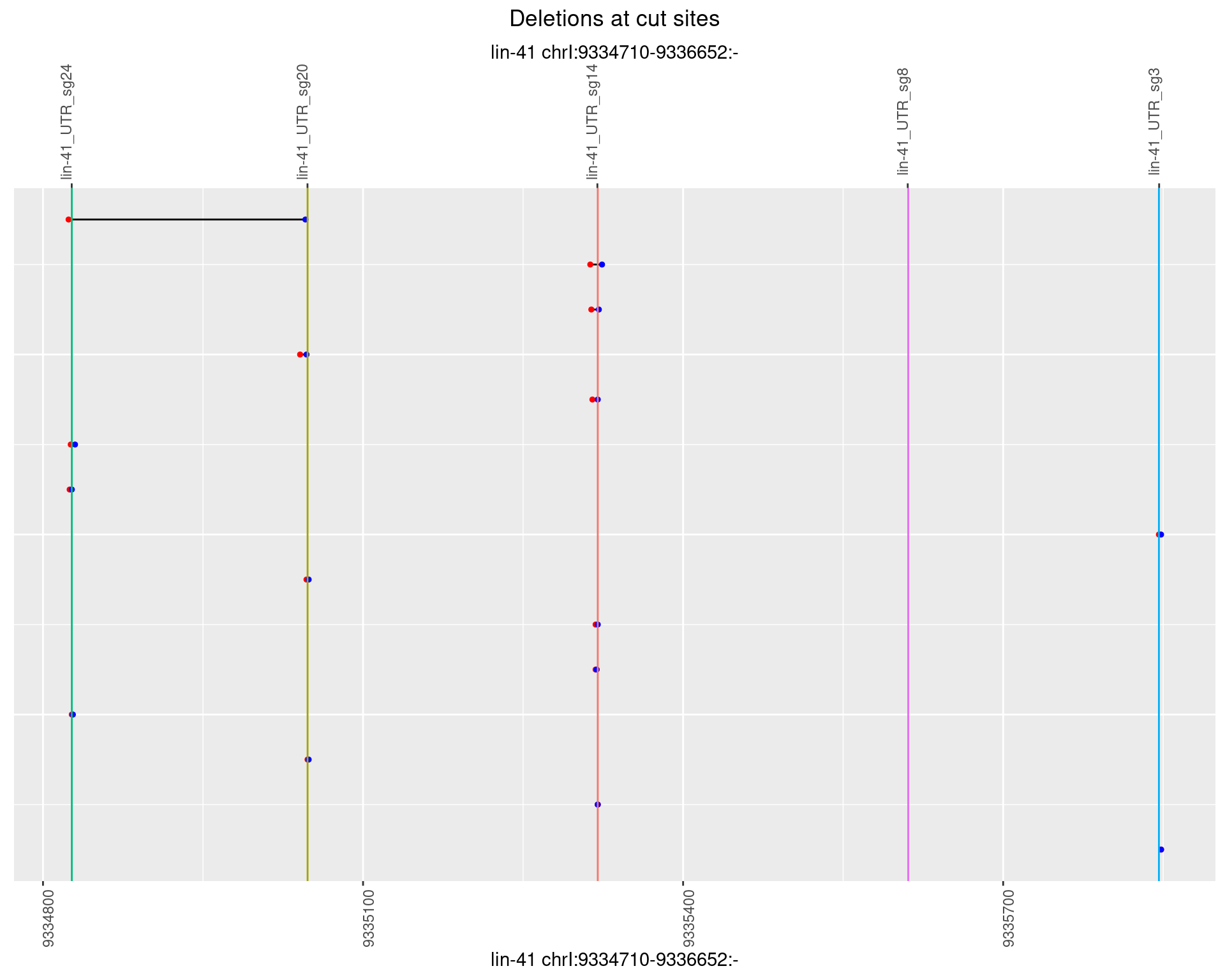
1.31.5 Freq: 0.01
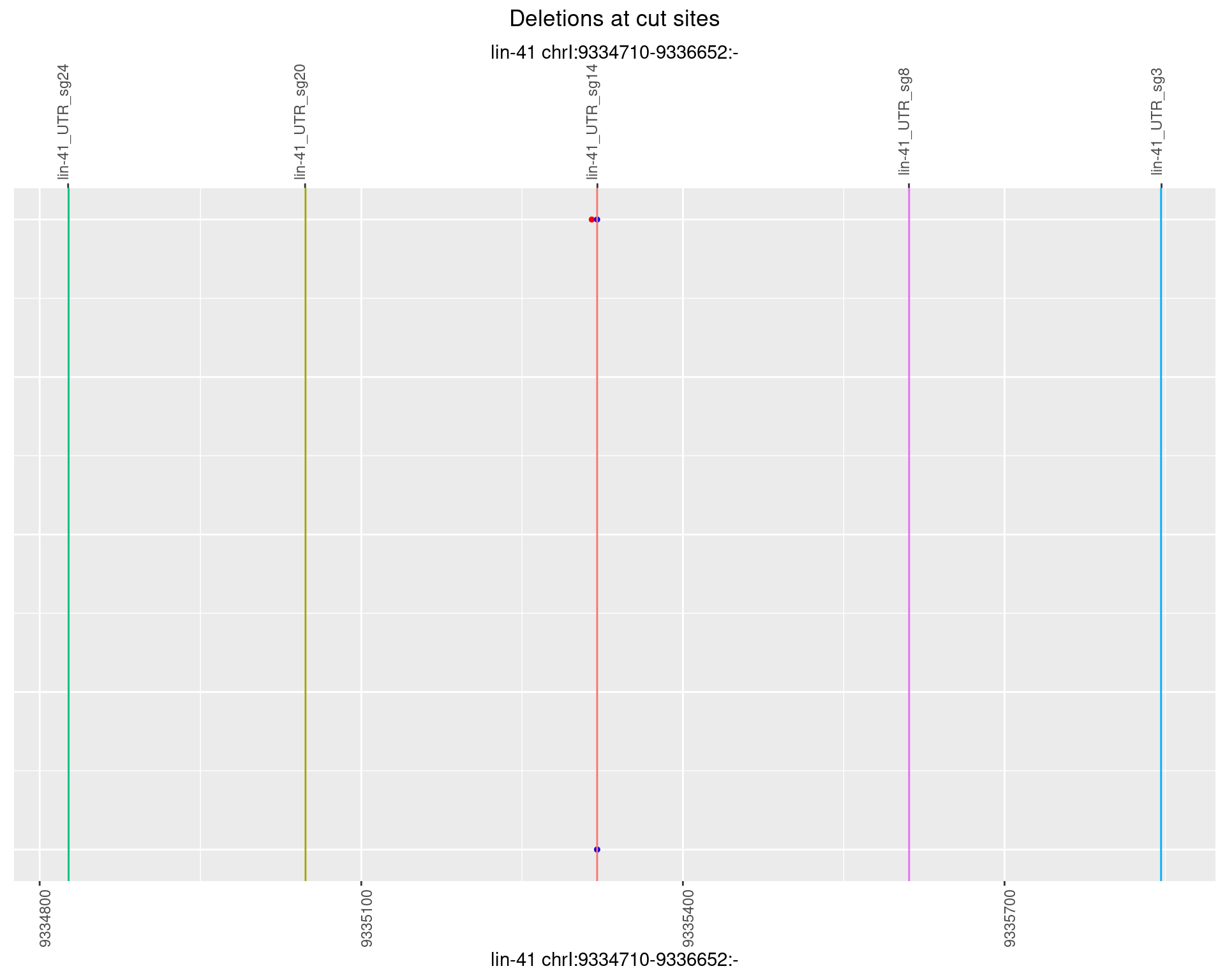
1.31.6 Freq: 0.1
No plot to show
1.32 gen_16C_F4_lin-41_sg26sg27
1.32.1 Freq: 0
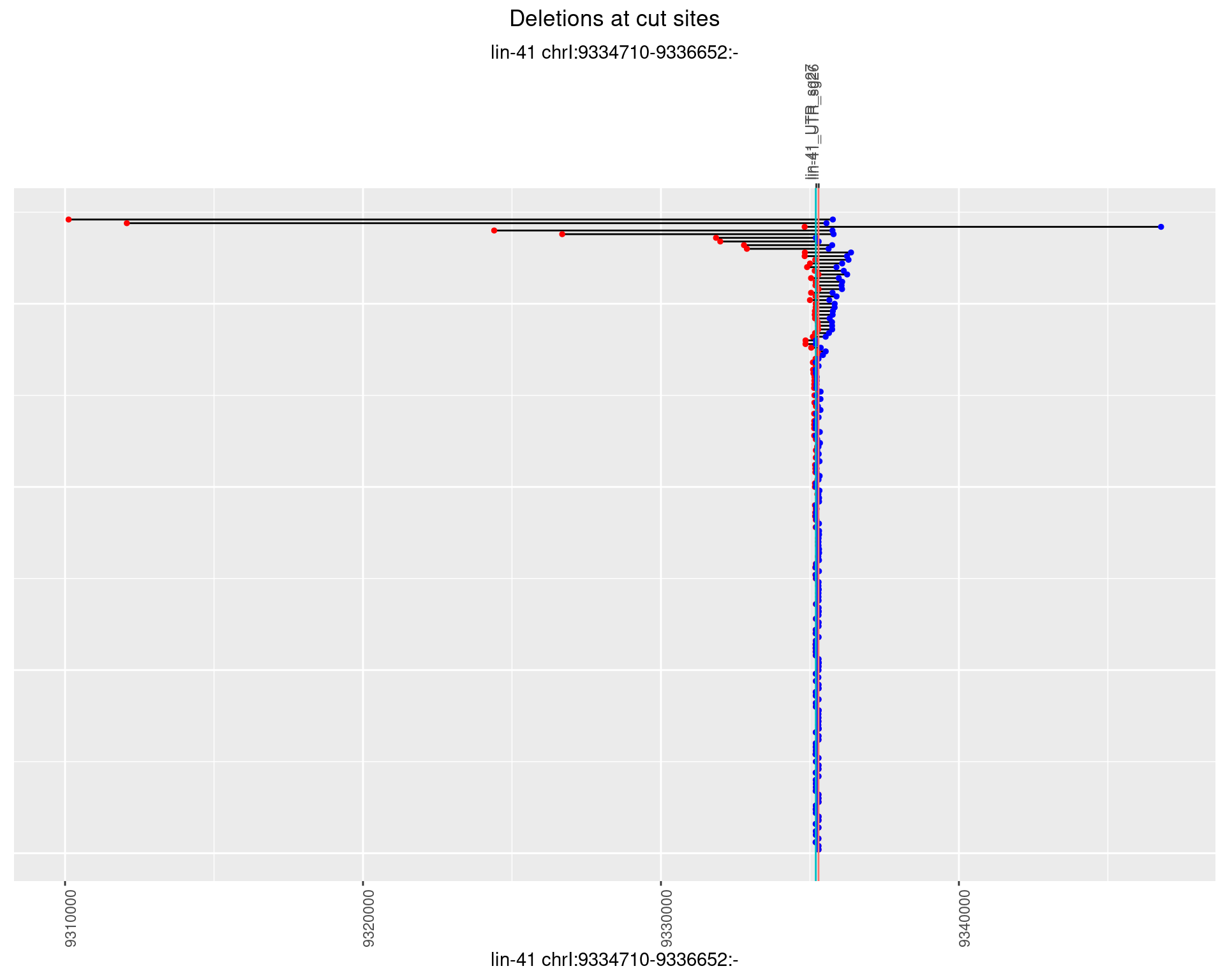
1.32.2 Freq: 1e-05
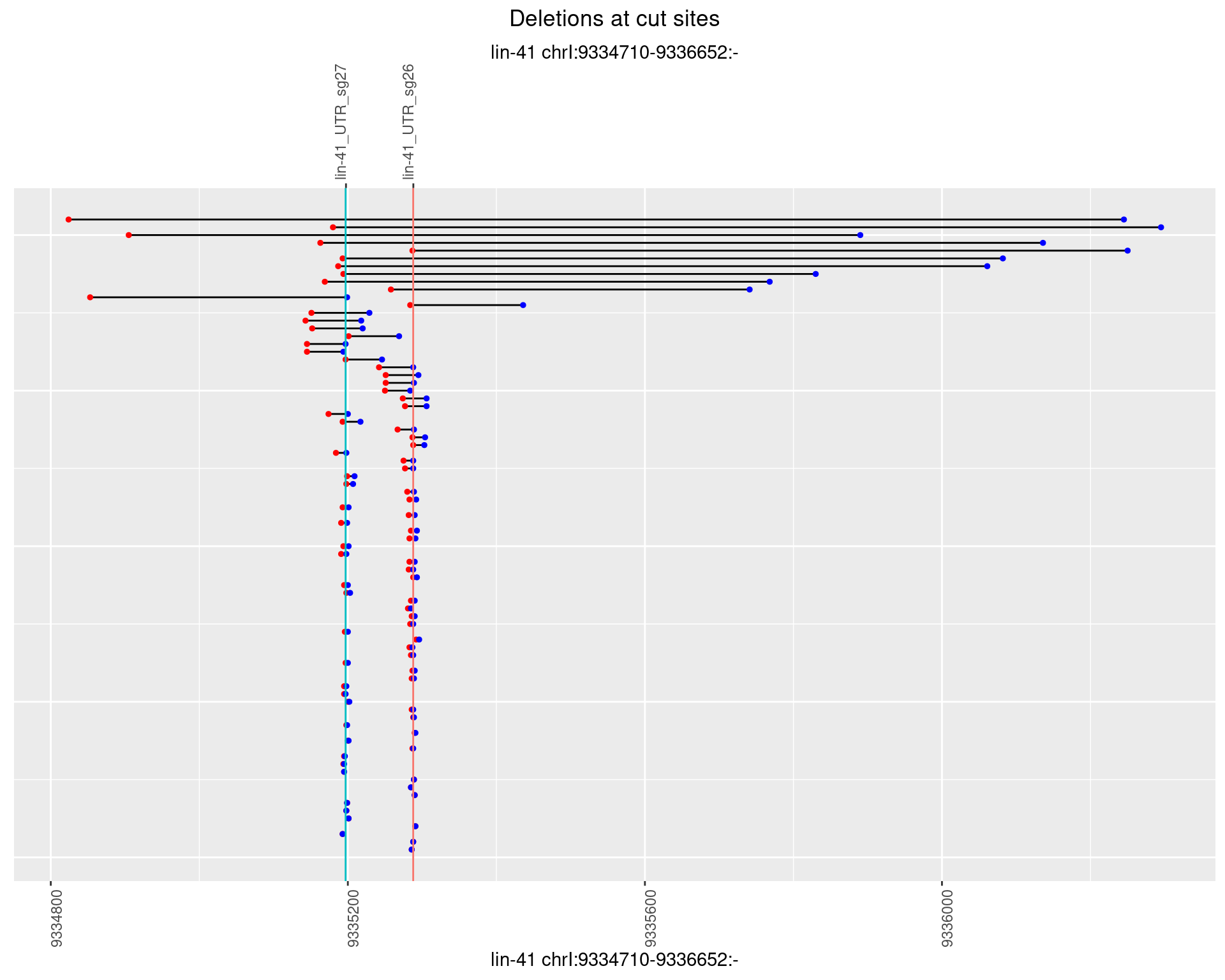
1.32.3 Freq: 1e-04
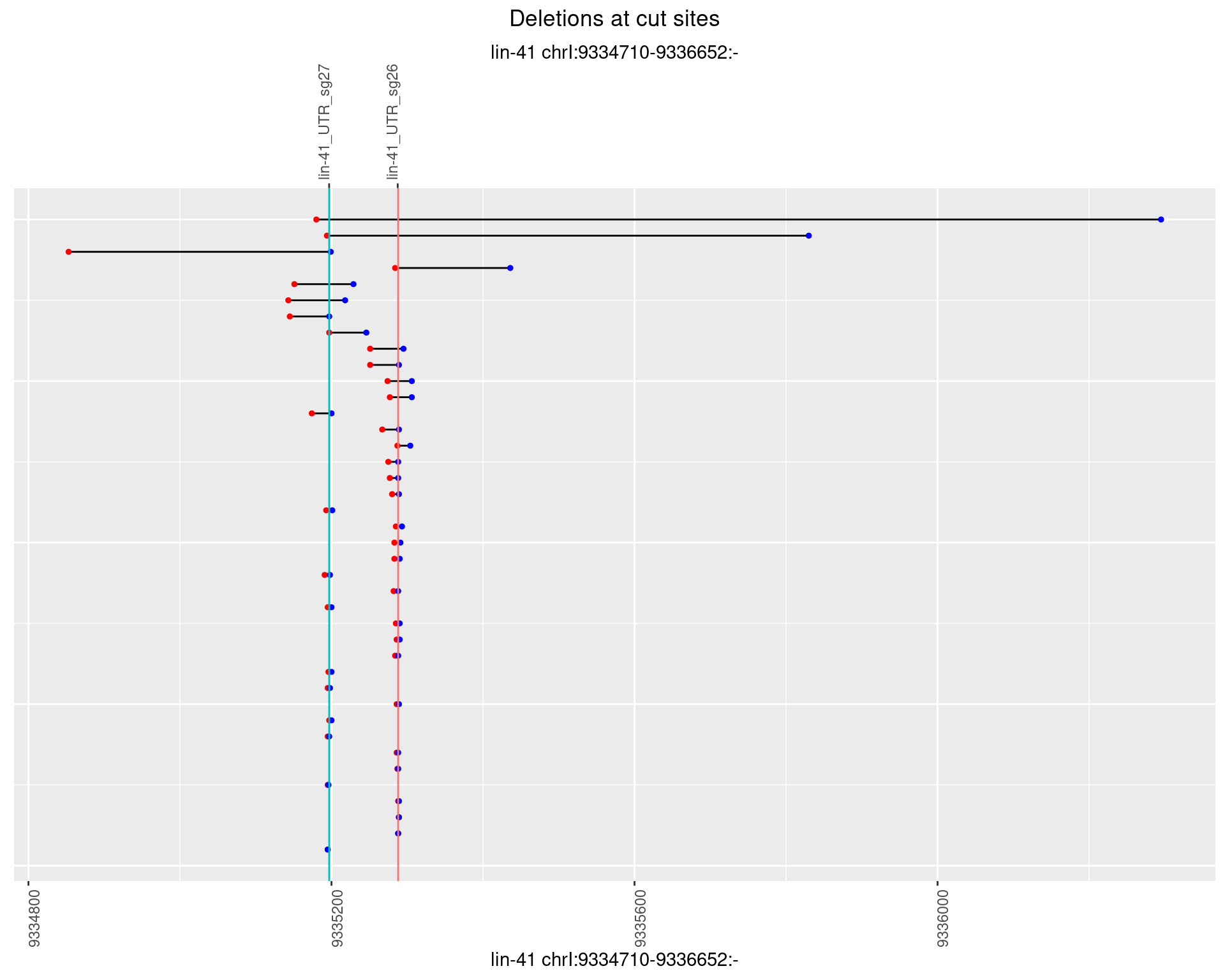
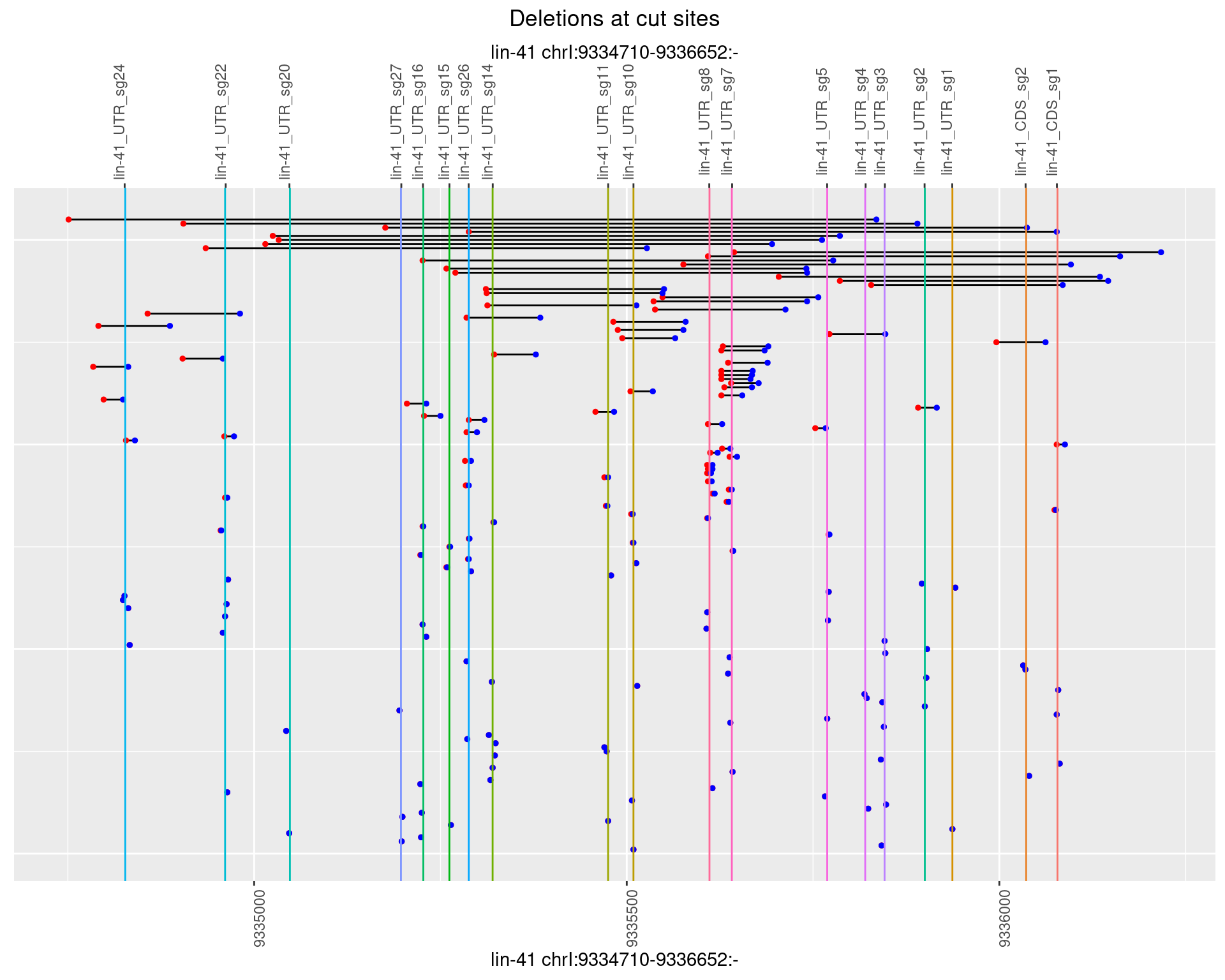
1.32.4 Freq: 0.001
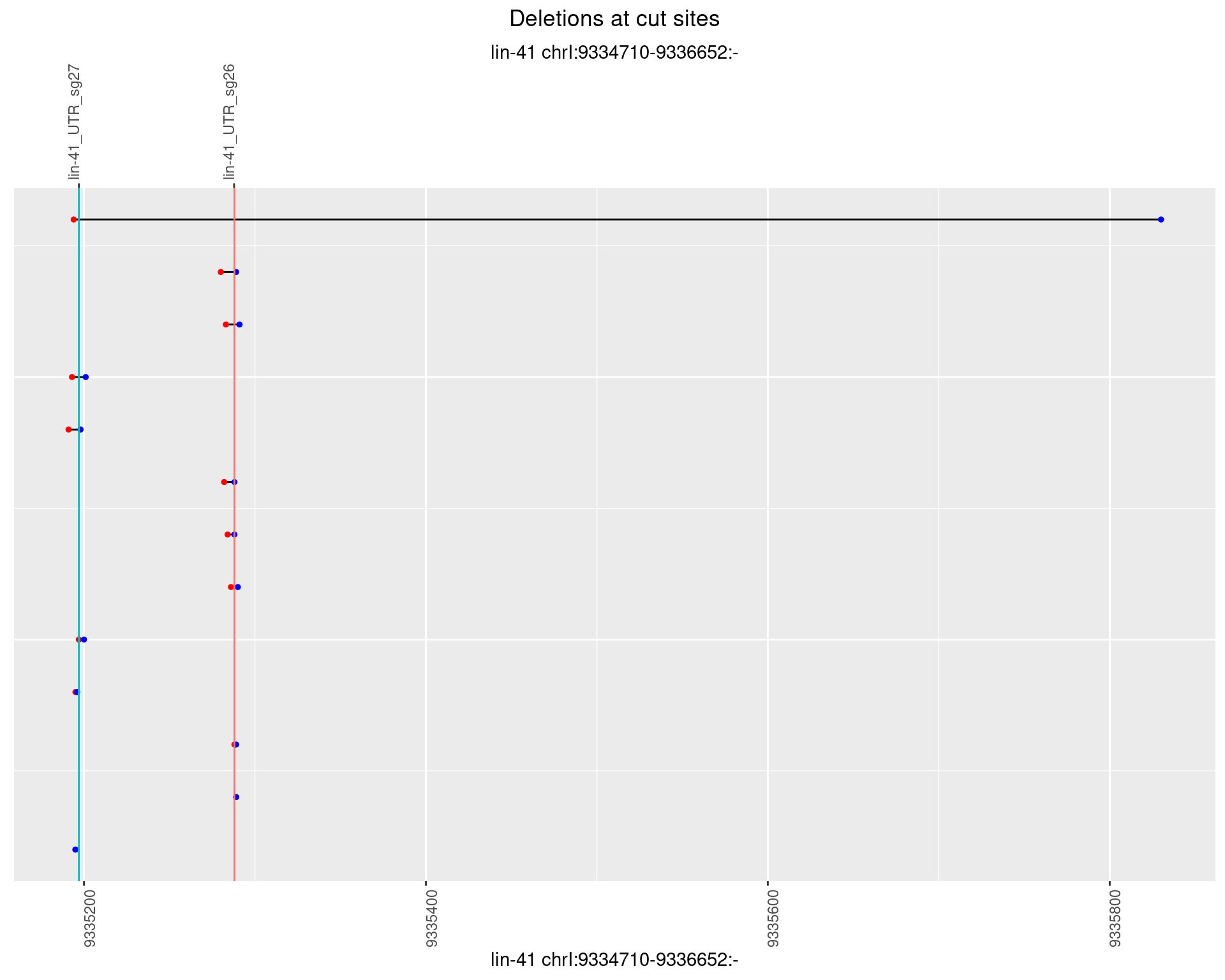
1.32.5 Freq: 0.01
1.32.6 Freq: 0.1
No plot to show
1.33 gen_16C_F5_lin-41_N2
1.33.1 Freq: 0
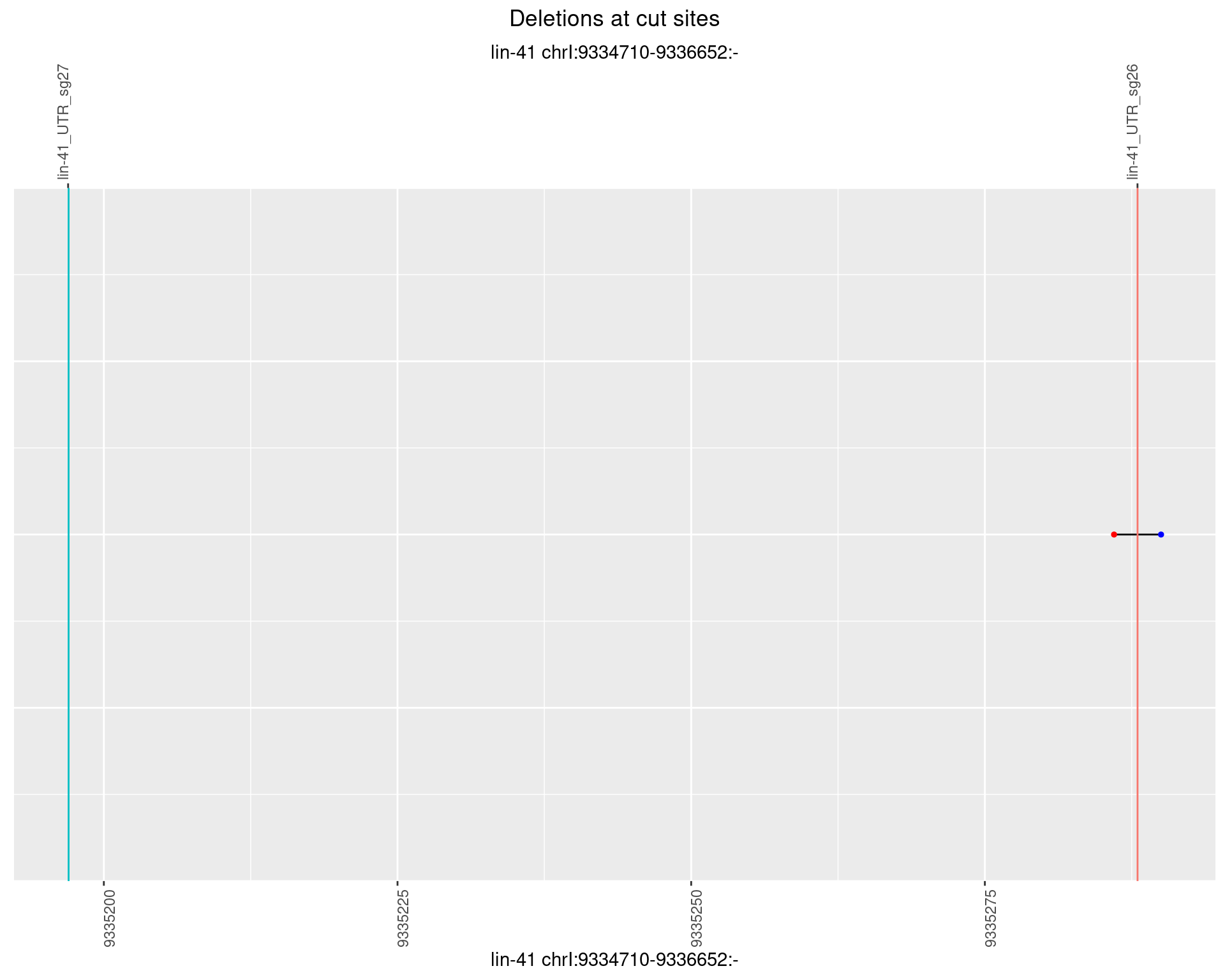
1.33.2 Freq: 1e-05
1.33.3 Freq: 1e-04
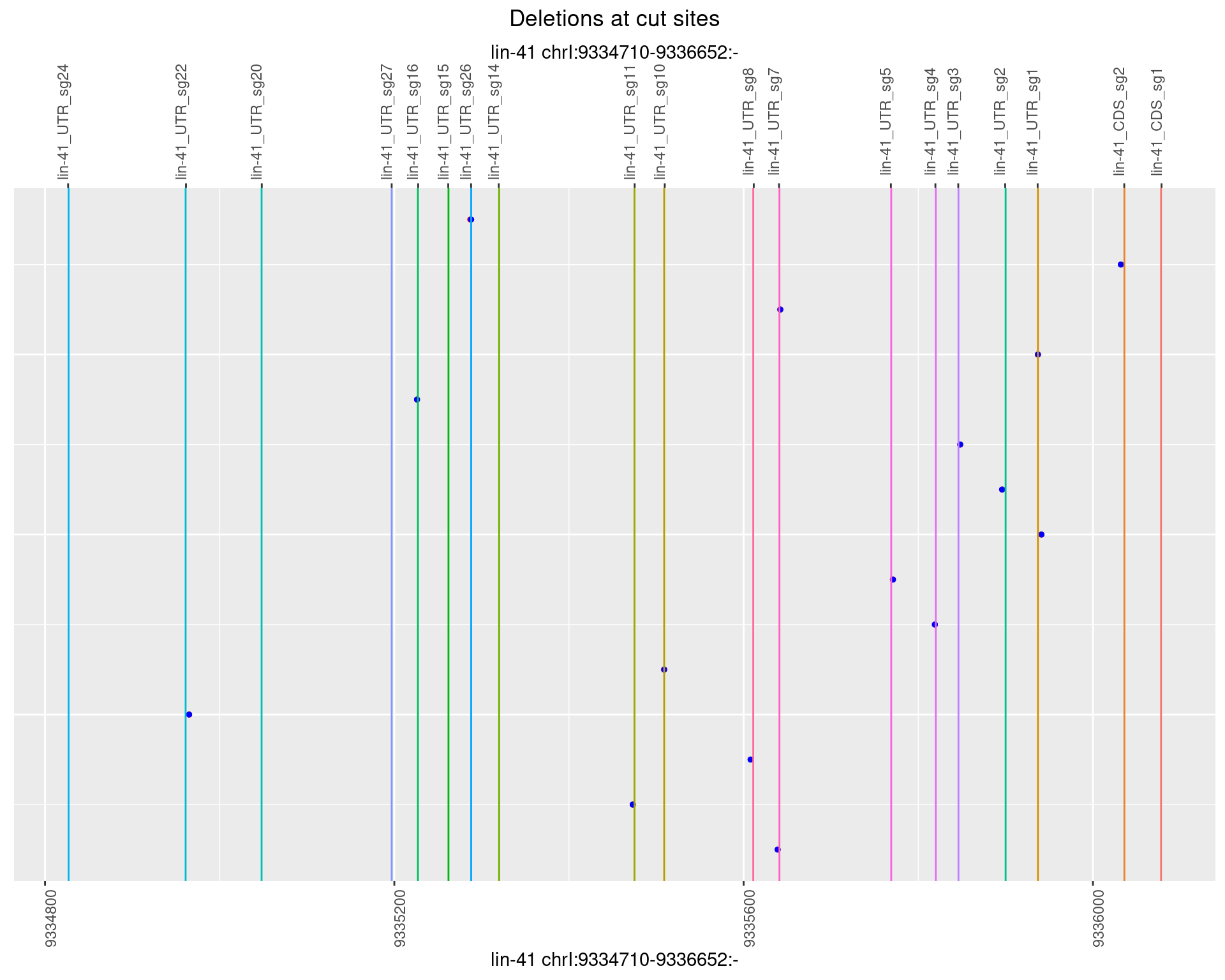
1.33.4 Freq: 0.001
1.33.5 Freq: 0.01
No plot to show
1.33.6 Freq: 0.1
No plot to show
1.34 gen_16C_F5_lin-41_sg15sg16
1.34.1 Freq: 0
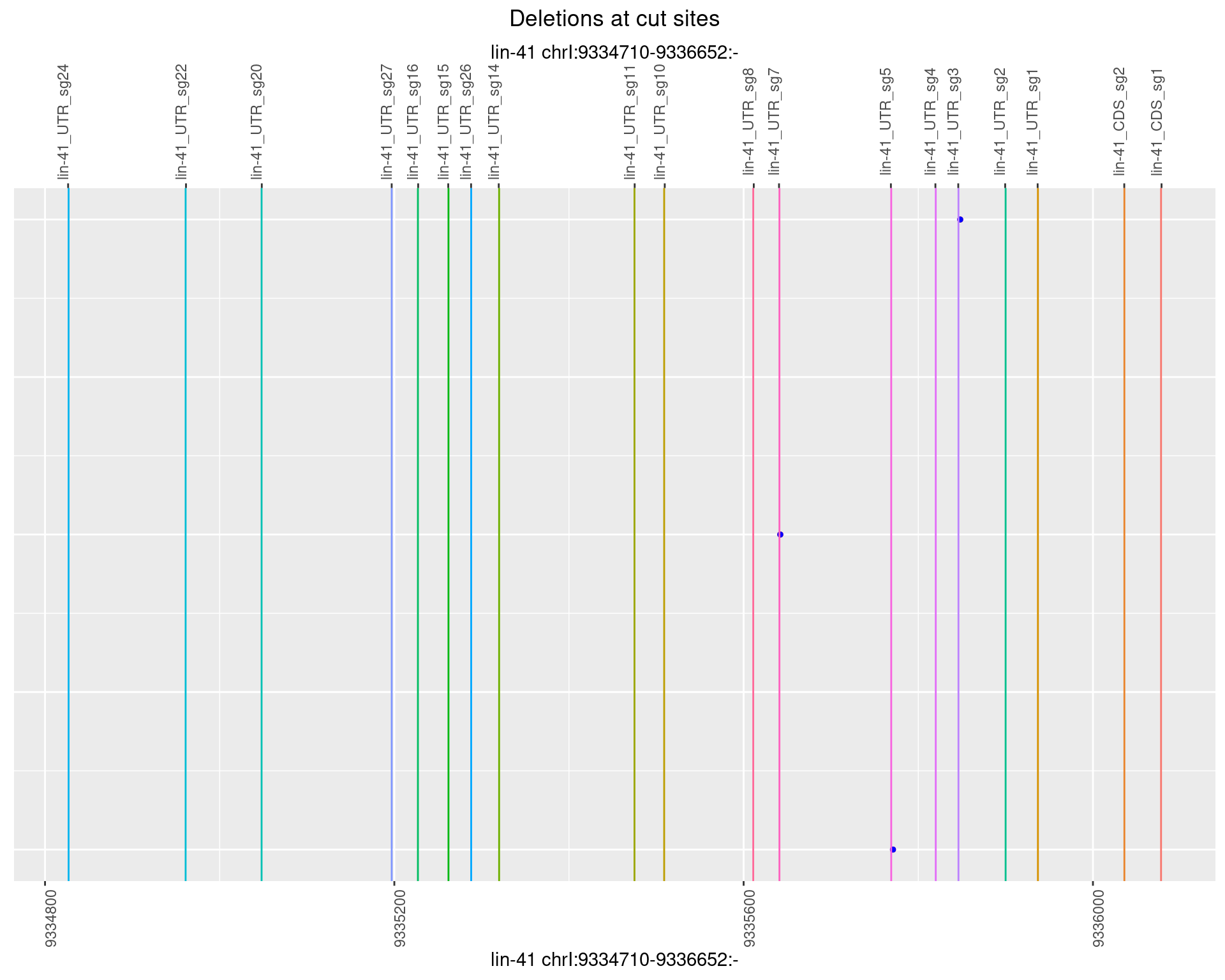
1.34.2 Freq: 1e-05
1.34.3 Freq: 1e-04
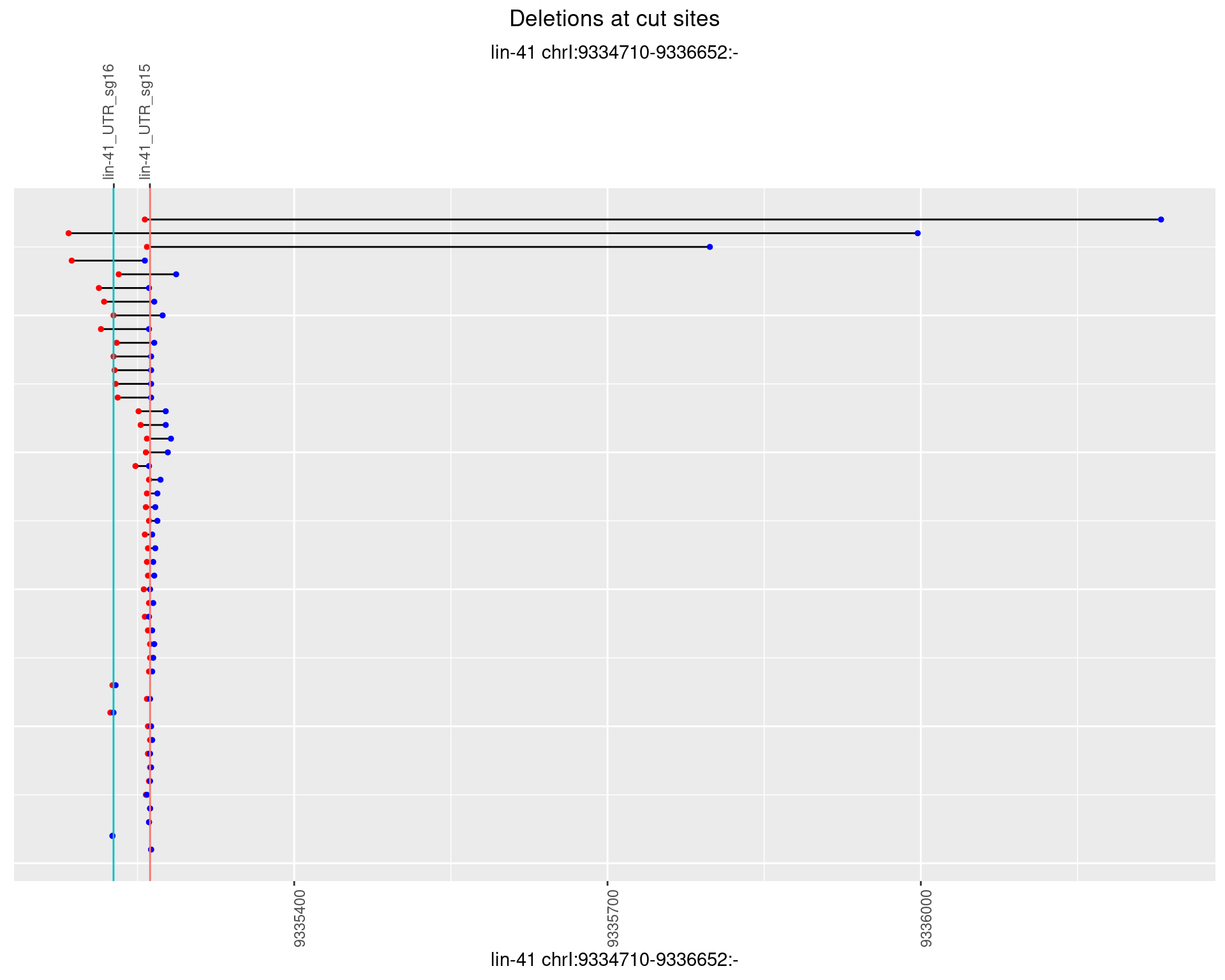
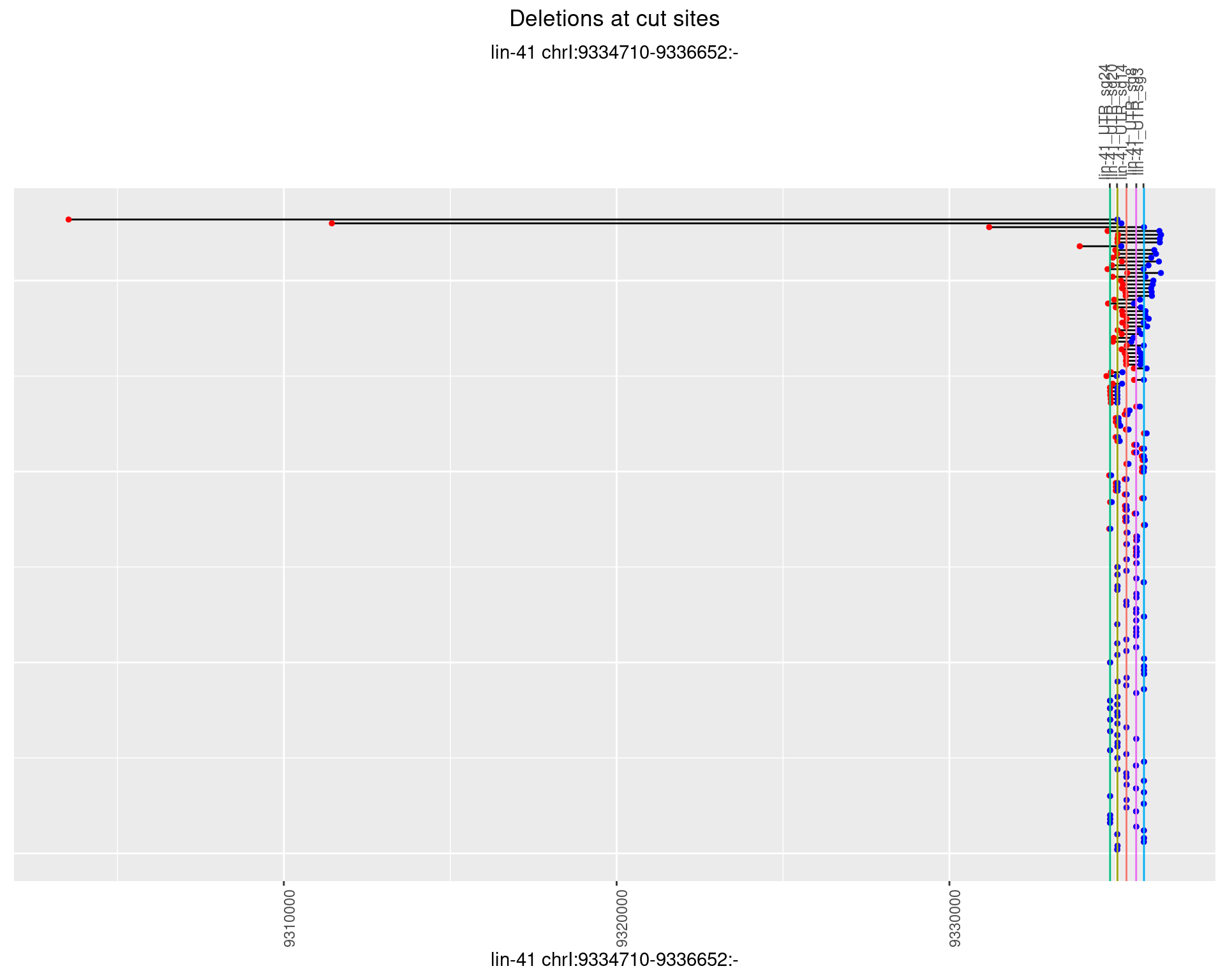
1.34.4 Freq: 0.001
1.34.5 Freq: 0.01
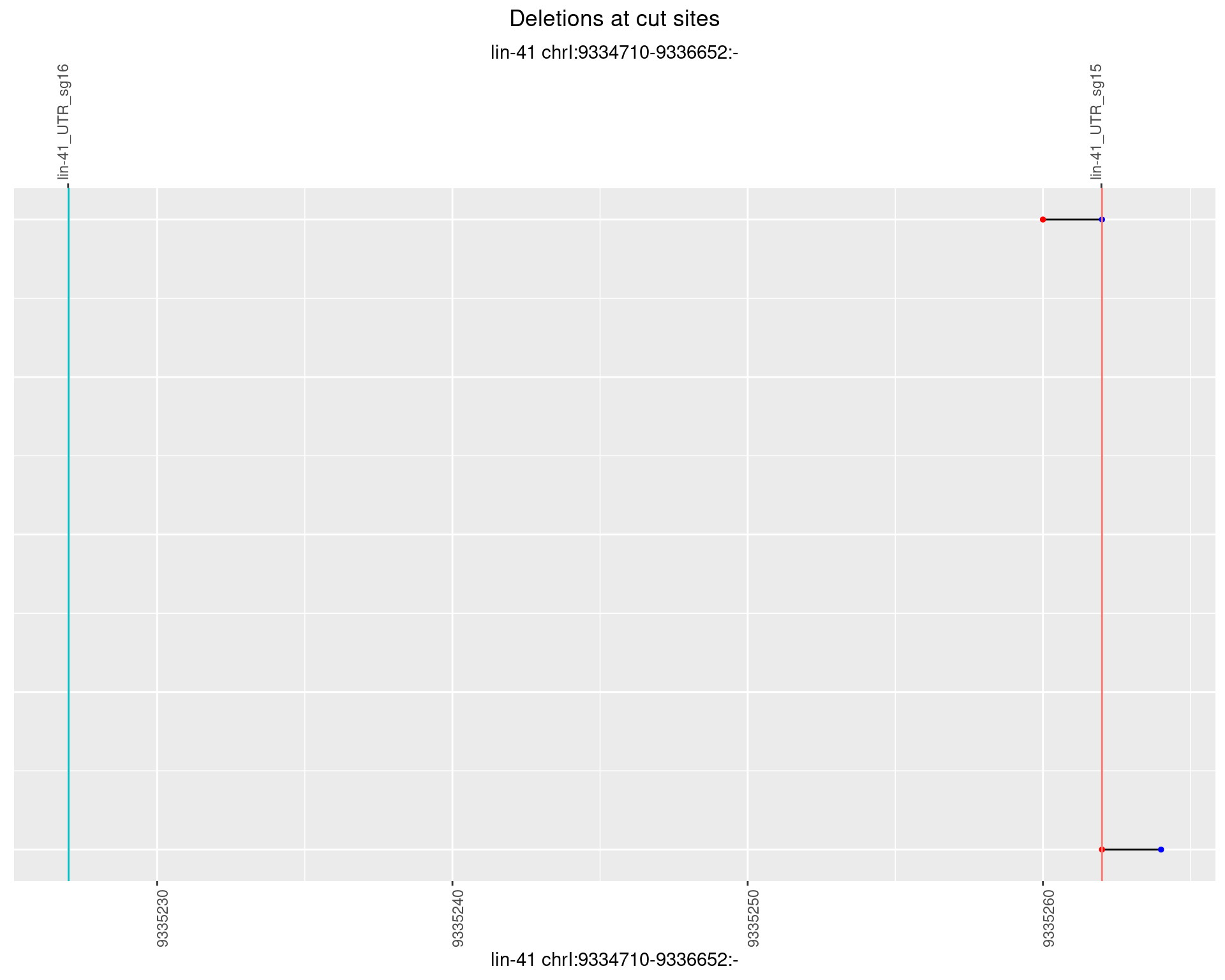
1.34.6 Freq: 0.1
No plot to show
1.35 gen_16C_F5_lin-41_pool
1.35.1 Freq: 0
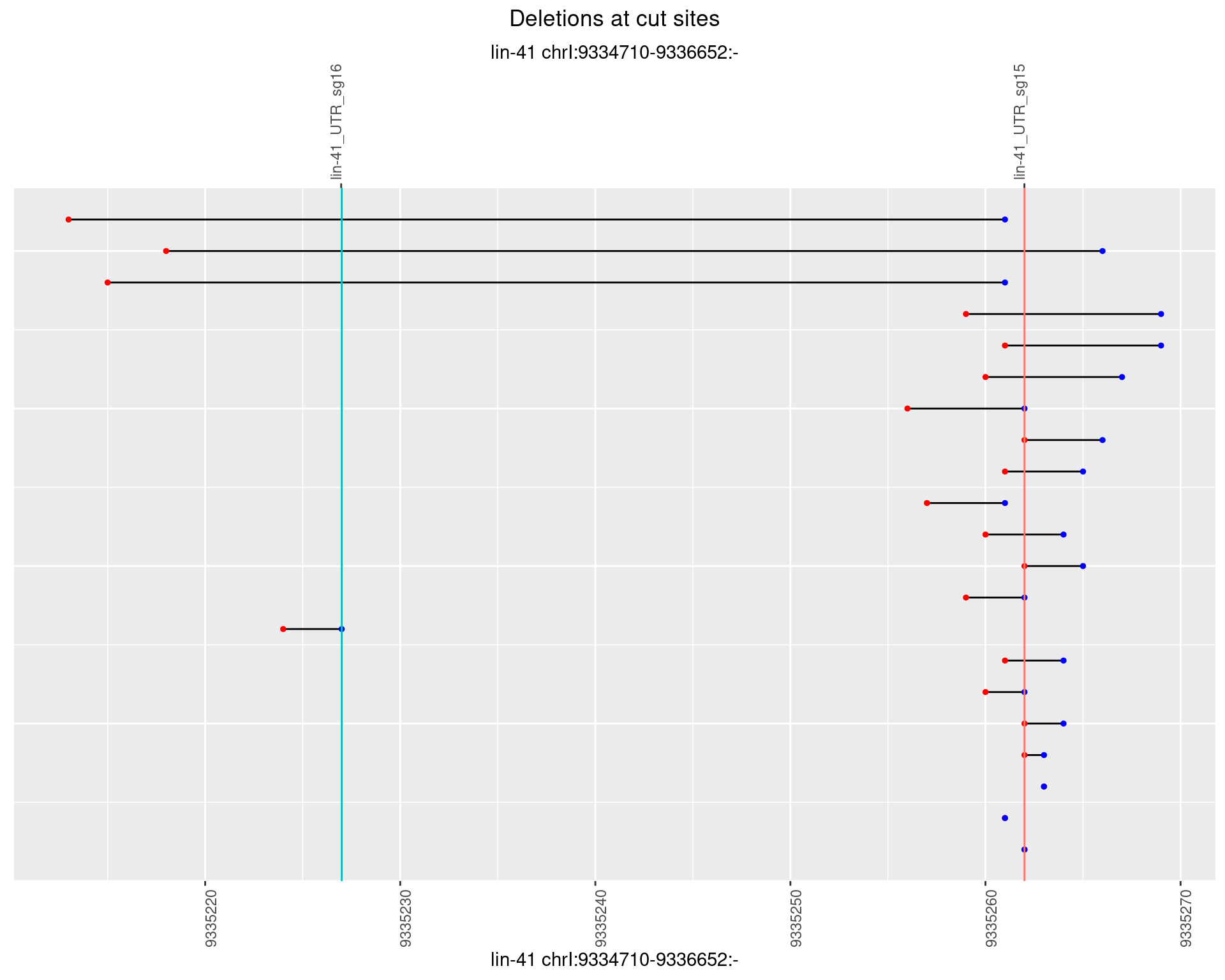
1.35.2 Freq: 1e-05
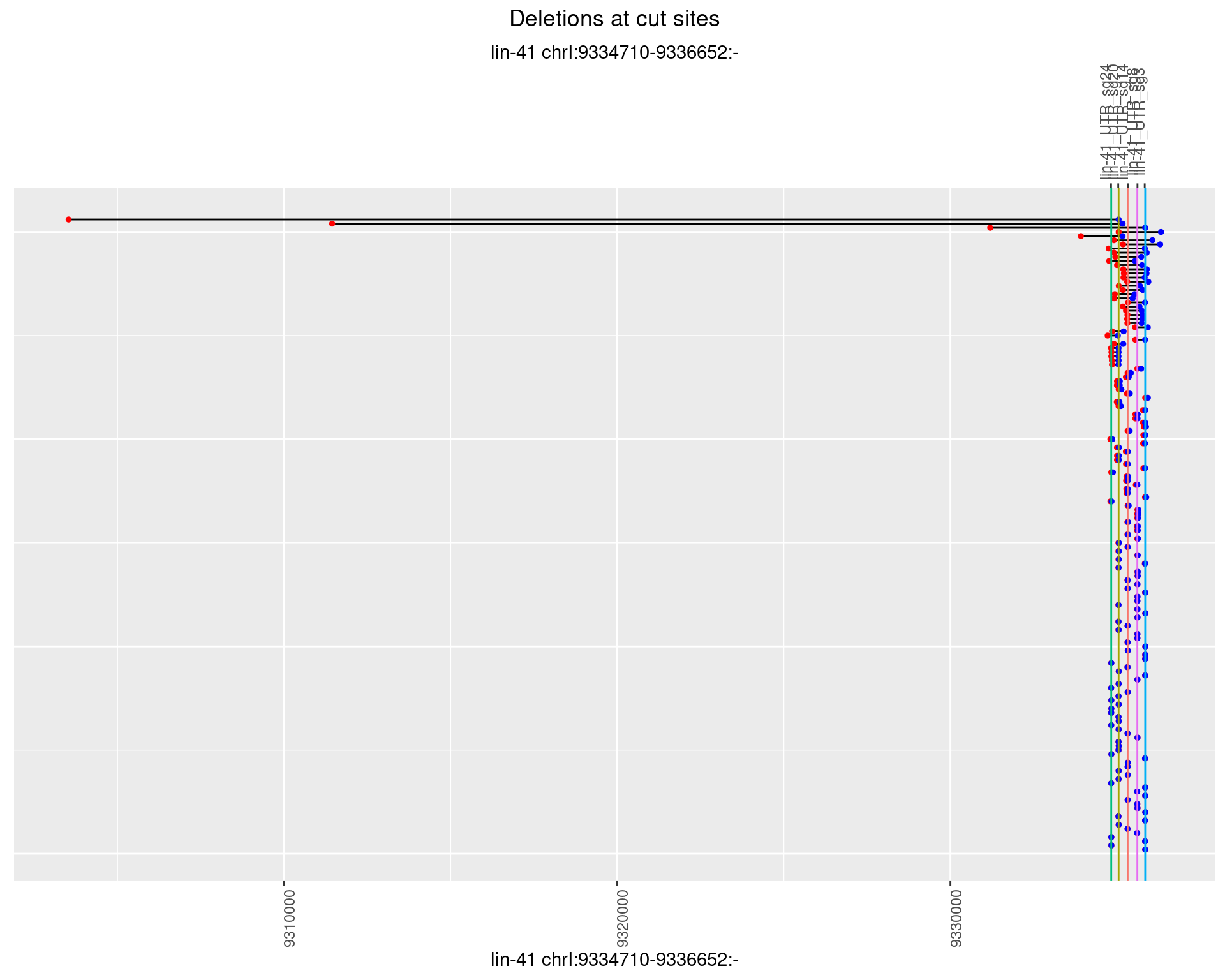
1.35.3 Freq: 1e-04
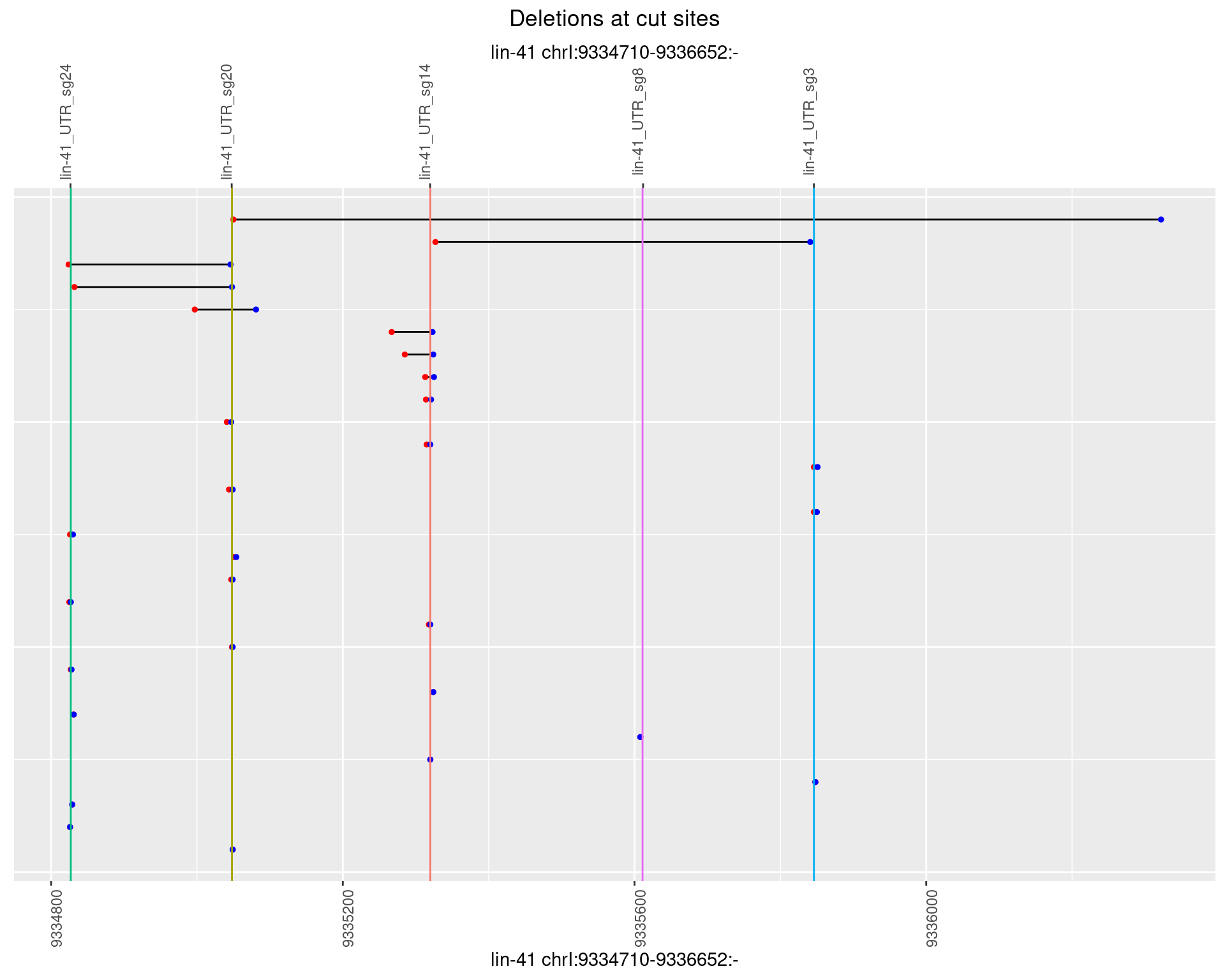
1.35.4 Freq: 0.001
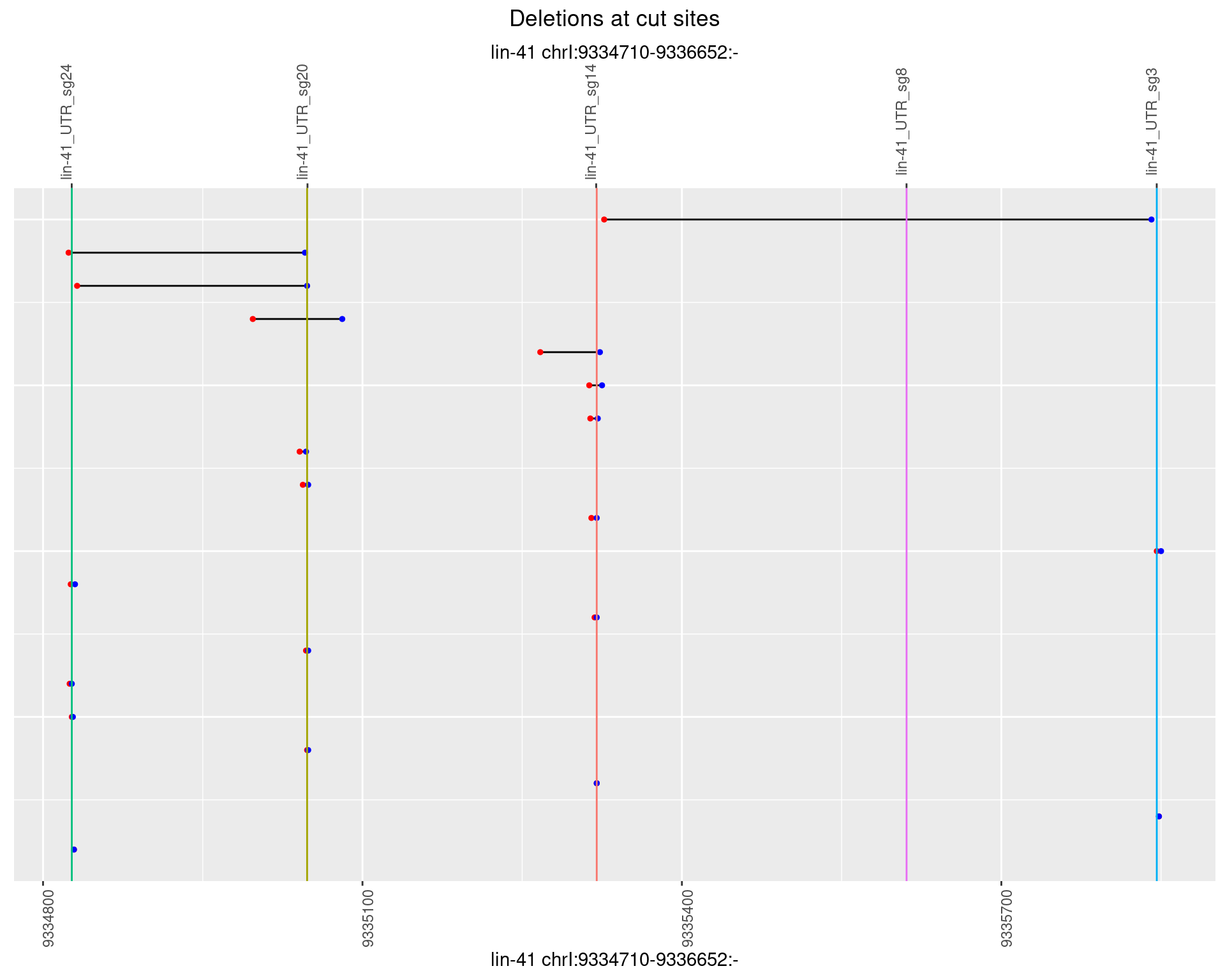
1.35.5 Freq: 0.01
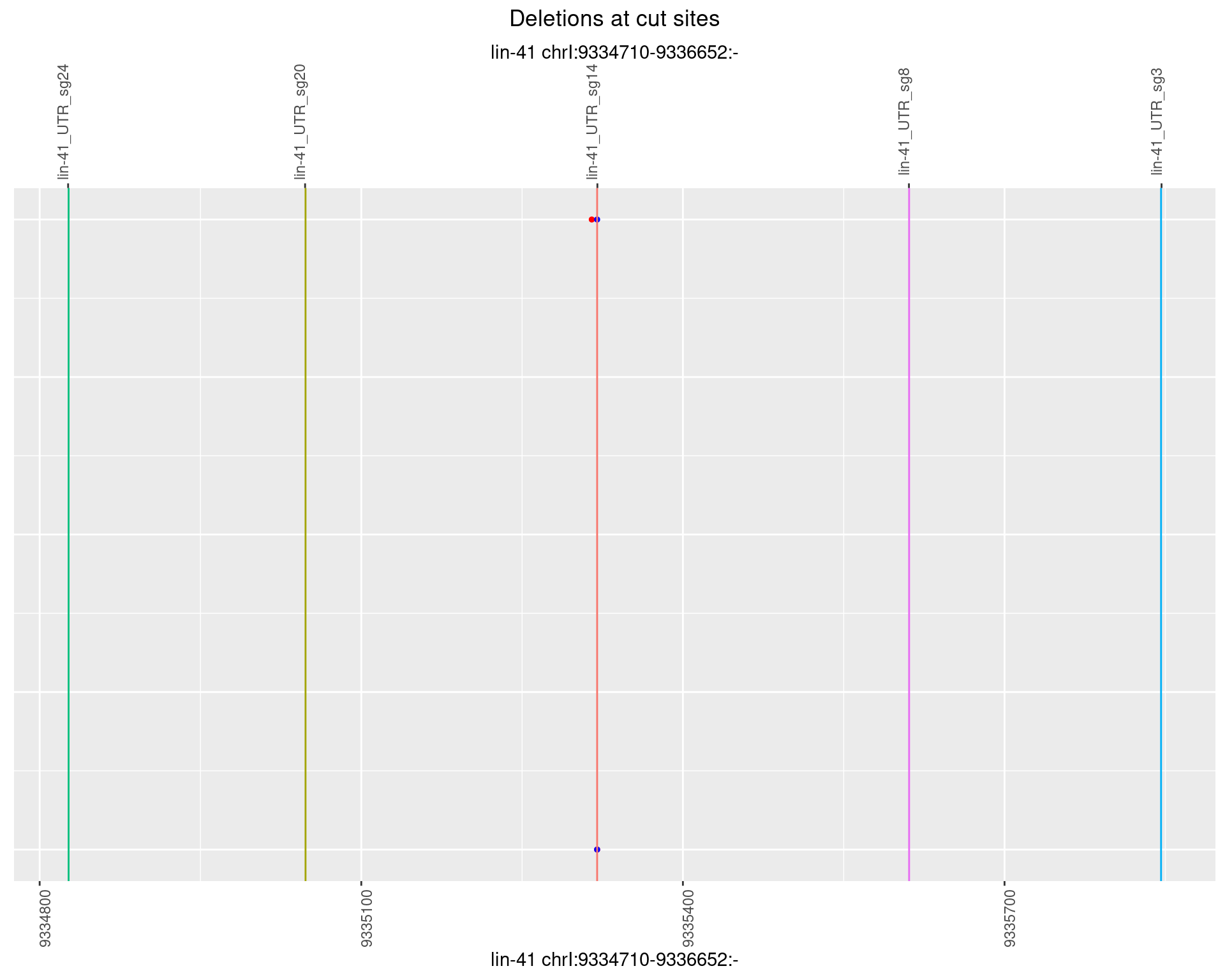
1.35.6 Freq: 0.1
No plot to show
1.36 gen_16C_F5_lin-41_sg26sg27
1.36.1 Freq: 0
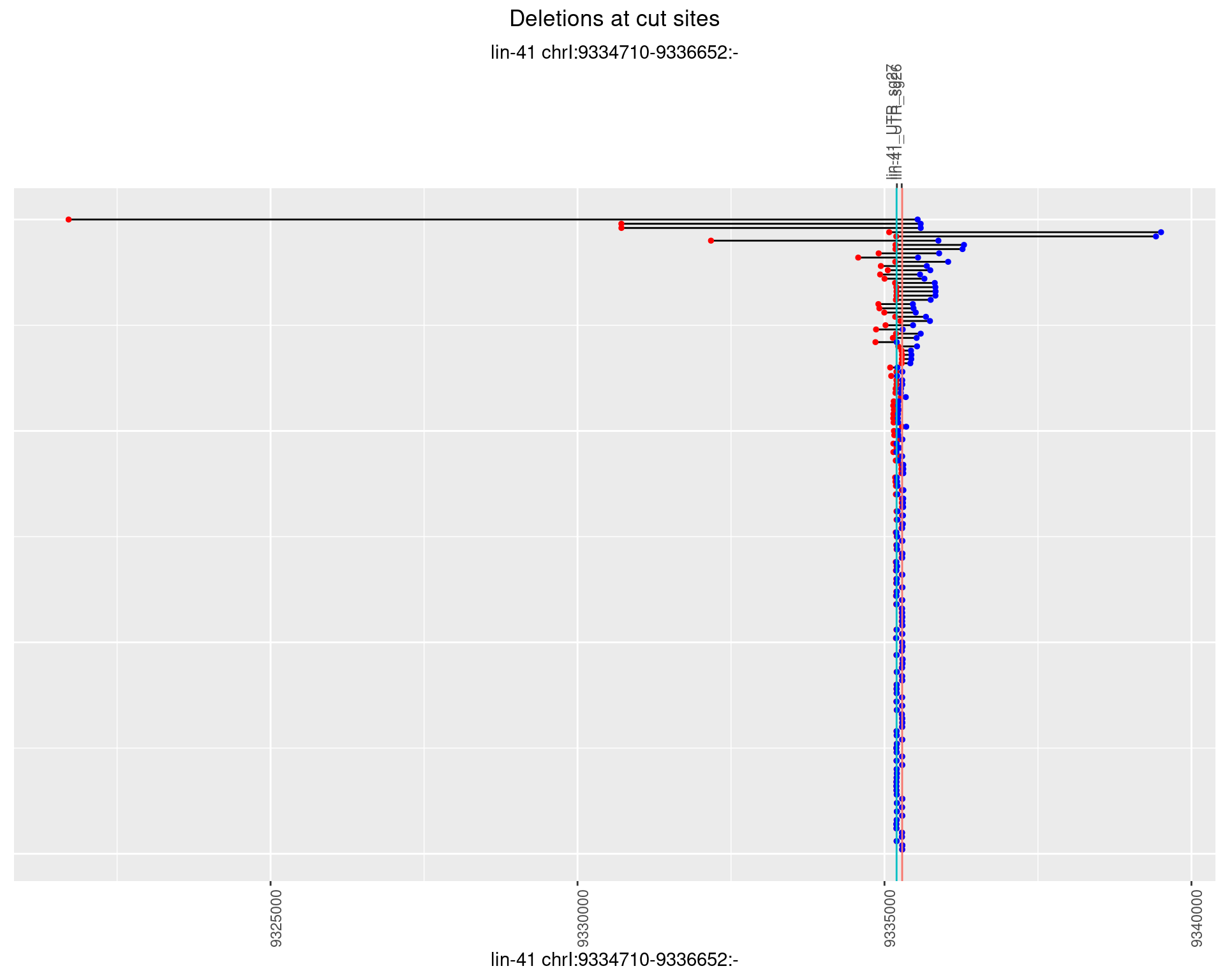
1.36.2 Freq: 1e-05
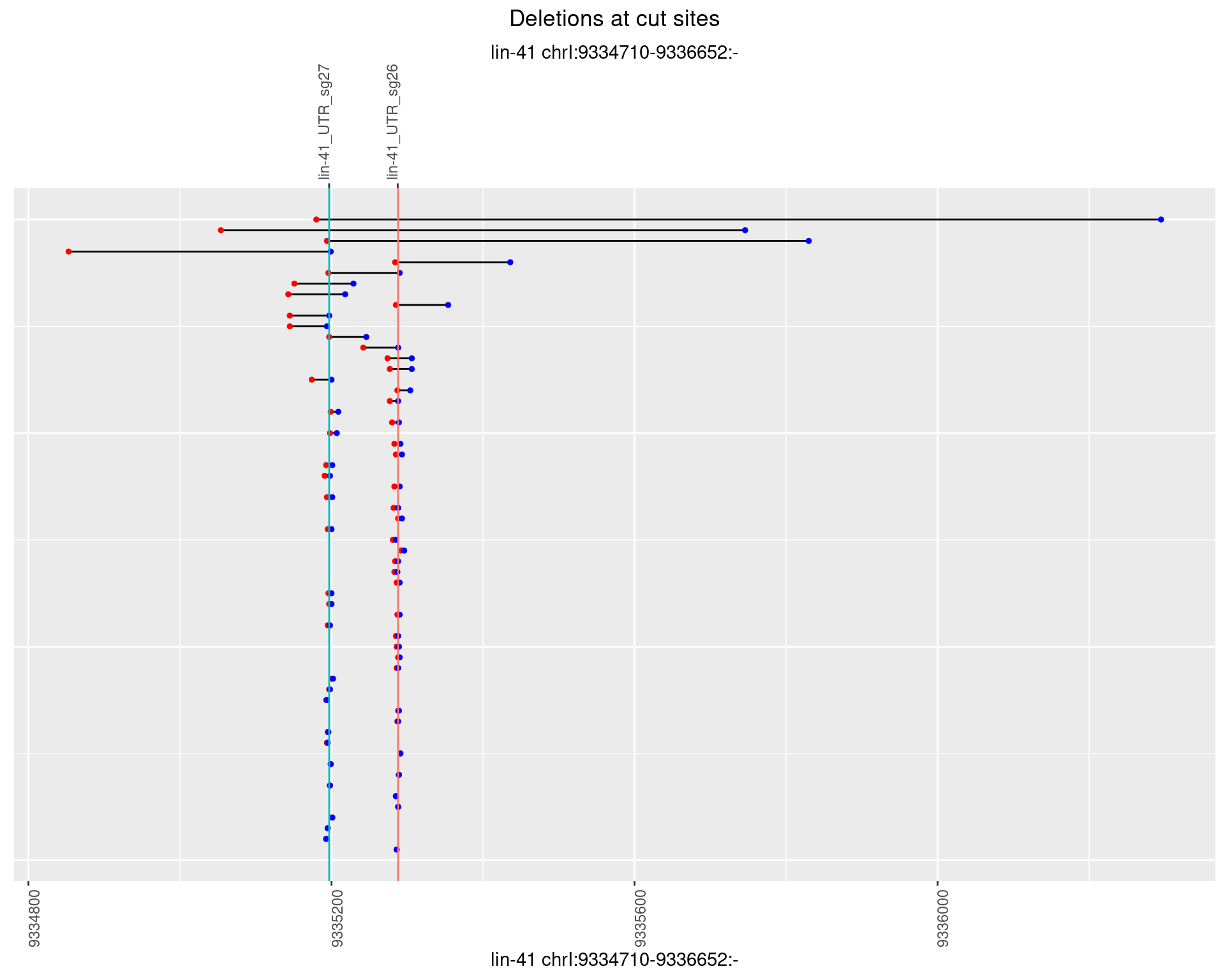
1.36.3 Freq: 1e-04
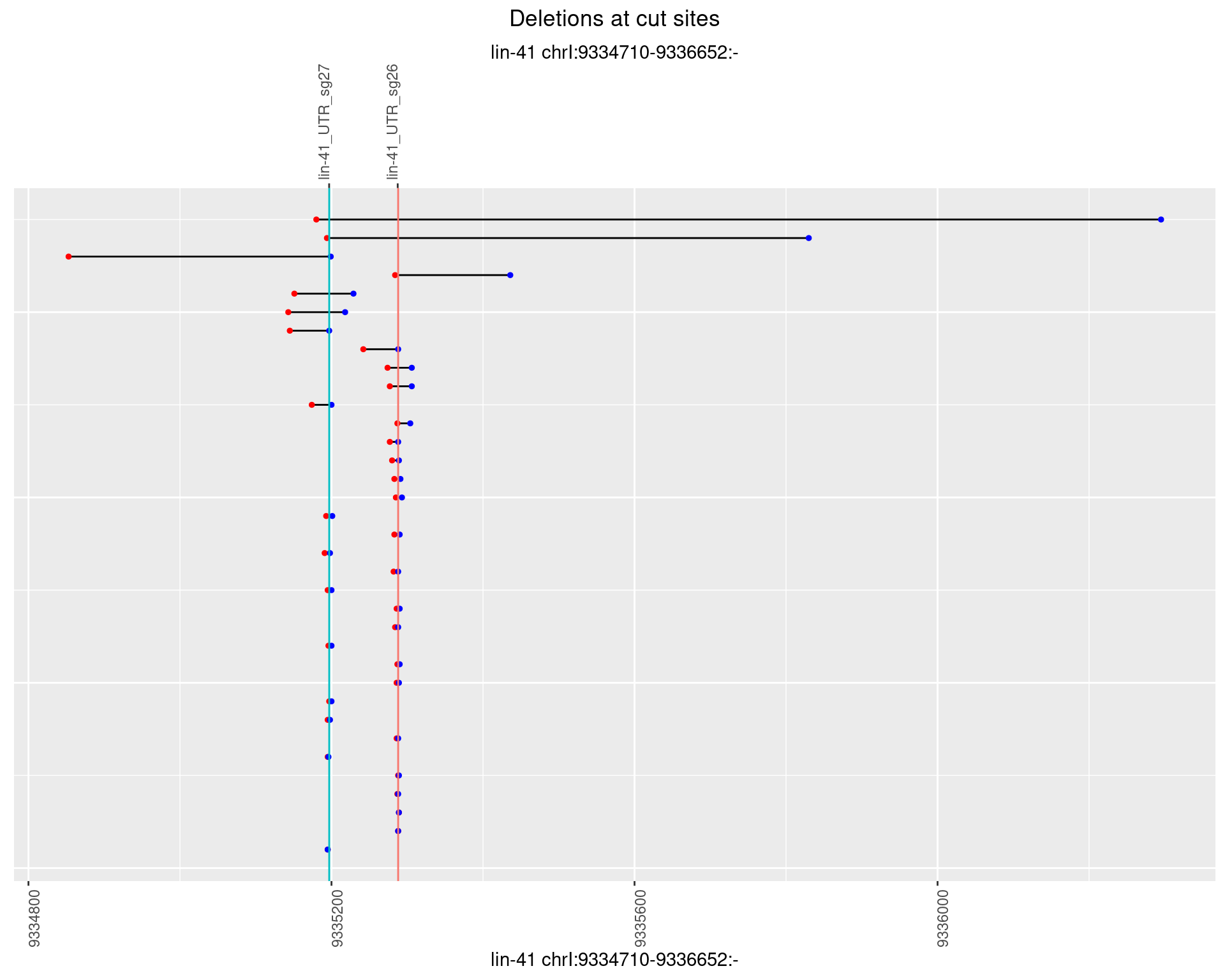
1.36.4 Freq: 0.001
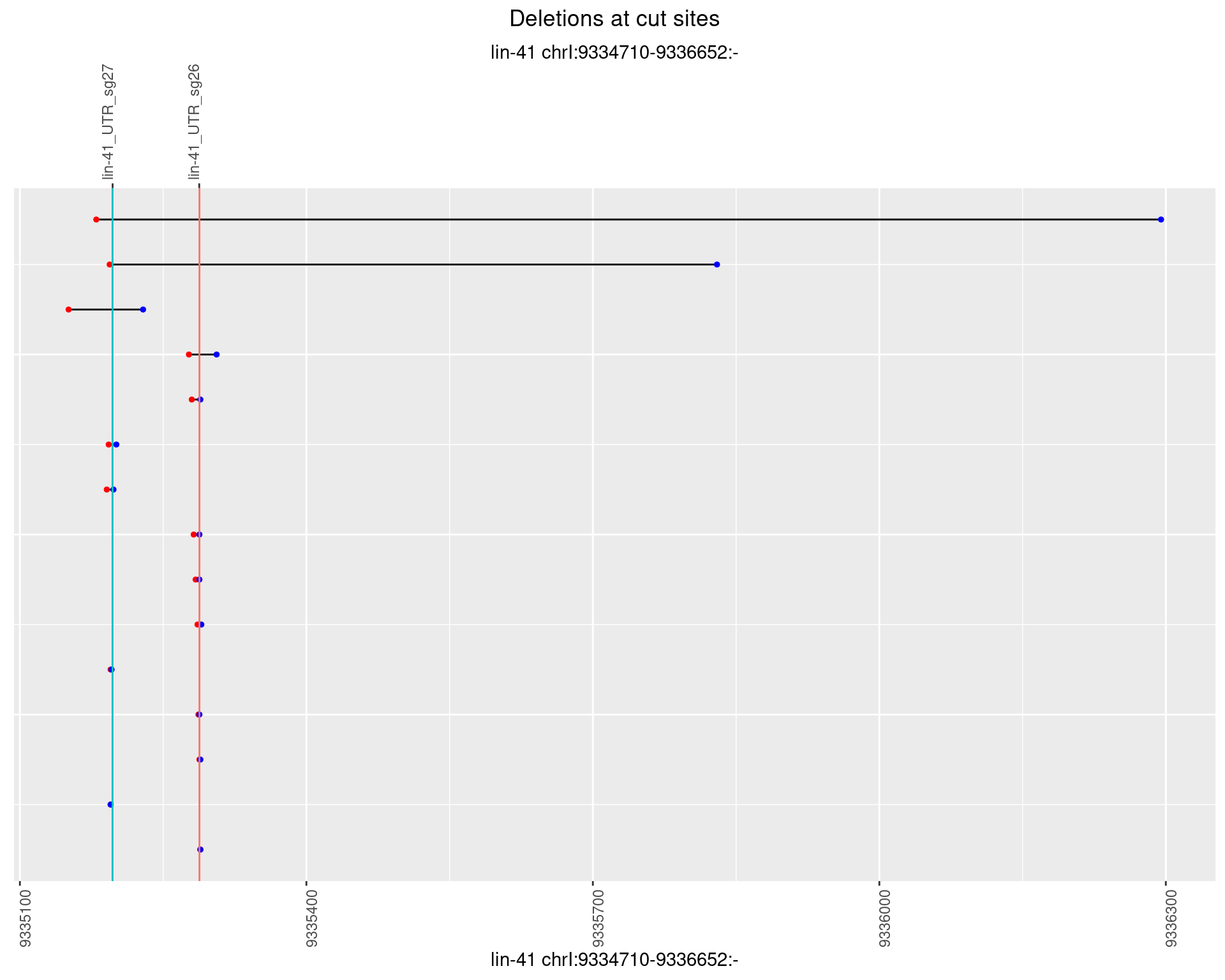
1.36.5 Freq: 0.01
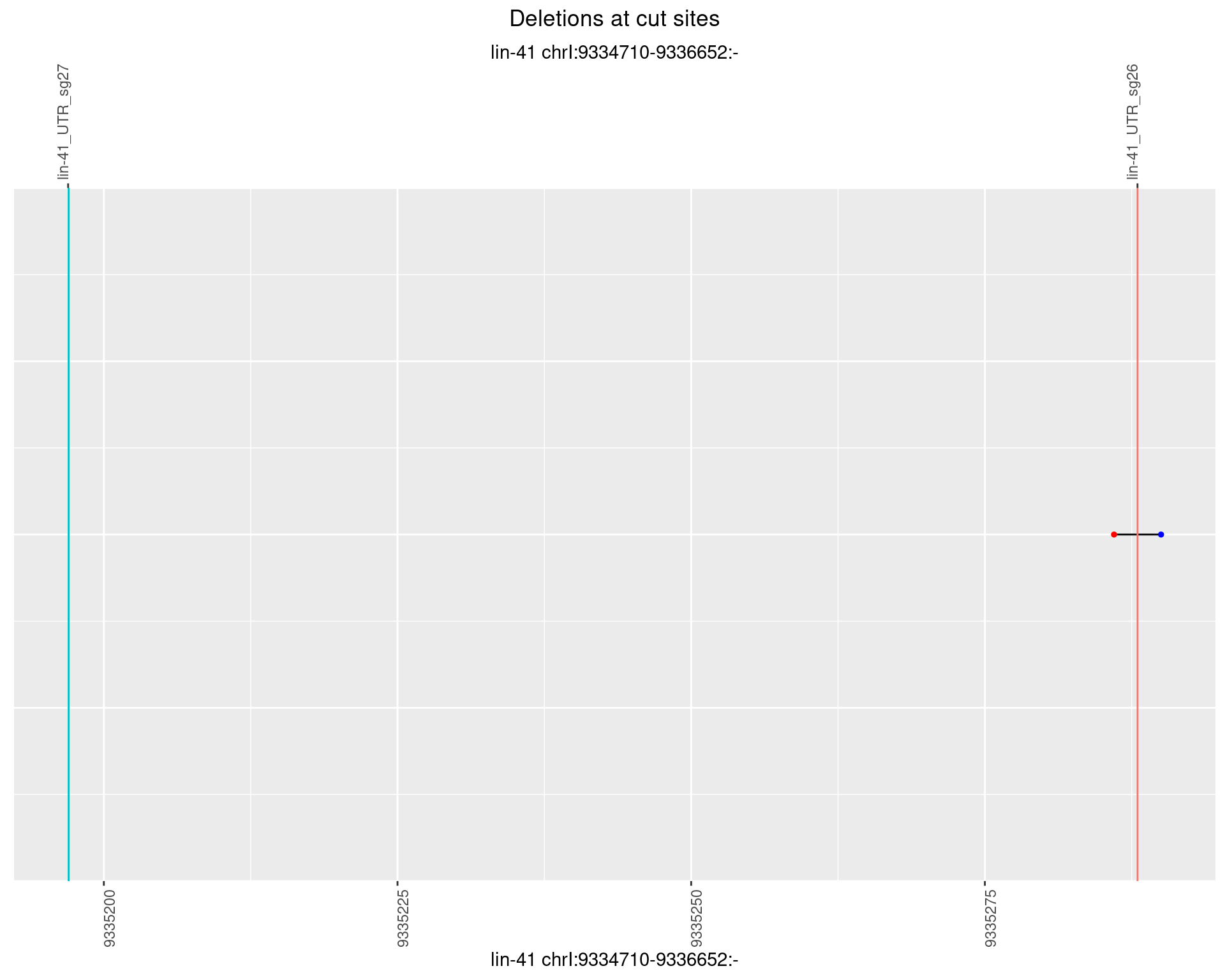
1.36.6 Freq: 0.1
No plot to show
1.37 lin41_DNA_N2
1.37.1 Freq: 0
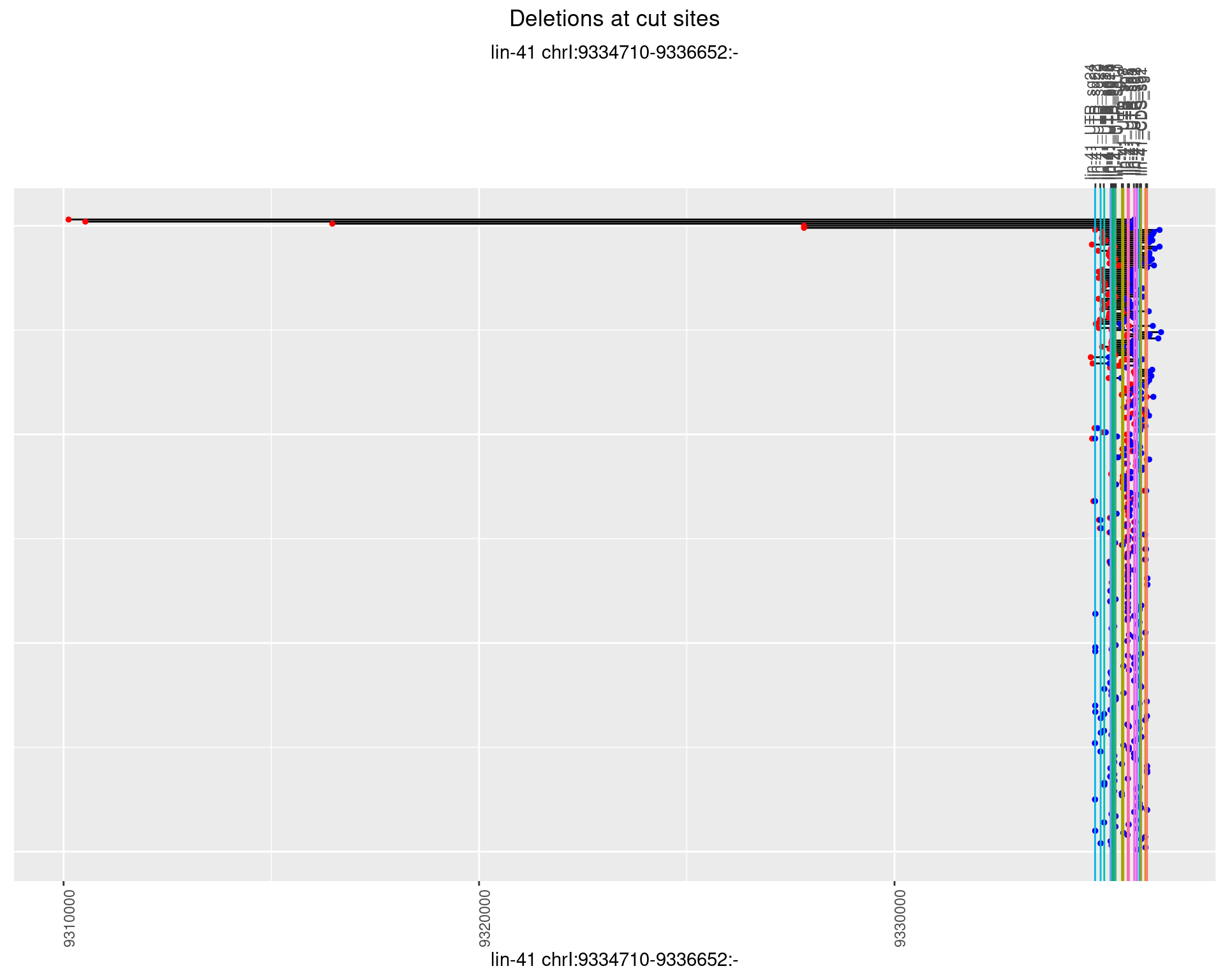
1.37.2 Freq: 1e-05
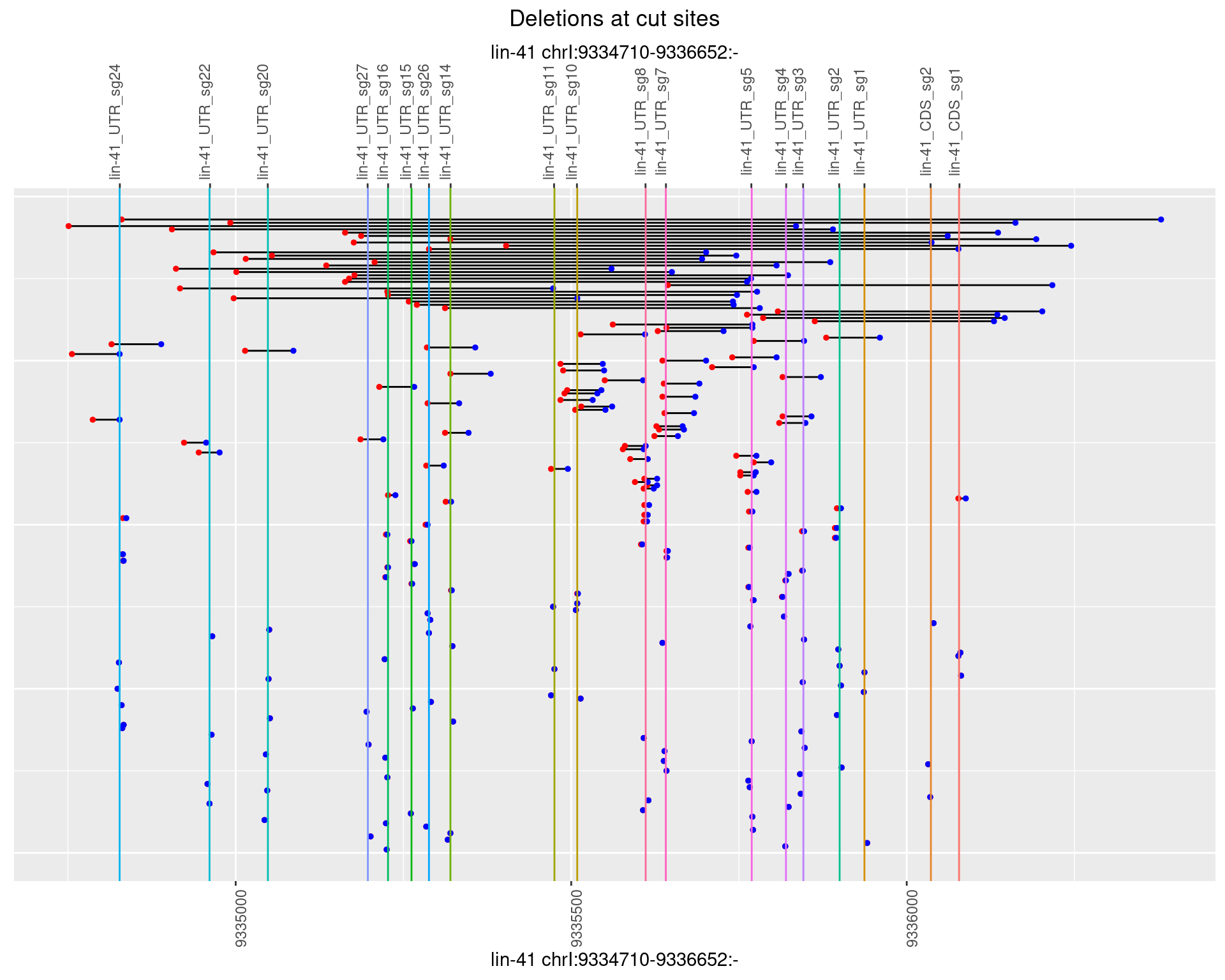
1.37.3 Freq: 1e-04
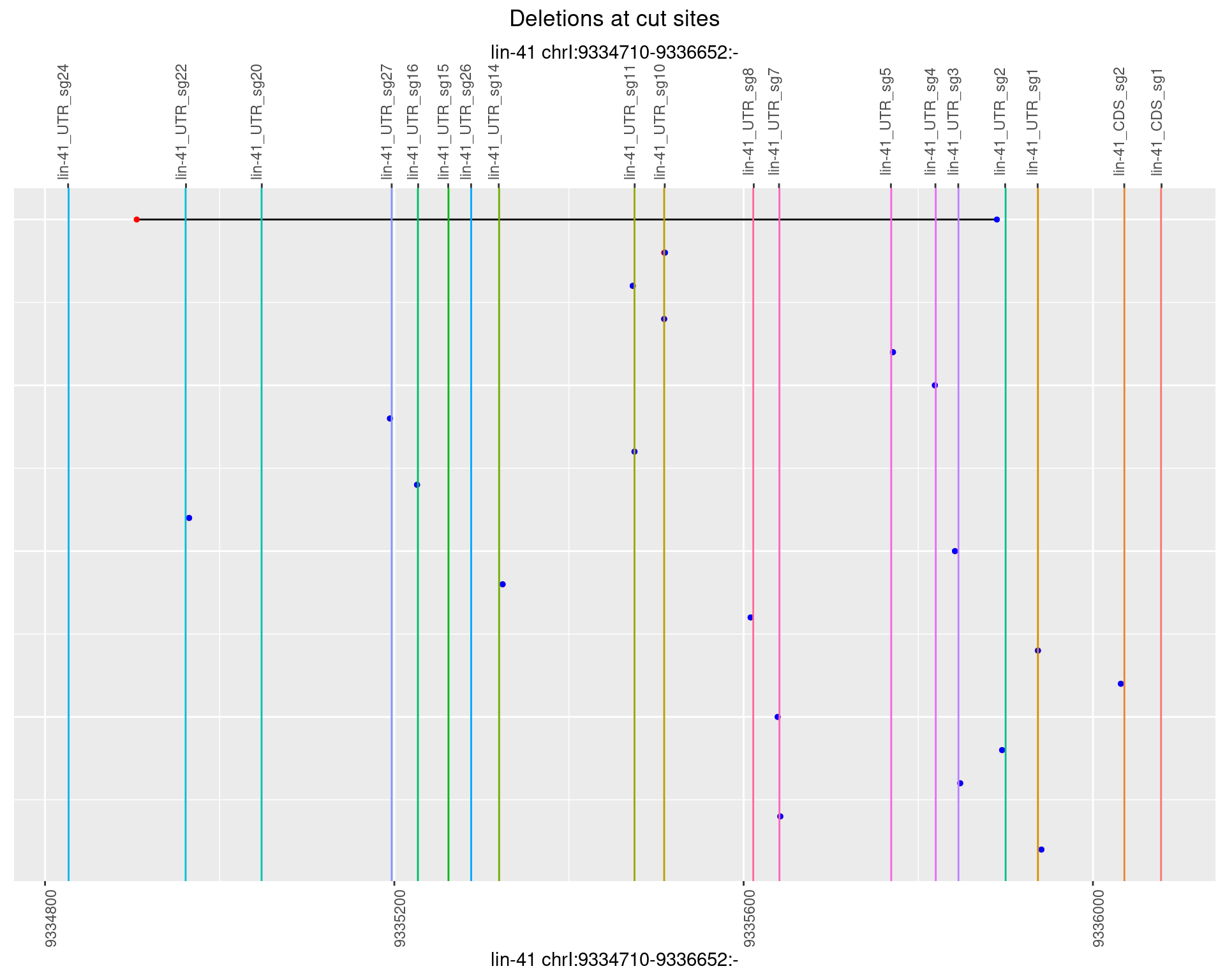
1.37.4 Freq: 0.001
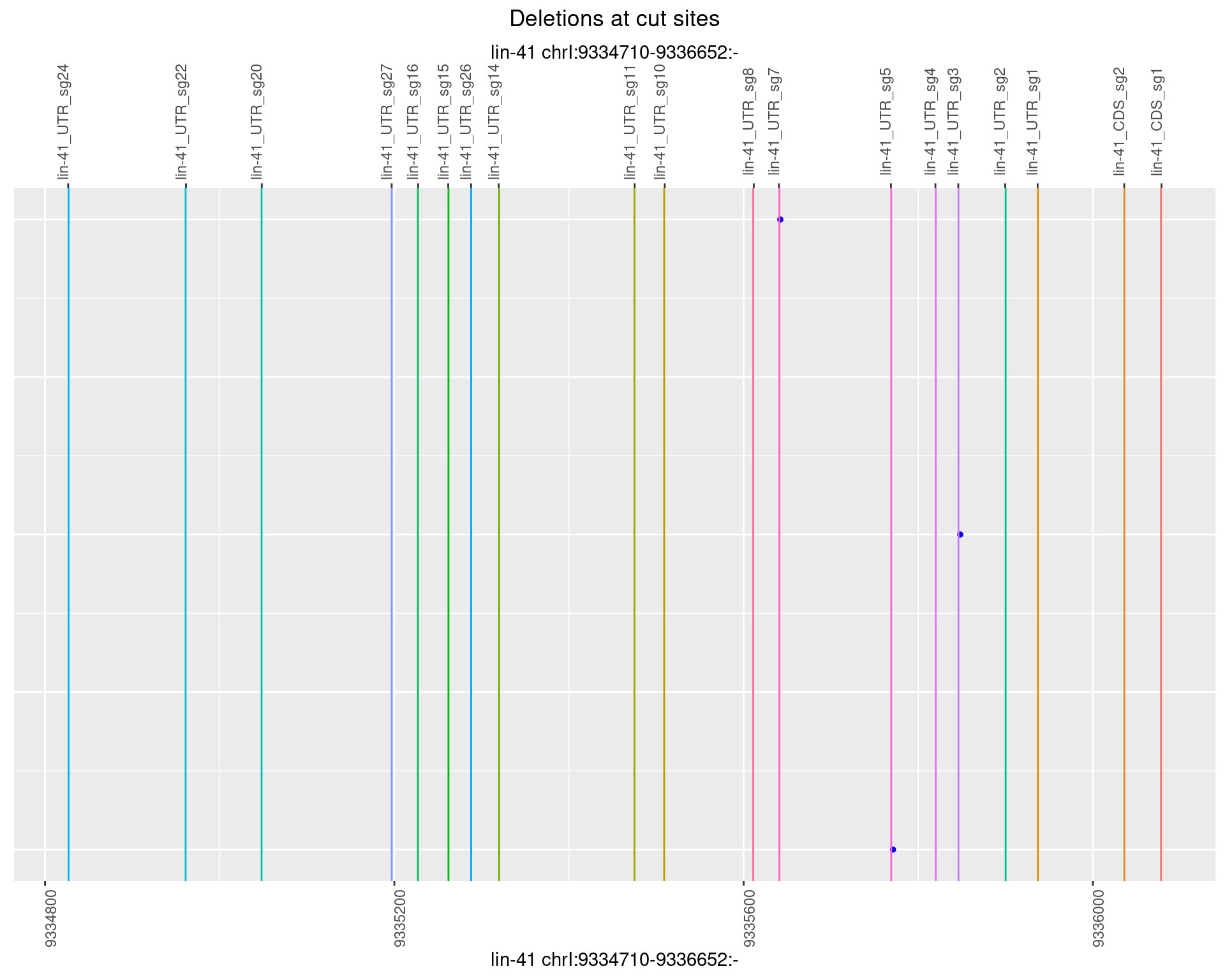
1.37.5 Freq: 0.01
No plot to show
1.37.6 Freq: 0.1
No plot to show
1.38 lin41_DNA_CDS
1.38.1 Freq: 0
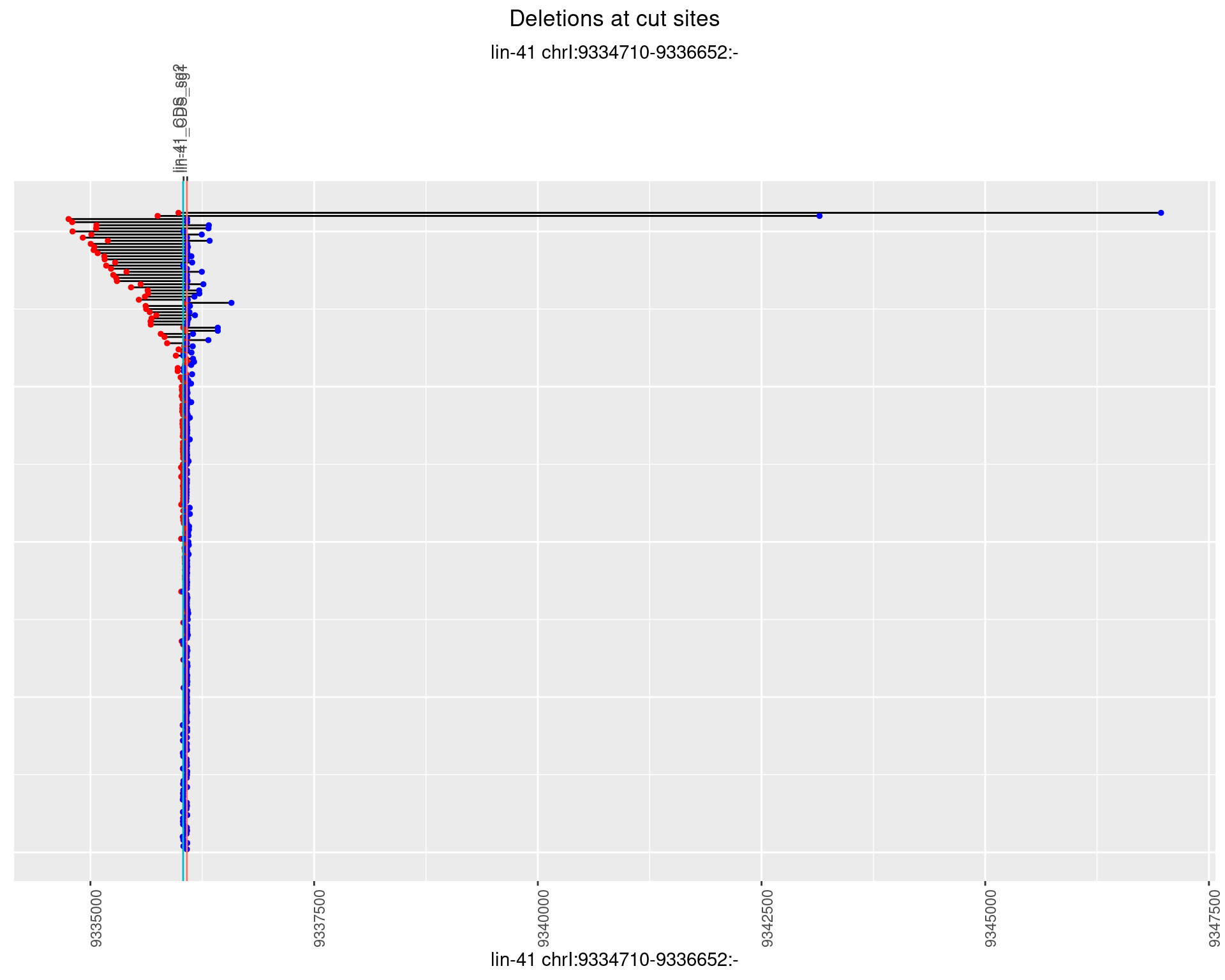
1.38.2 Freq: 1e-05
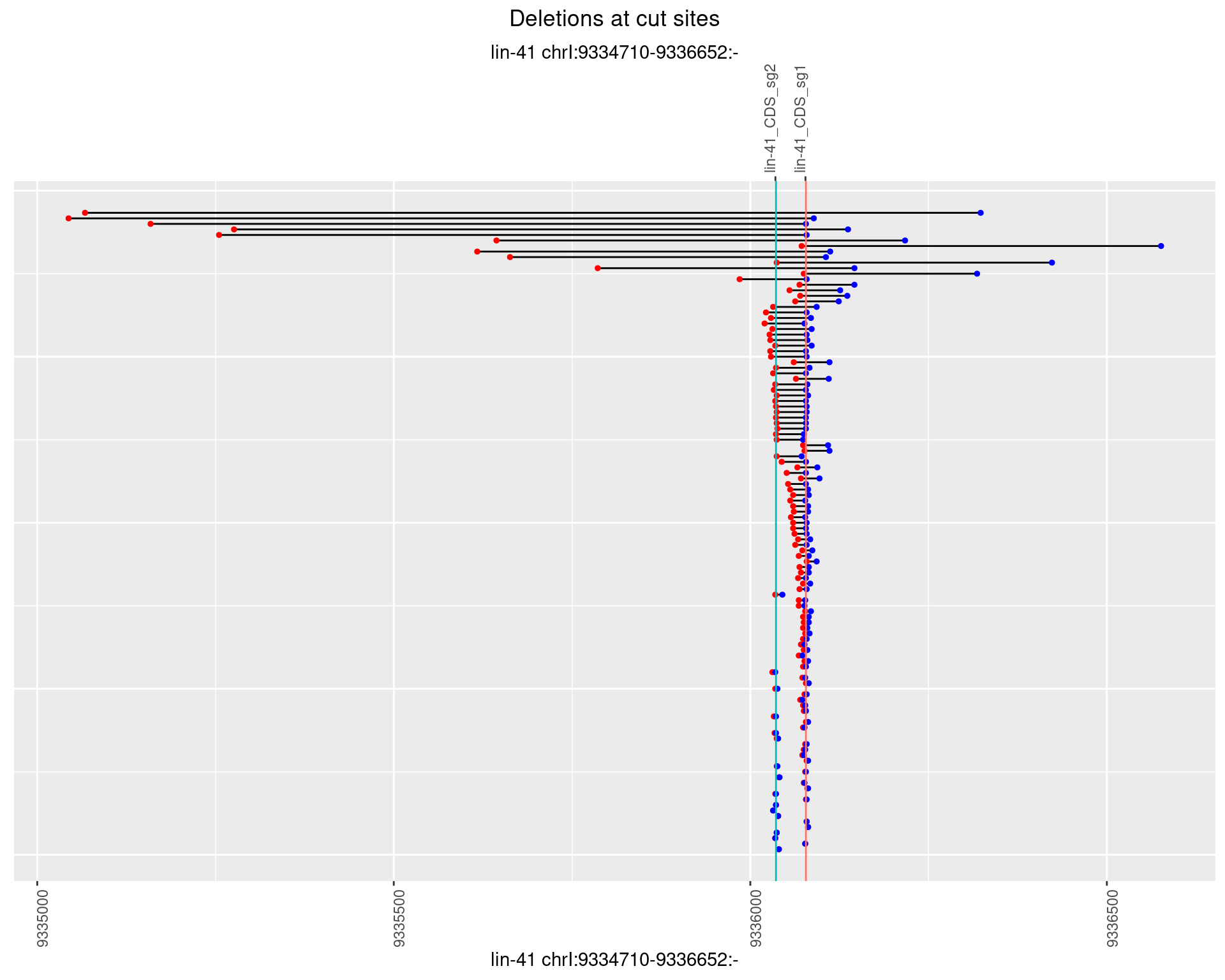
1.38.3 Freq: 1e-04
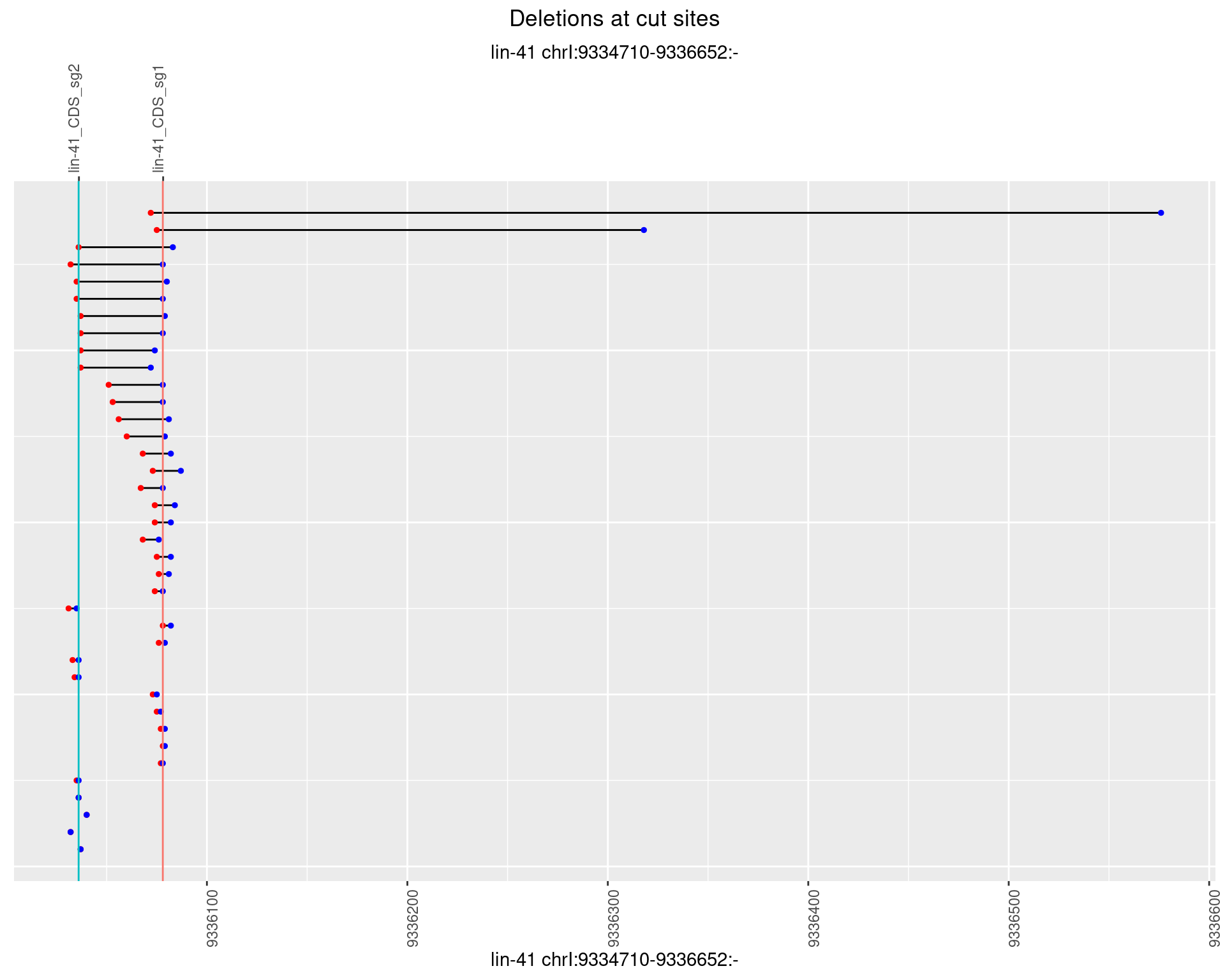
1.38.4 Freq: 0.001
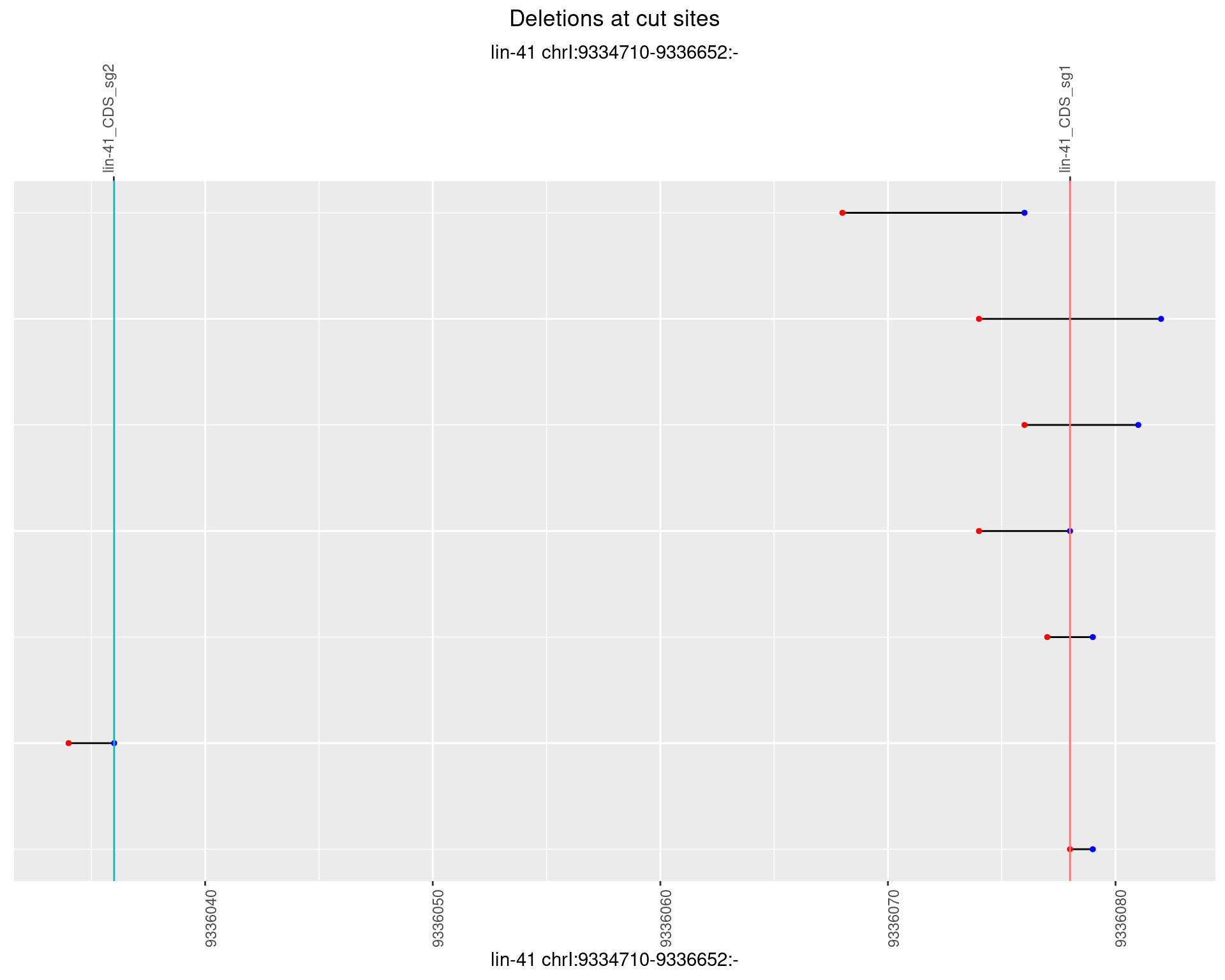
1.38.5 Freq: 0.01
No plot to show
1.38.6 Freq: 0.1
No plot to show
1.39 lin41_DNA_1516
1.39.1 Freq: 0
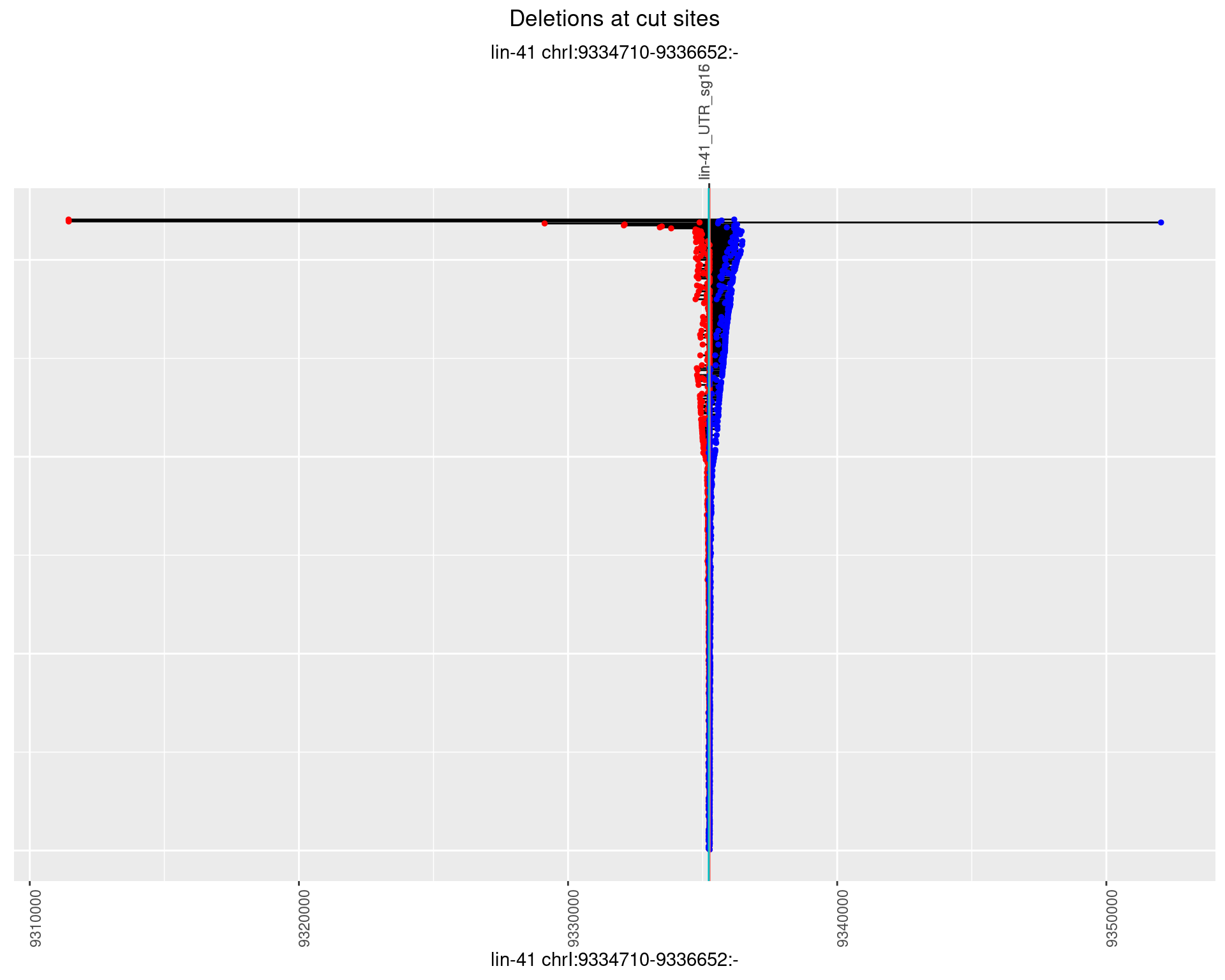
1.39.2 Freq: 1e-05
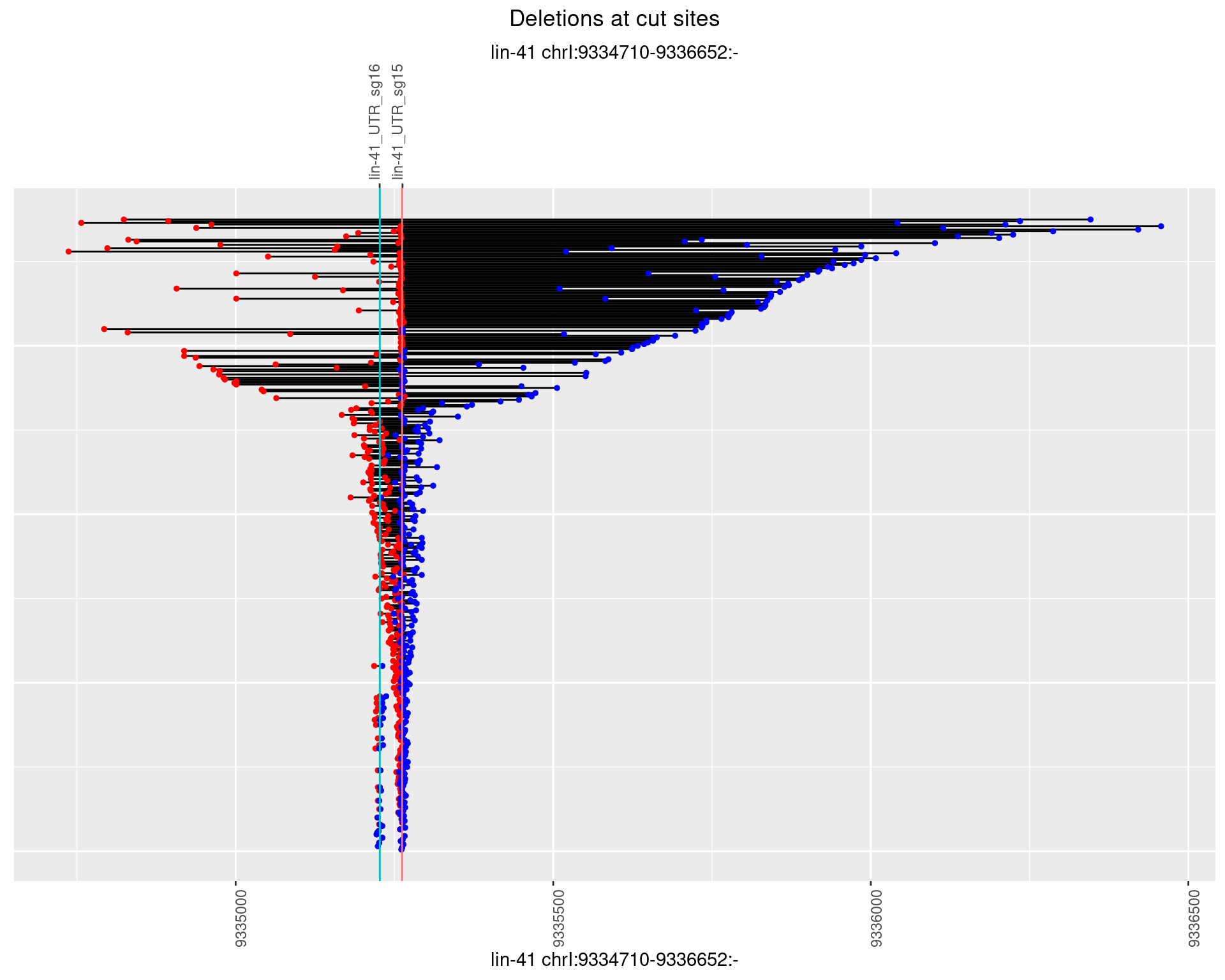
1.39.3 Freq: 1e-04
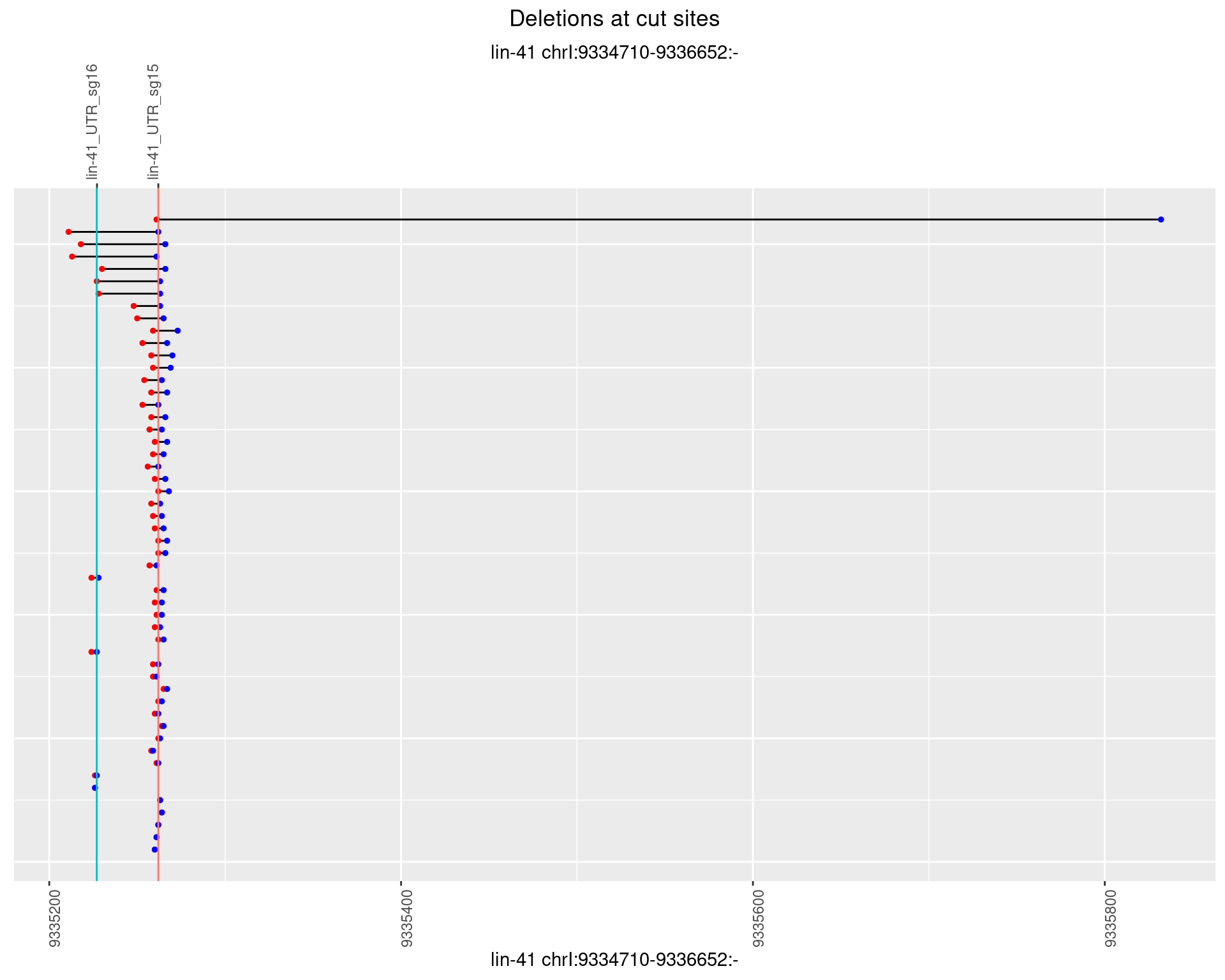
1.39.4 Freq: 0.001
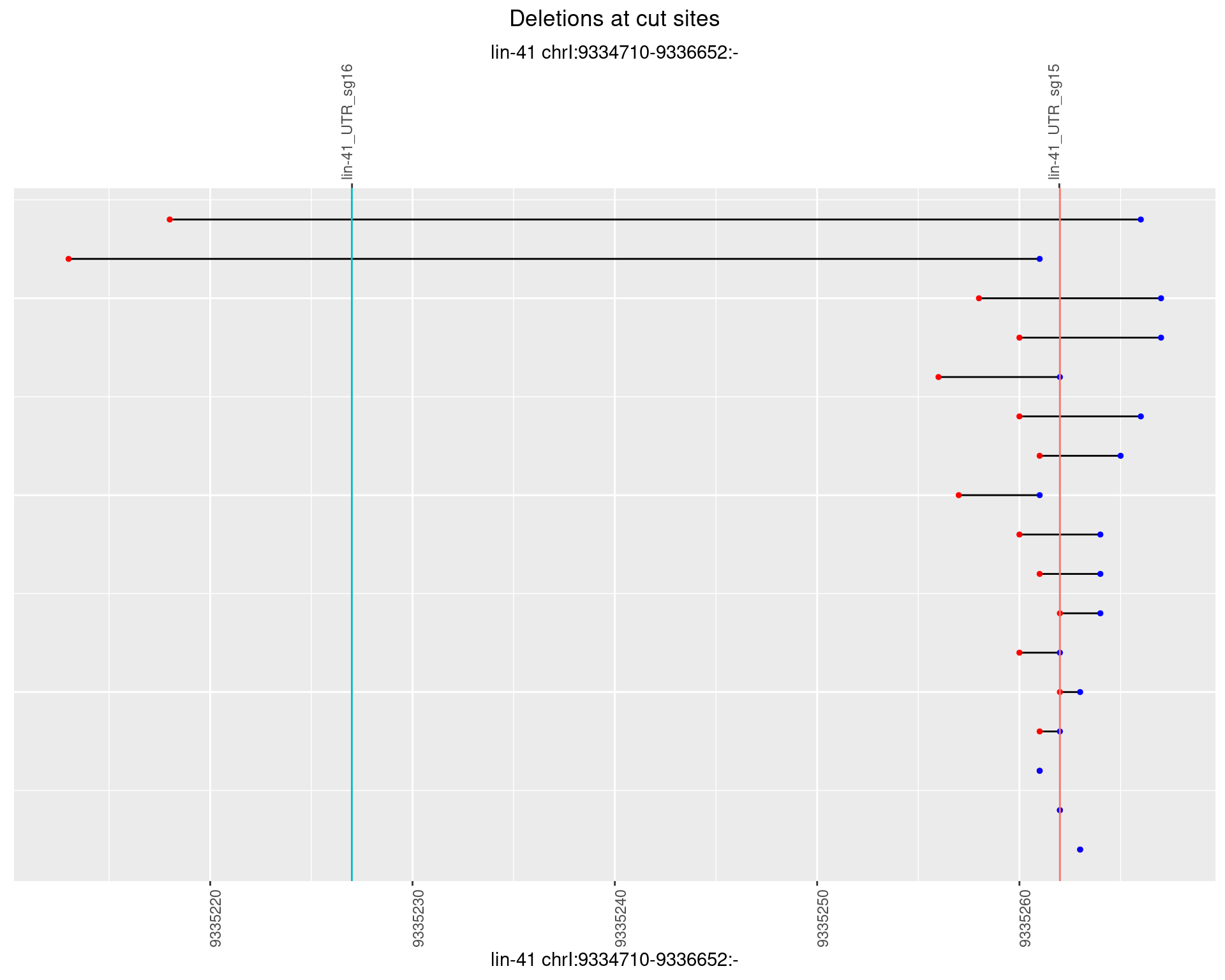
1.39.5 Freq: 0.01
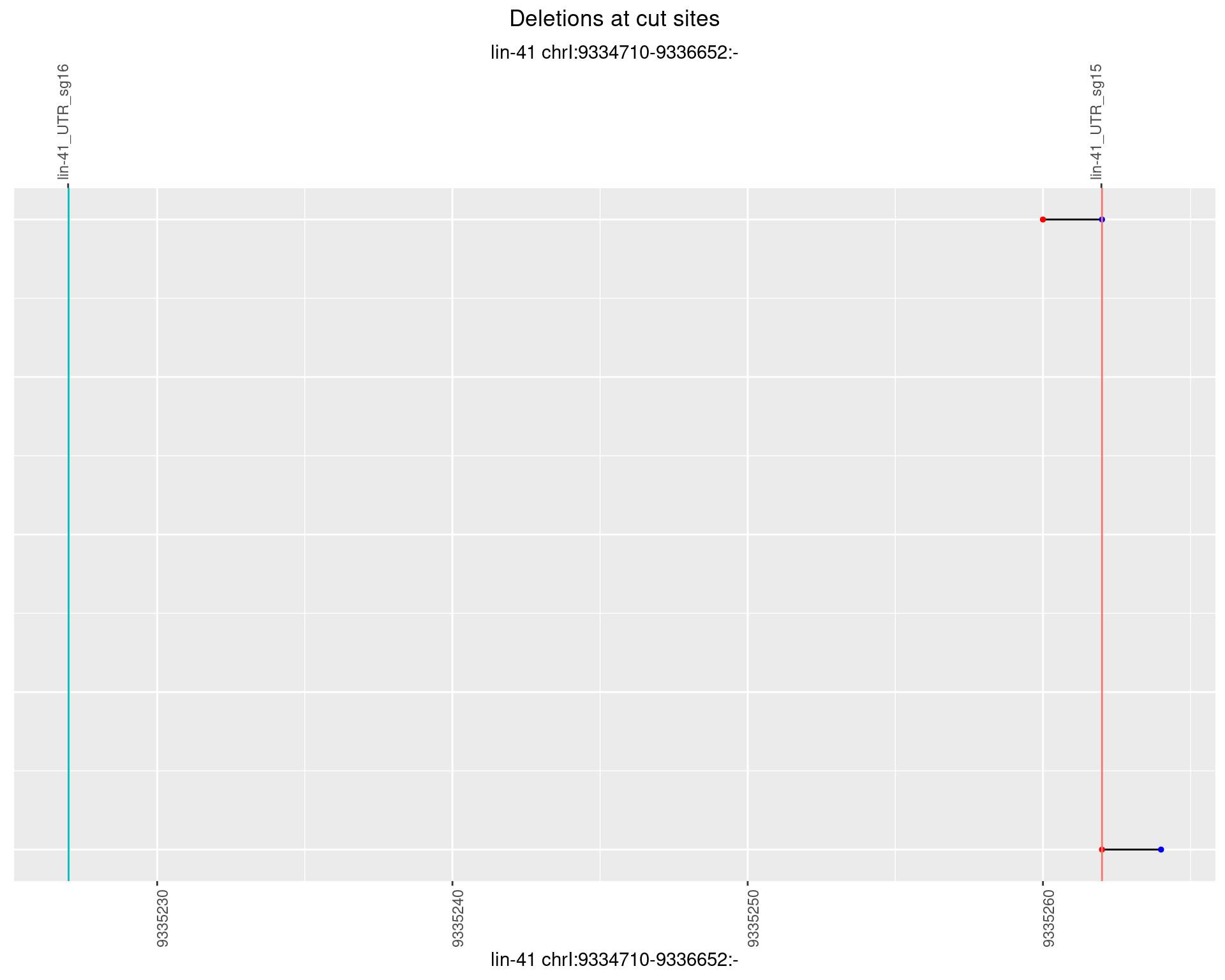
1.39.6 Freq: 0.1
No plot to show
1.40 lin41_DNA_pool1
1.40.1 Freq: 0
1.40.2 Freq: 1e-05
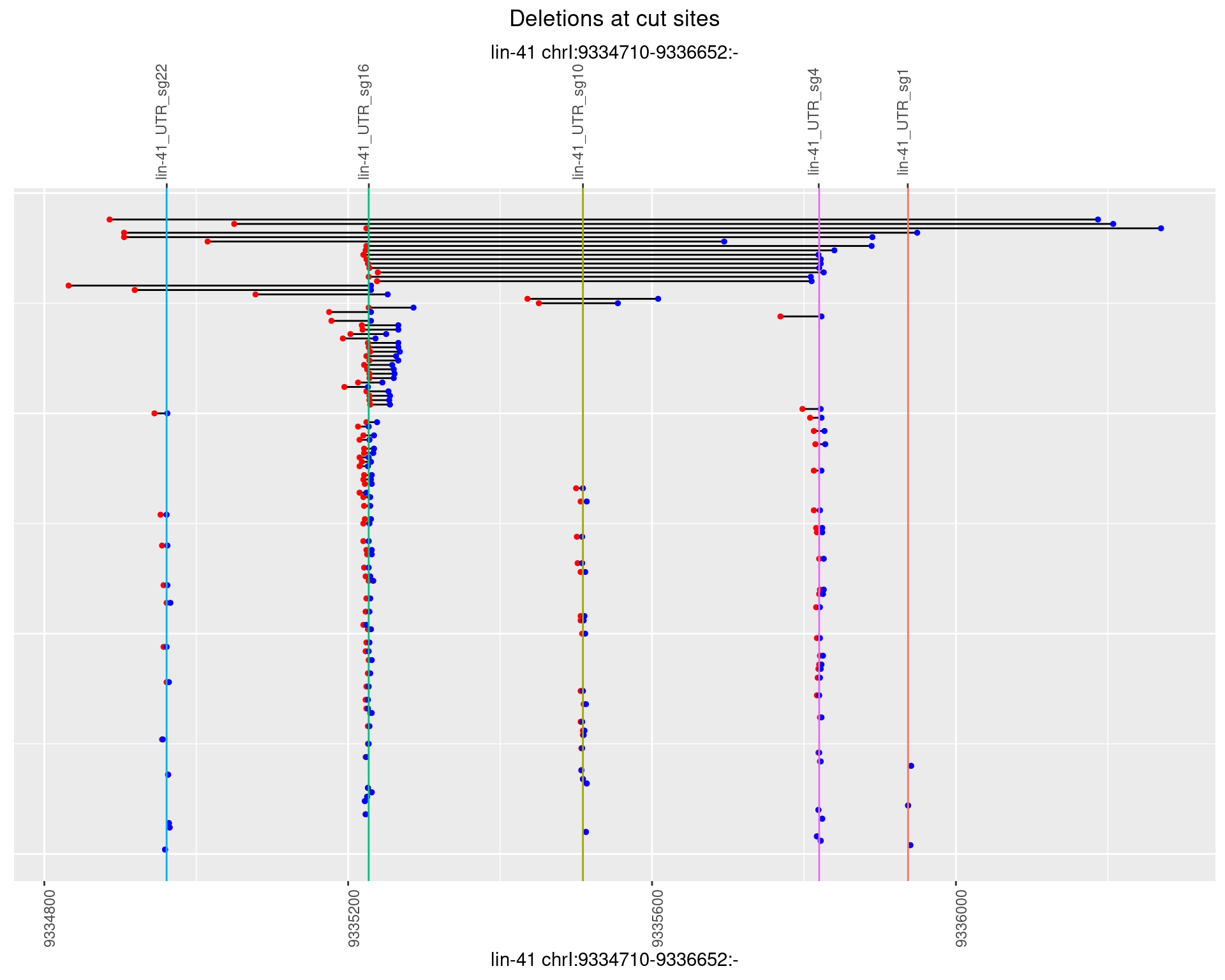
1.40.3 Freq: 1e-04
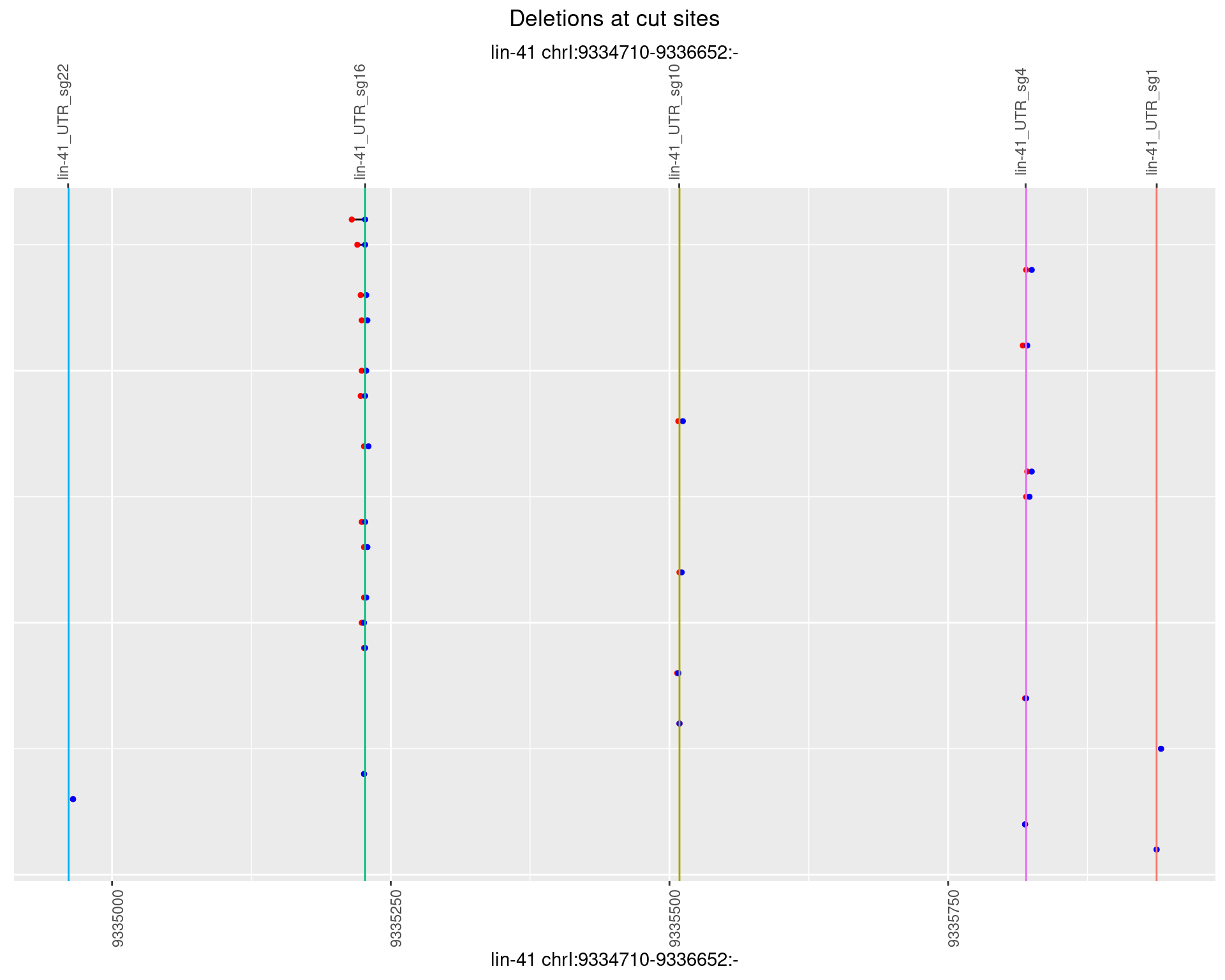
1.40.4 Freq: 0.001
1.40.5 Freq: 0.01
No plot to show
1.40.6 Freq: 0.1
No plot to show
1.41 lin41_DNA_pool2
1.41.1 Freq: 0
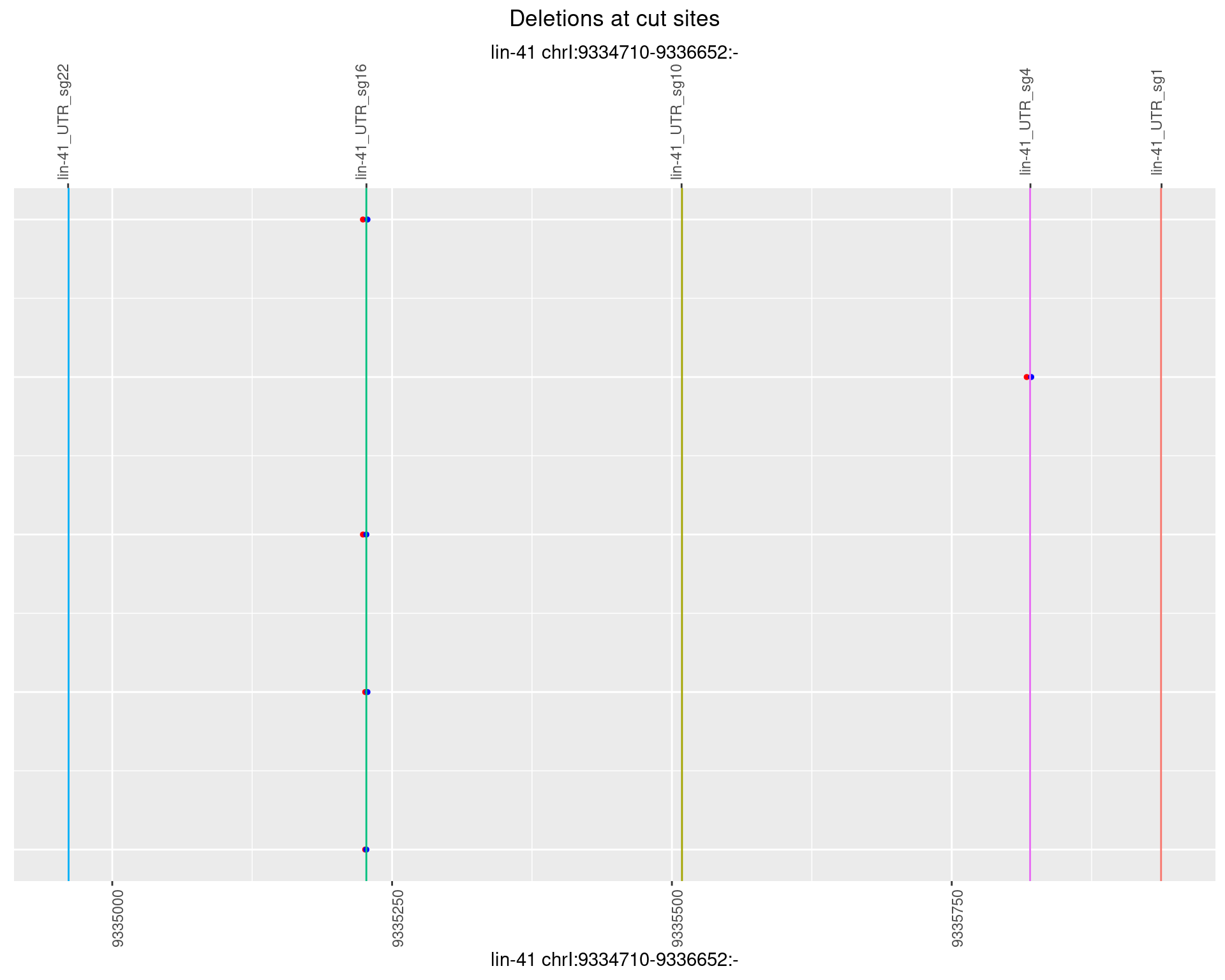
1.41.2 Freq: 1e-05
1.41.3 Freq: 1e-04
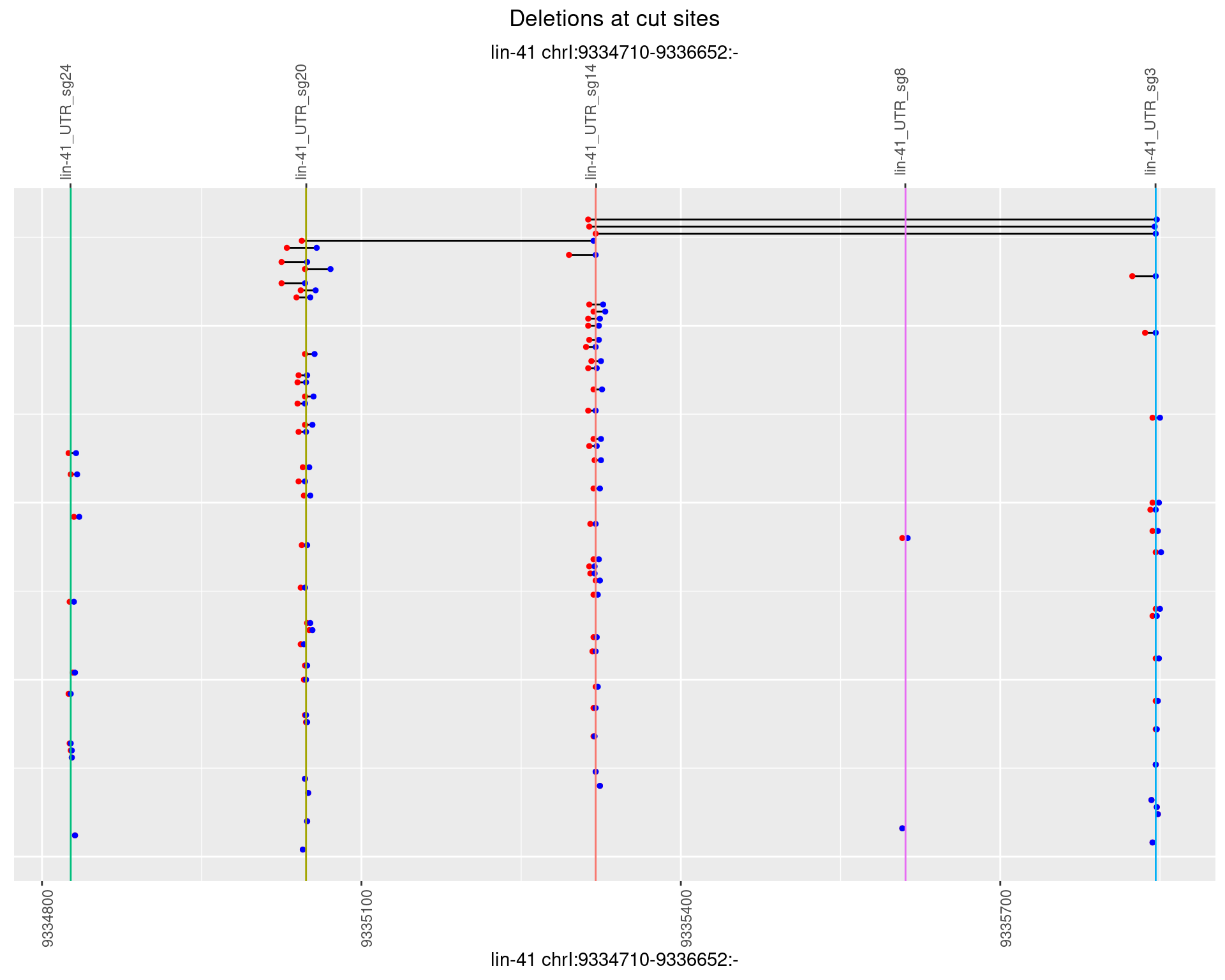
1.41.4 Freq: 0.001
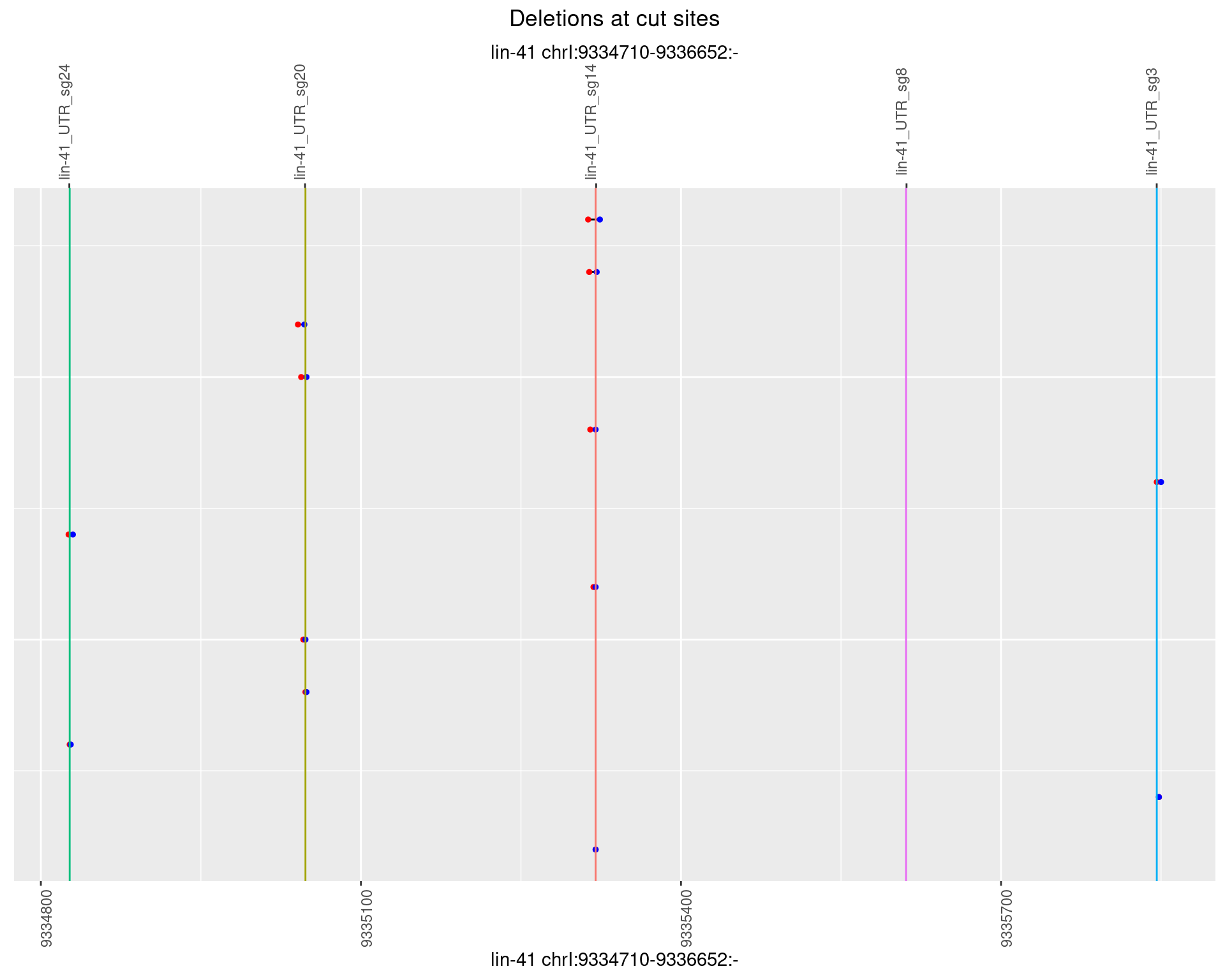
1.41.5 Freq: 0.01
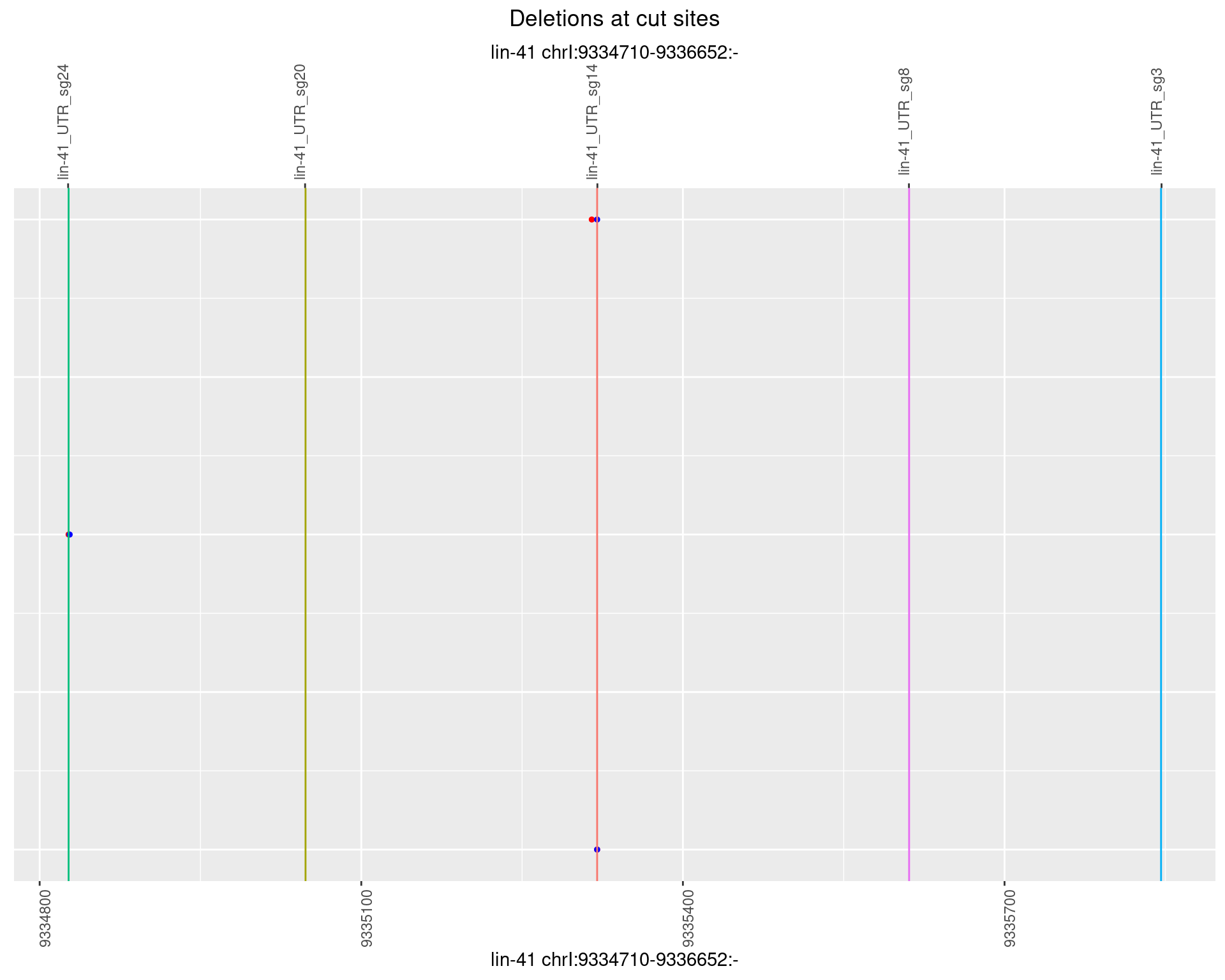
1.41.6 Freq: 0.1
No plot to show
1.42 lin41_DNA_pool3
1.42.1 Freq: 0
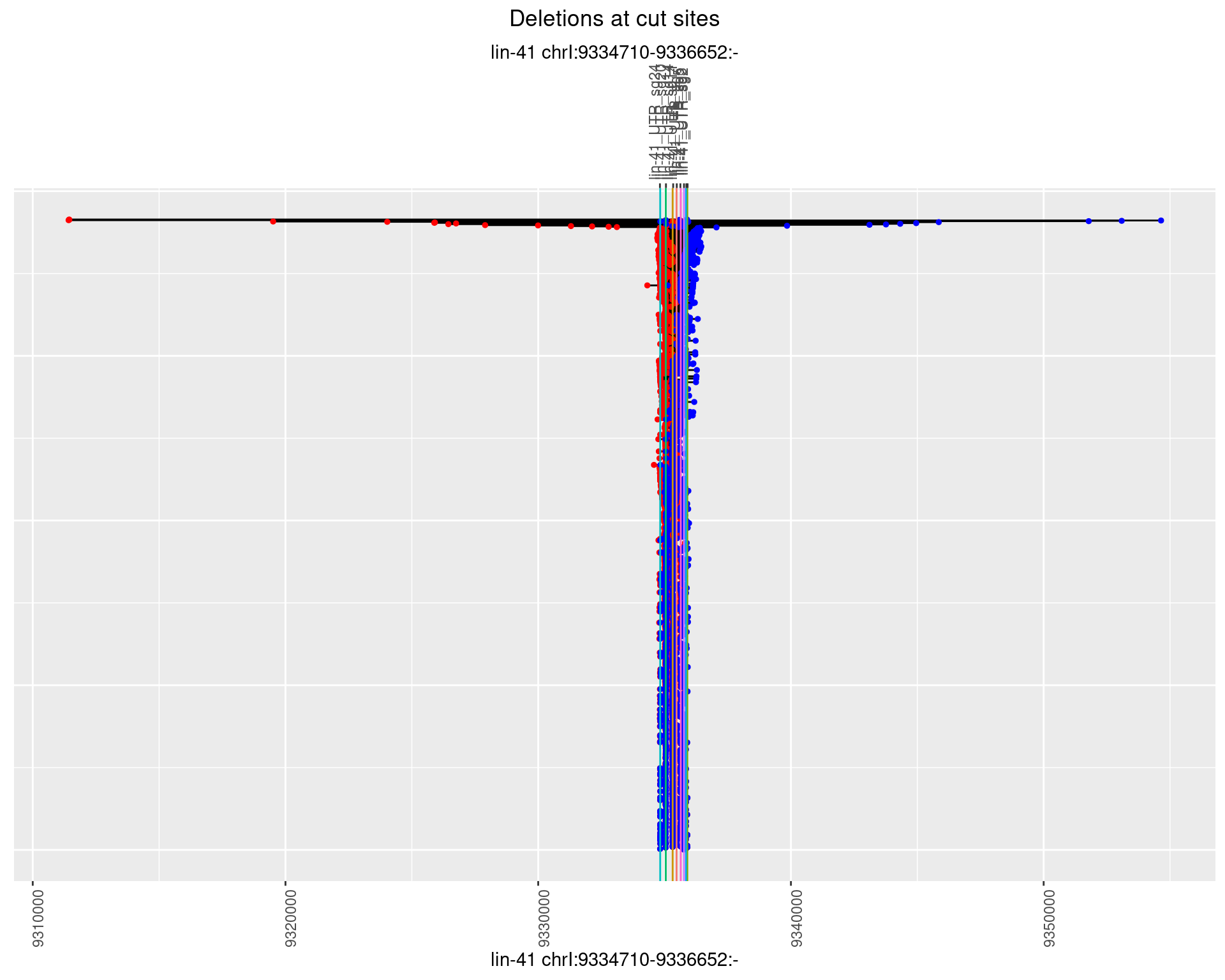
1.42.2 Freq: 1e-05
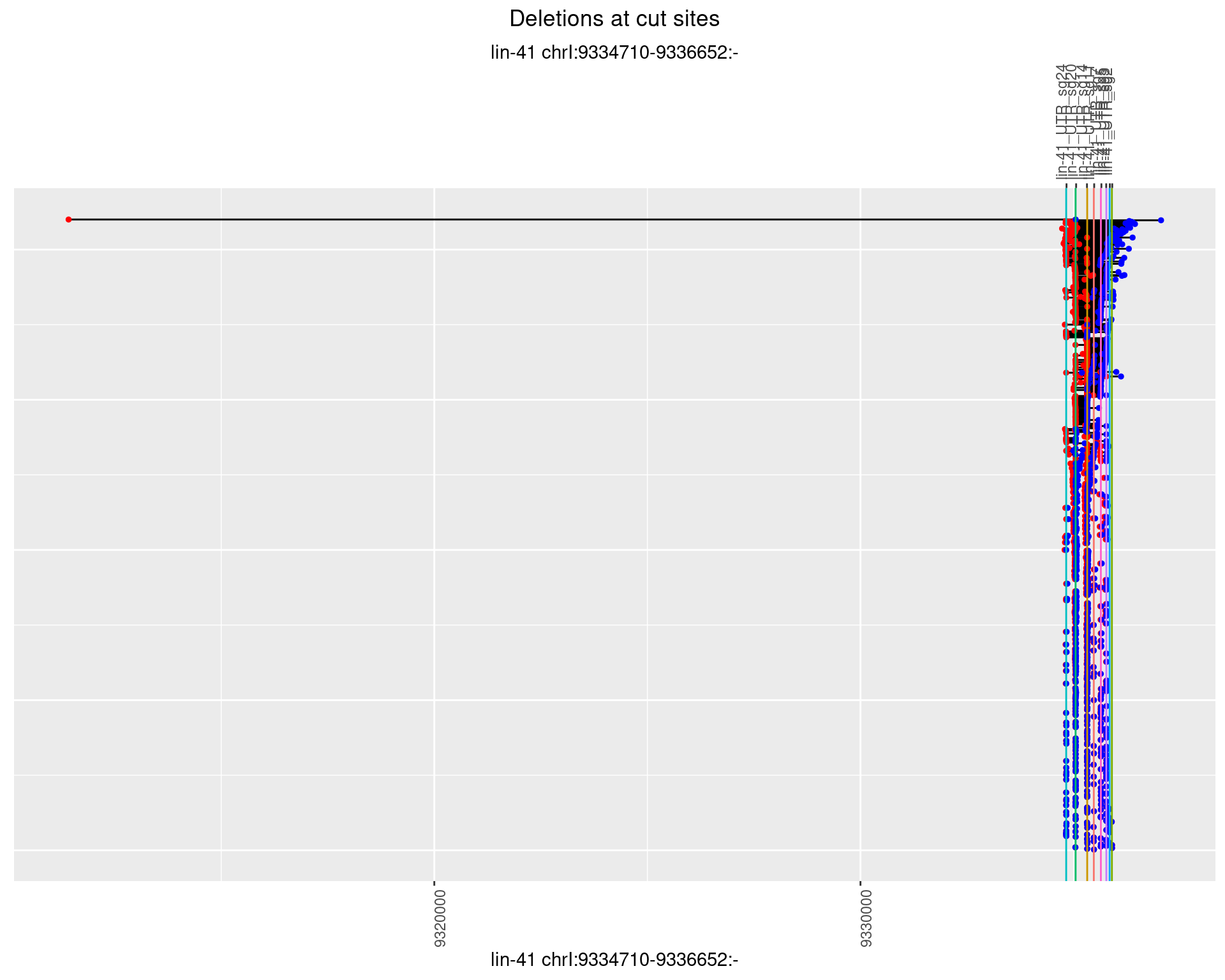
1.42.3 Freq: 1e-04
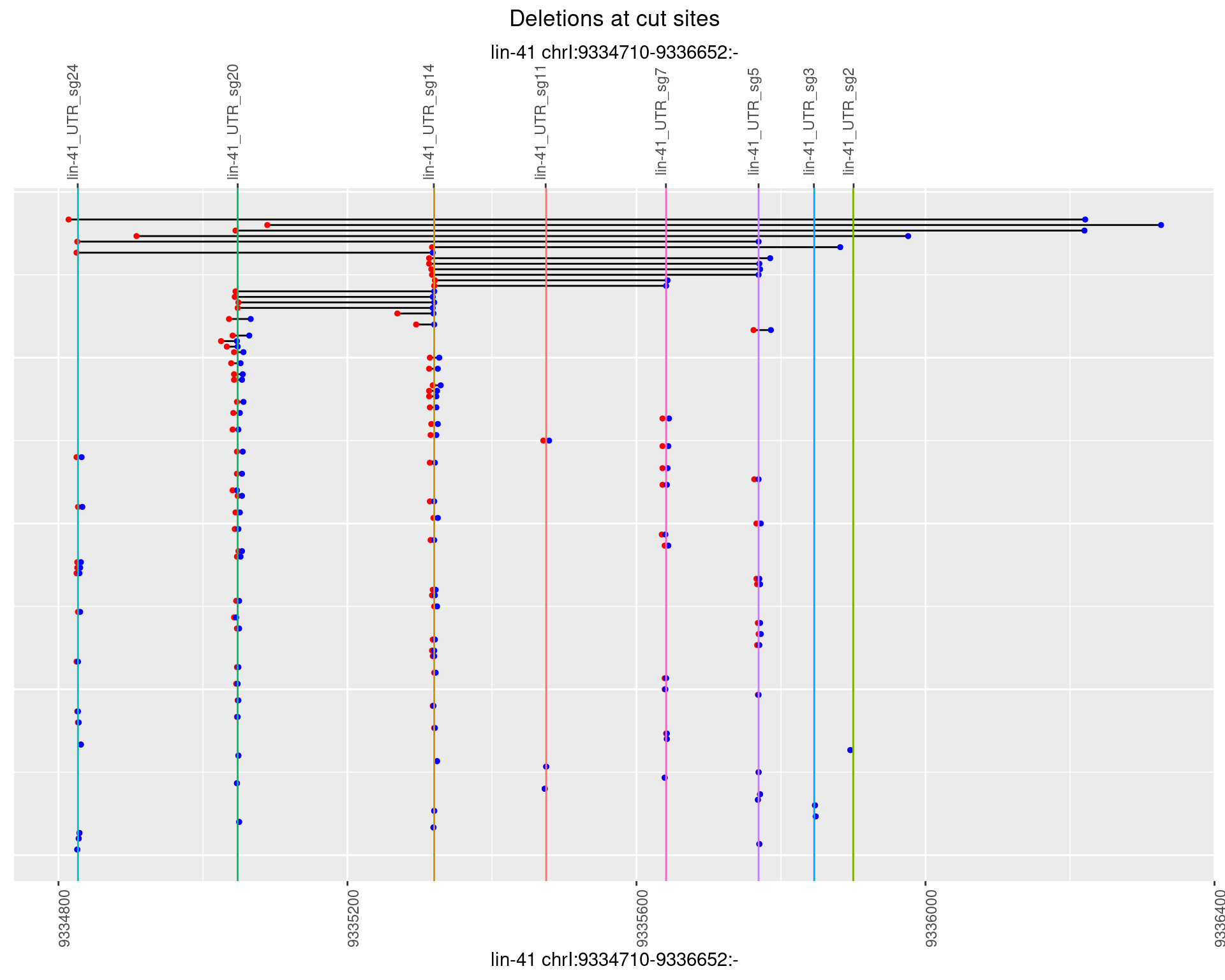
1.42.4 Freq: 0.001
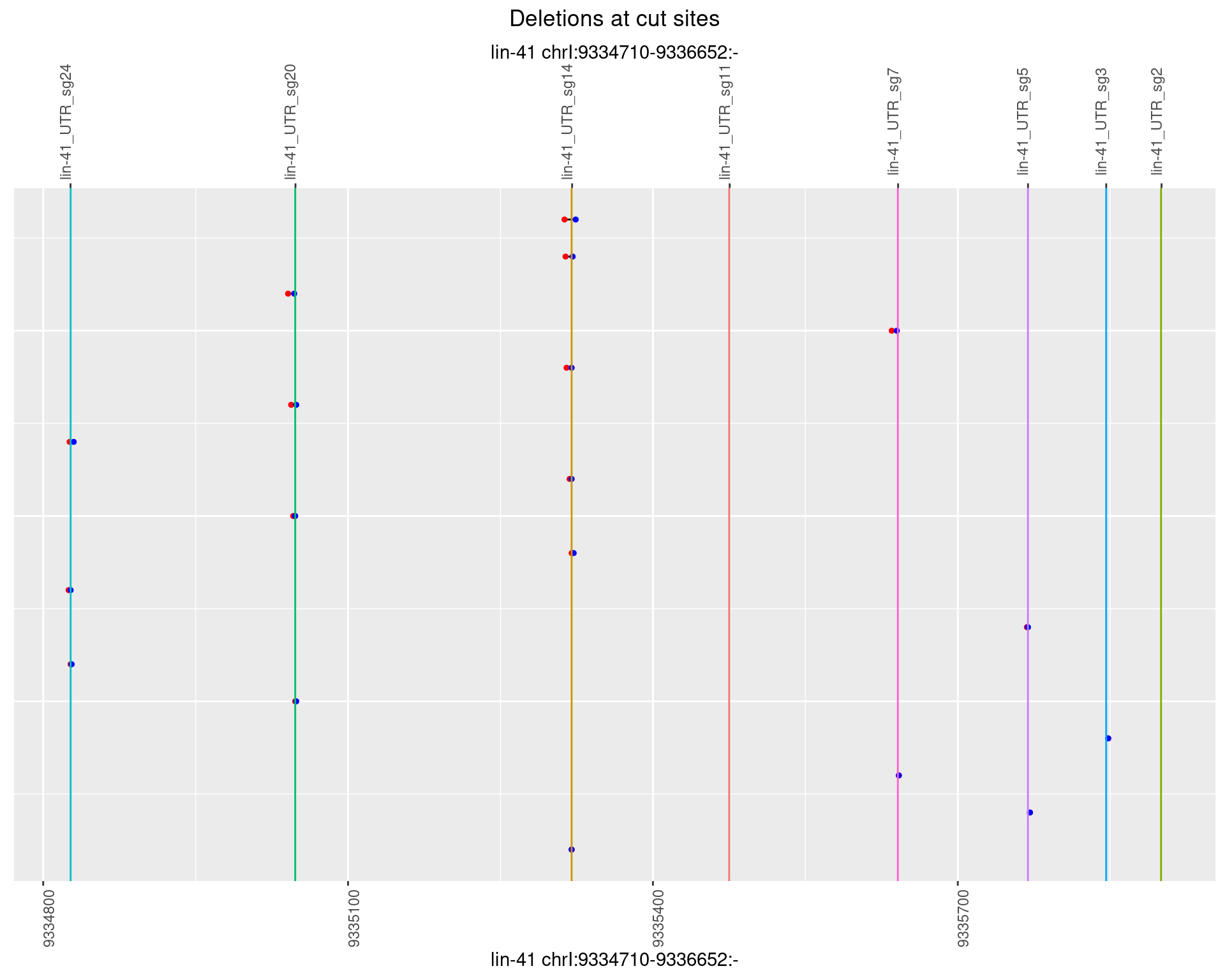
1.42.5 Freq: 0.01
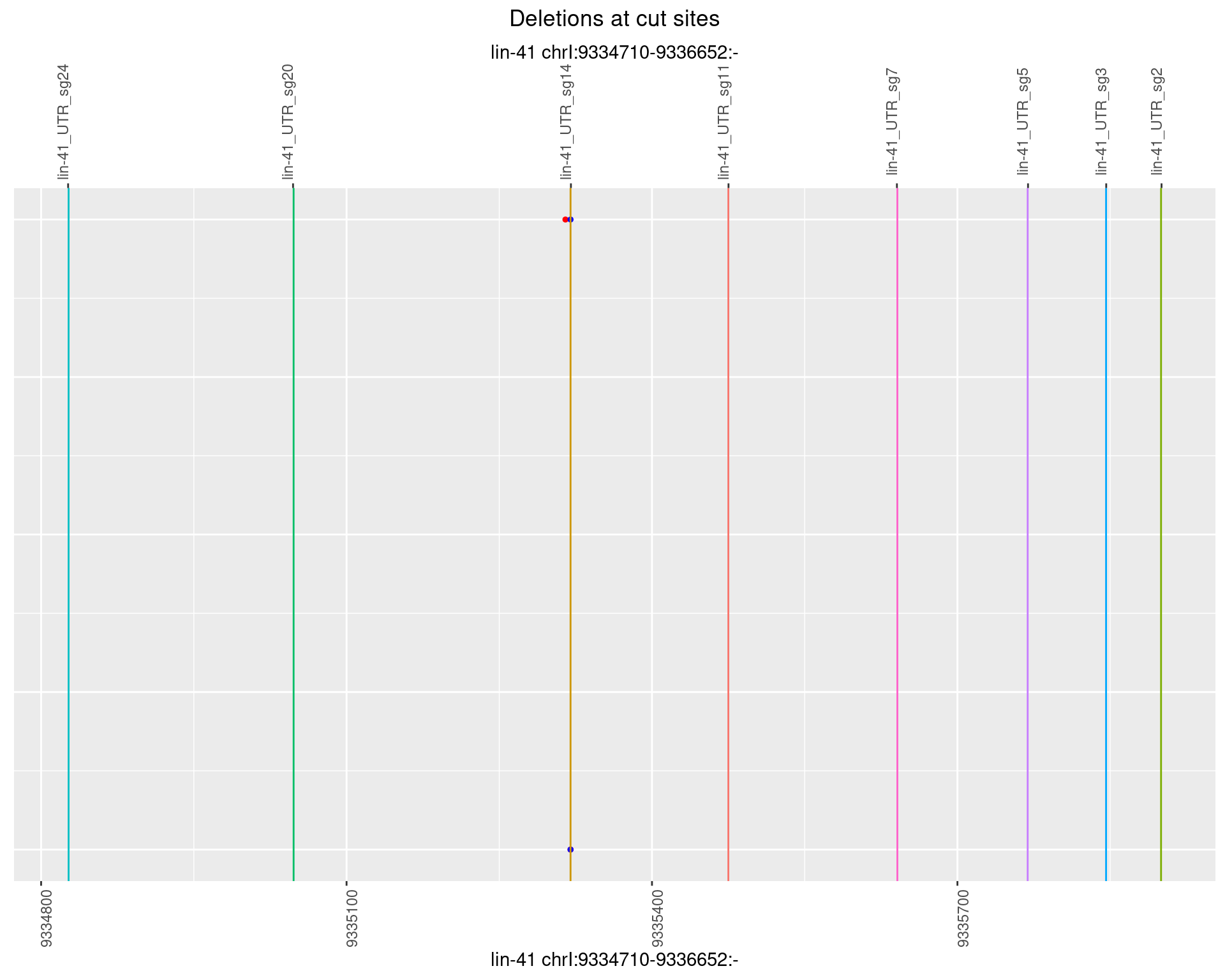
1.42.6 Freq: 0.1
No plot to show
1.43 lin41_RNA_L1_N2_3end
1.43.1 Freq: 0
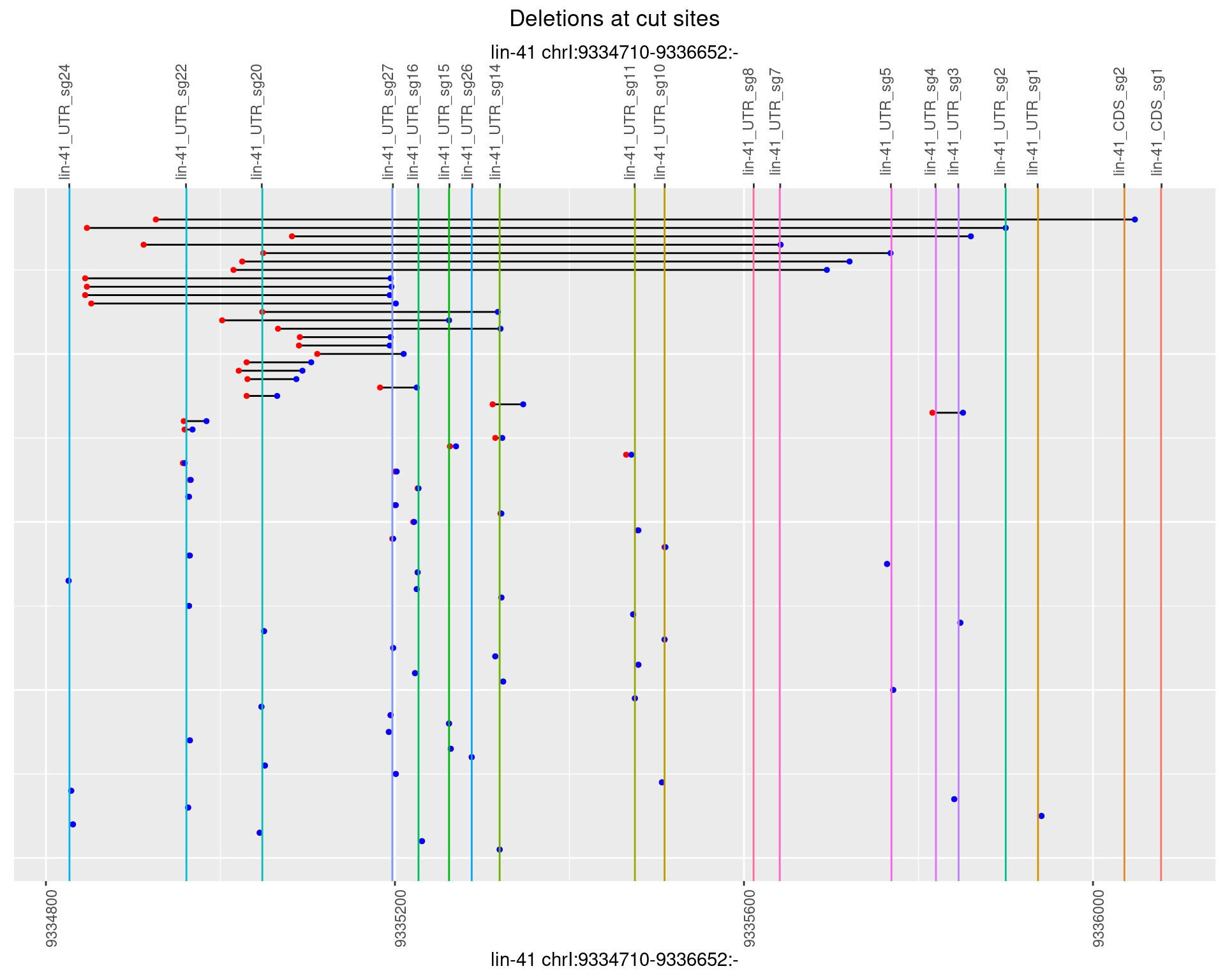
1.43.2 Freq: 1e-05
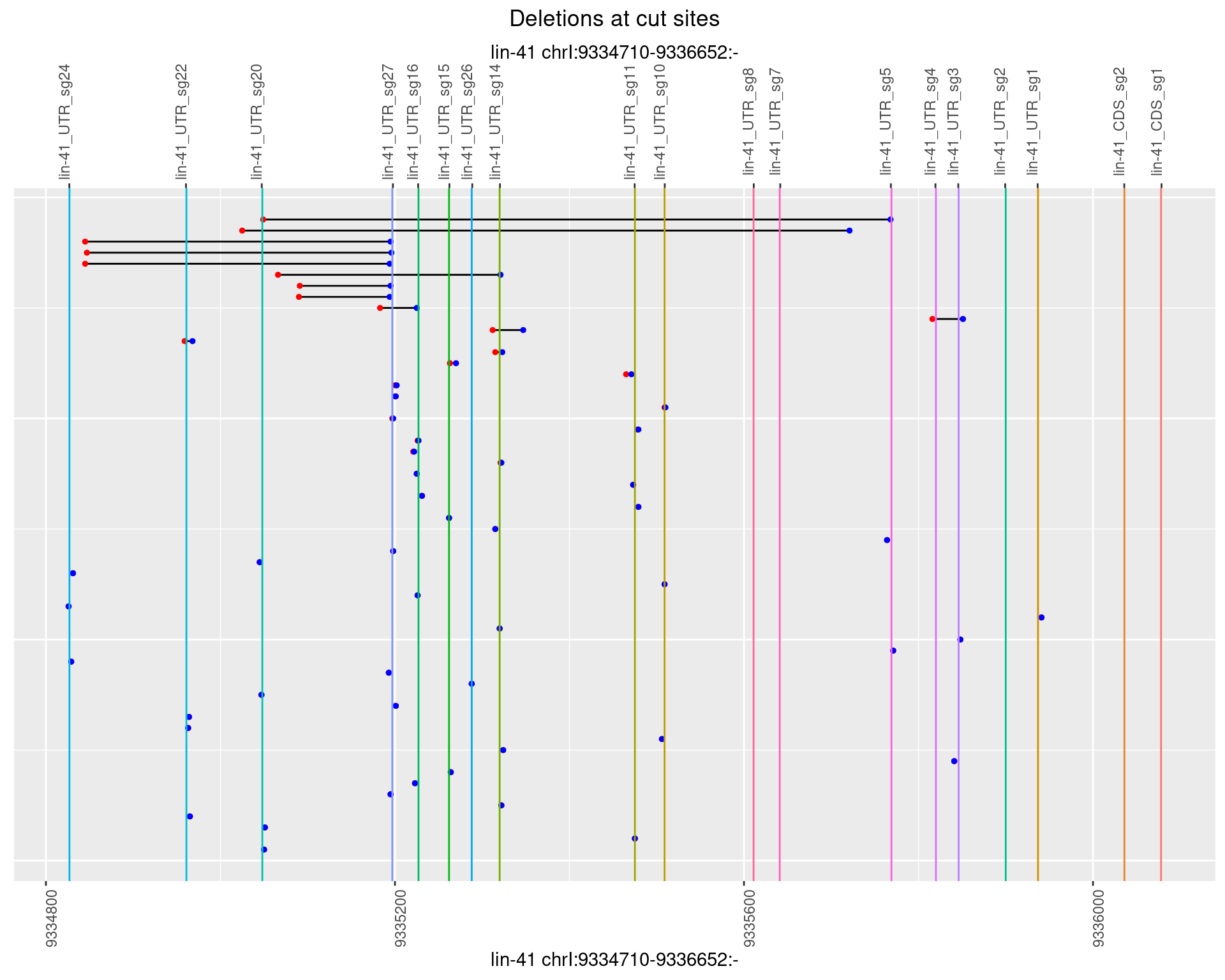
1.43.3 Freq: 1e-04
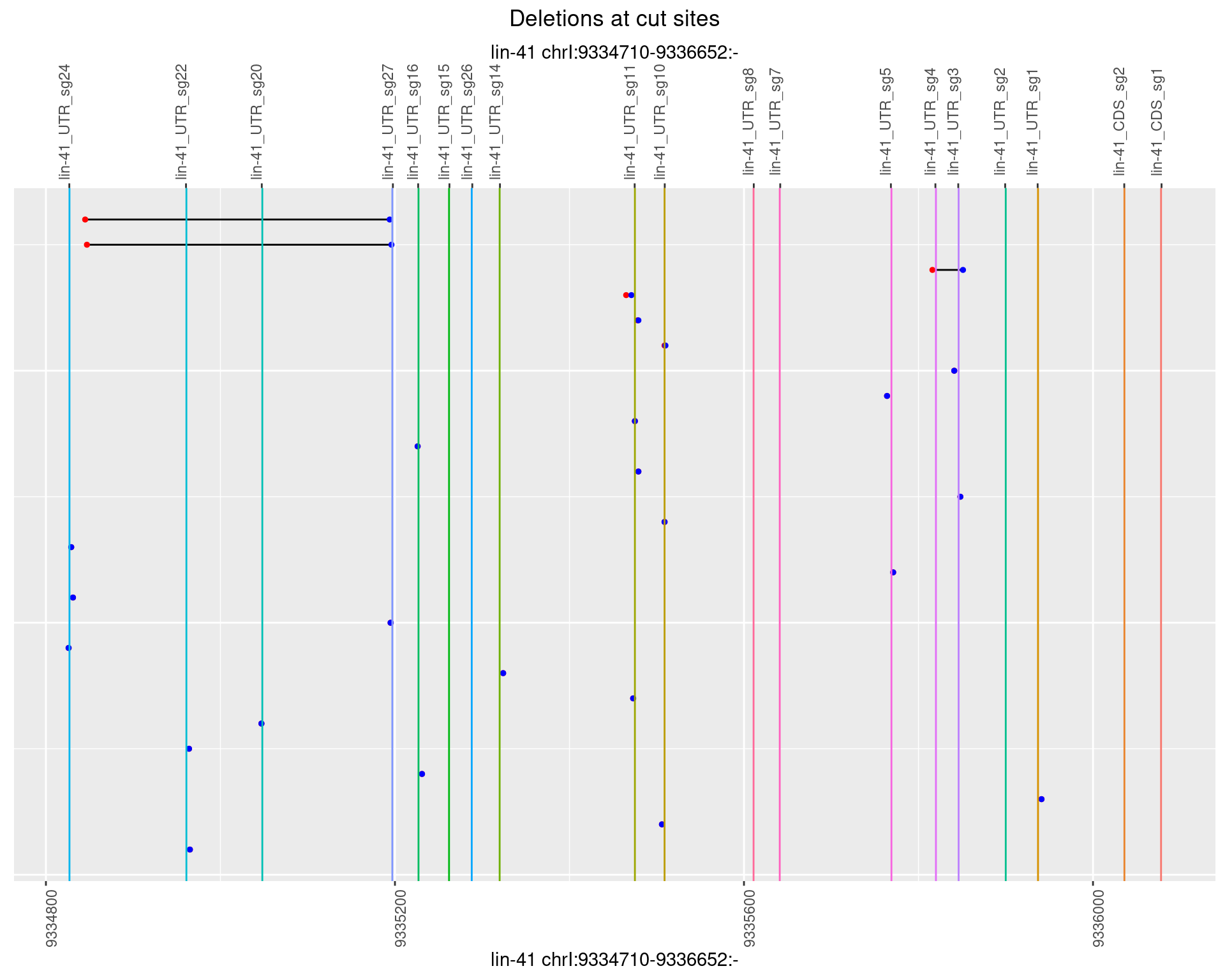
1.43.4 Freq: 0.001
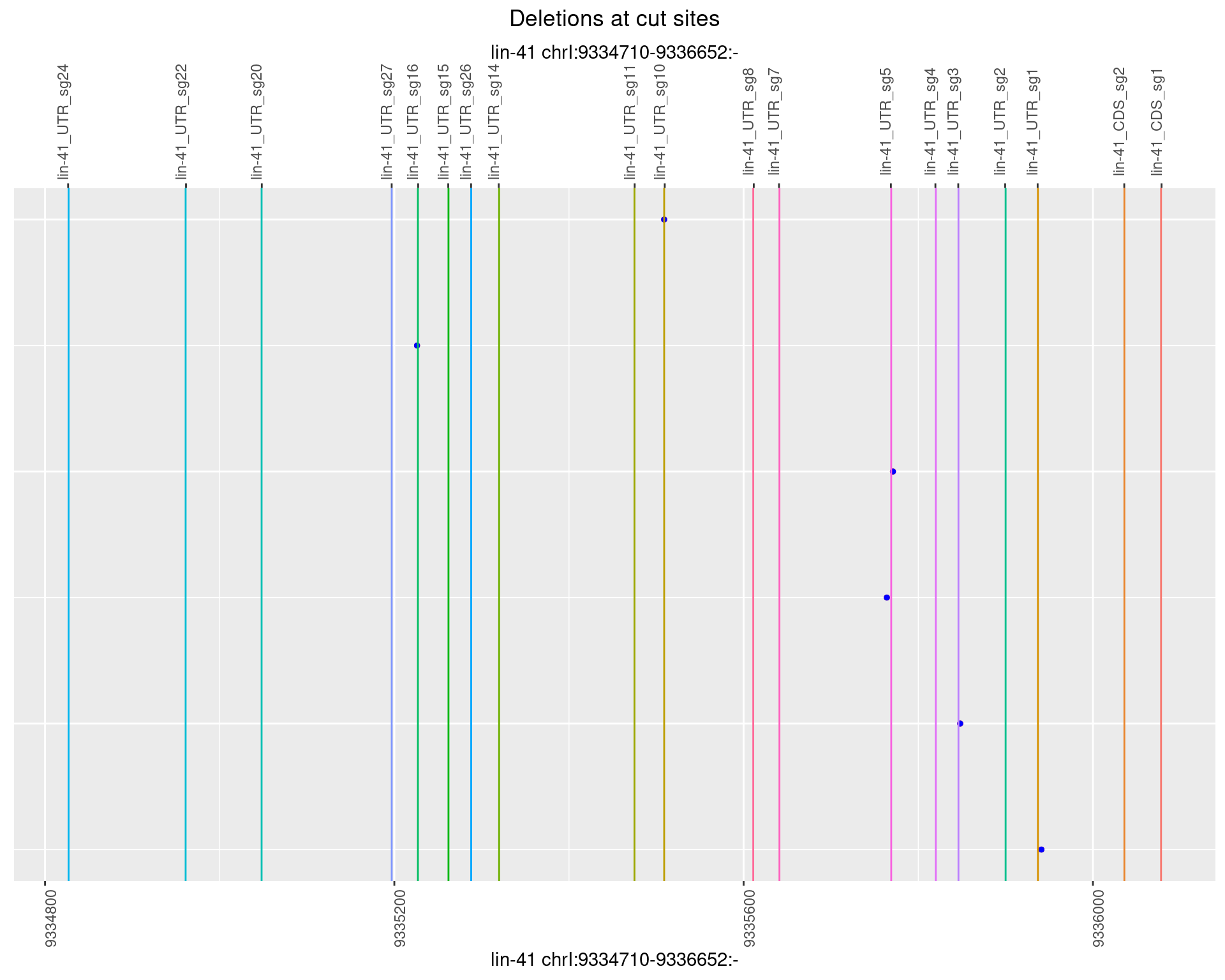
1.43.5 Freq: 0.01
No plot to show
1.43.6 Freq: 0.1
No plot to show
1.44 lin41_RNA_L1_1516_3end
1.44.1 Freq: 0
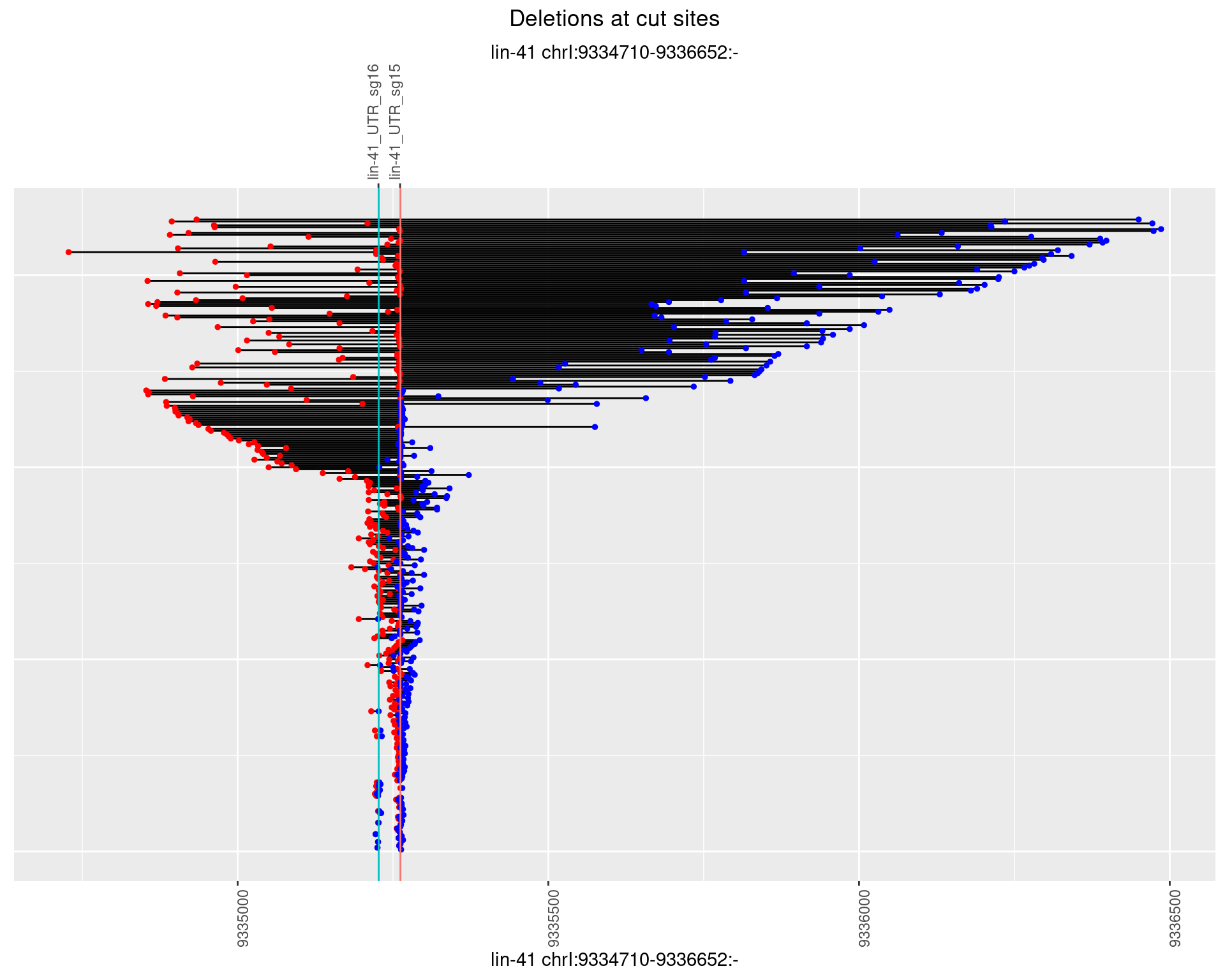
1.44.2 Freq: 1e-05
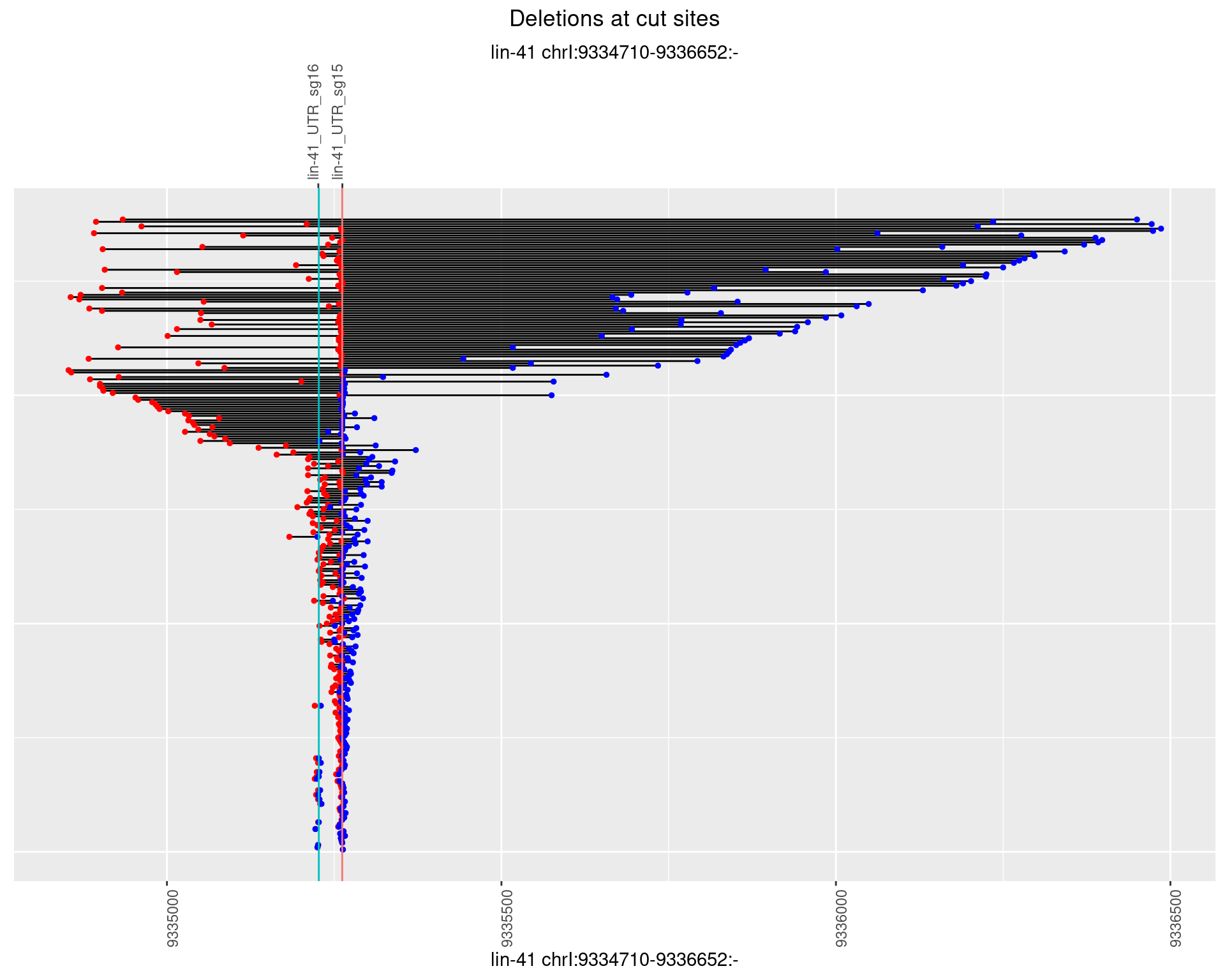
1.44.3 Freq: 1e-04
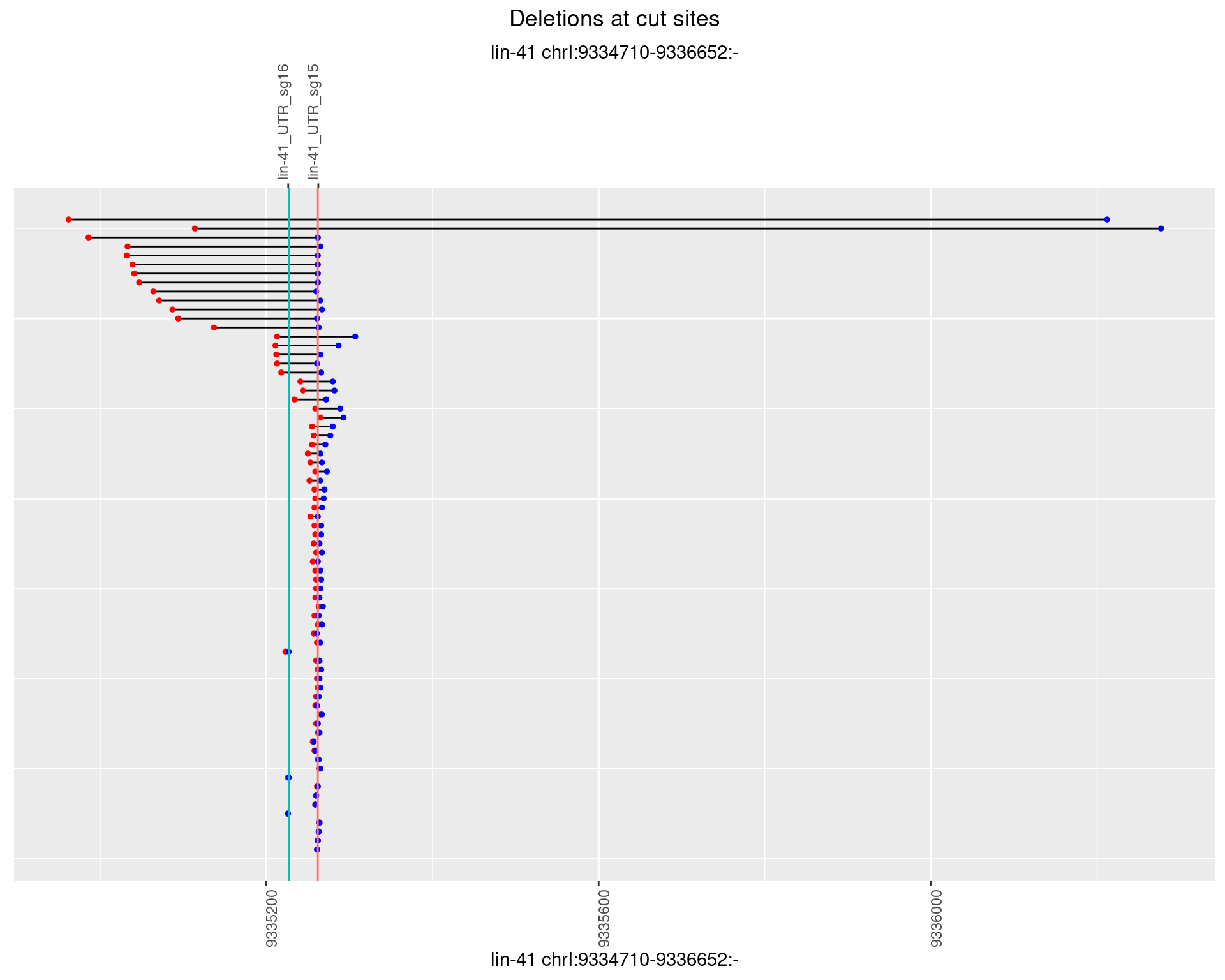
1.44.4 Freq: 0.001
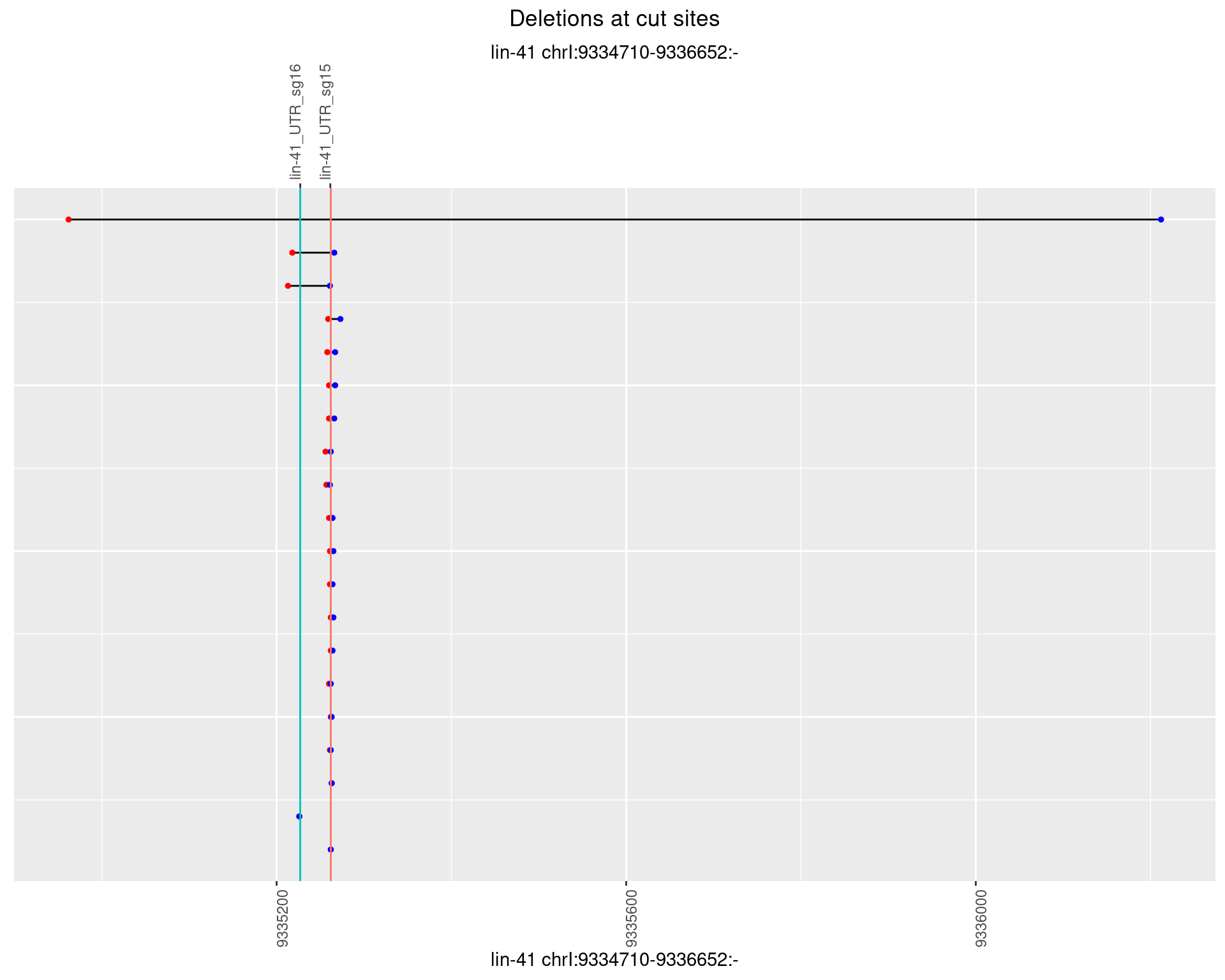
1.44.5 Freq: 0.01
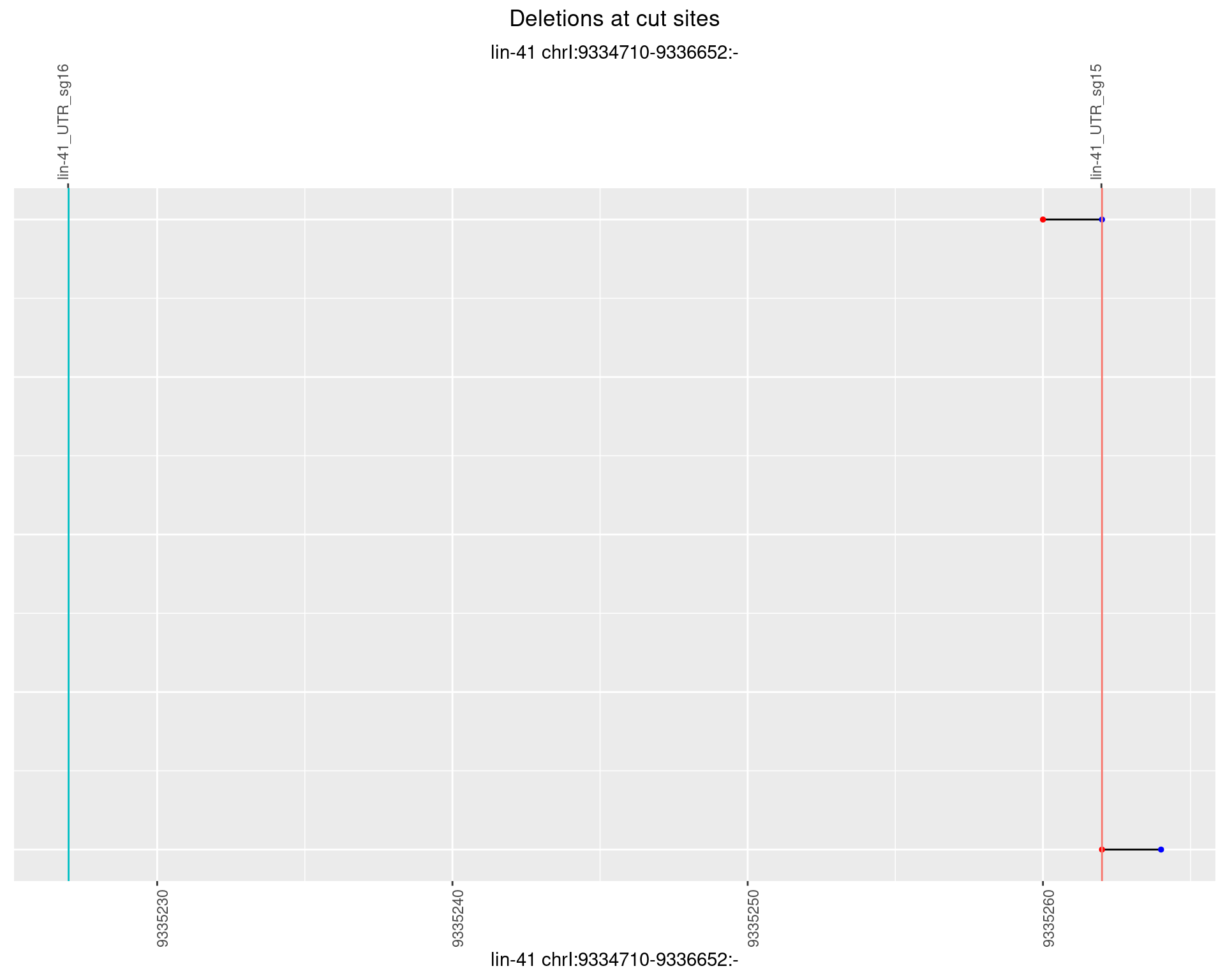
1.44.6 Freq: 0.1
No plot to show
1.45 lin41_RNA_L1_2627_3end
1.45.1 Freq: 0
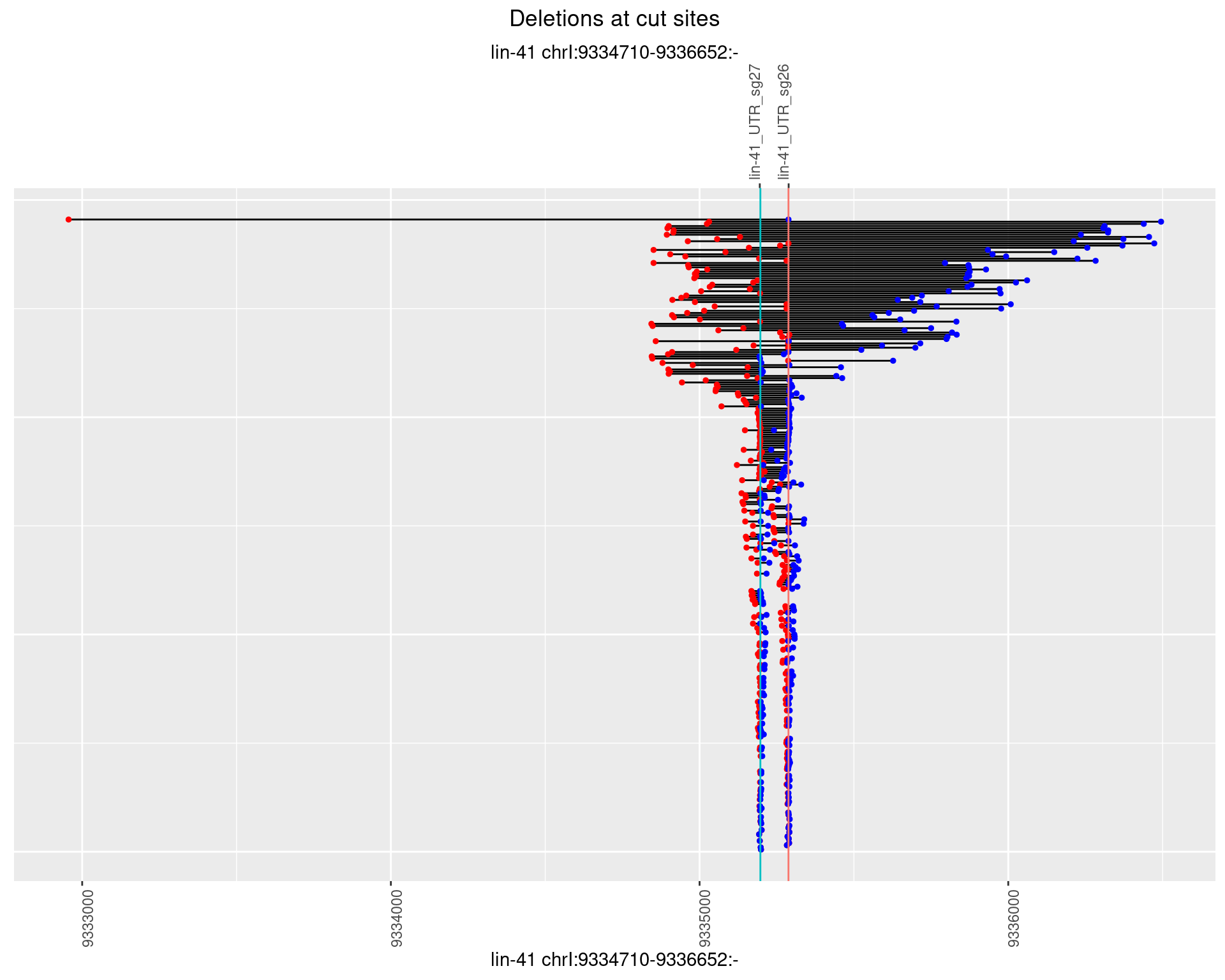
1.45.2 Freq: 1e-05
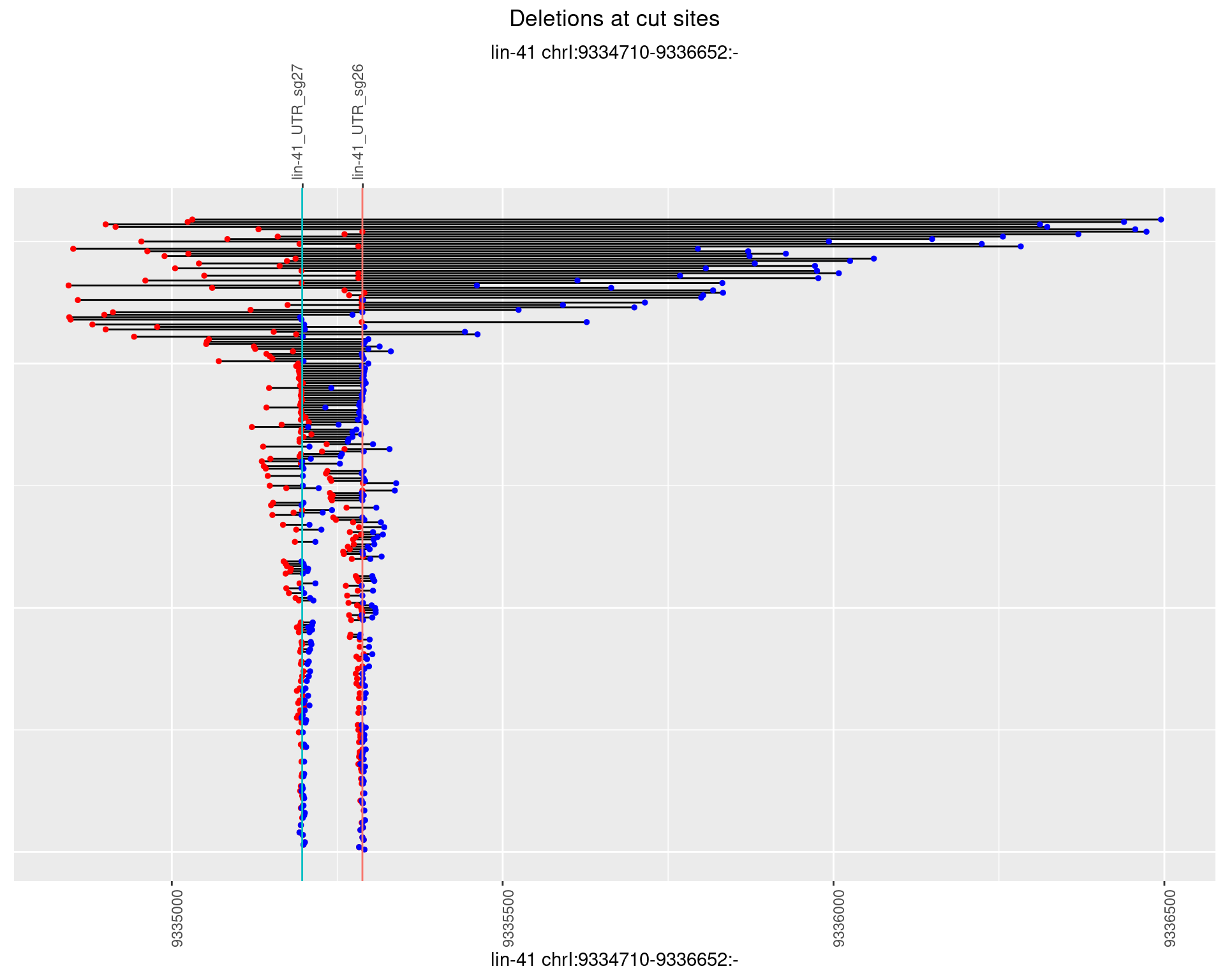
1.45.3 Freq: 1e-04
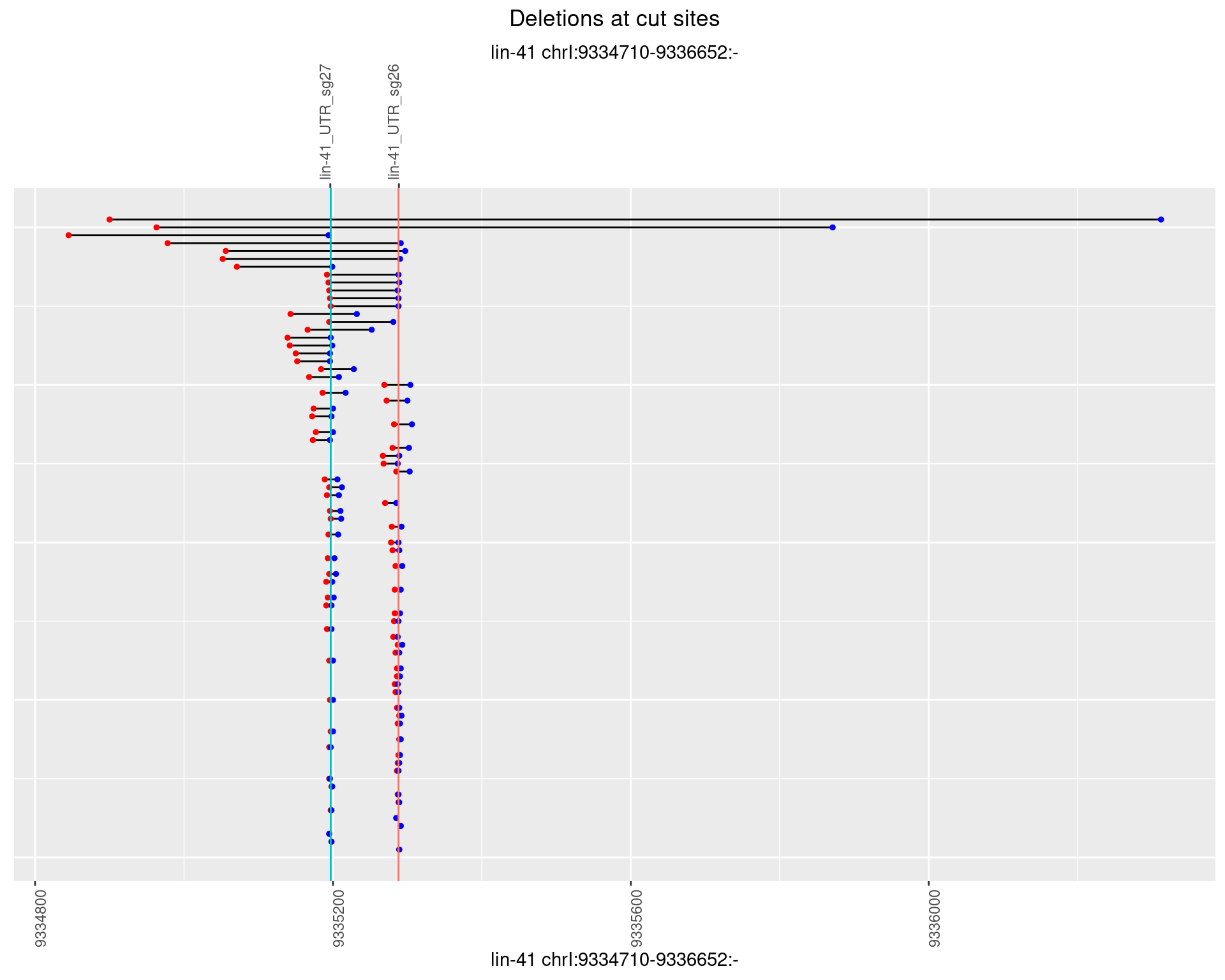
1.45.4 Freq: 0.001
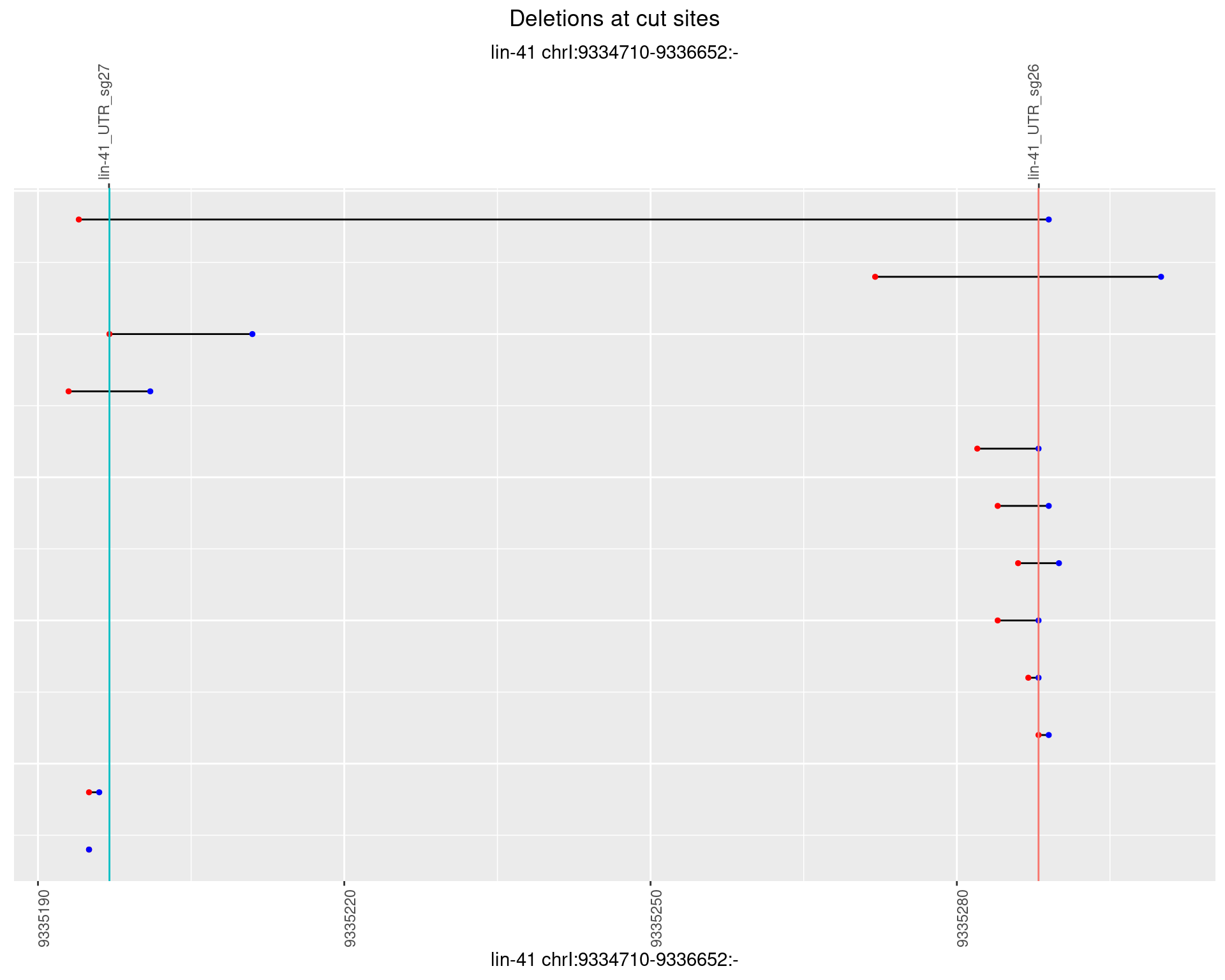
1.45.5 Freq: 0.01
No plot to show
1.45.6 Freq: 0.1
No plot to show
1.46 lin41_RNA_L1_pool2_3end
1.46.1 Freq: 0
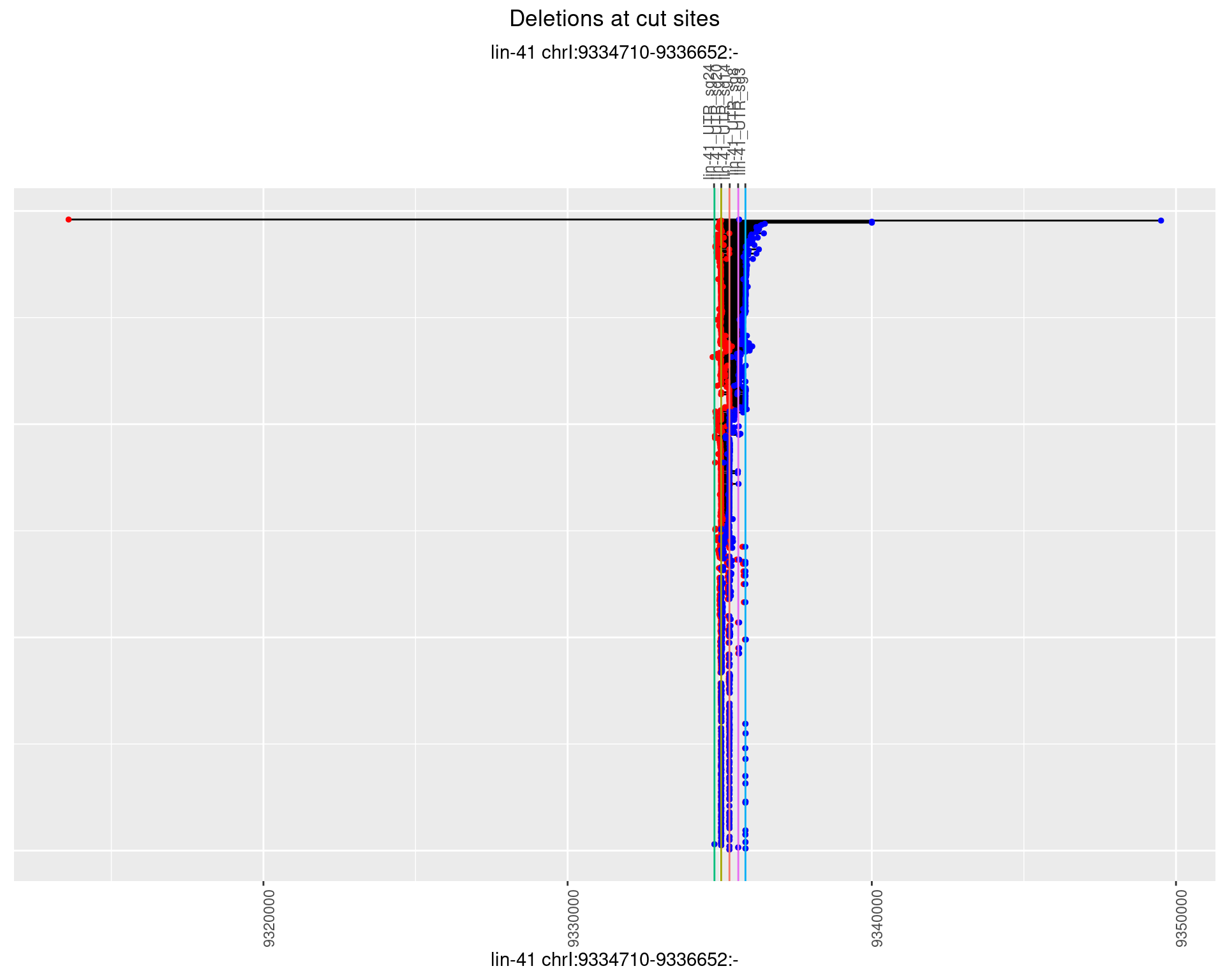
1.46.2 Freq: 1e-05
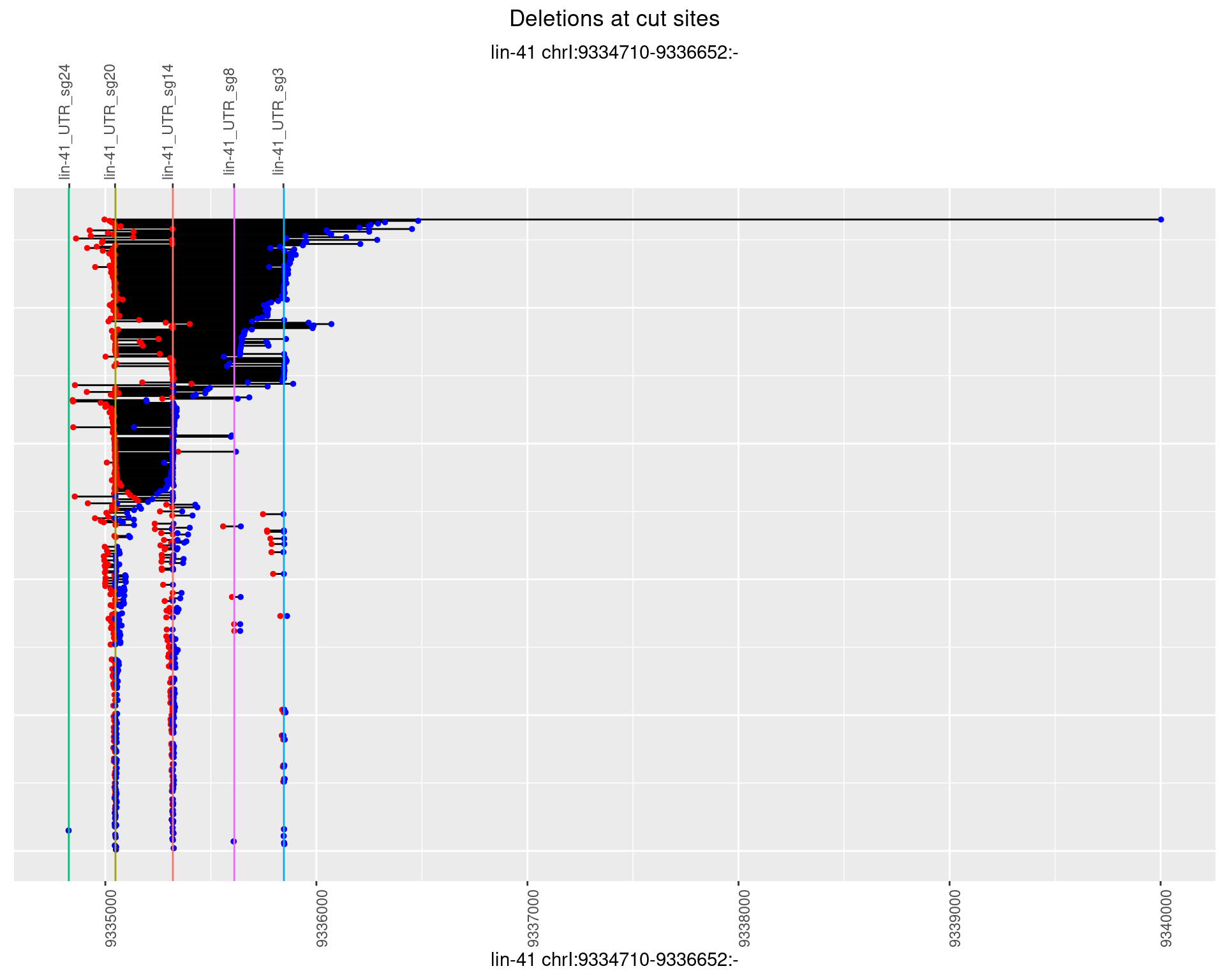
1.46.3 Freq: 1e-04
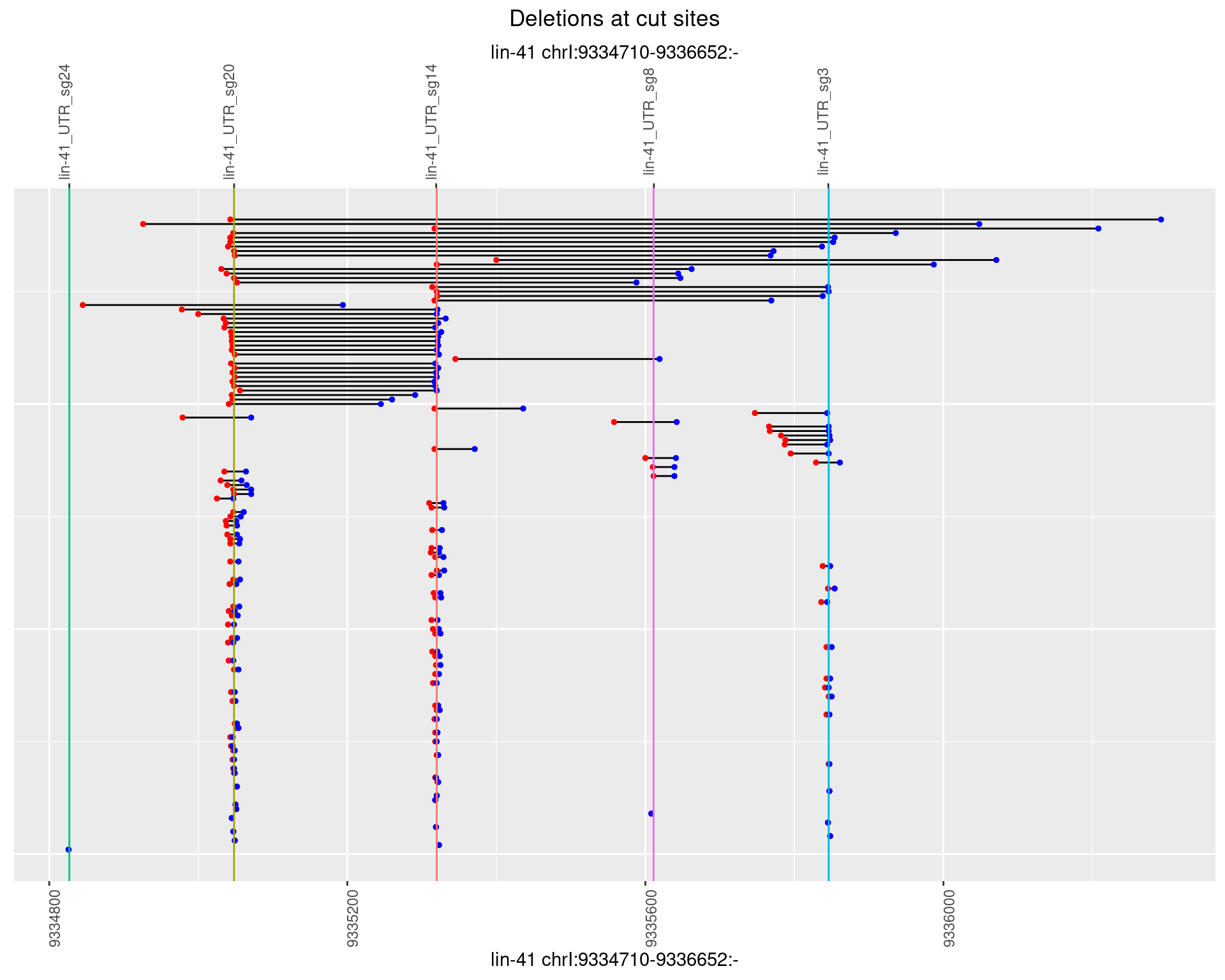
1.46.4 Freq: 0.001
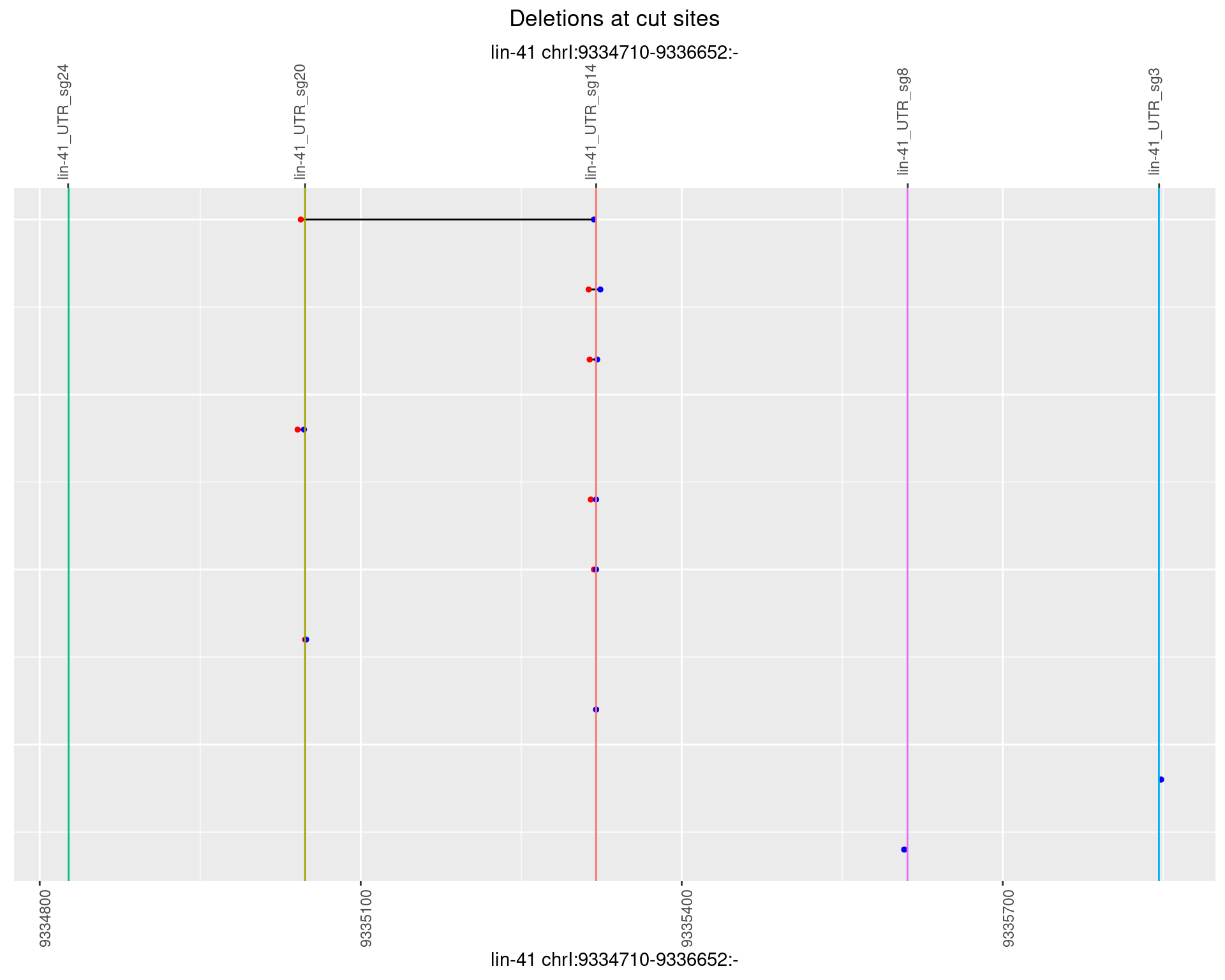
1.46.5 Freq: 0.01
1.46.6 Freq: 0.1
No plot to show
1.47 lin41_RNA_L1_pool3_3end
1.47.1 Freq: 0
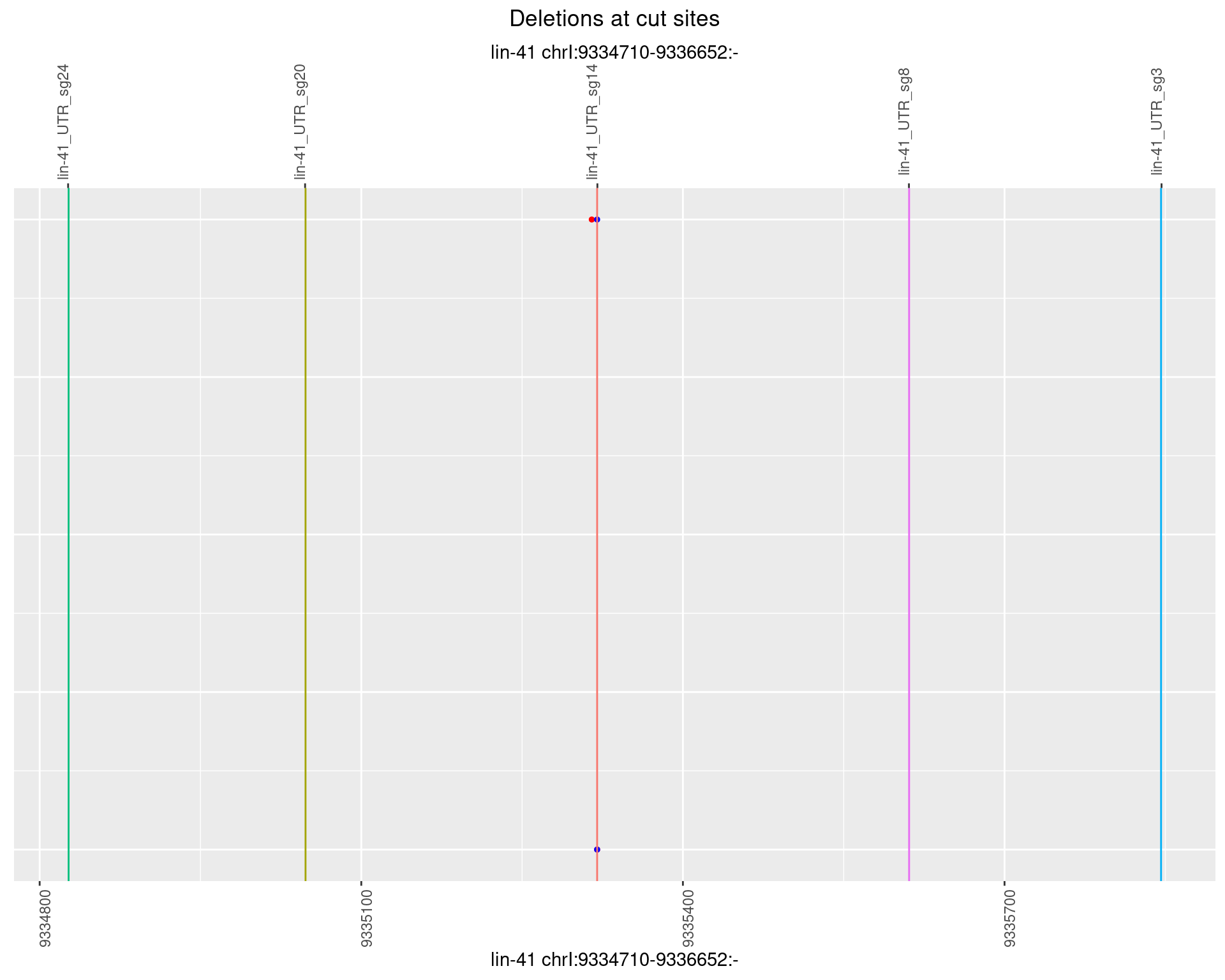
1.47.2 Freq: 1e-05
1.47.3 Freq: 1e-04
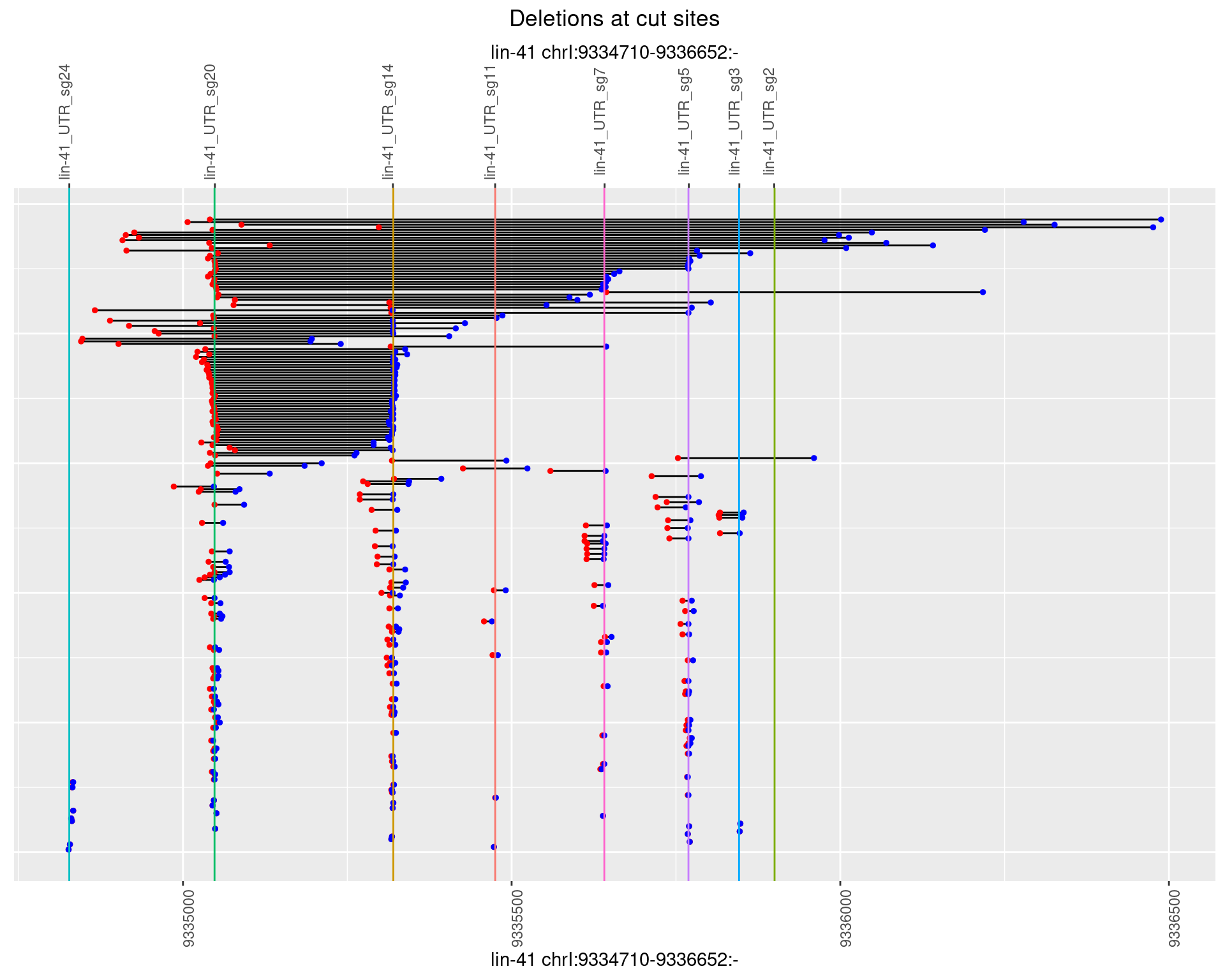
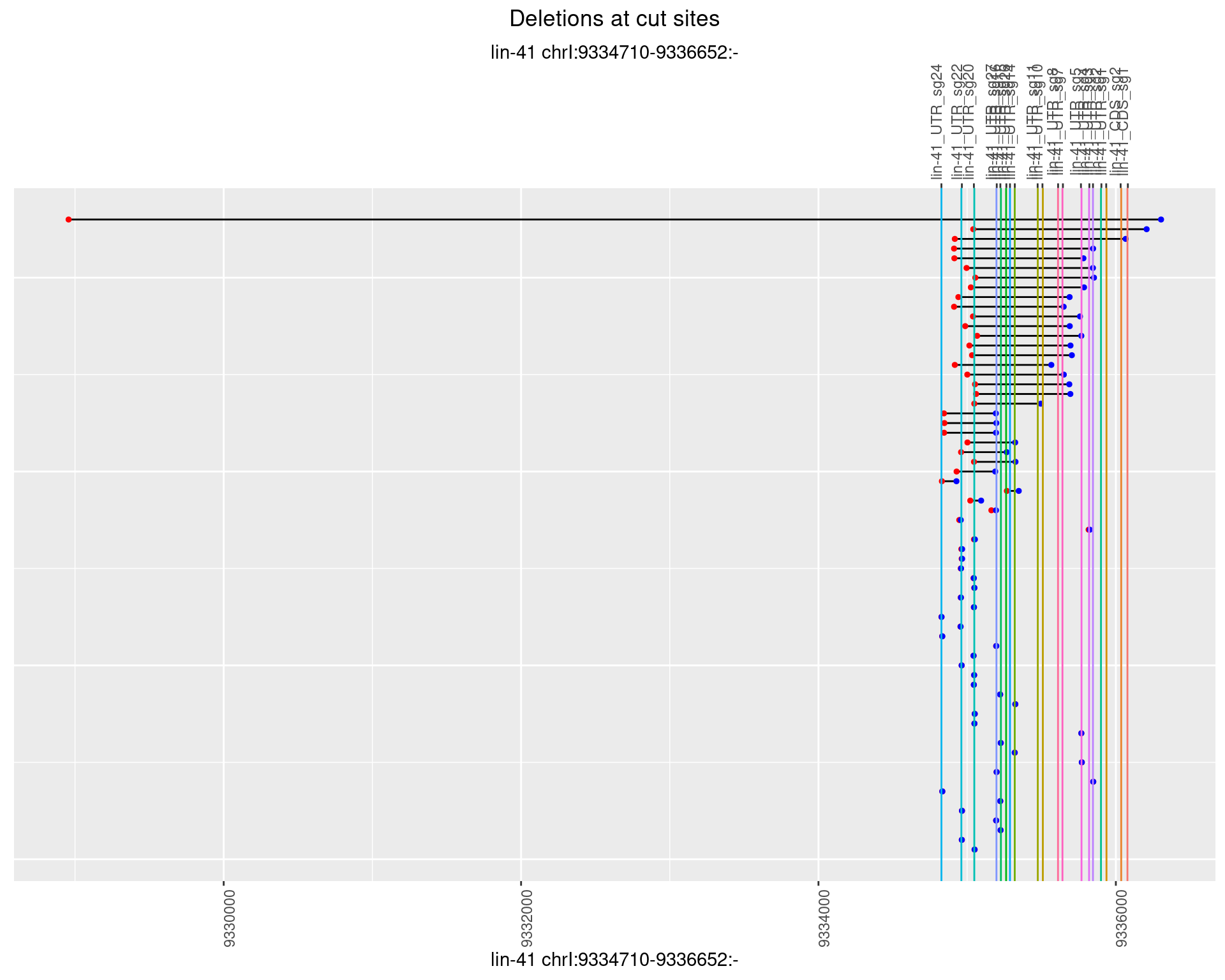
1.47.4 Freq: 0.001
1.47.5 Freq: 0.01
1.47.6 Freq: 0.1
No plot to show
1.48 lin41_RNA_L4_N2_3end
1.48.1 Freq: 0
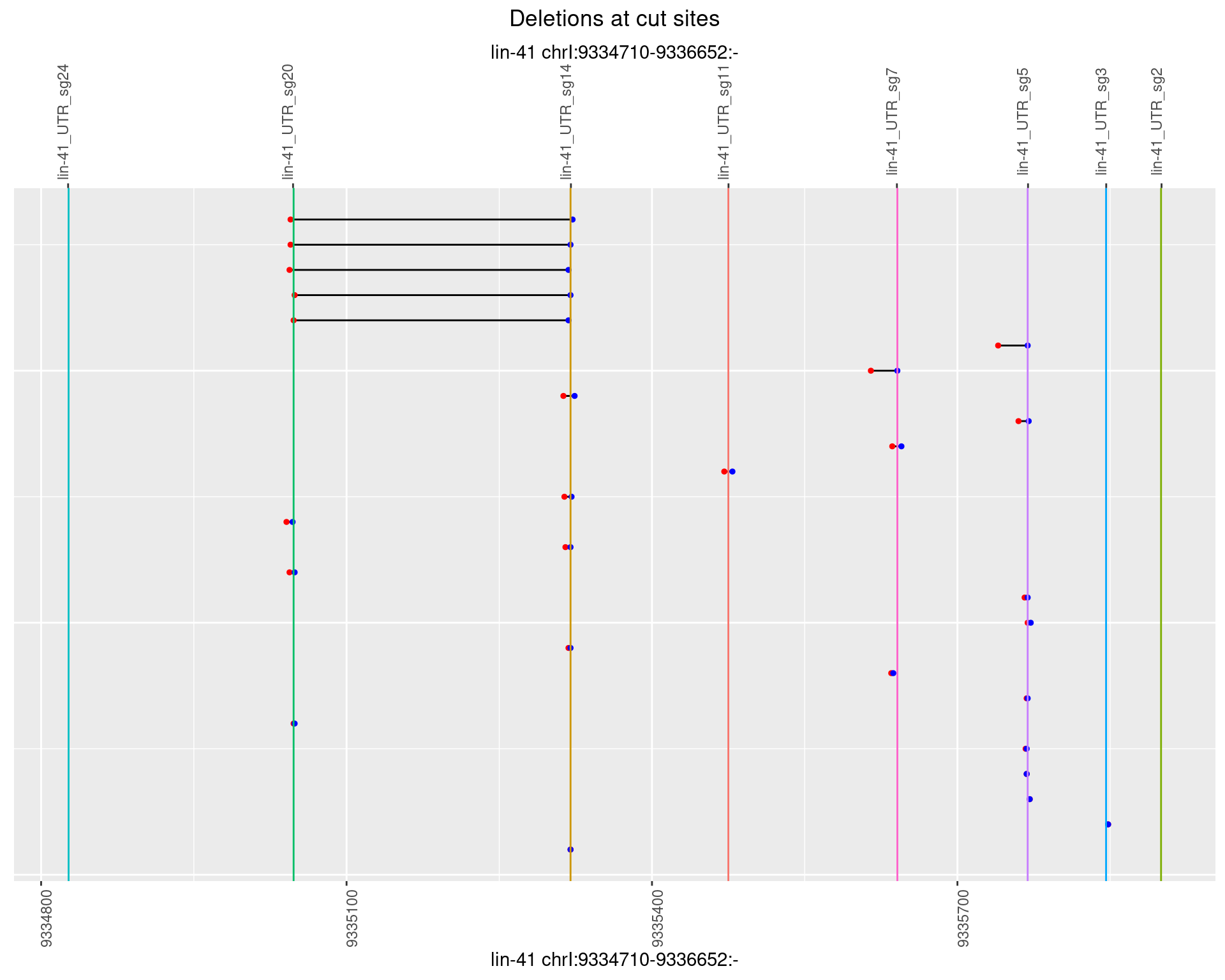
1.48.2 Freq: 1e-05
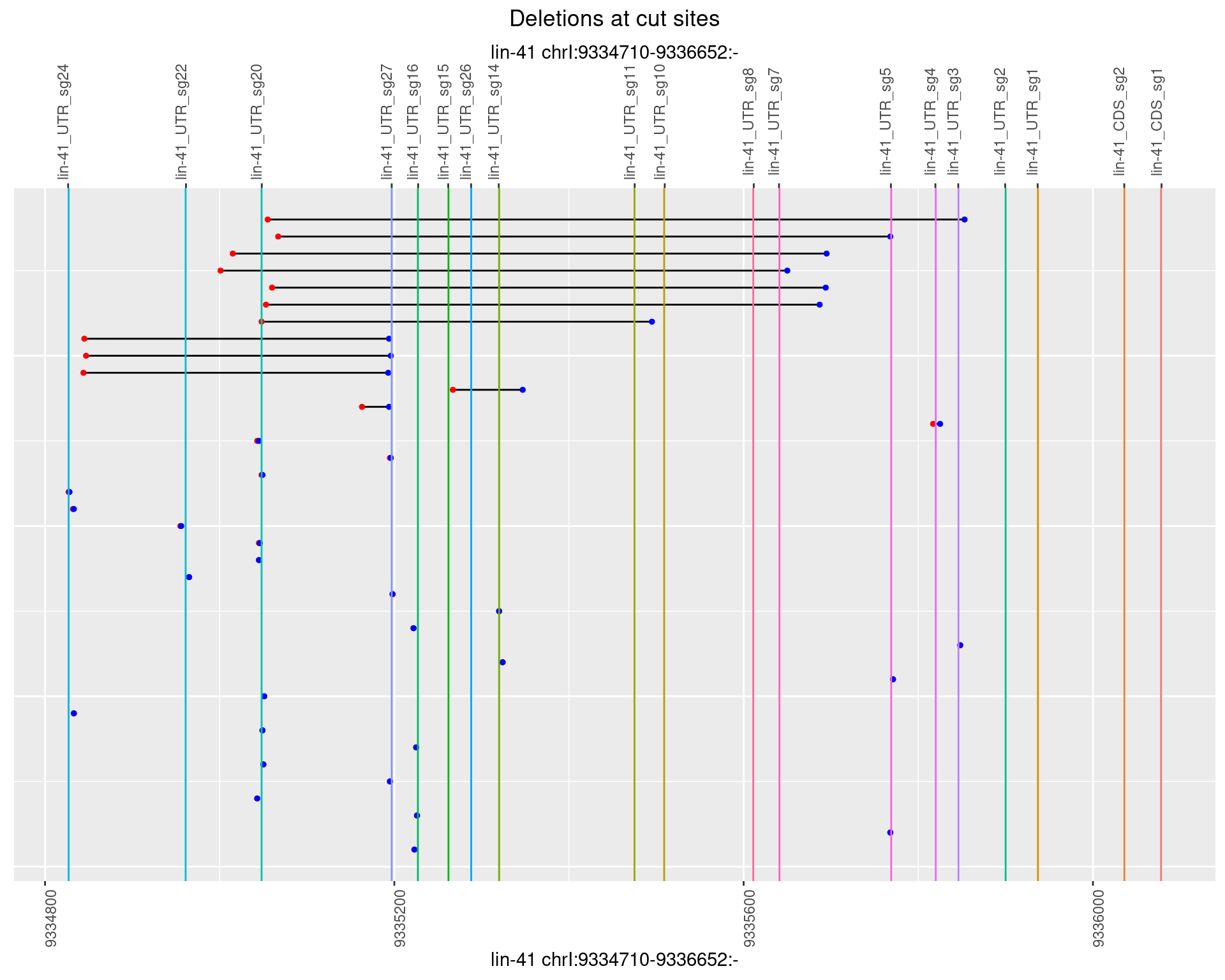
1.48.3 Freq: 1e-04
1.48.4 Freq: 0.001
1.48.5 Freq: 0.01
No plot to show
1.48.6 Freq: 0.1
No plot to show
1.49 lin41_RNA_L4_1516_3end
1.49.1 Freq: 0
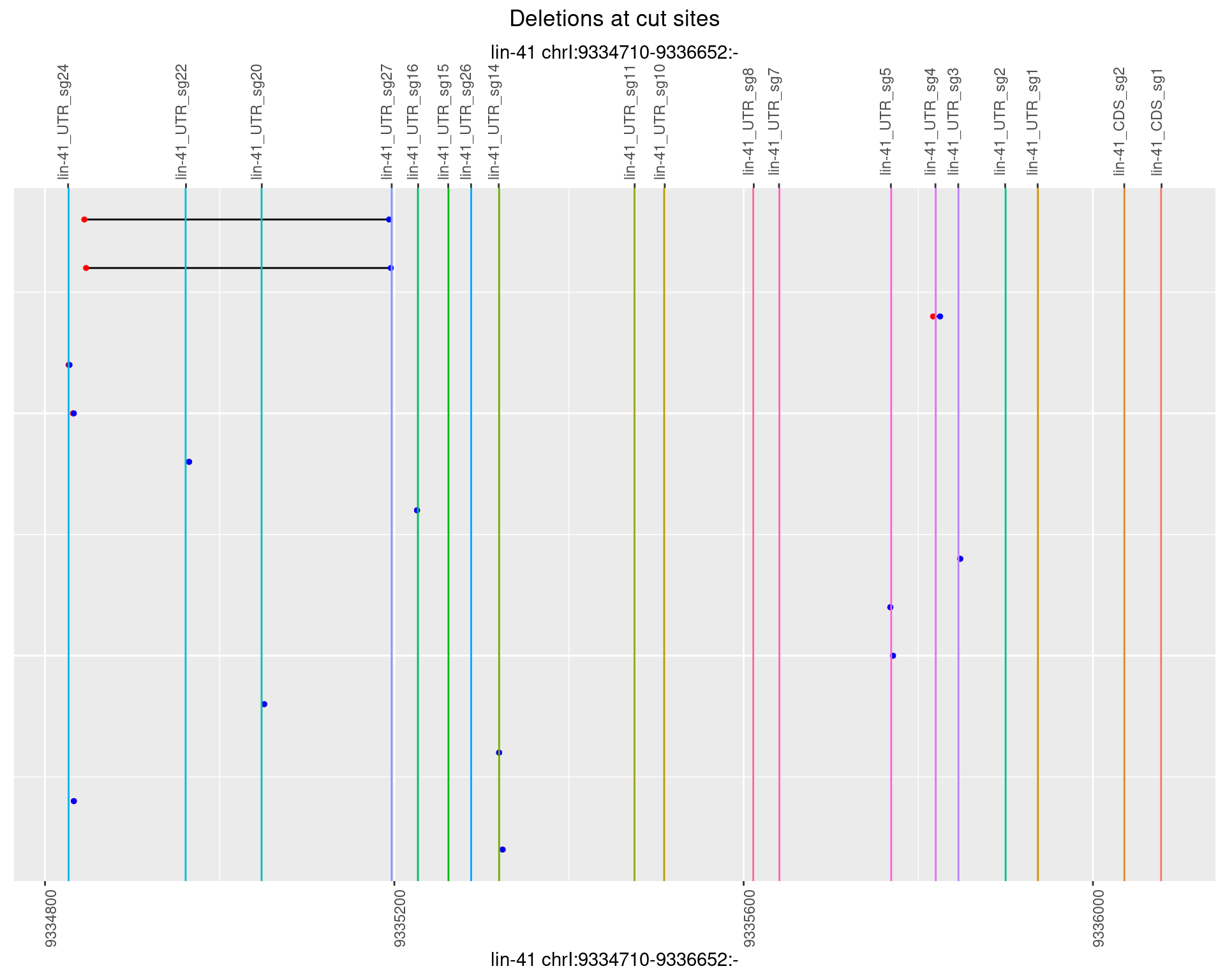
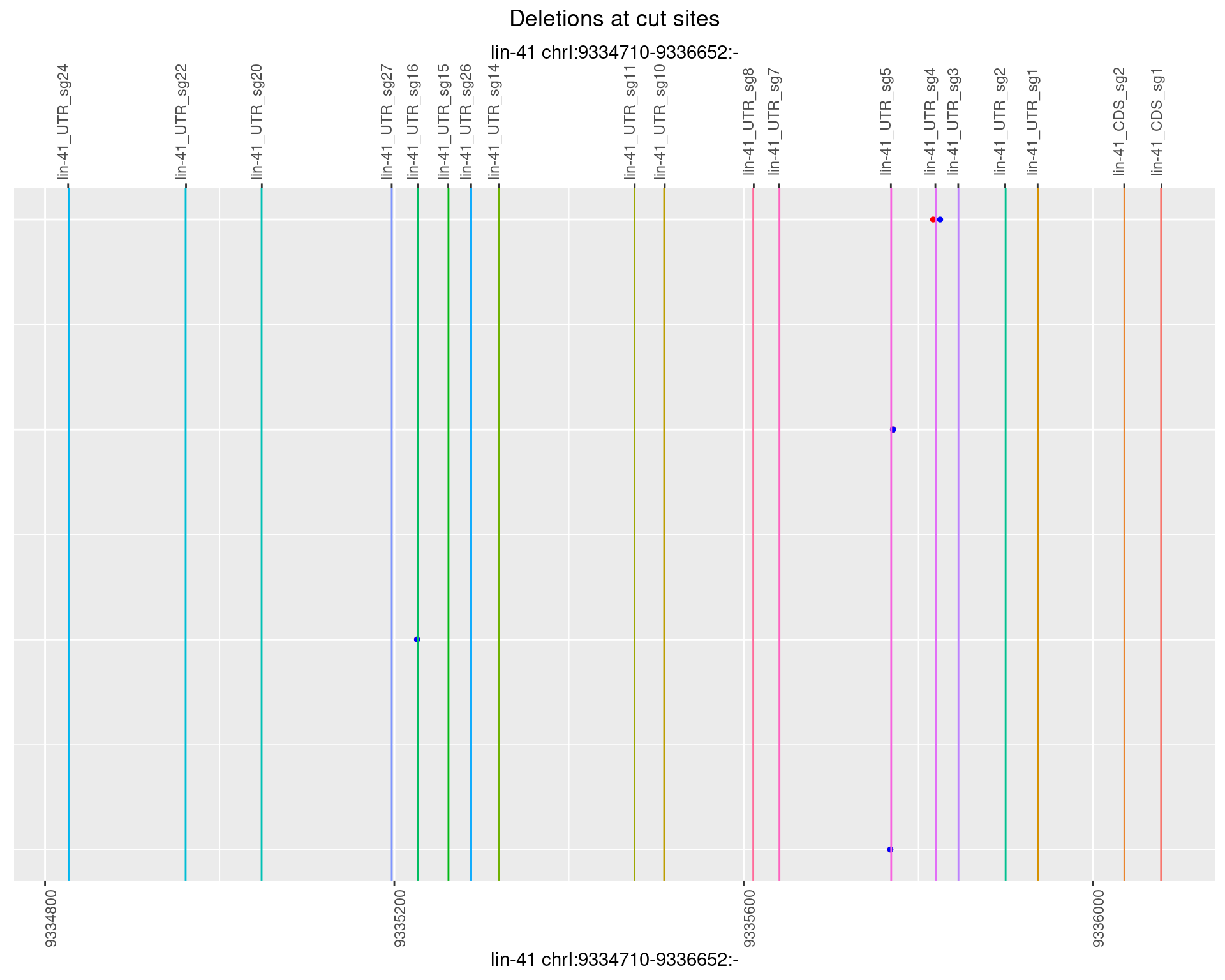
1.49.2 Freq: 1e-05
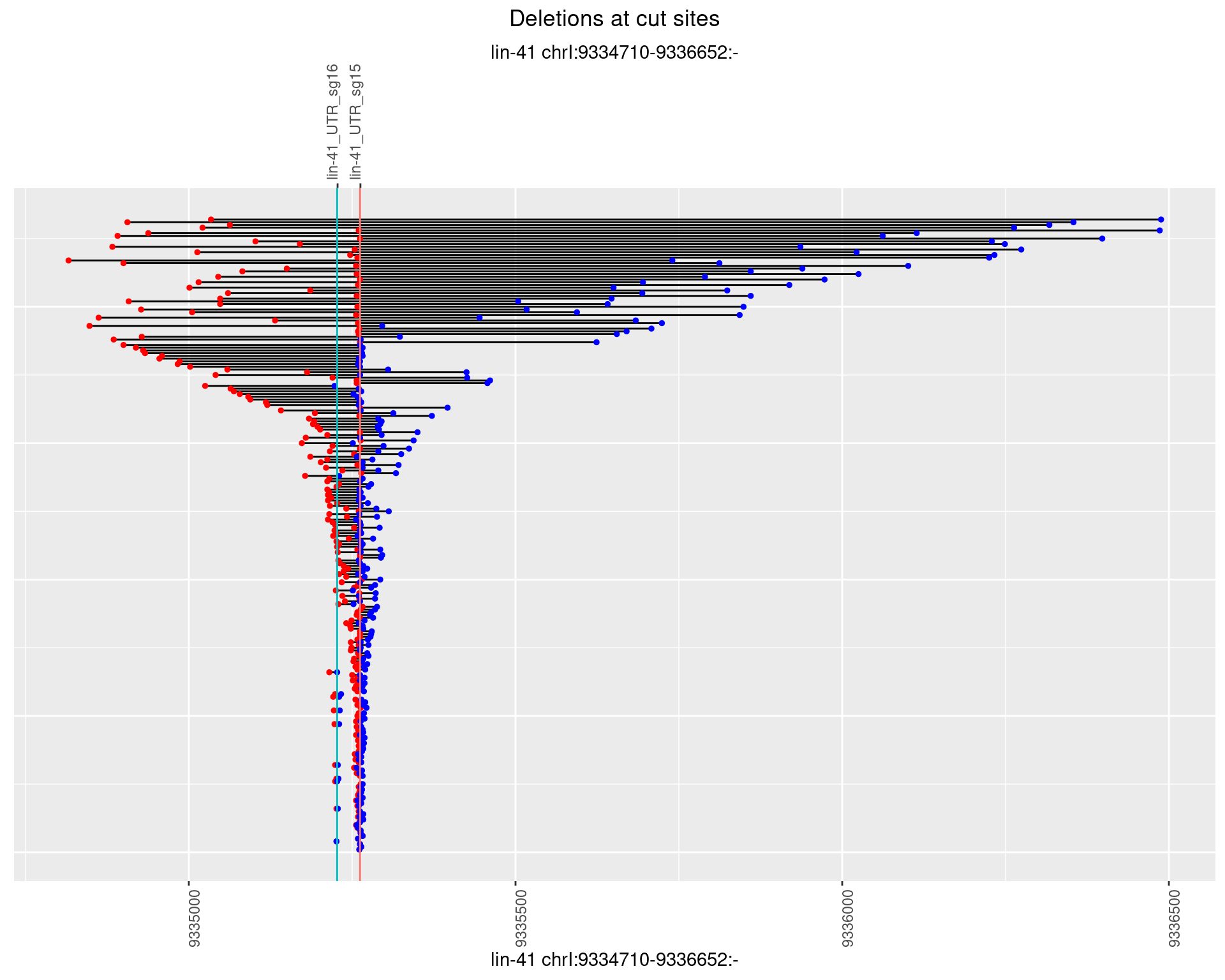
1.49.3 Freq: 1e-04
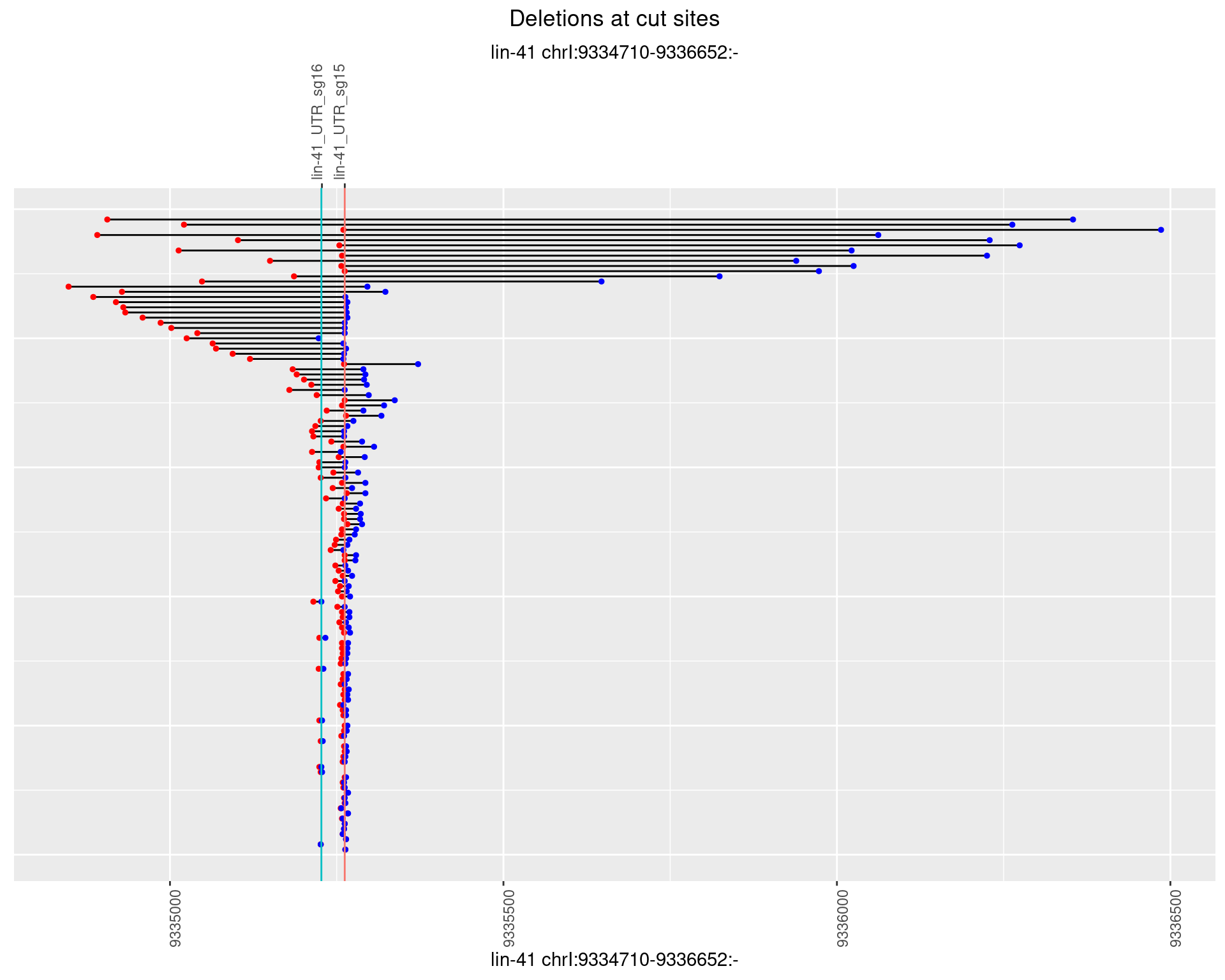
1.49.4 Freq: 0.001
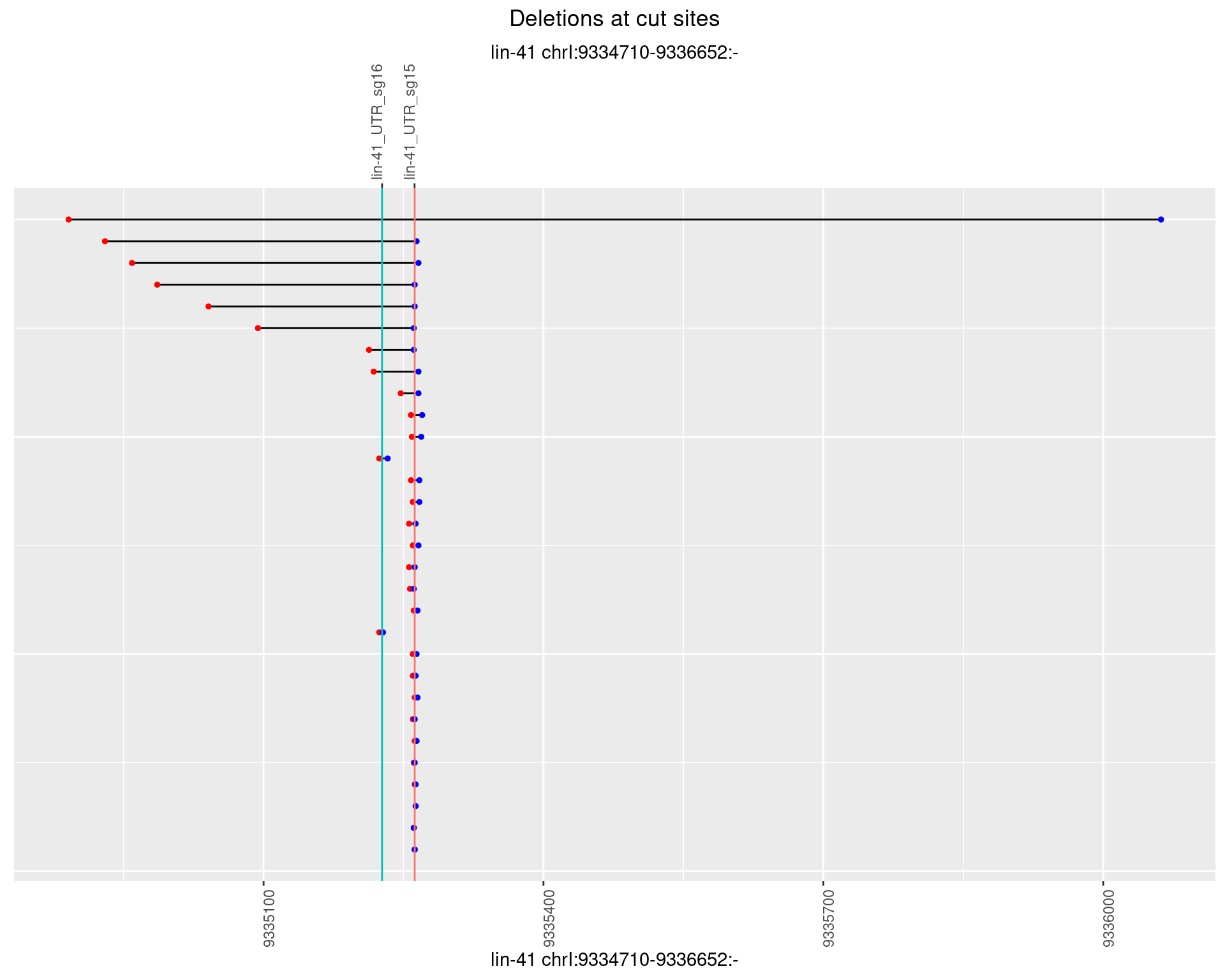
1.49.5 Freq: 0.01
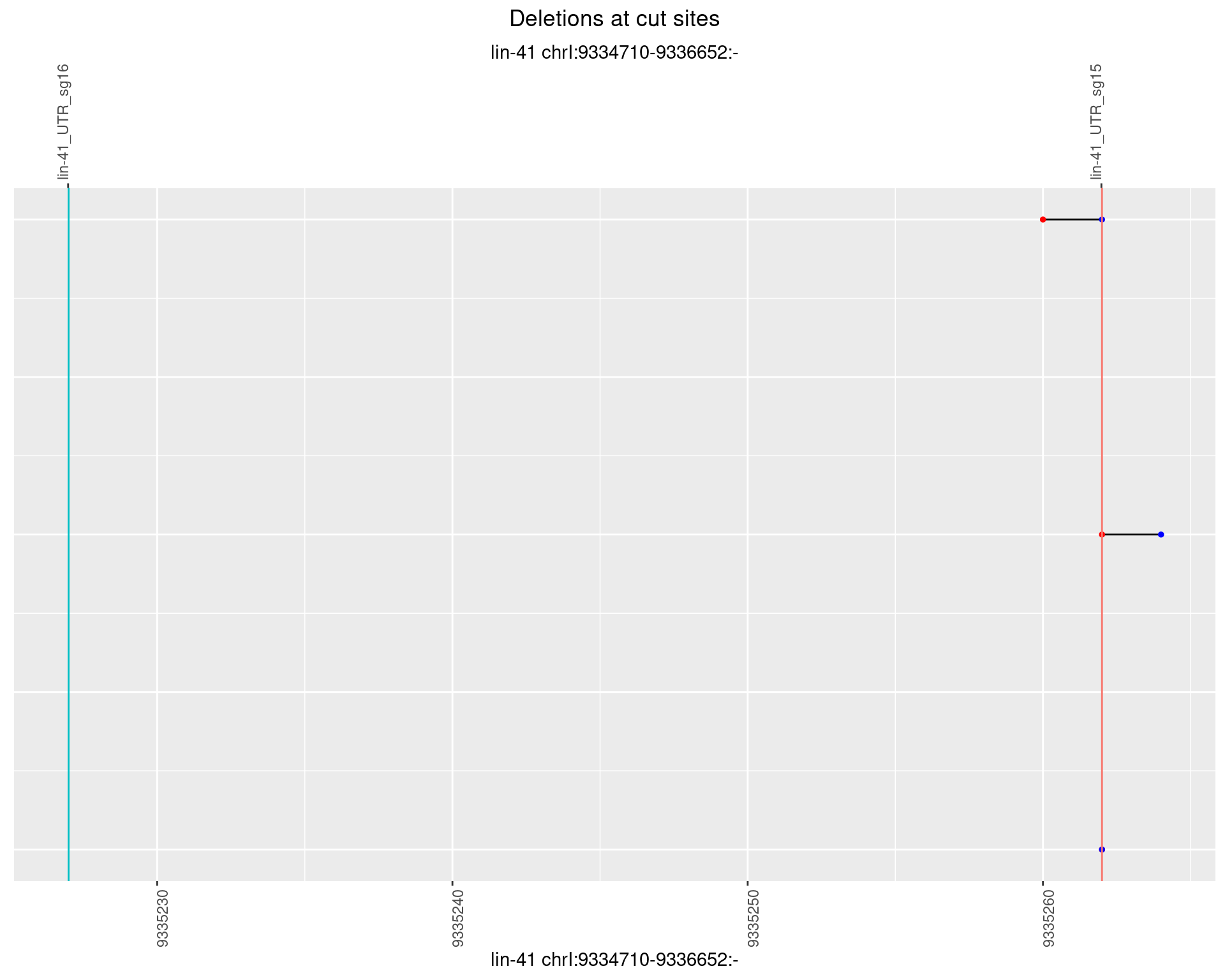
1.49.6 Freq: 0.1
No plot to show
1.50 lin41_RNA_L4_2627_3end
1.50.1 Freq: 0
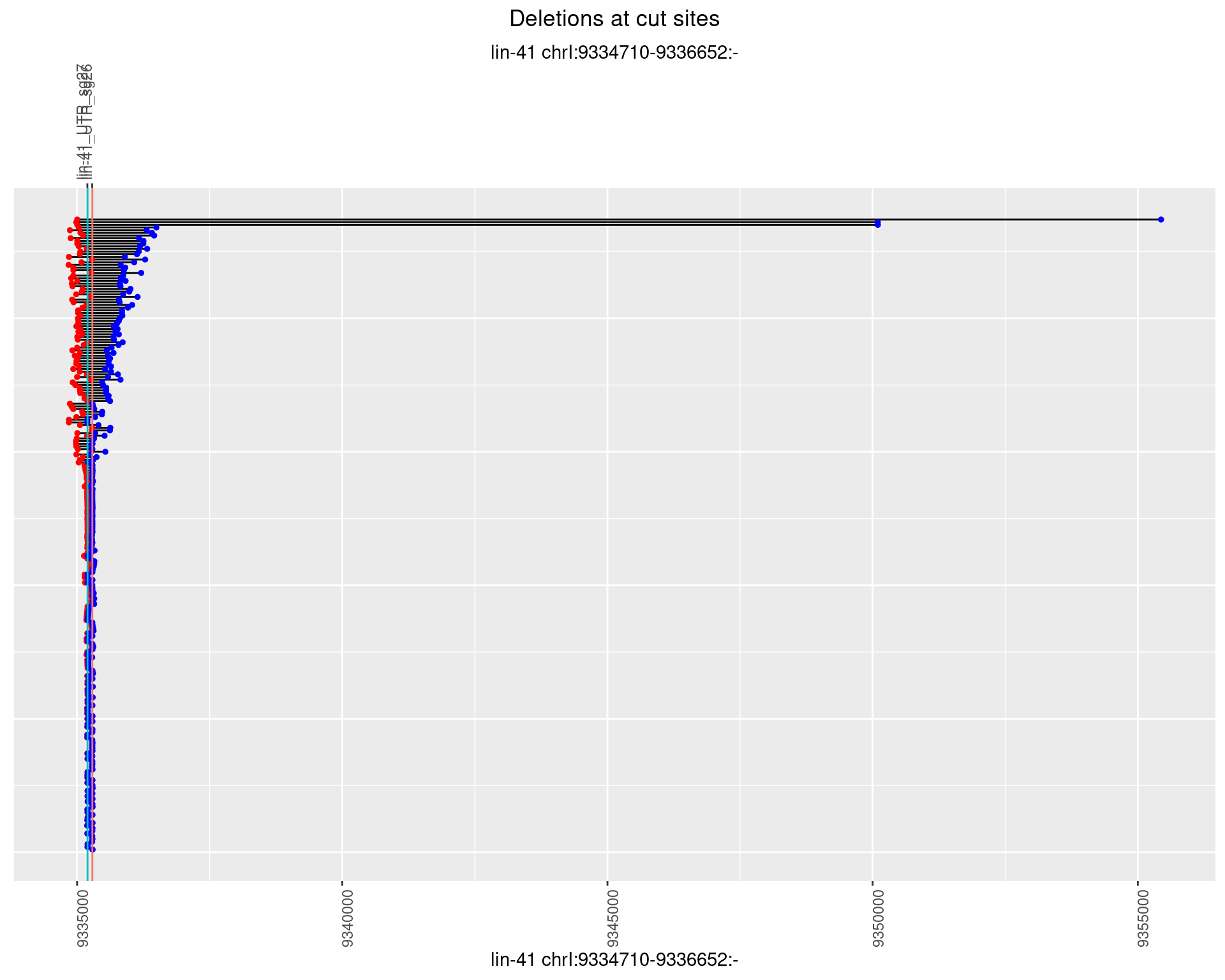
1.50.2 Freq: 1e-05
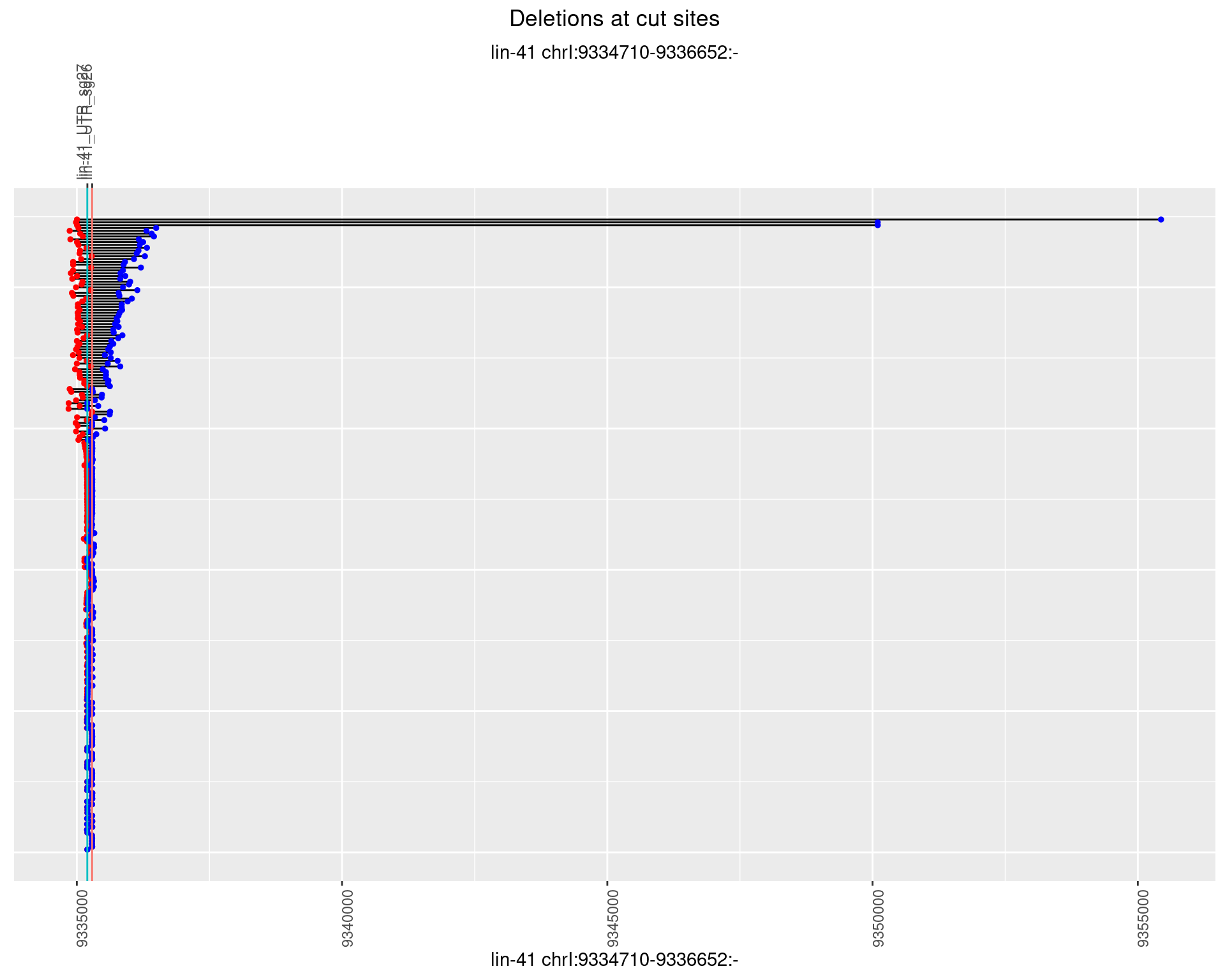
1.50.3 Freq: 1e-04
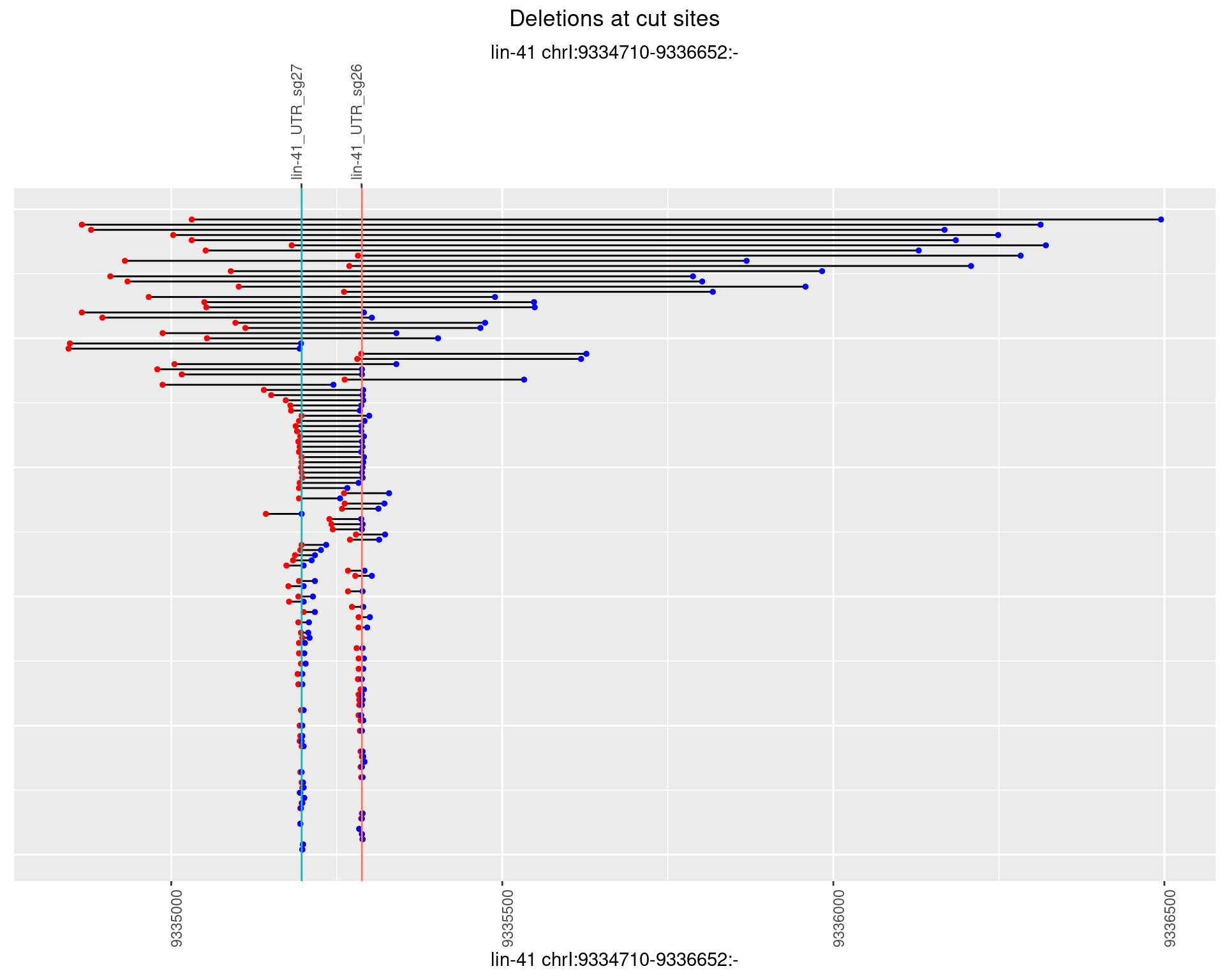
1.50.4 Freq: 0.001
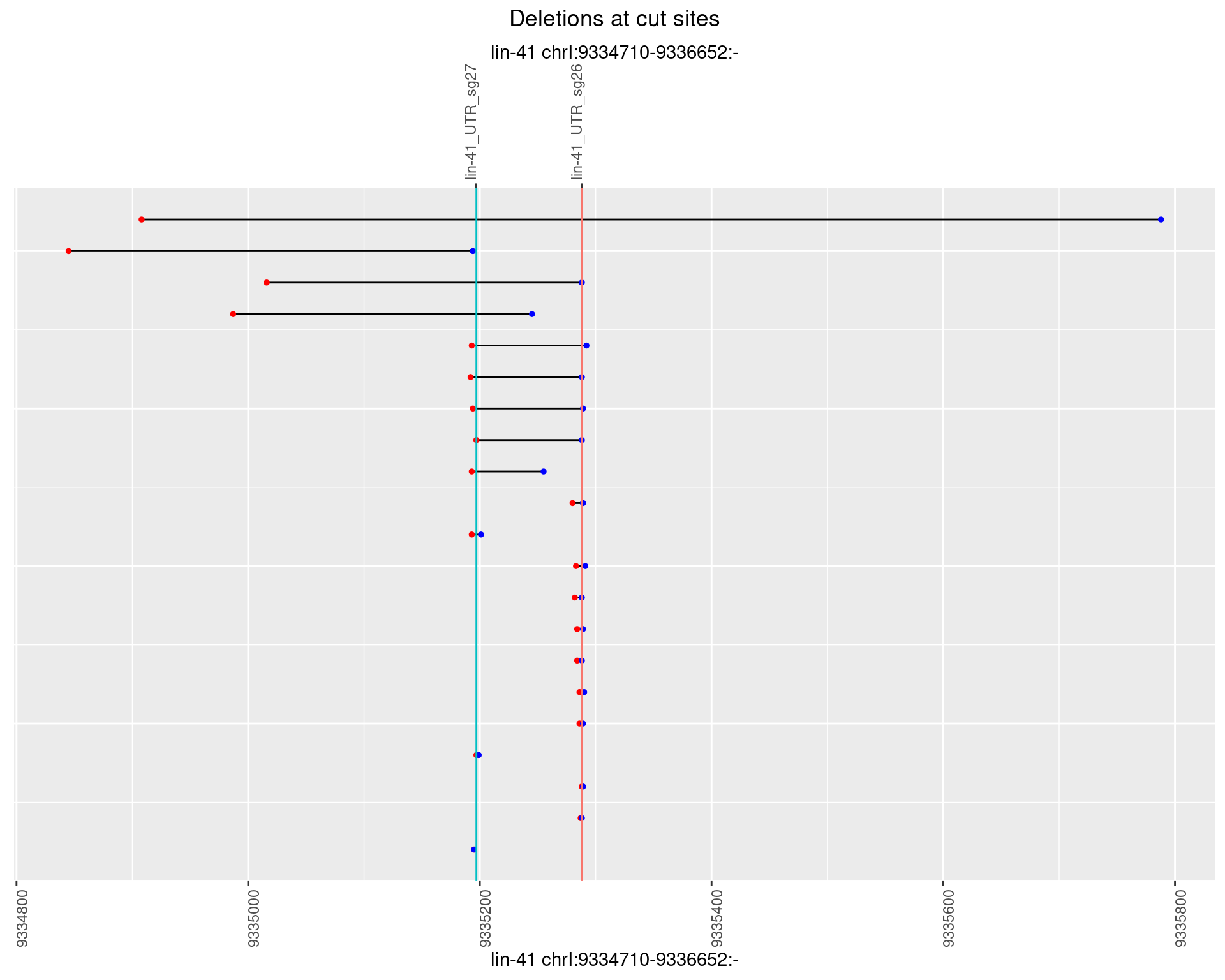
1.50.5 Freq: 0.01
No plot to show
1.50.6 Freq: 0.1
No plot to show
1.51 lin41_RNA_L4_pool2_3end
1.51.1 Freq: 0
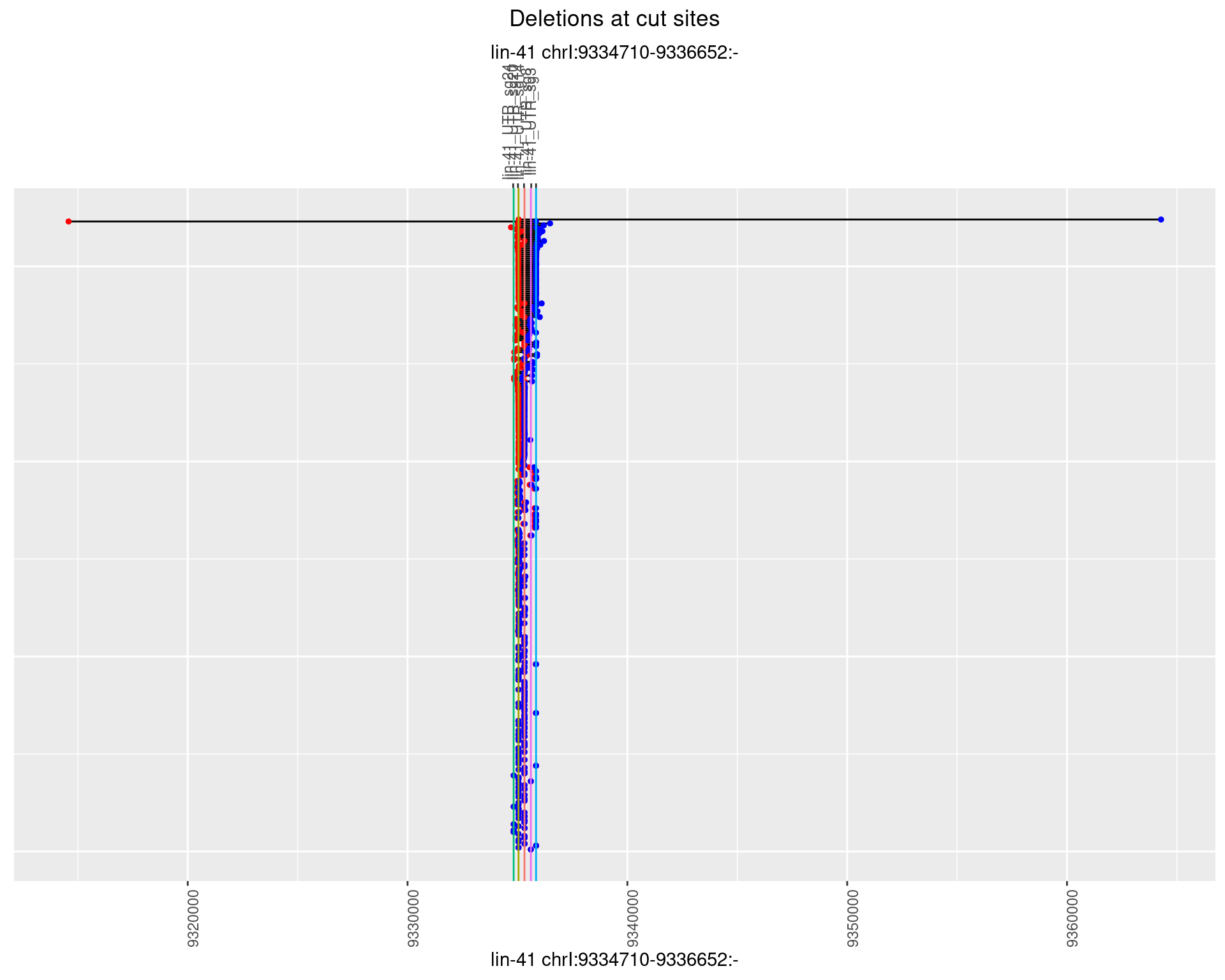
1.51.2 Freq: 1e-05
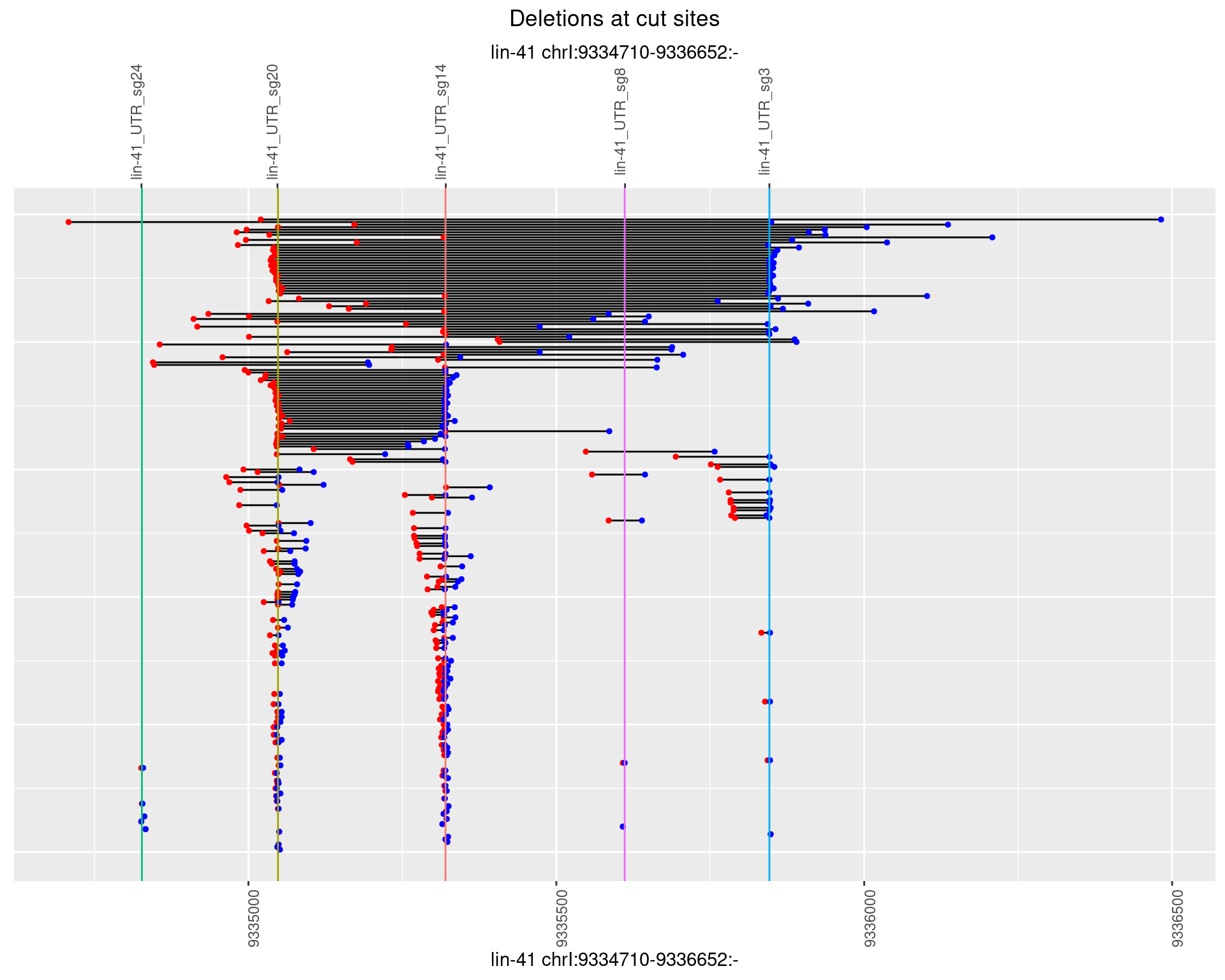
1.51.3 Freq: 1e-04
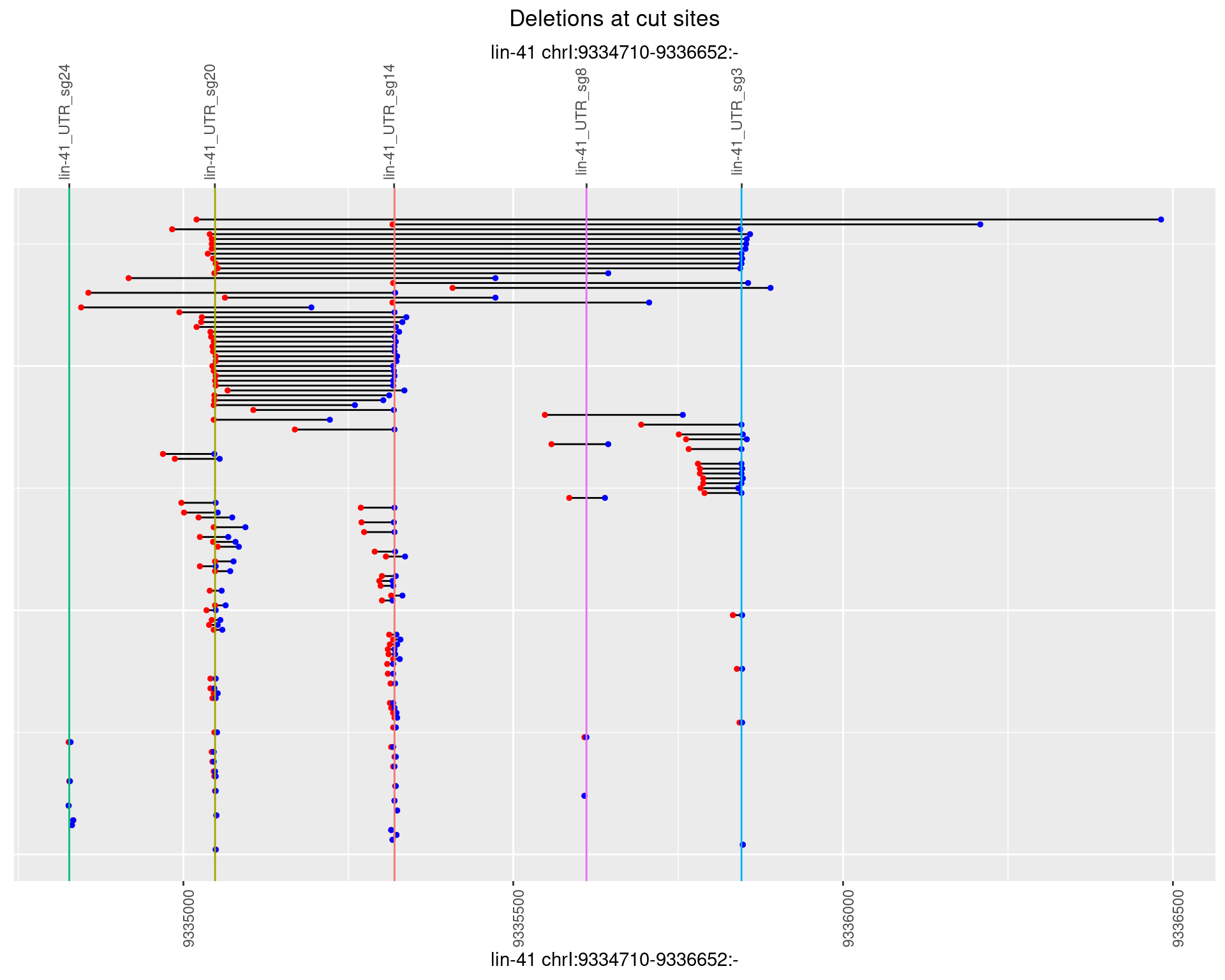
1.51.4 Freq: 0.001
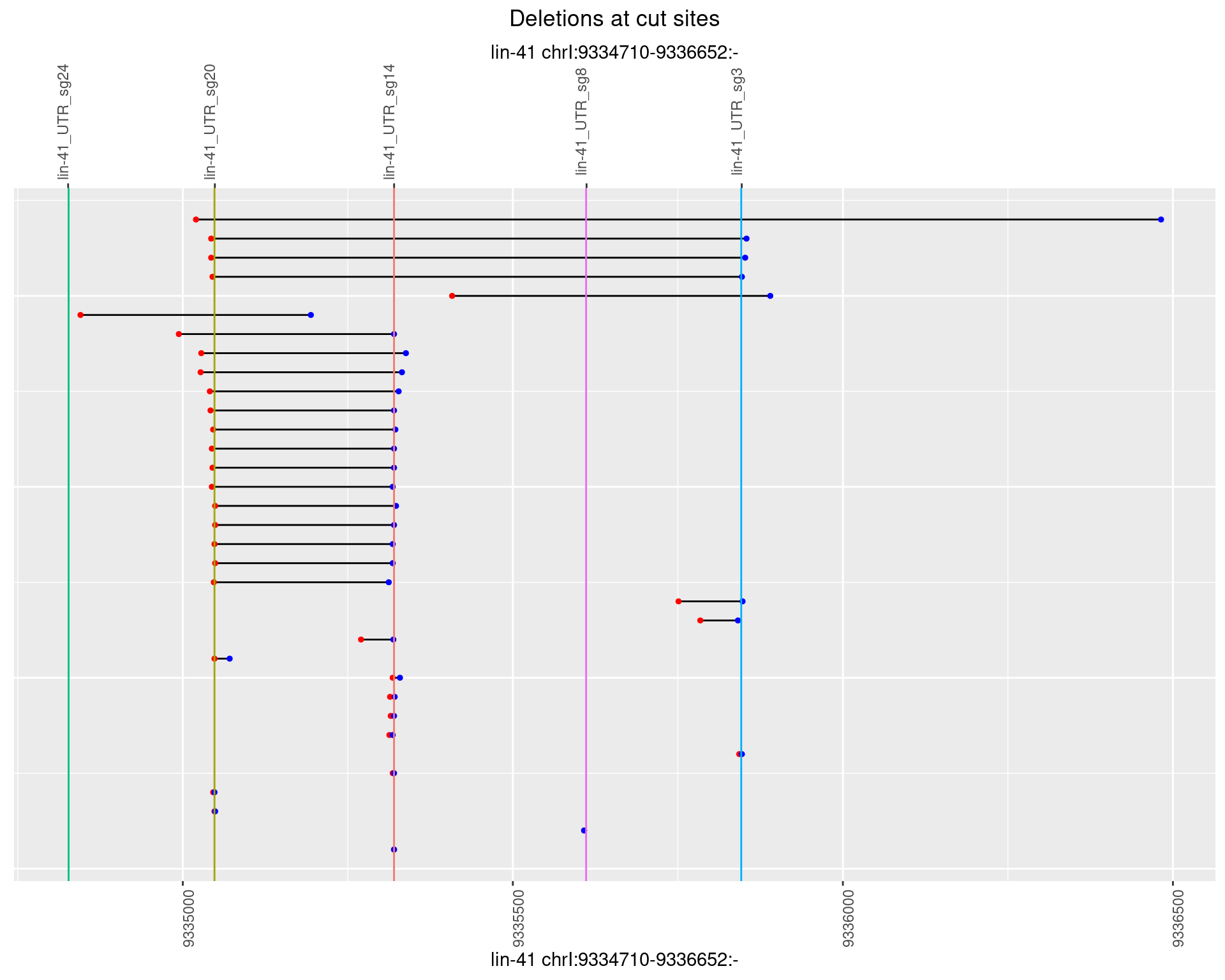
1.51.5 Freq: 0.01
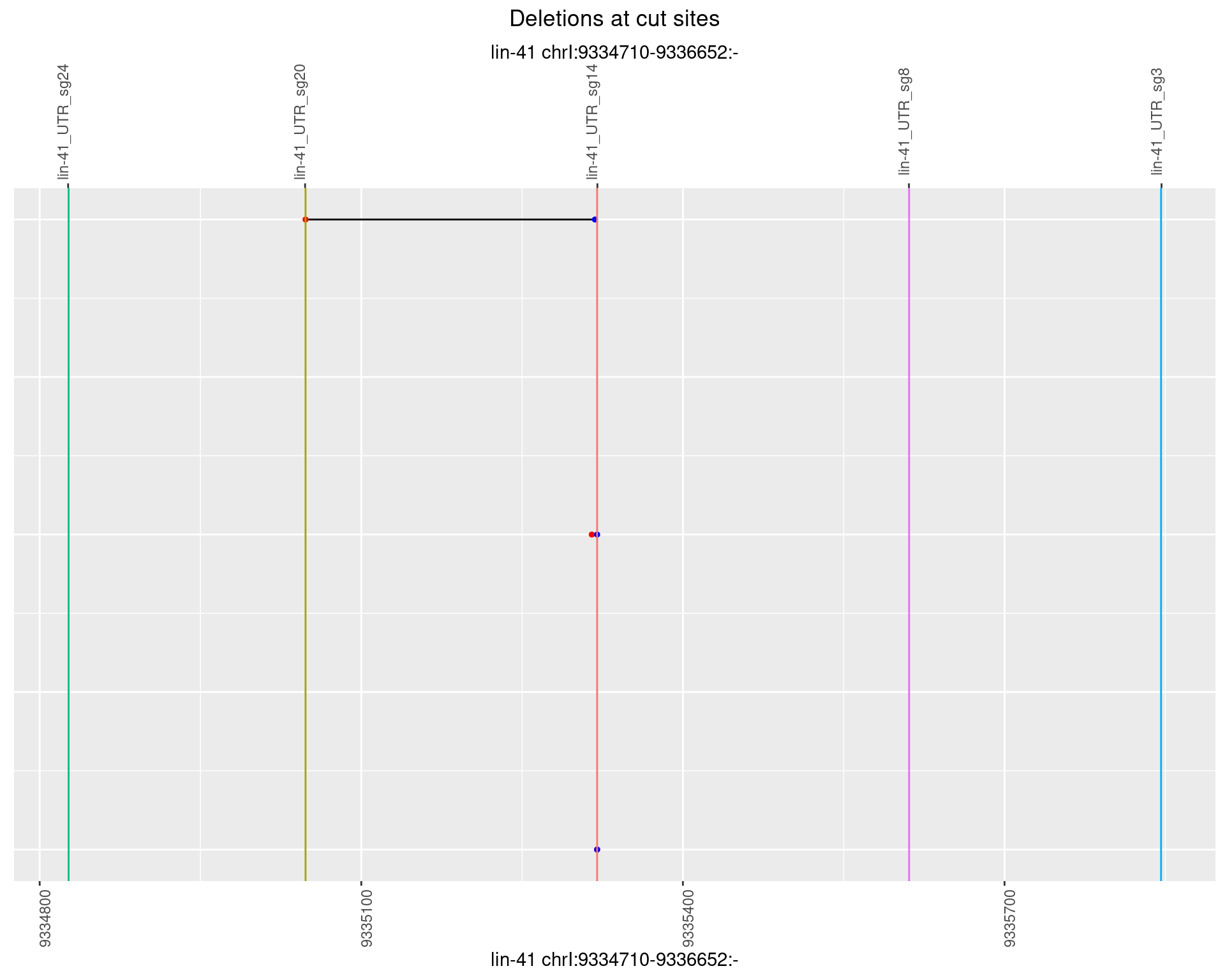
1.51.6 Freq: 0.1
No plot to show
1.52 lin41_RNA_L1_N2_middle
1.52.1 Freq: 0
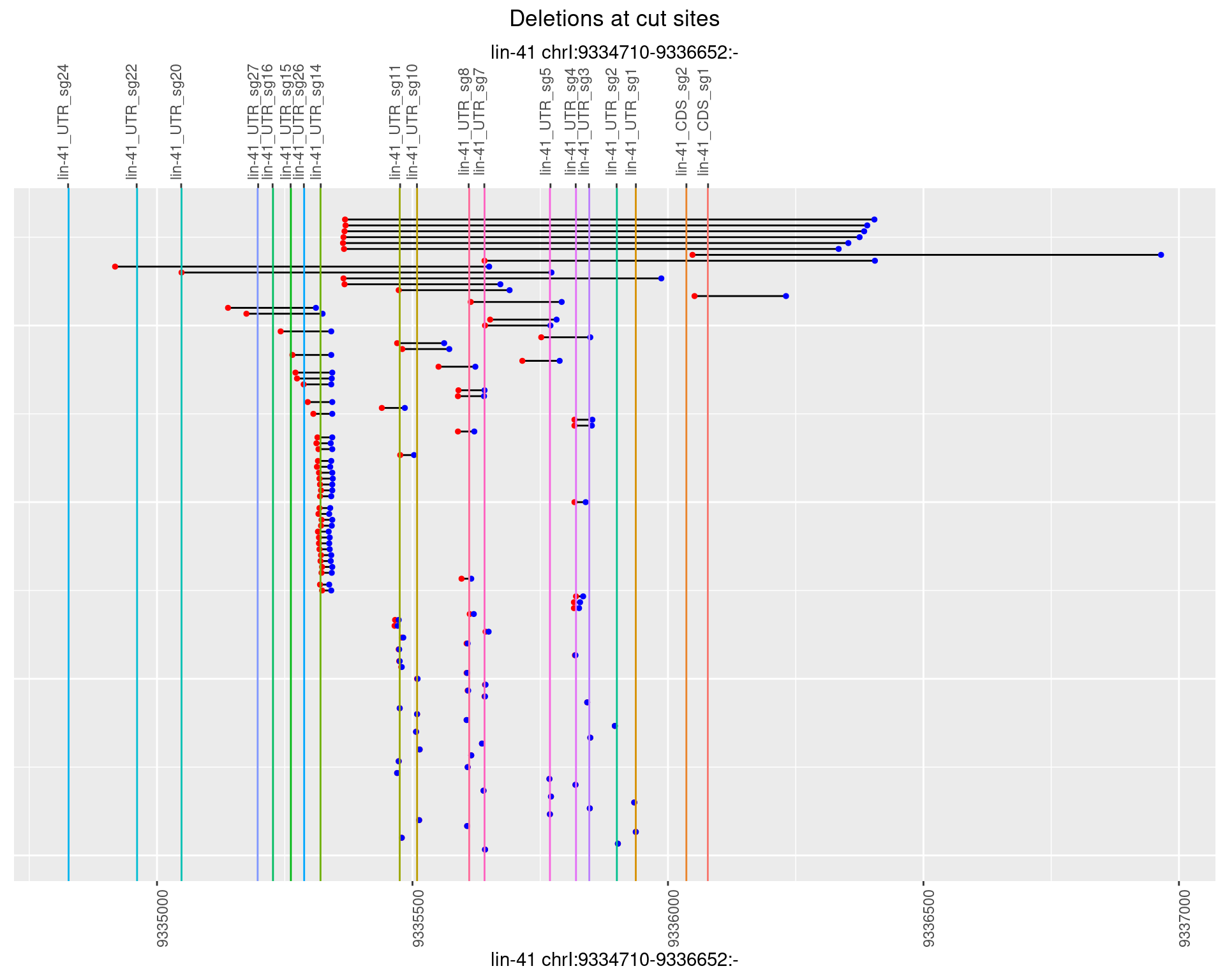
1.52.2 Freq: 1e-05
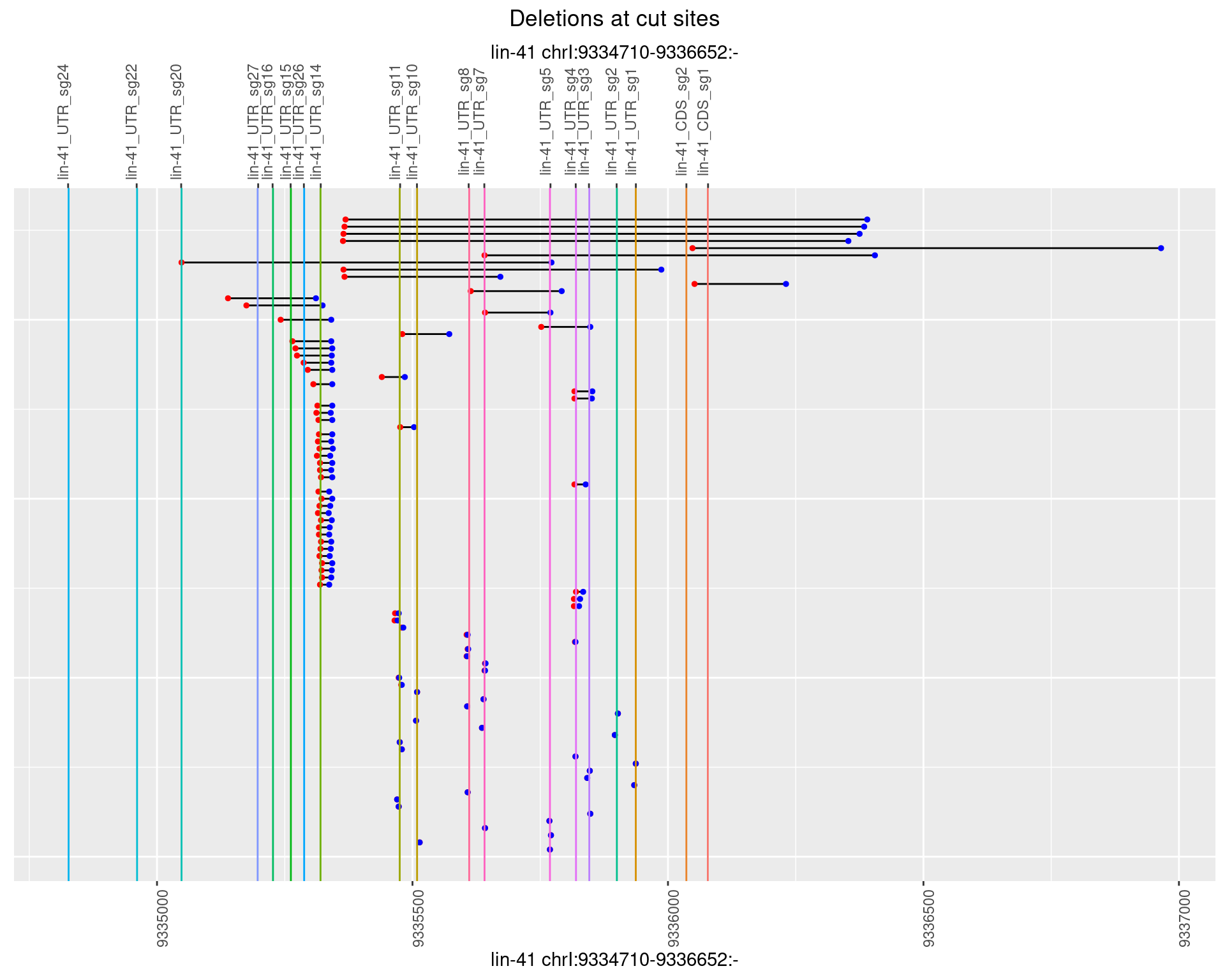
1.52.3 Freq: 1e-04
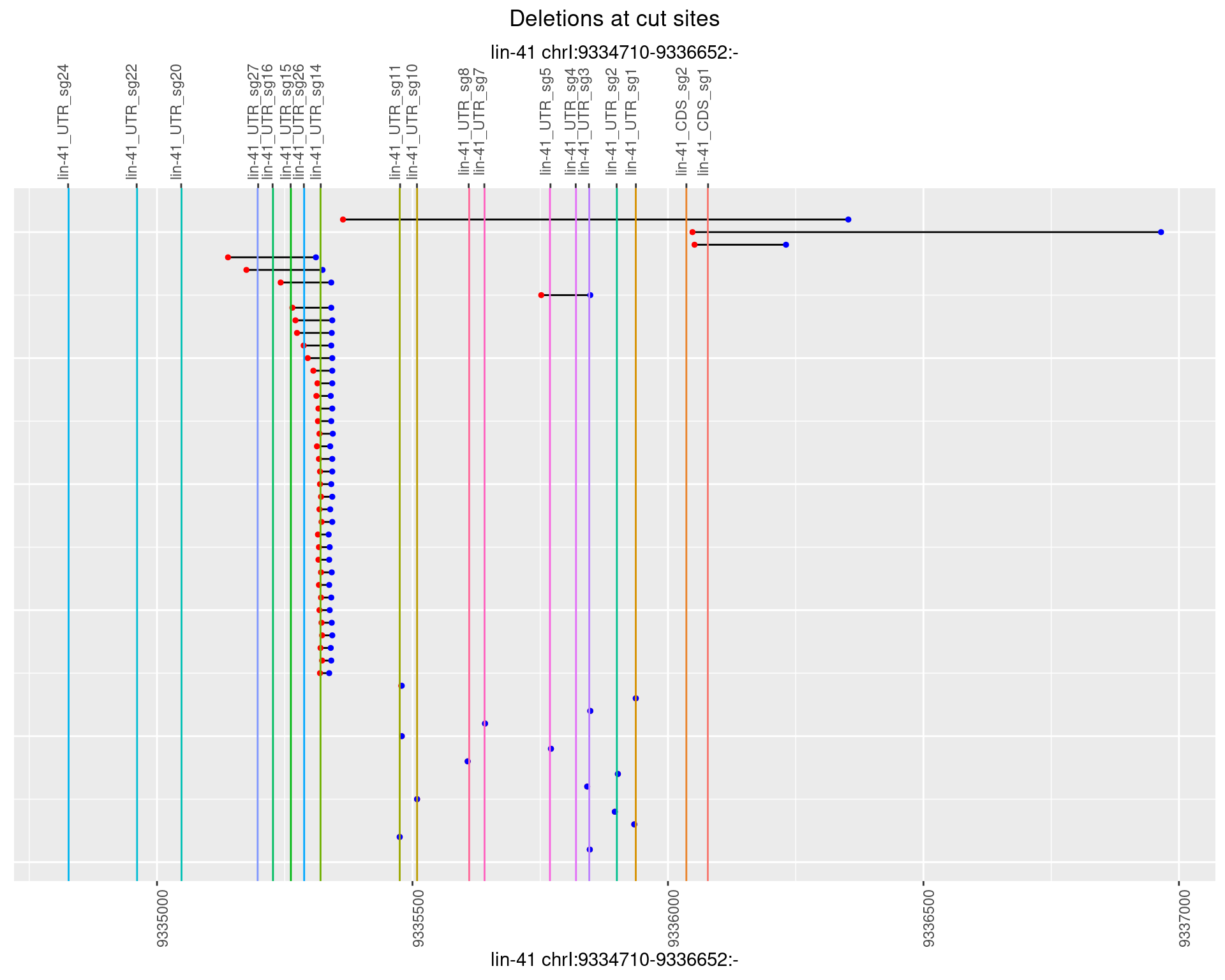
1.52.4 Freq: 0.001
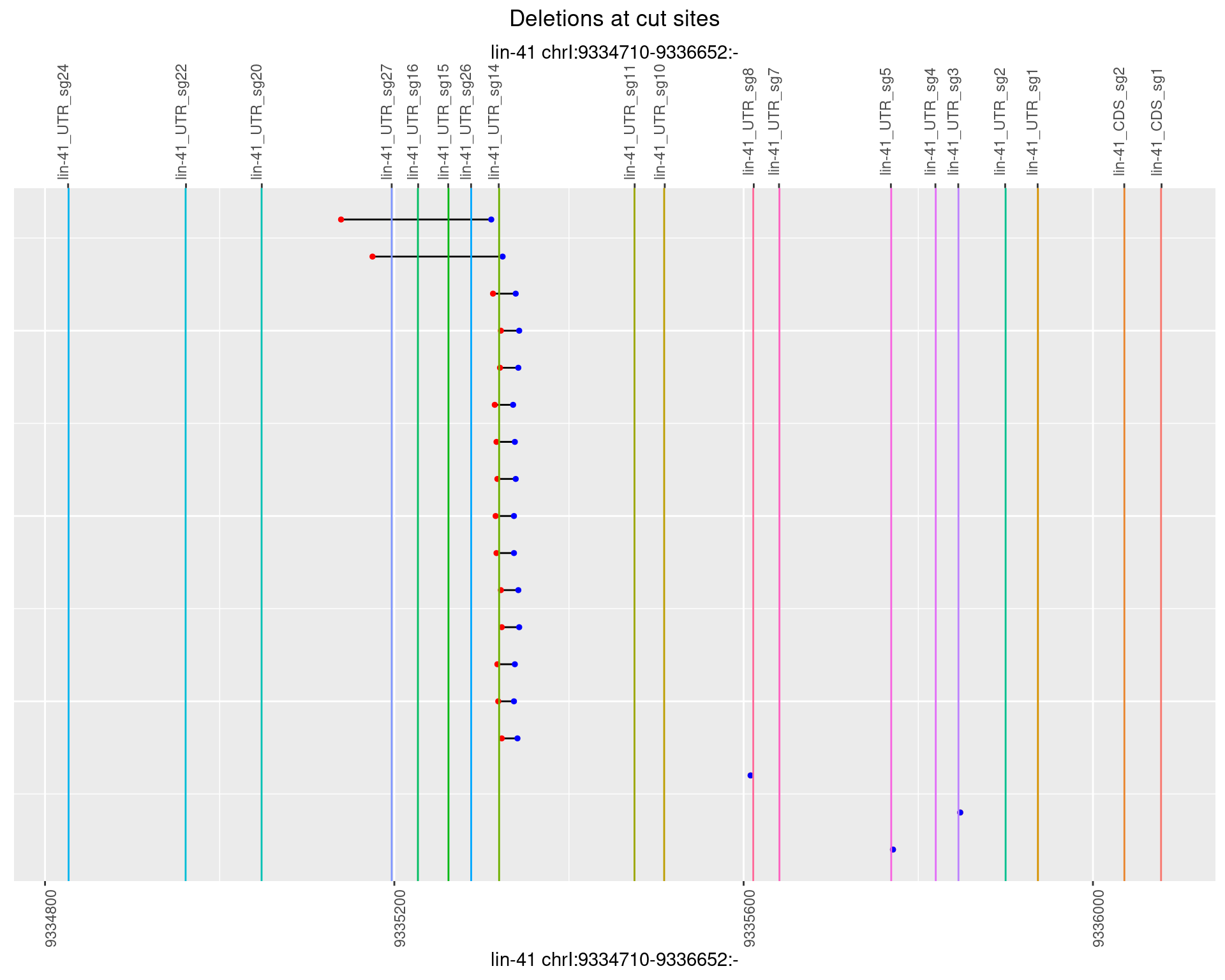
1.52.5 Freq: 0.01
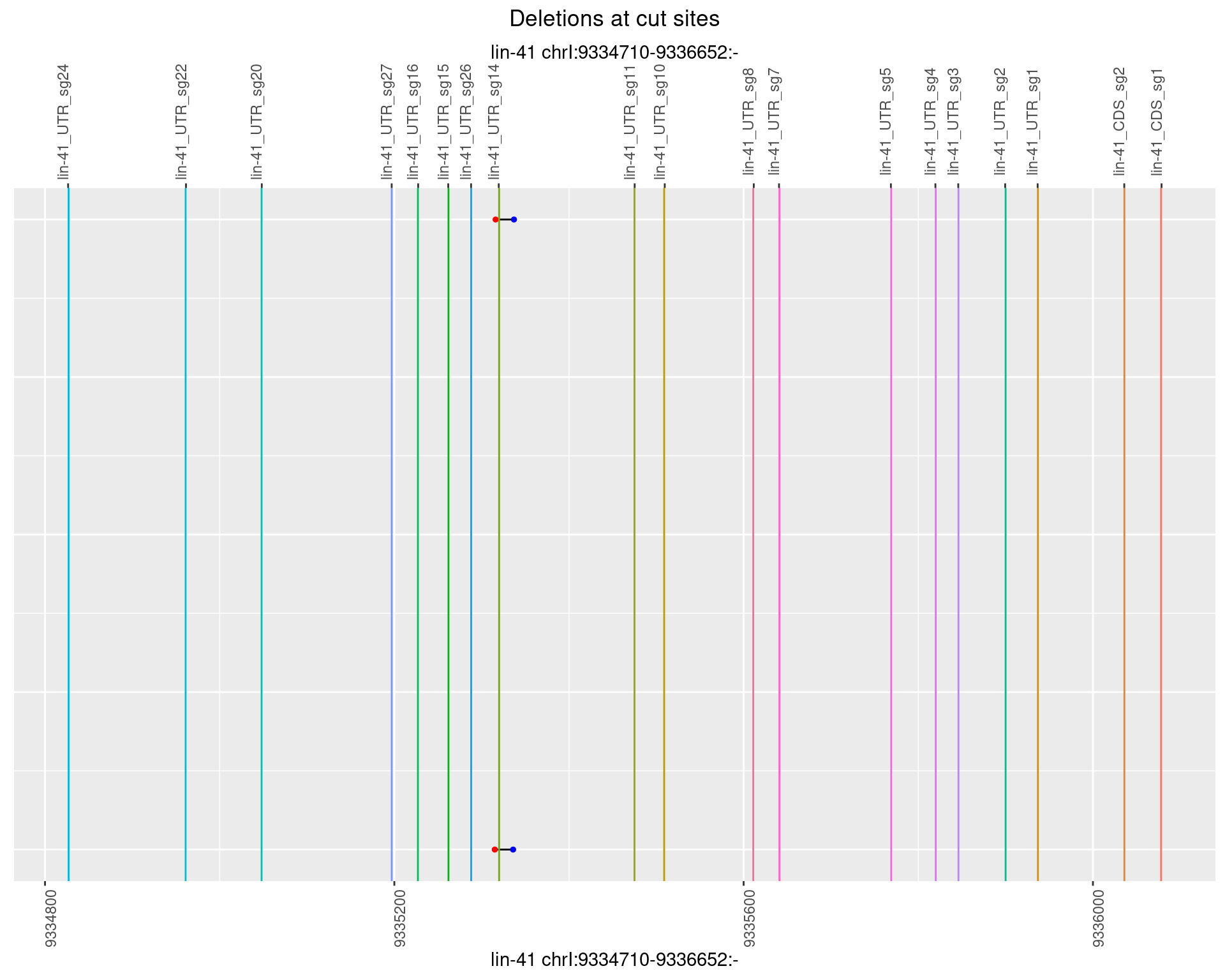
1.52.6 Freq: 0.1
No plot to show
1.53 lin41_RNA_L1_1516_middle
1.53.1 Freq: 0
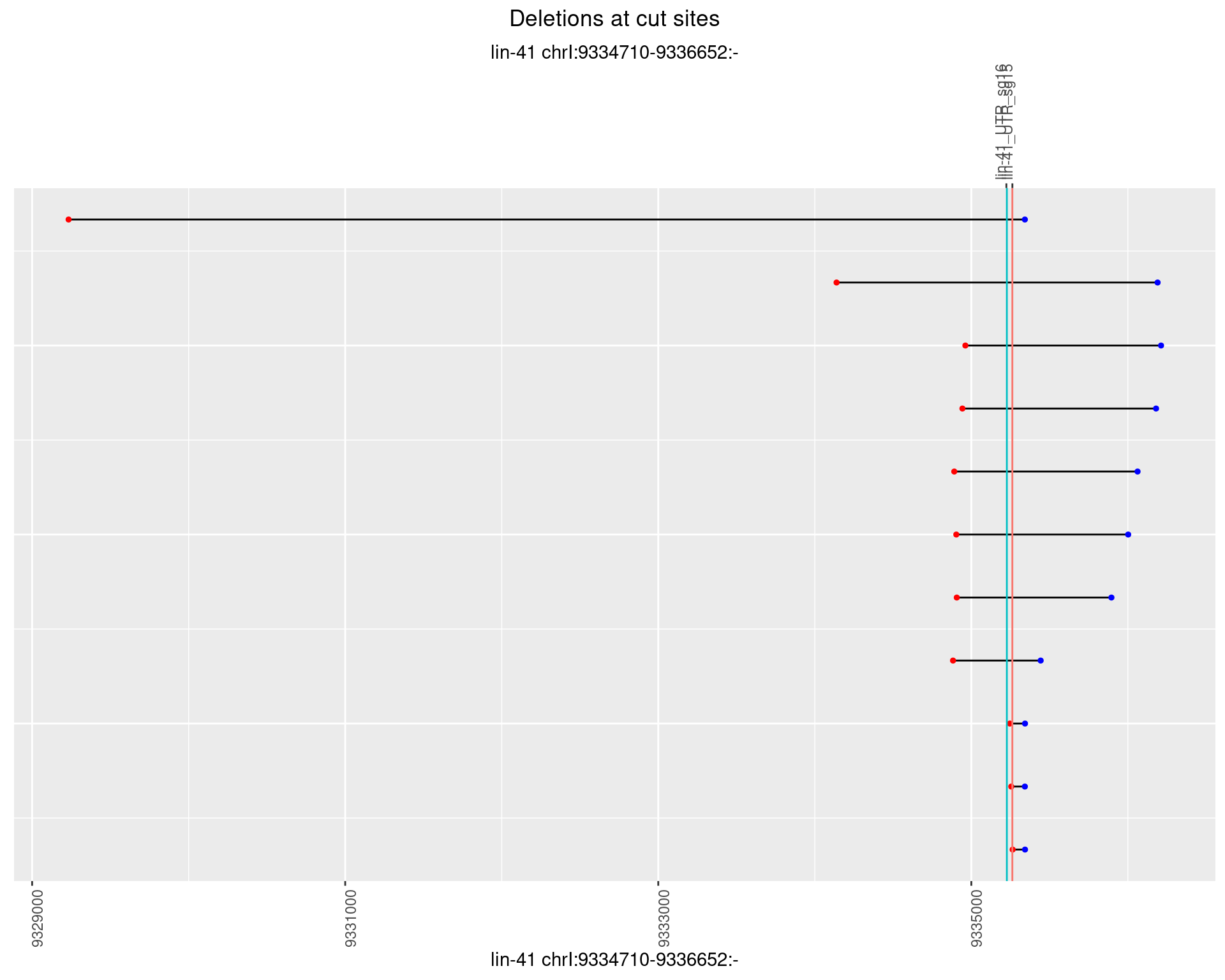
1.53.2 Freq: 1e-05
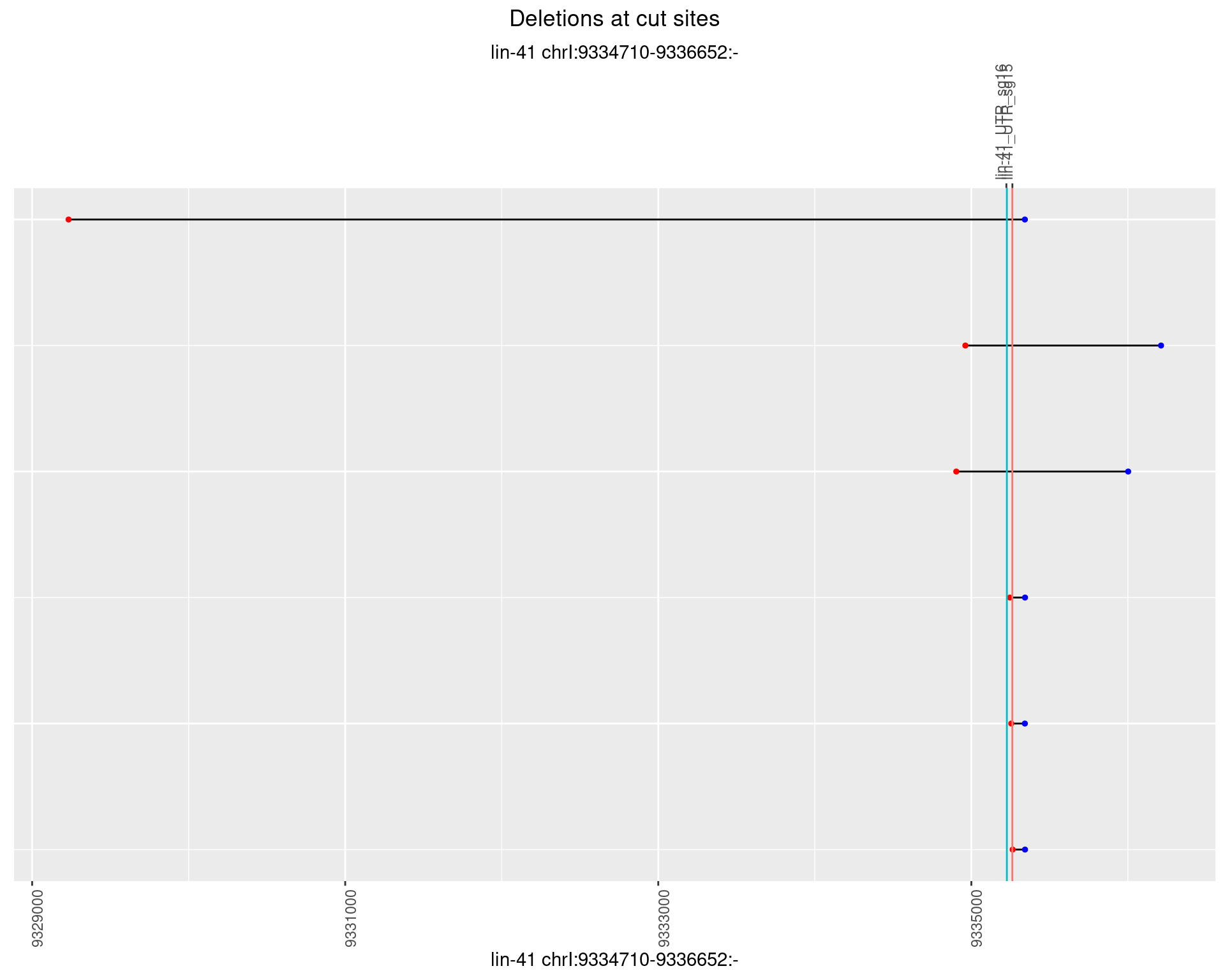
1.53.3 Freq: 1e-04
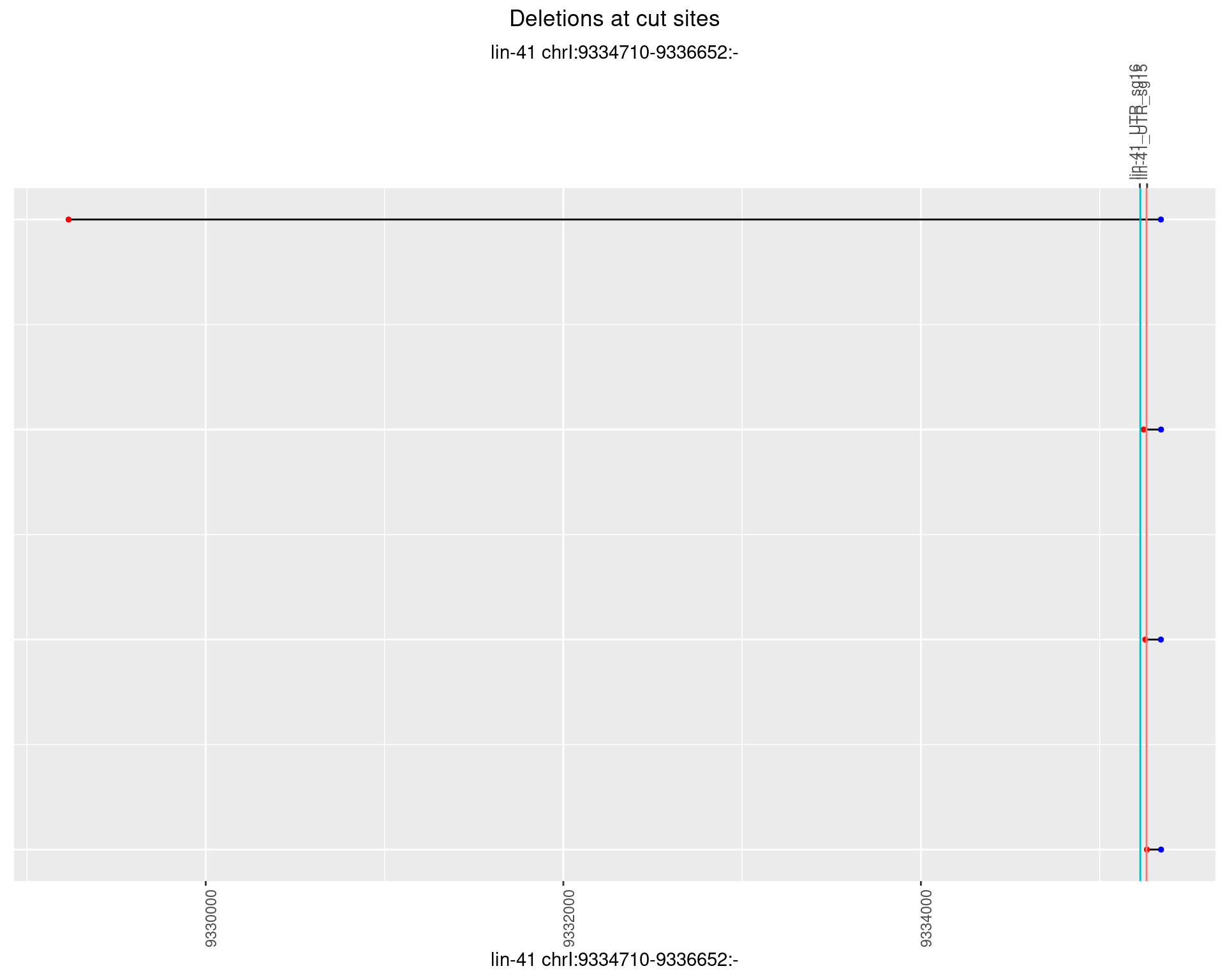
1.53.4 Freq: 0.001
No plot to show
1.53.5 Freq: 0.01
No plot to show
1.53.6 Freq: 0.1
No plot to show
1.54 lin41_RNA_L1_2627_middle
1.54.1 Freq: 0
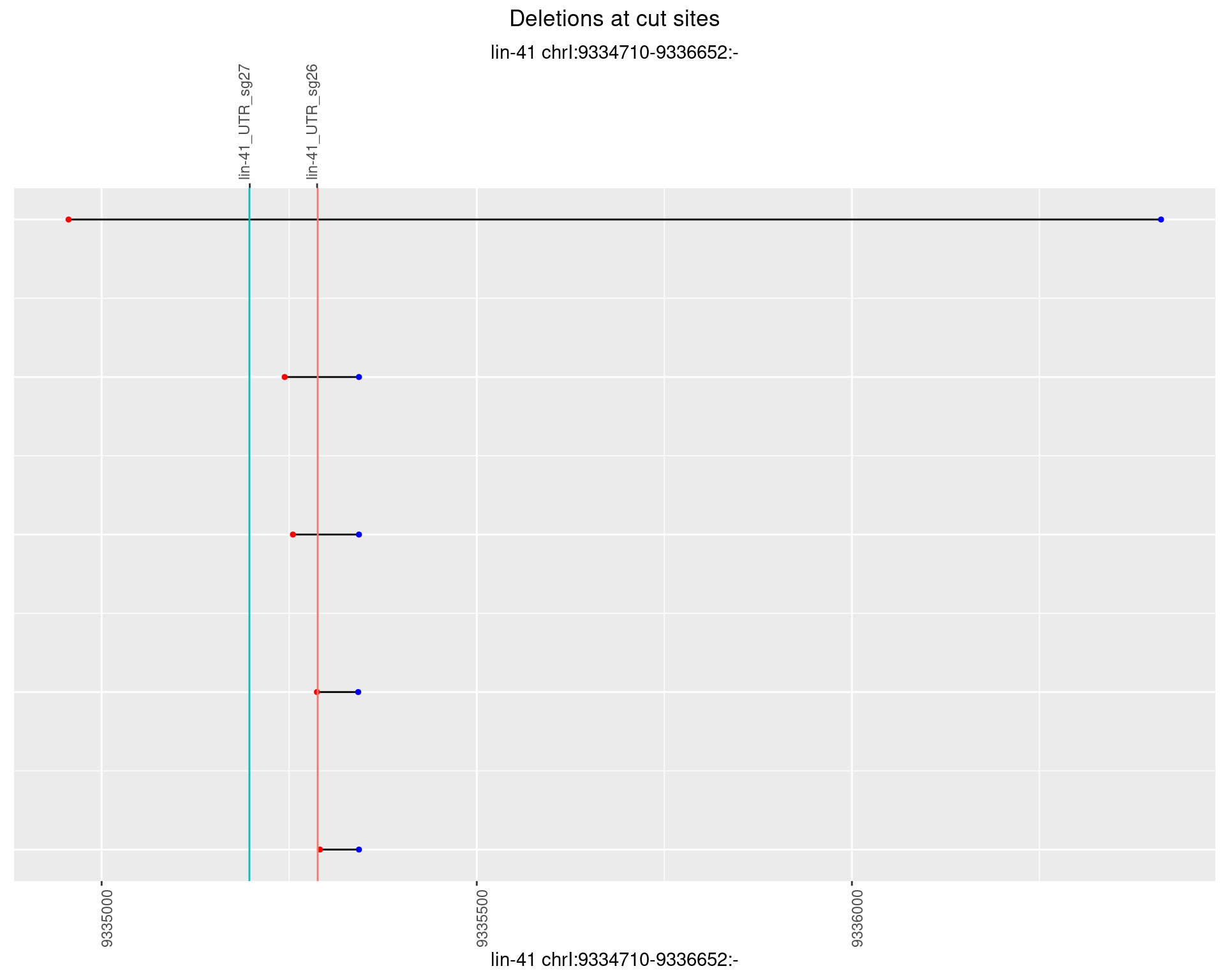
1.54.2 Freq: 1e-05
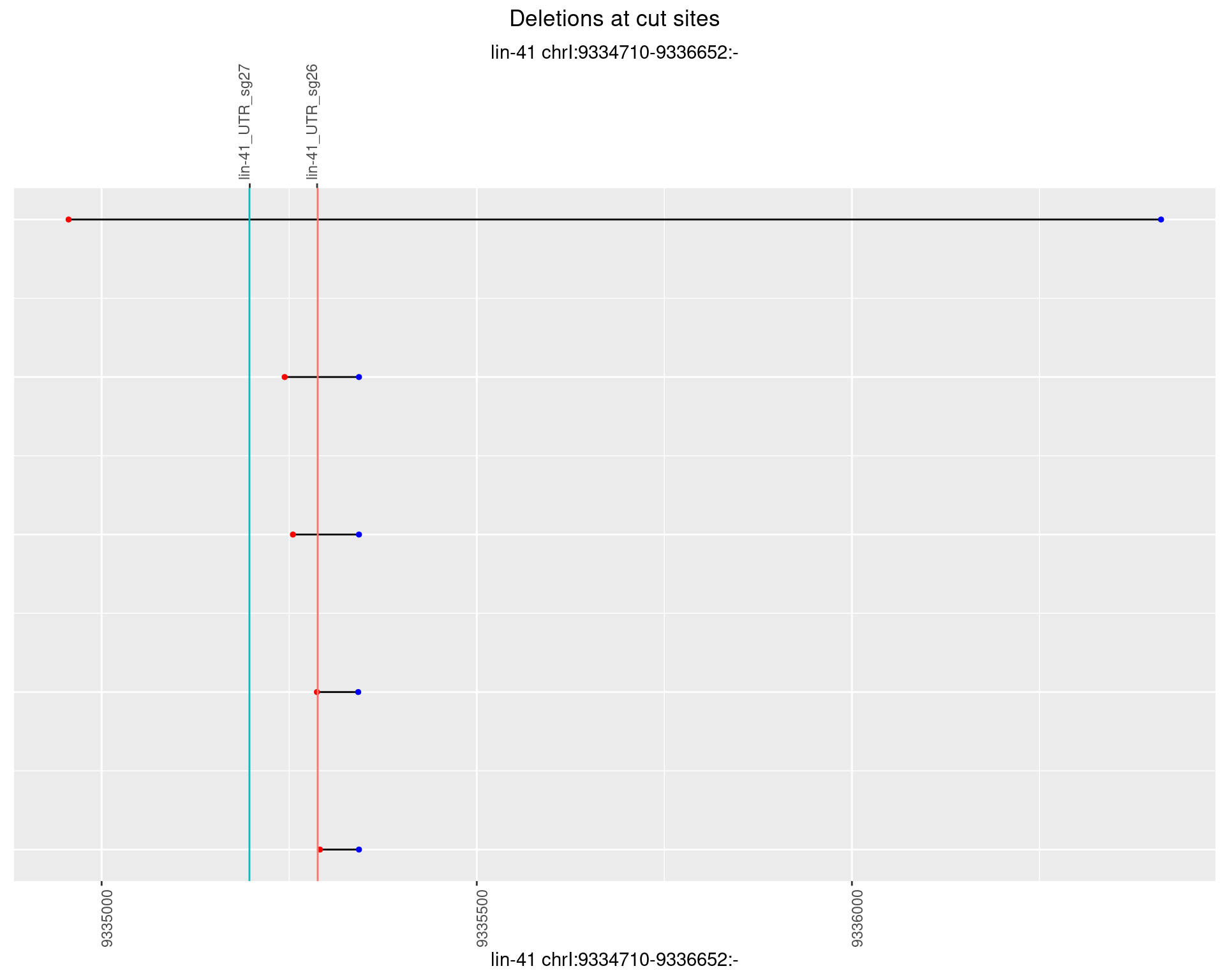
1.54.3 Freq: 1e-04
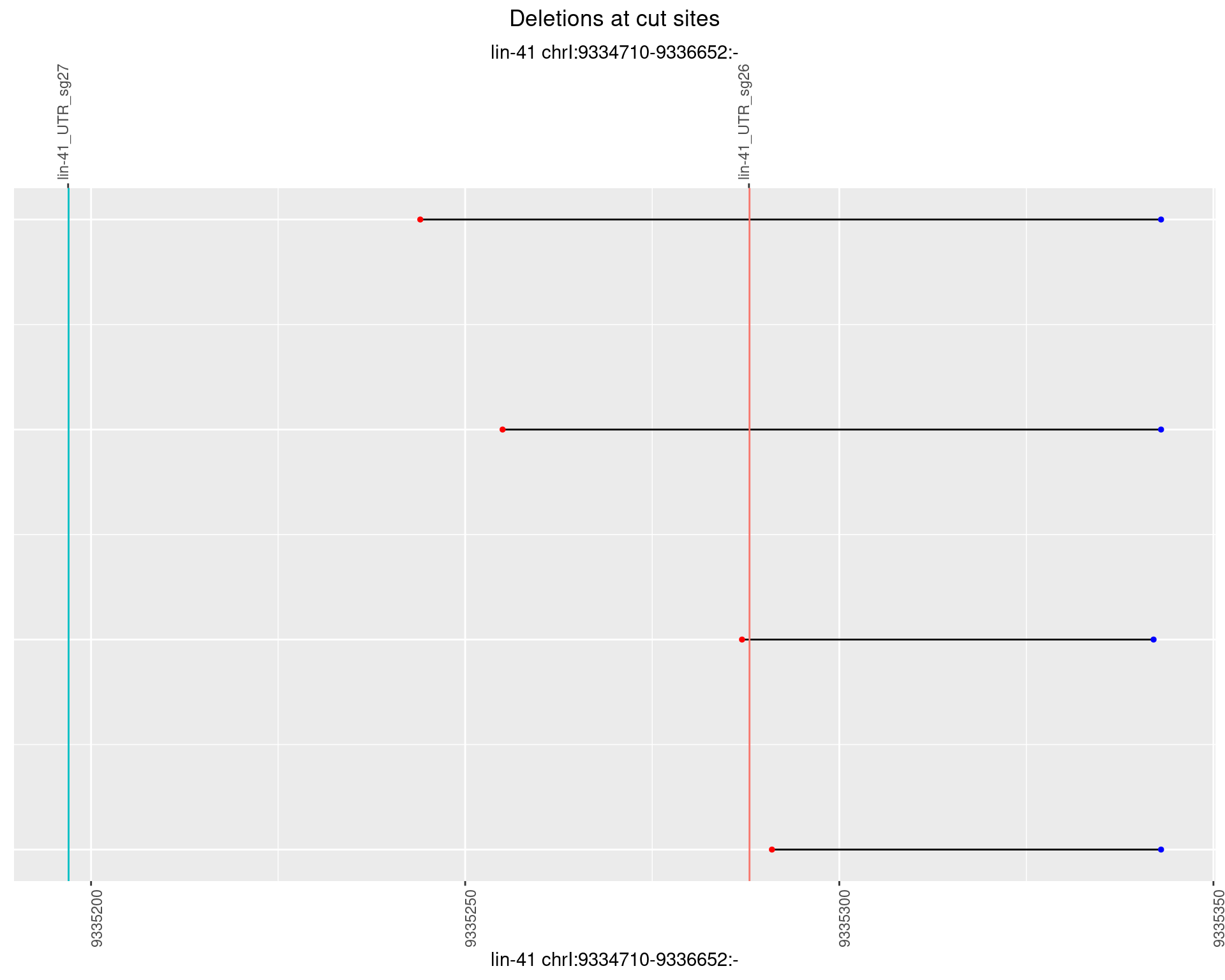
1.54.4 Freq: 0.001
No plot to show
1.54.5 Freq: 0.01
No plot to show
1.54.6 Freq: 0.1
No plot to show
1.55 lin41_RNA_L1_pool2_middle
1.55.1 Freq: 0
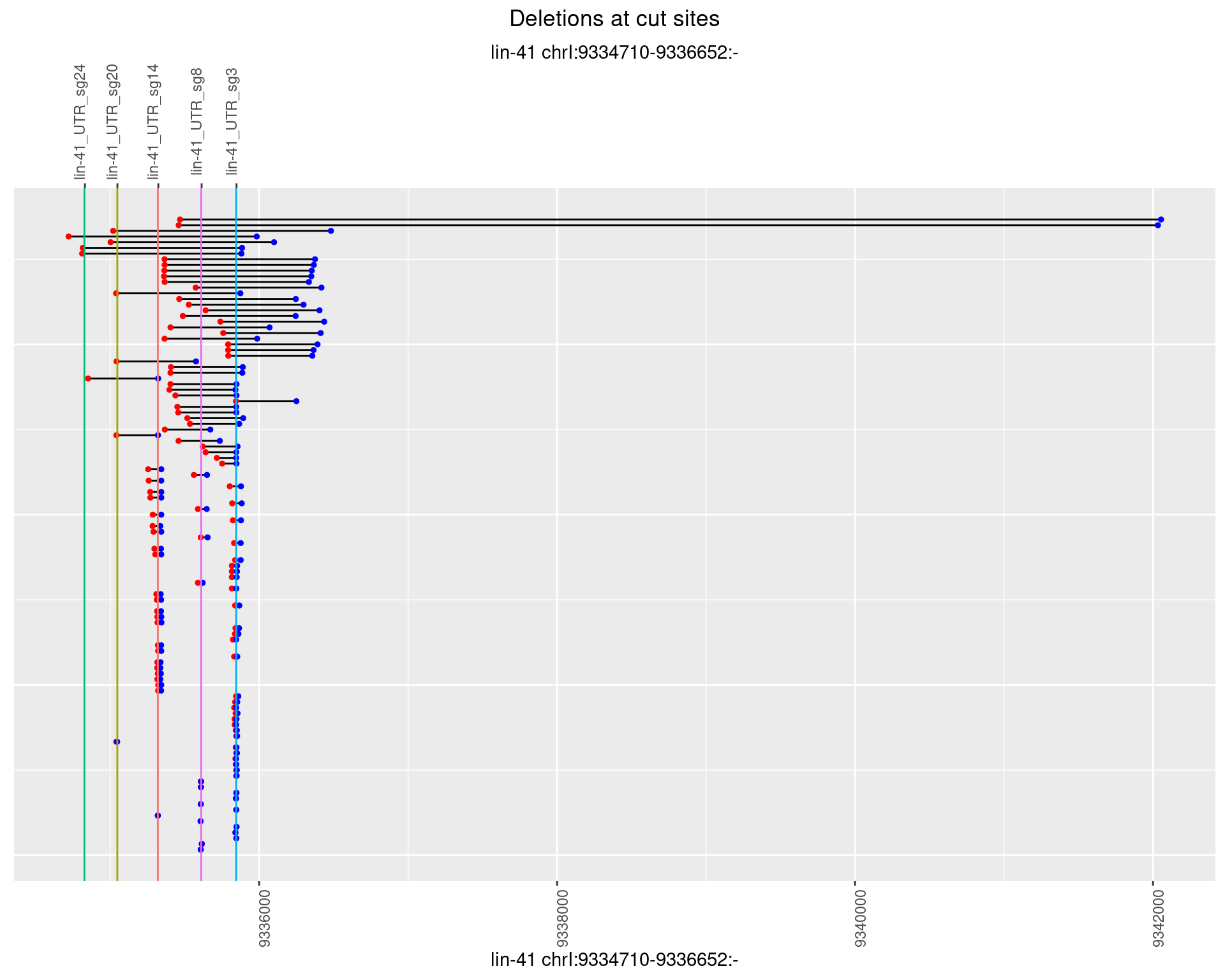
1.55.2 Freq: 1e-05
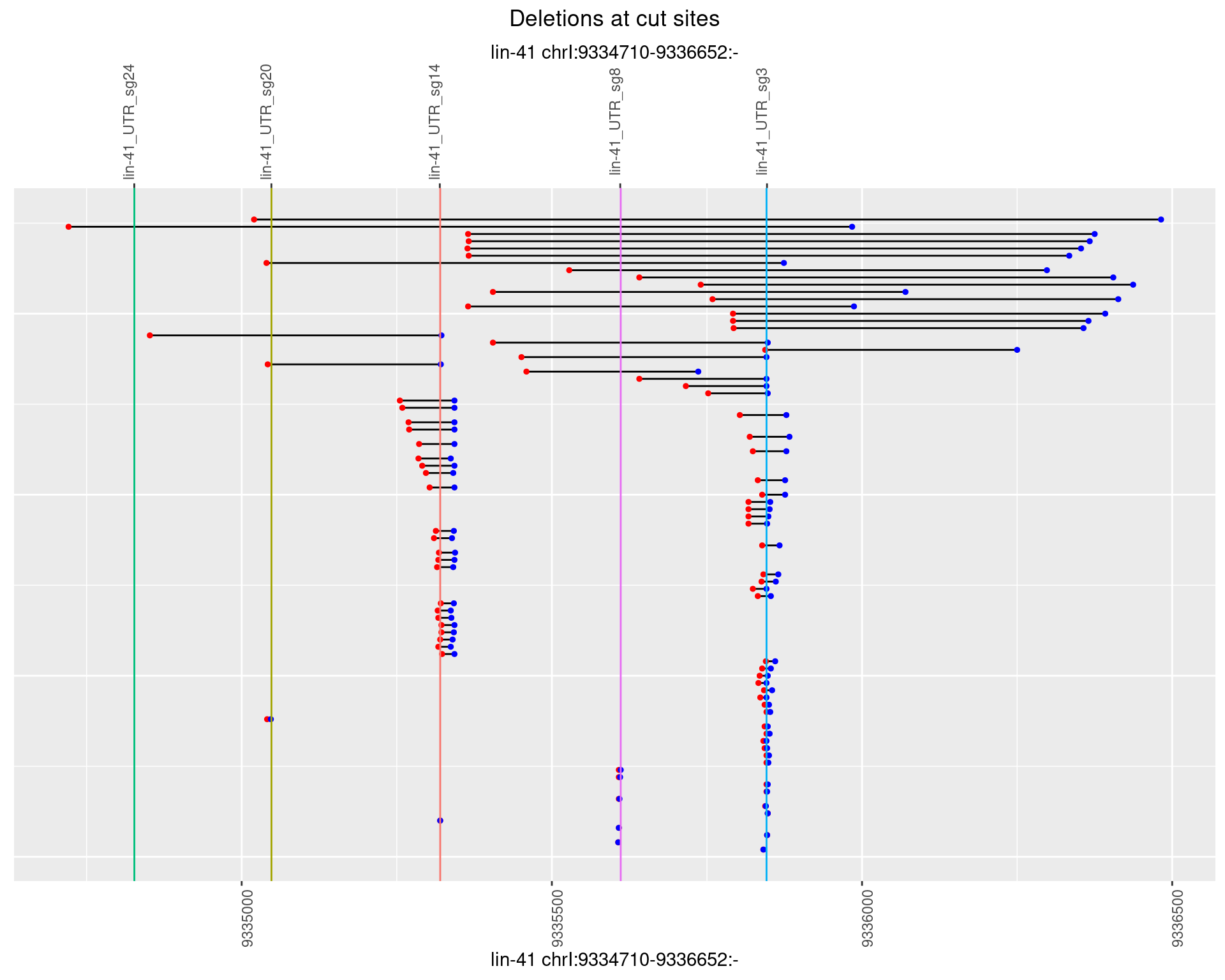
1.55.3 Freq: 1e-04
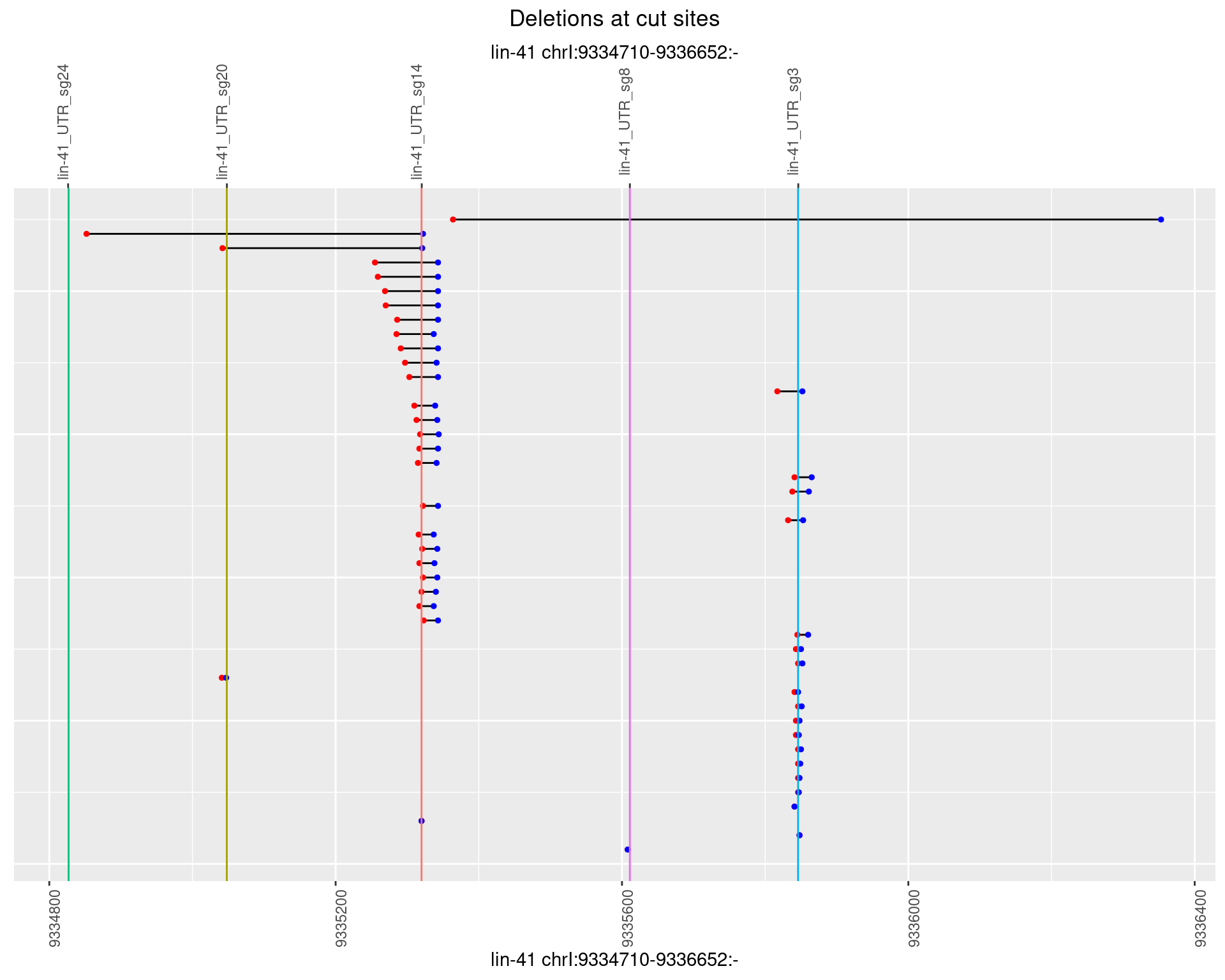
1.55.4 Freq: 0.001
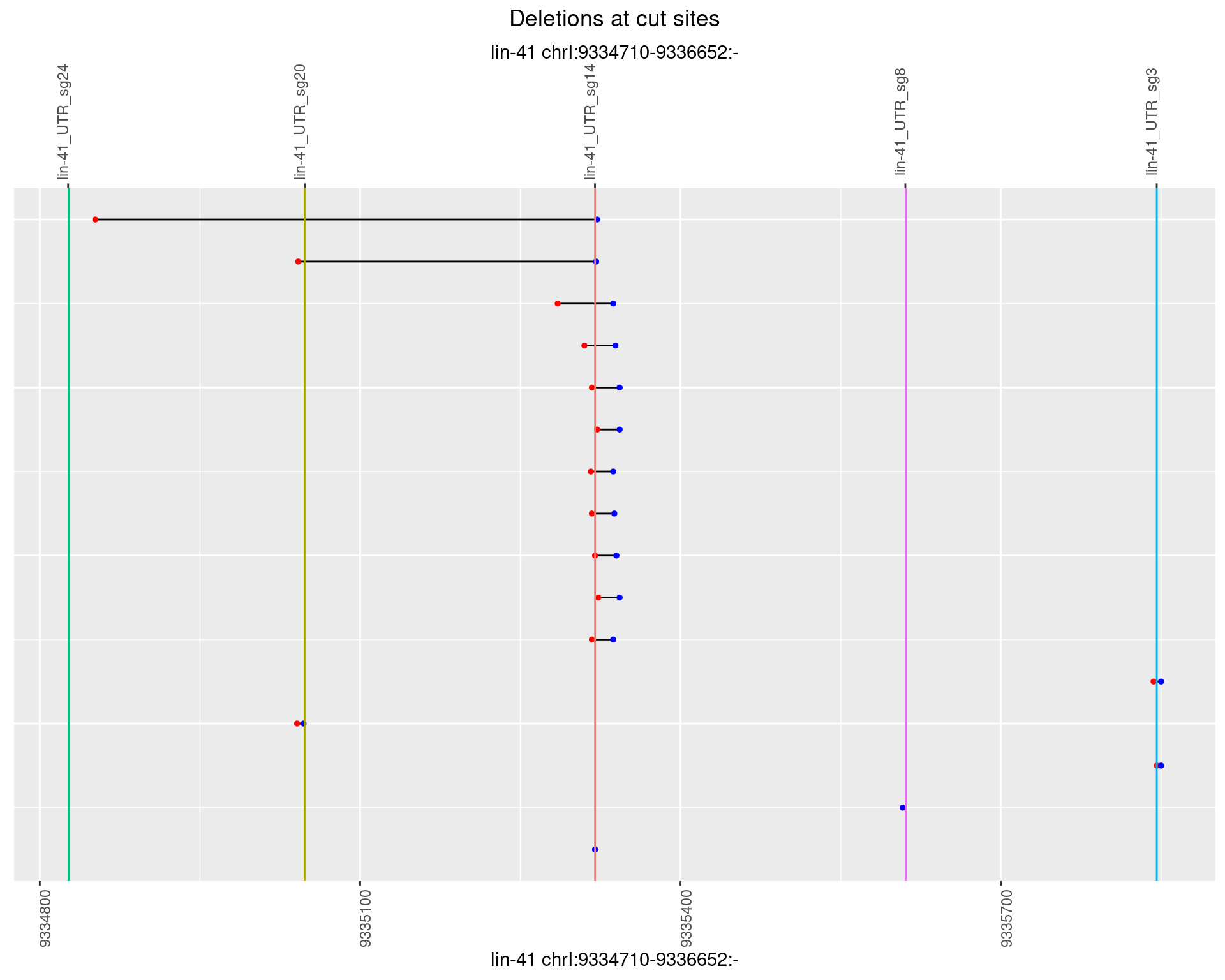
1.55.5 Freq: 0.01
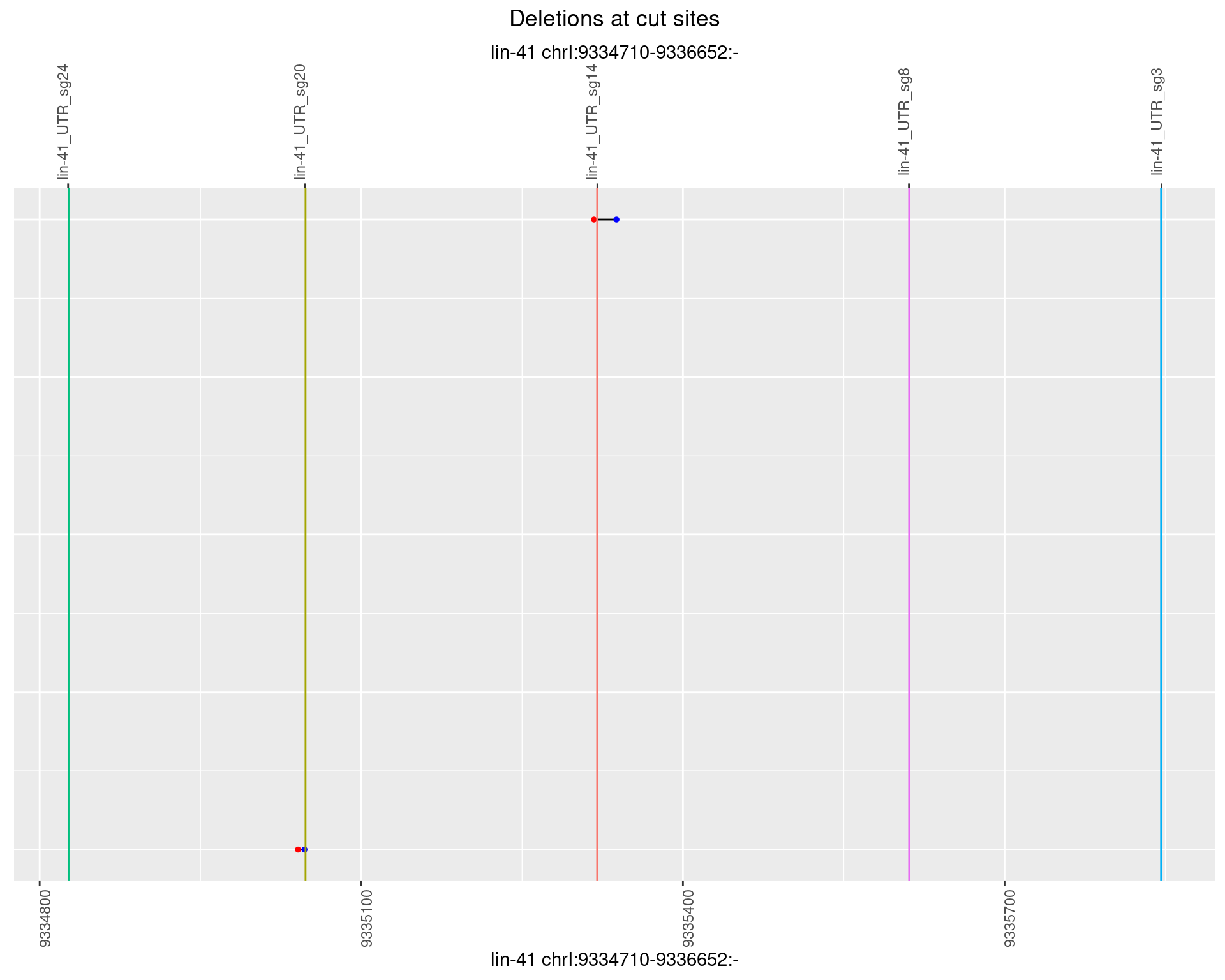
1.55.6 Freq: 0.1
No plot to show
1.56 lin41_RNA_L1_pool3_middle
1.56.1 Freq: 0
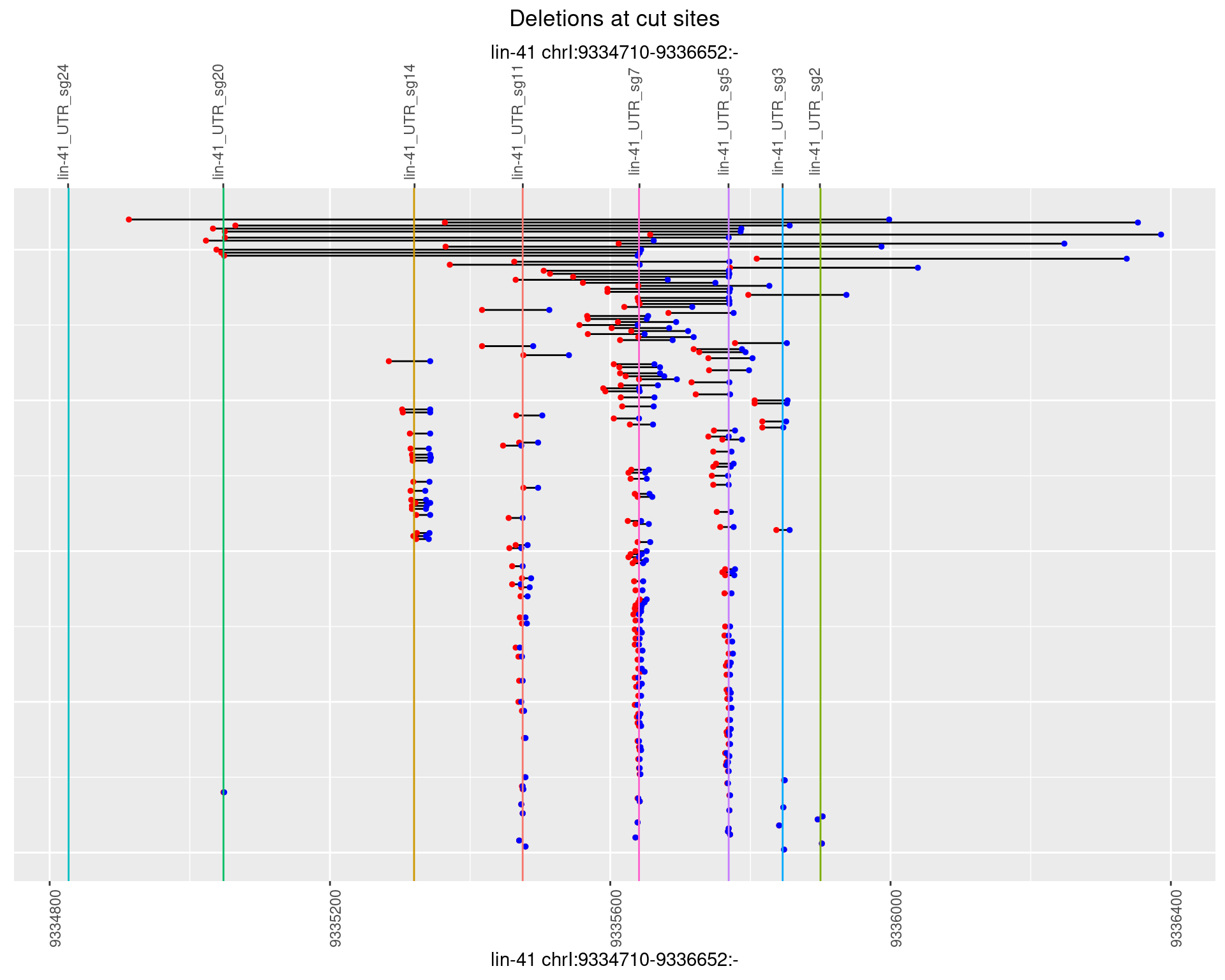
1.56.2 Freq: 1e-05
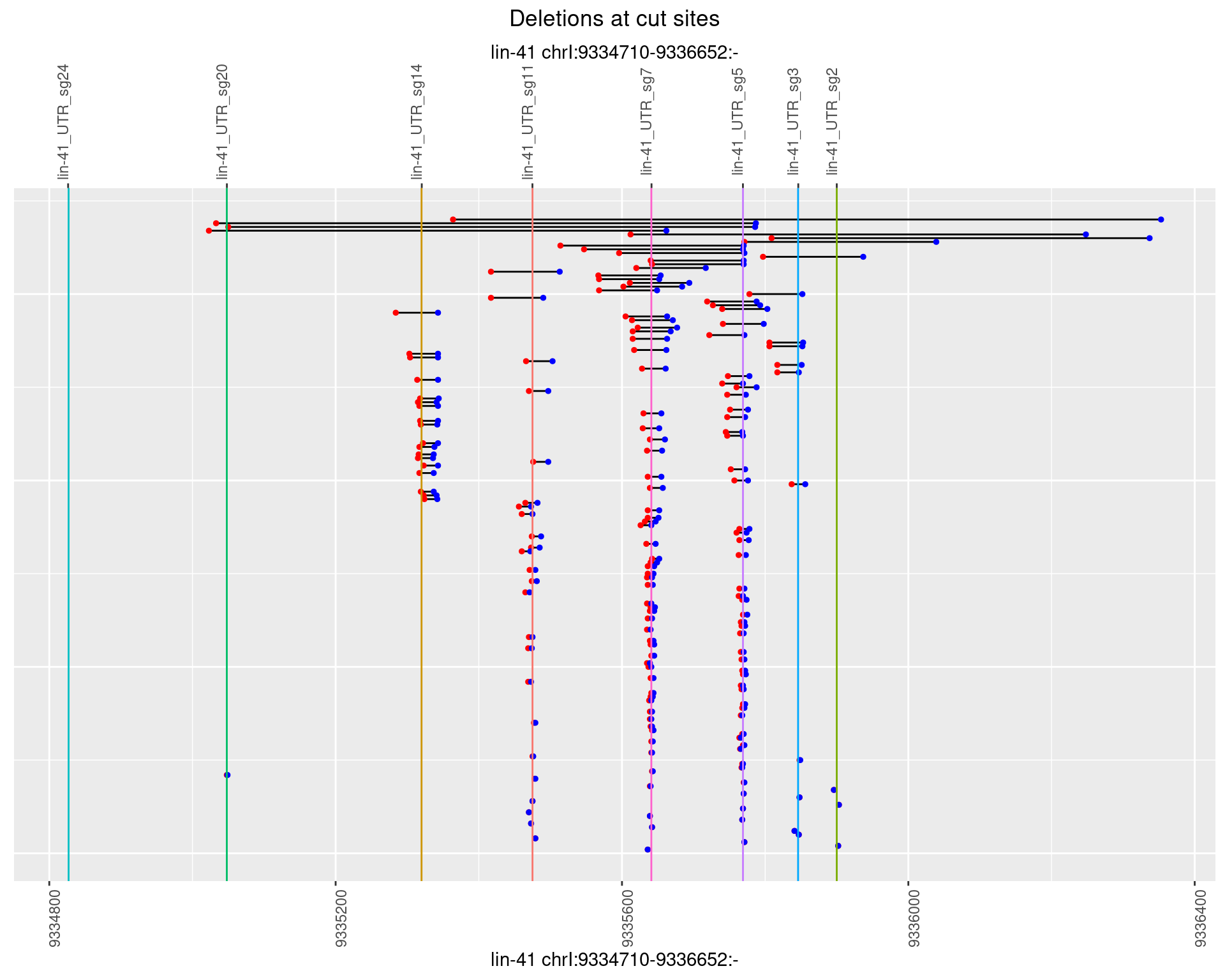
1.56.3 Freq: 1e-04
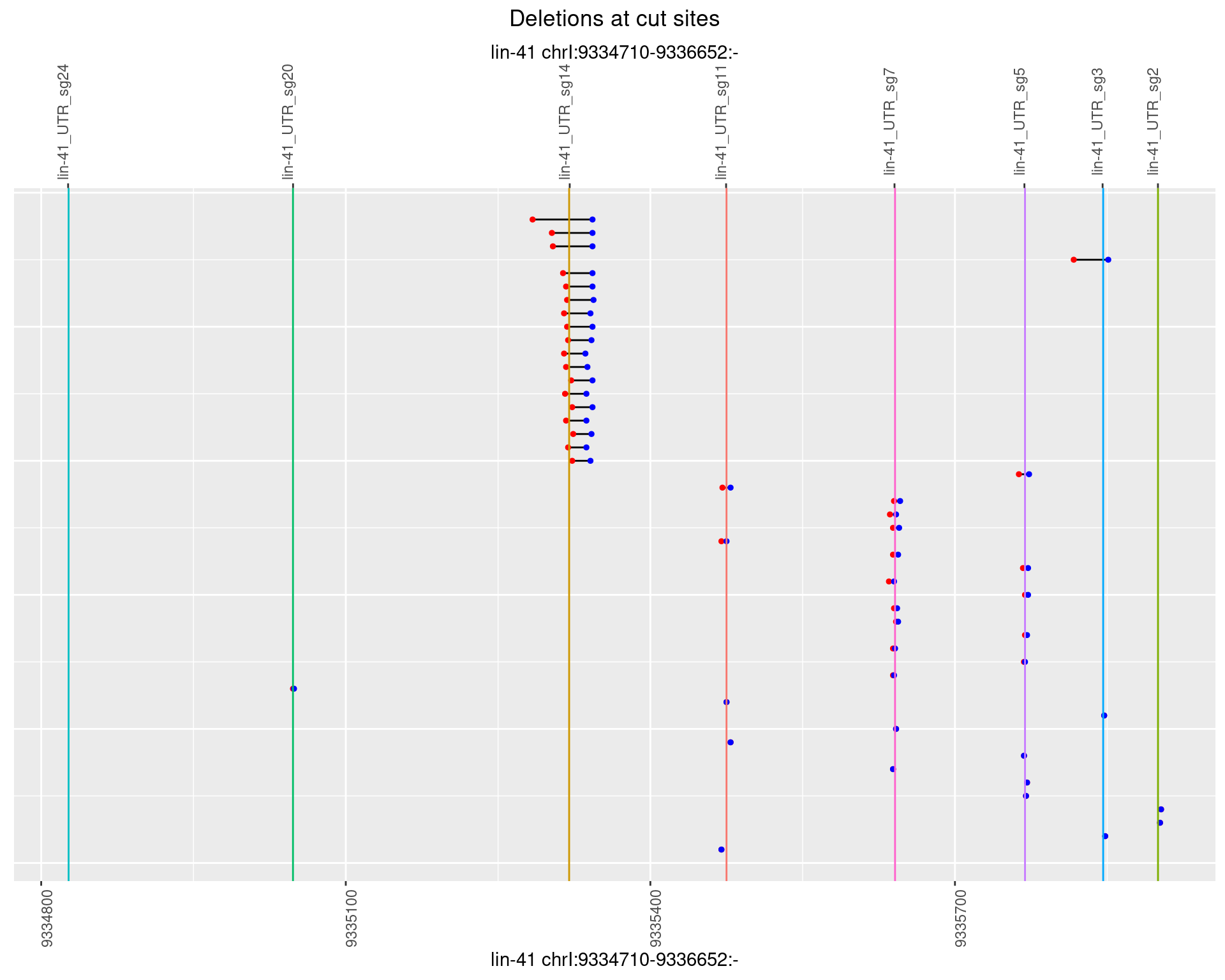
1.56.4 Freq: 0.001
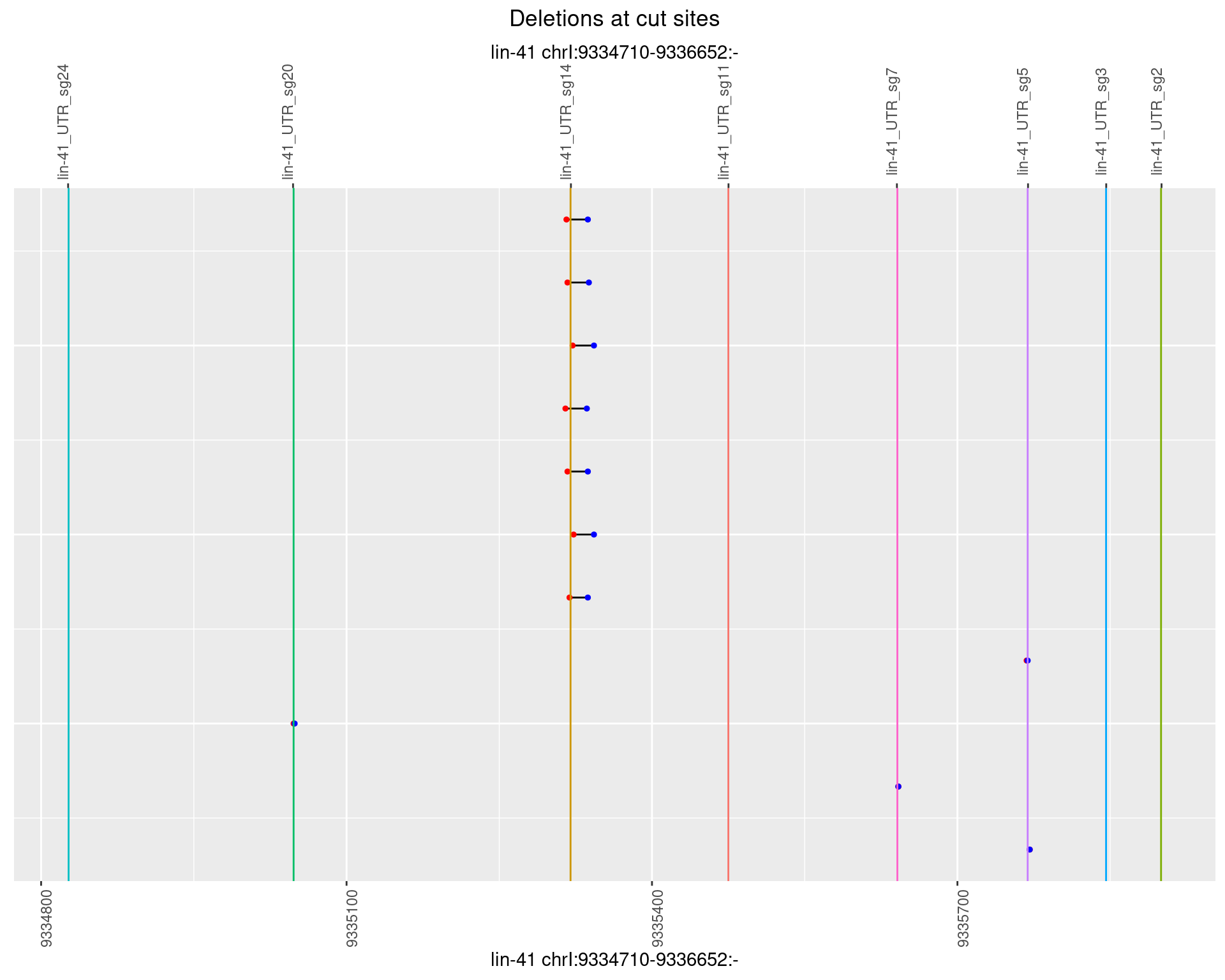
1.56.5 Freq: 0.01
1.56.6 Freq: 0.1
No plot to show
1.57 lin41_RNA_L4_N2_middle
1.57.1 Freq: 0
1.57.2 Freq: 1e-05
1.57.3 Freq: 1e-04
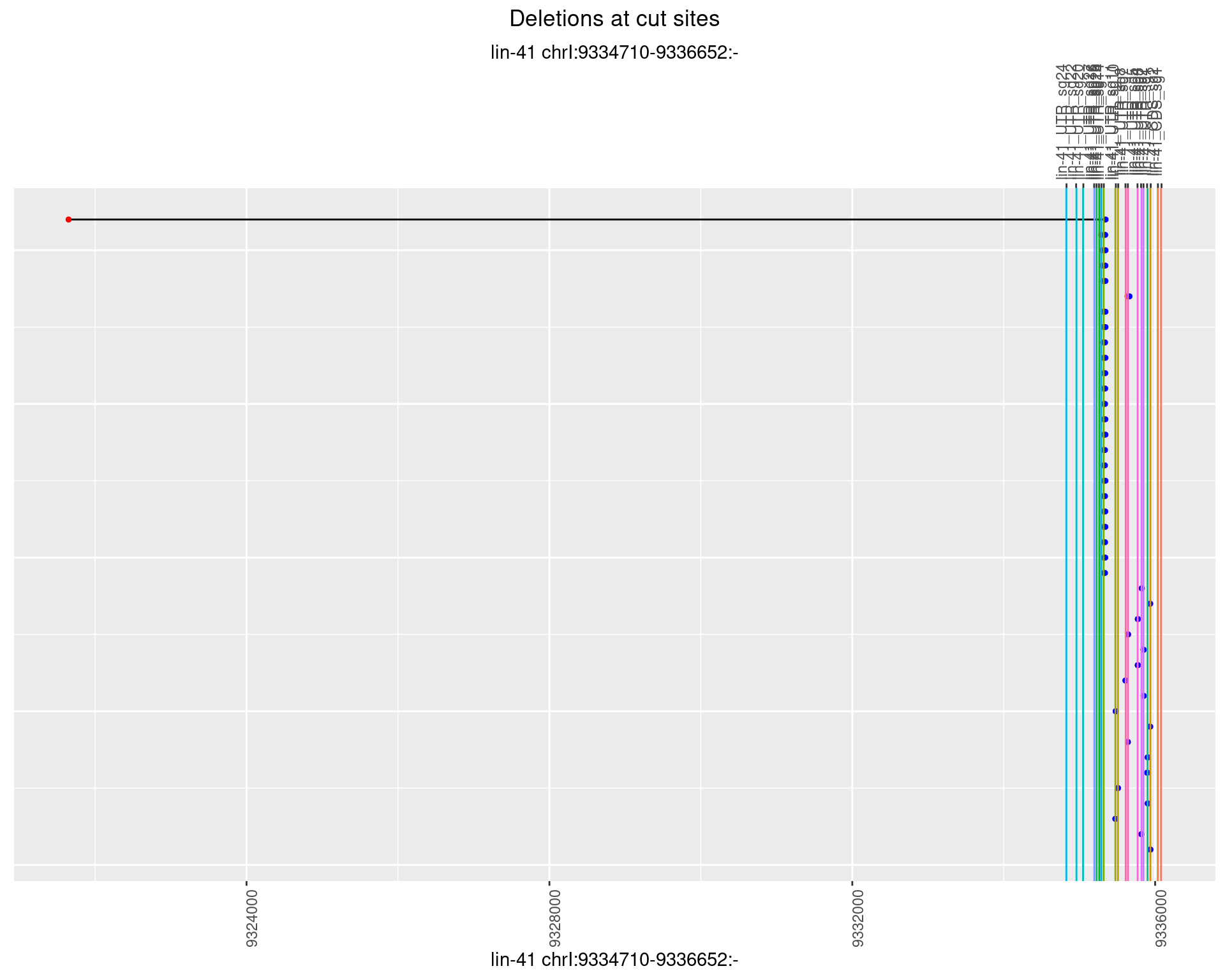
1.57.4 Freq: 0.001
1.57.5 Freq: 0.01
1.57.6 Freq: 0.1
No plot to show
1.58 lin41_RNA_L4_1516_middle
1.58.1 Freq: 0
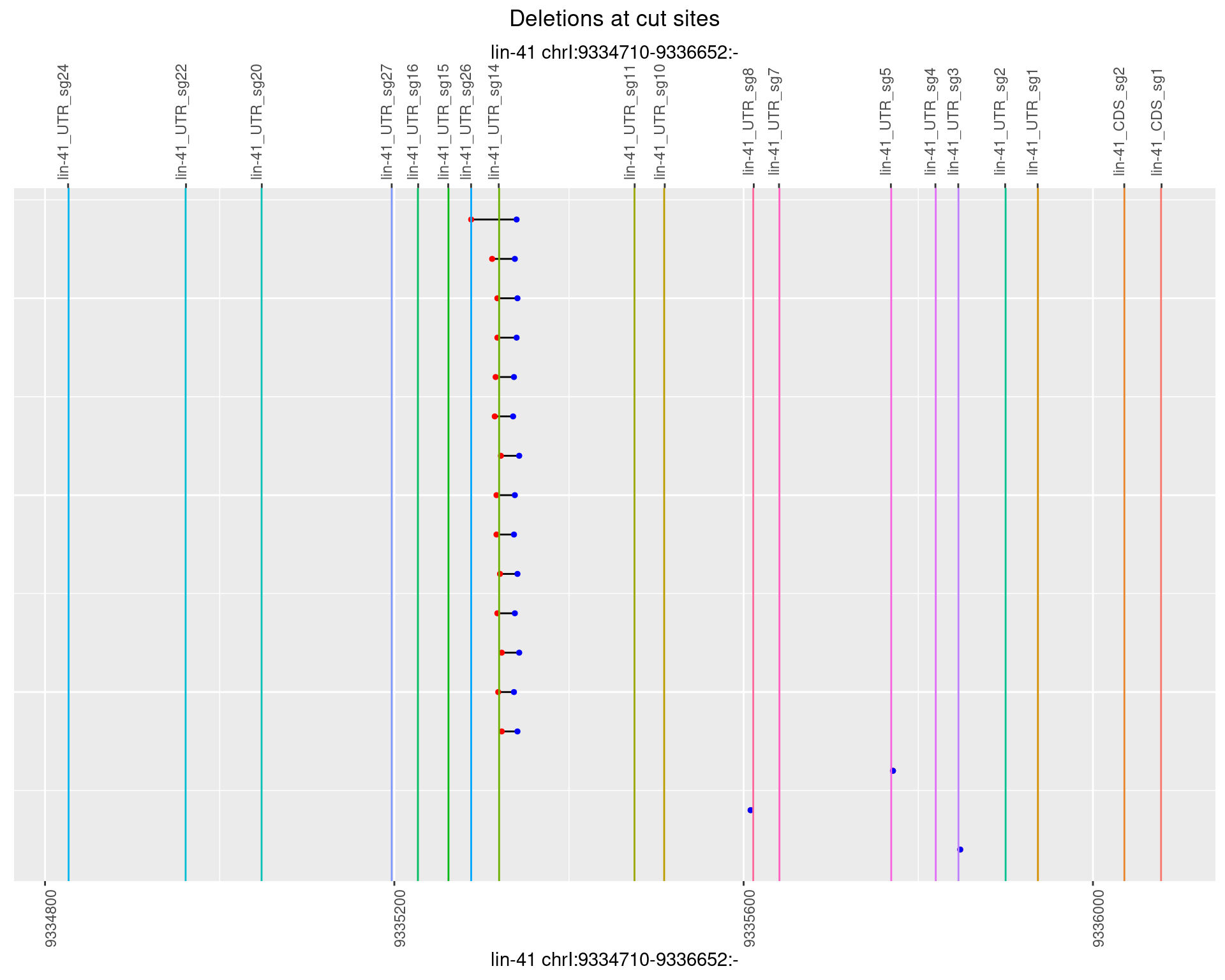
1.58.2 Freq: 1e-05
No plot to show
1.58.3 Freq: 1e-04
No plot to show
1.58.4 Freq: 0.001
No plot to show
1.58.5 Freq: 0.01
No plot to show
1.58.6 Freq: 0.1
No plot to show
1.59 lin41_RNA_L4_2627_middle
1.59.1 Freq: 0
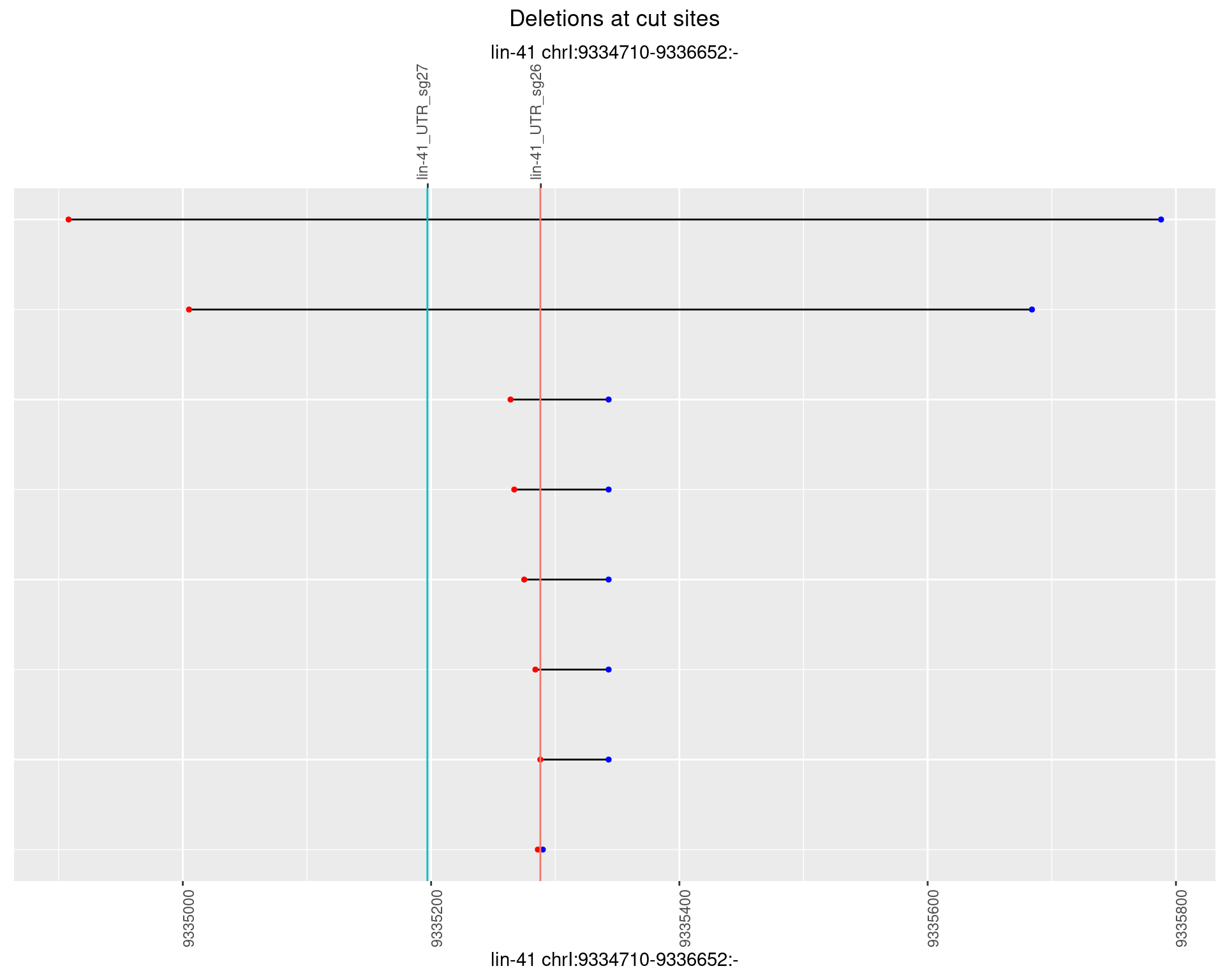
1.59.2 Freq: 1e-05
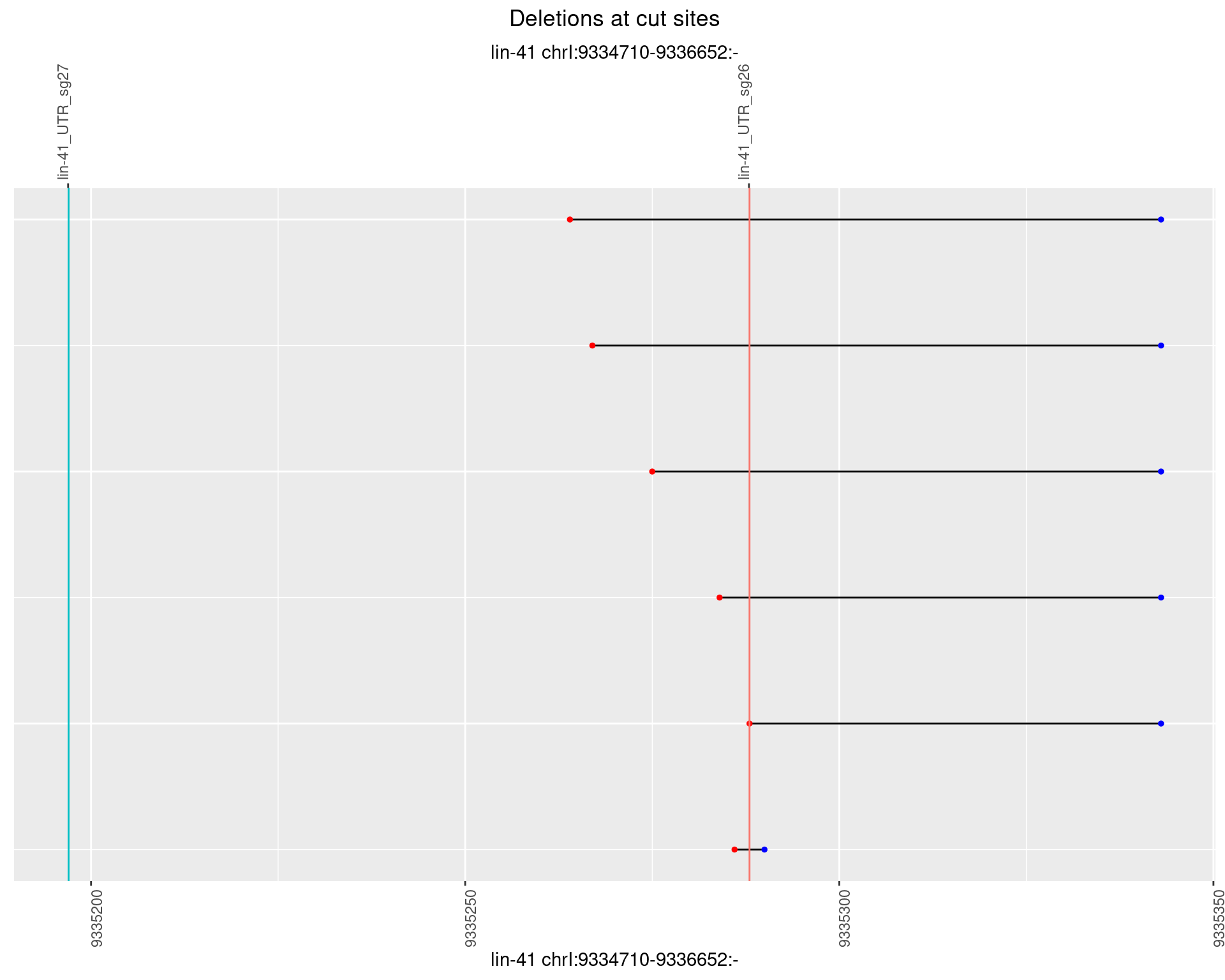
1.59.3 Freq: 1e-04
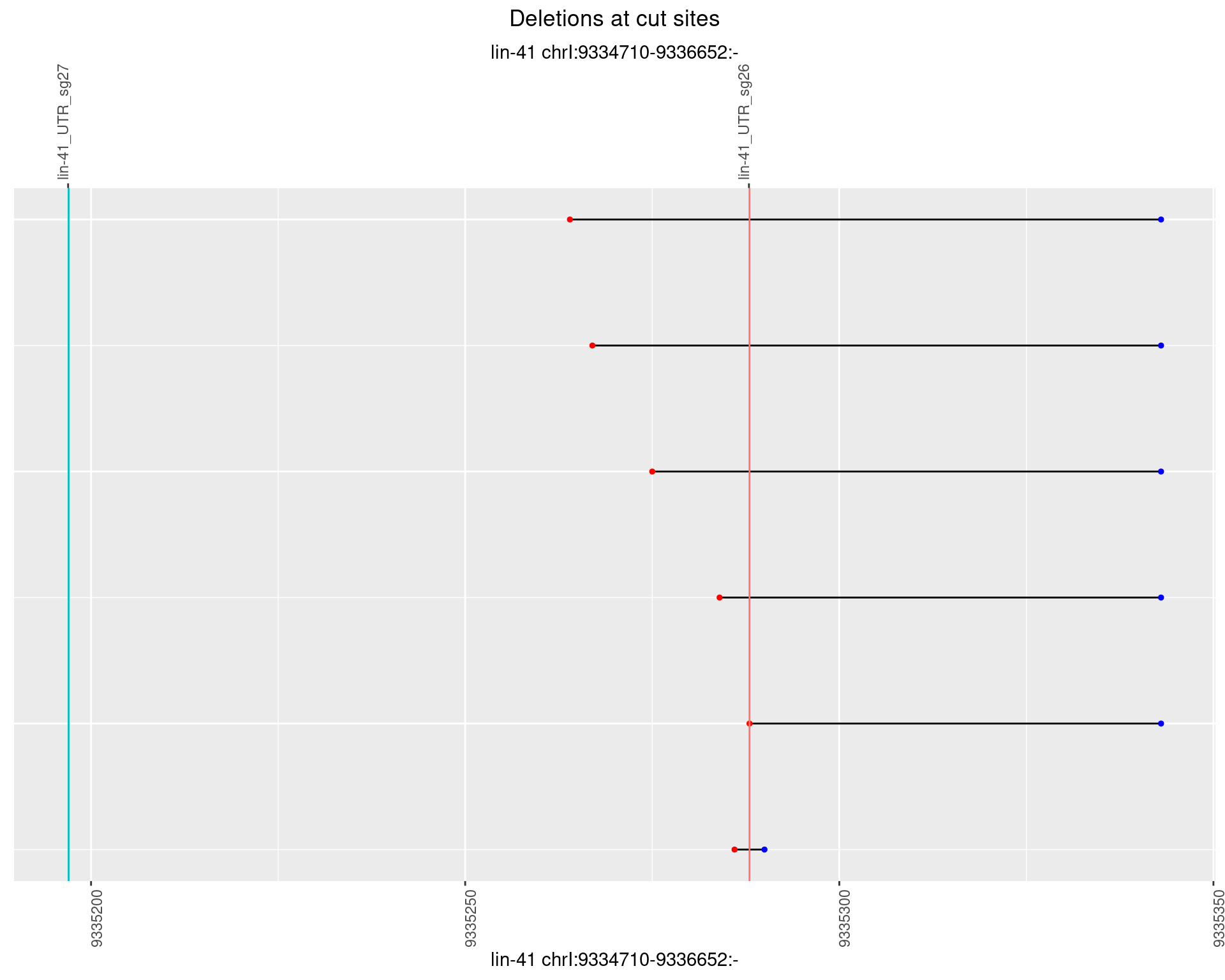
1.59.4 Freq: 0.001
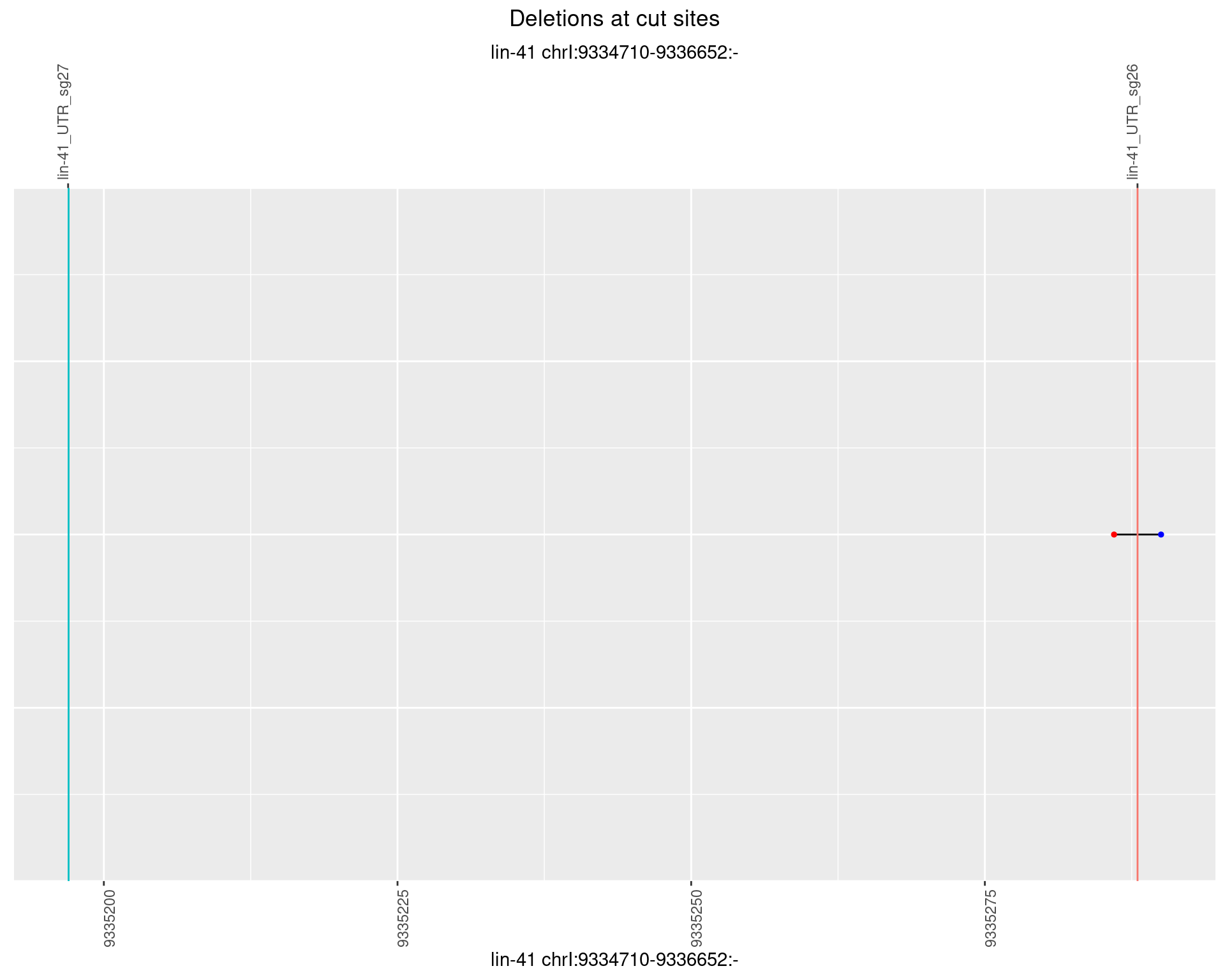
1.59.5 Freq: 0.01
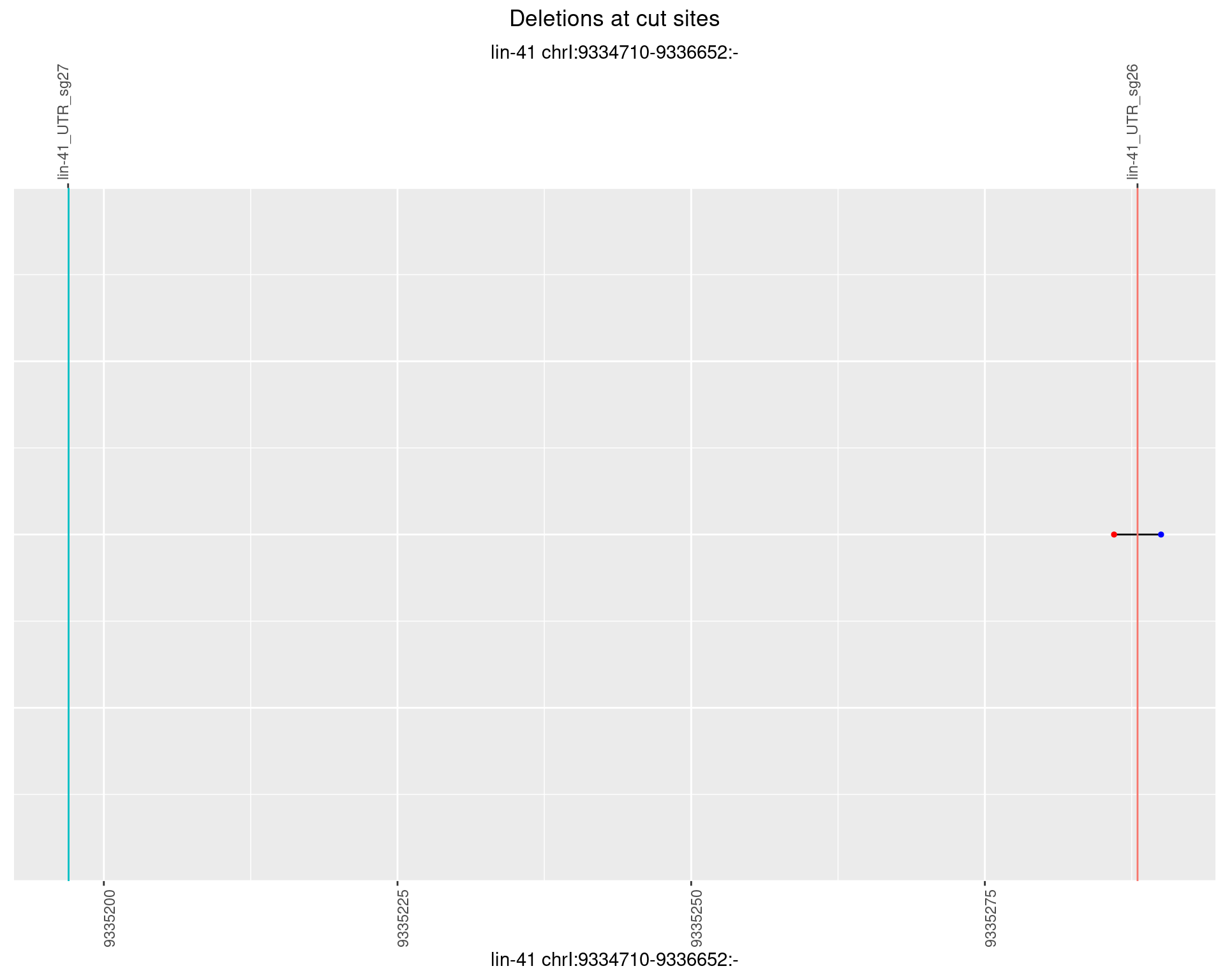
1.59.6 Freq: 0.1
No plot to show
1.60 lin41_RNA_L4_pool2_middle
1.60.1 Freq: 0
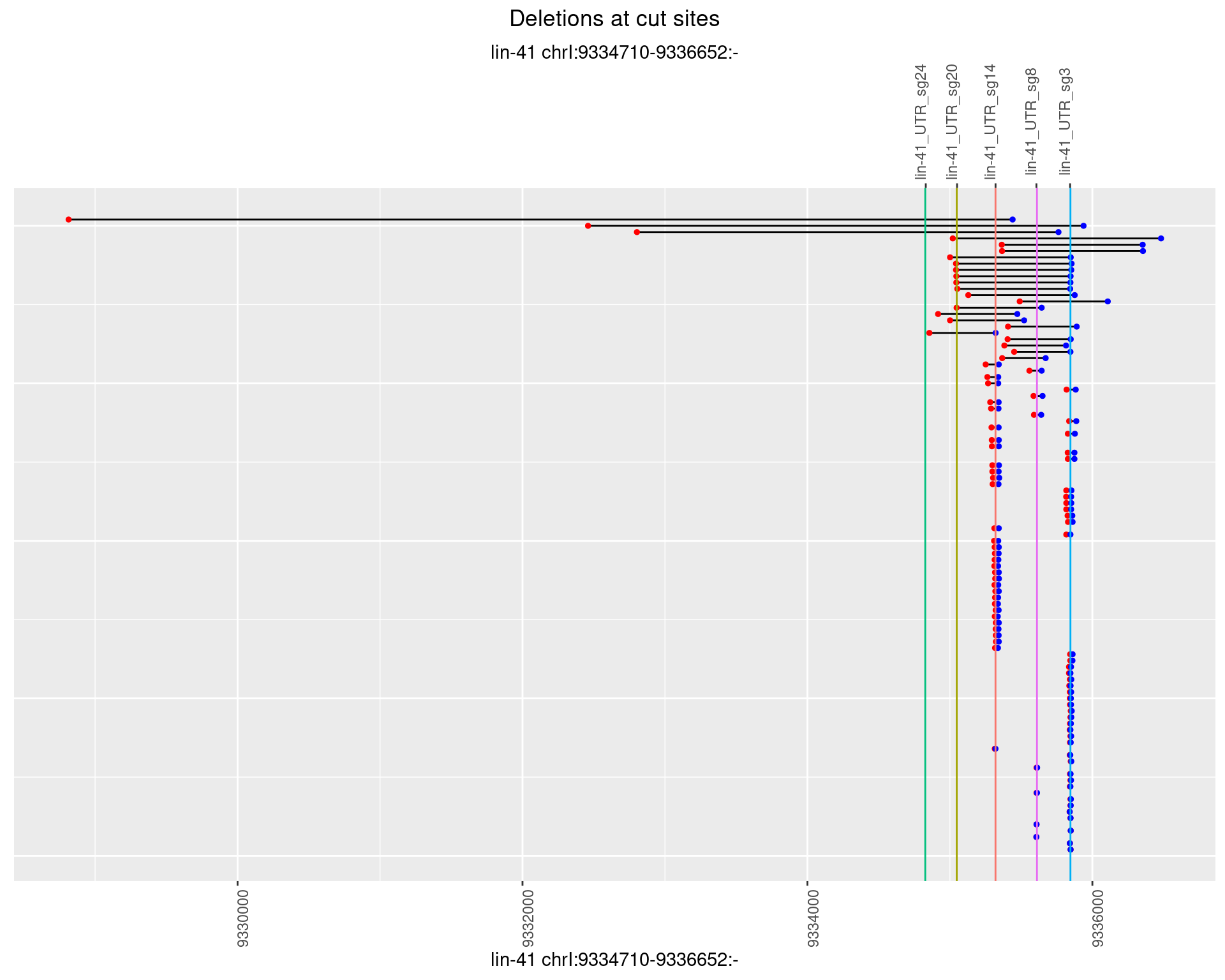
1.60.2 Freq: 1e-05
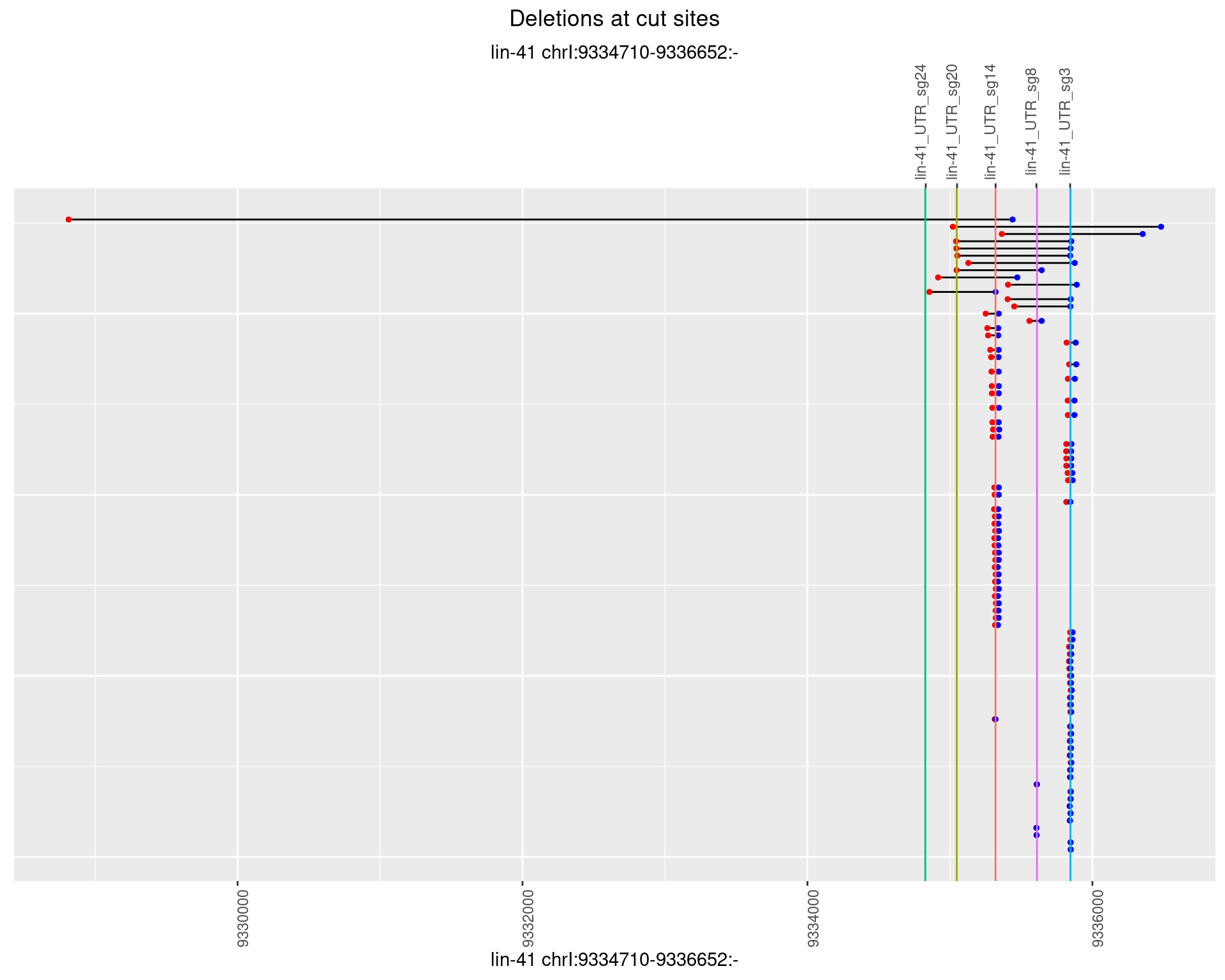
1.60.3 Freq: 1e-04
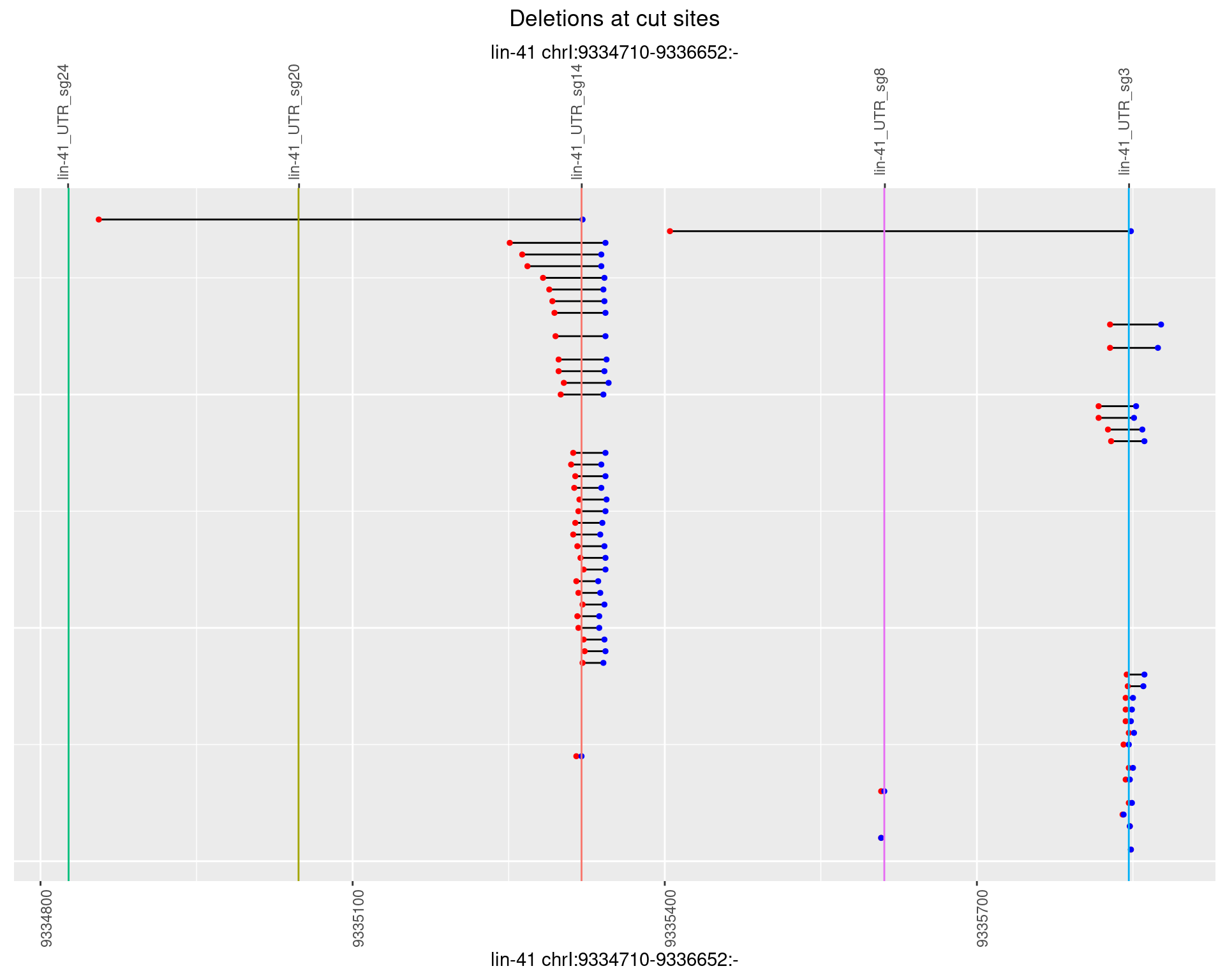
1.60.4 Freq: 0.001
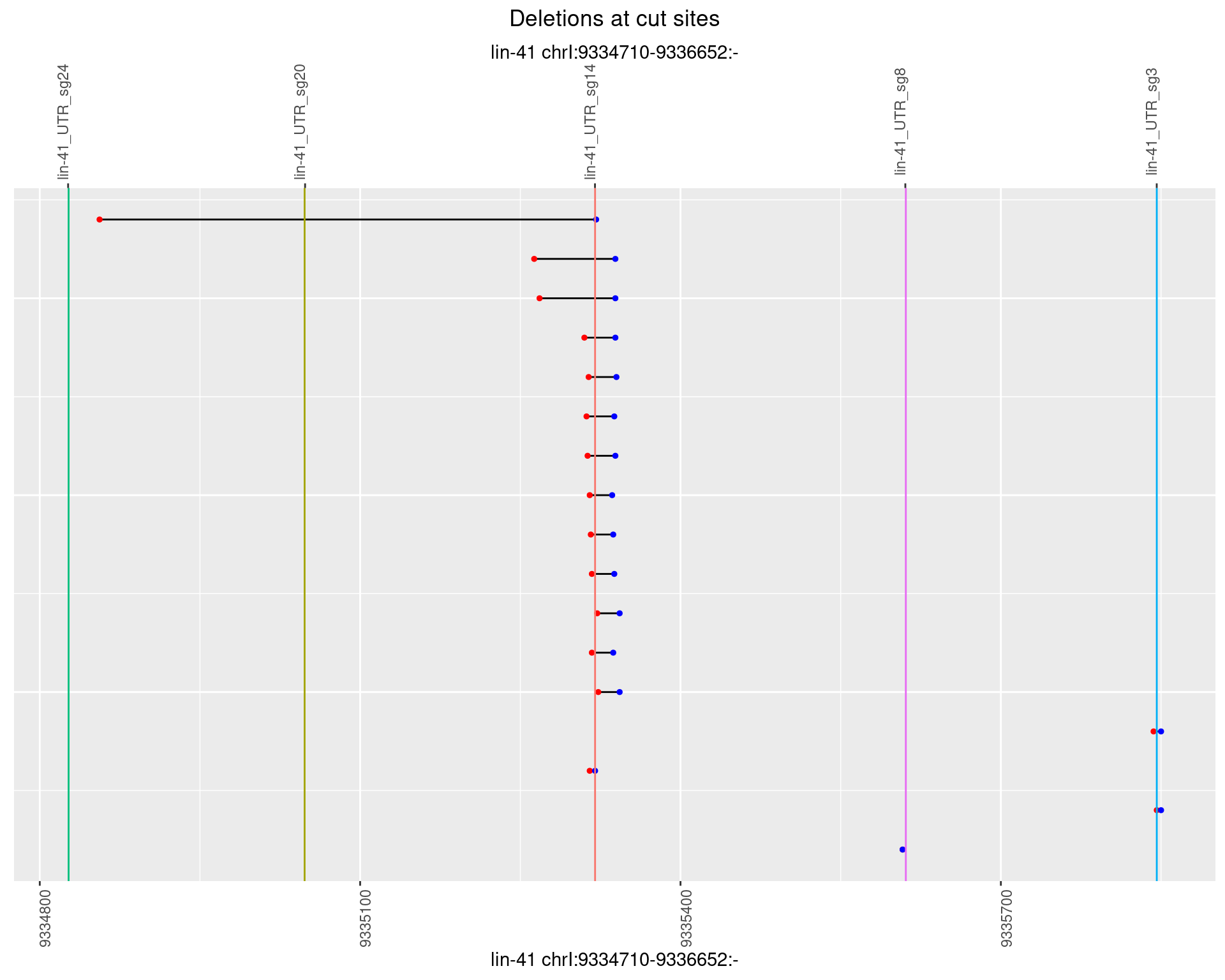
1.60.5 Freq: 0.01
No plot to show
1.60.6 Freq: 0.1
No plot to show
1.61 lin41_RNA_L4_pool3_middle
1.61.1 Freq: 0
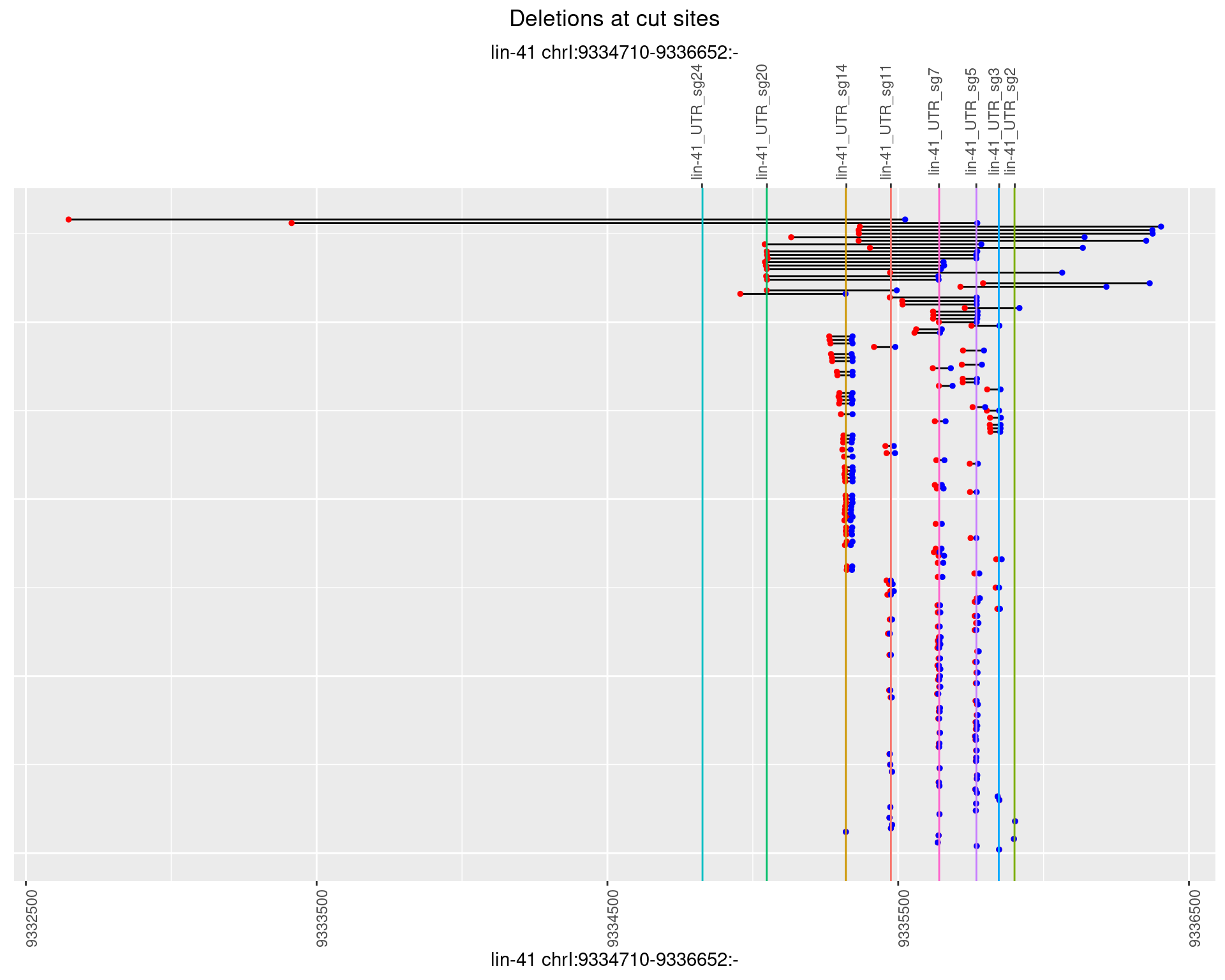
1.61.2 Freq: 1e-05
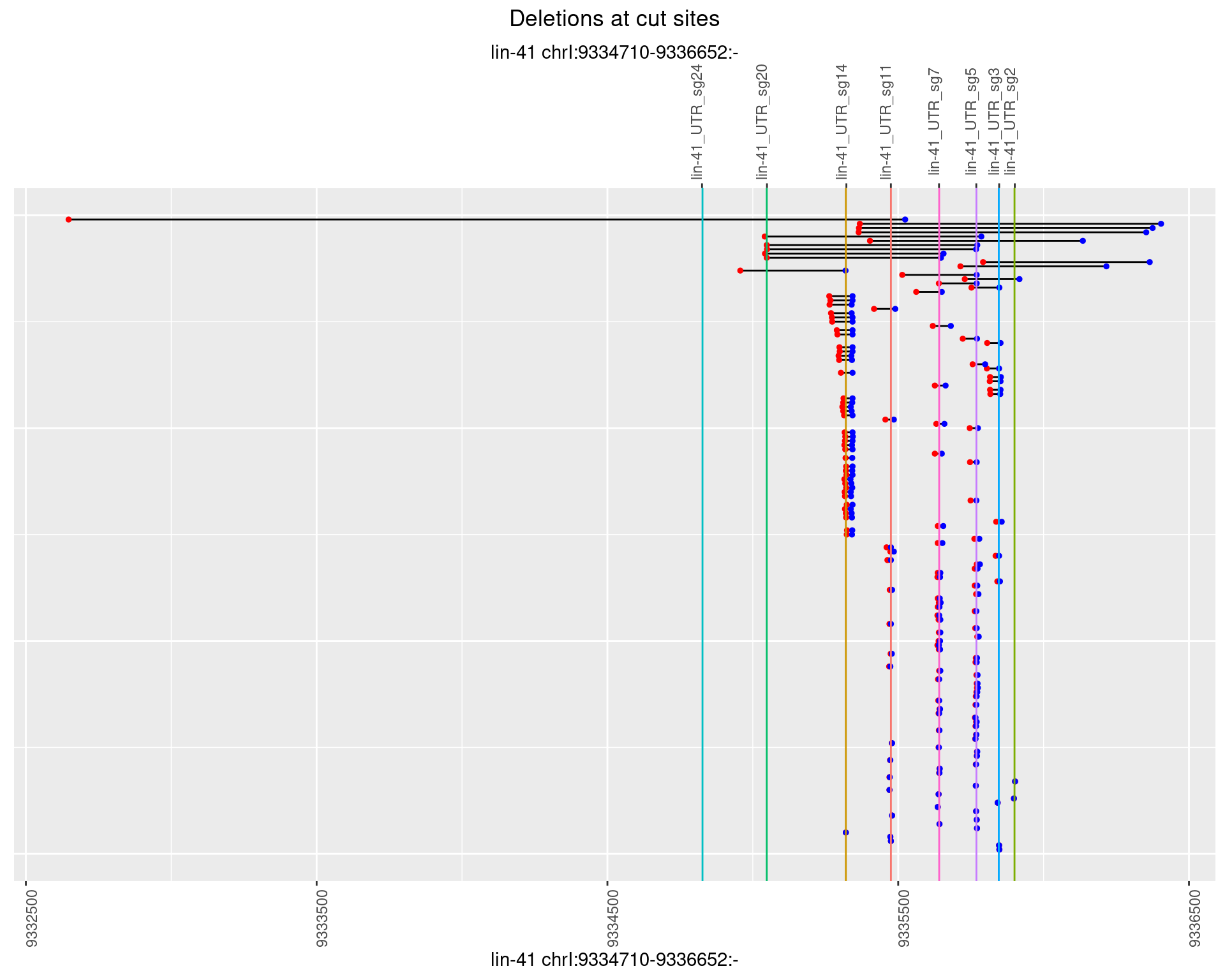
1.61.3 Freq: 1e-04
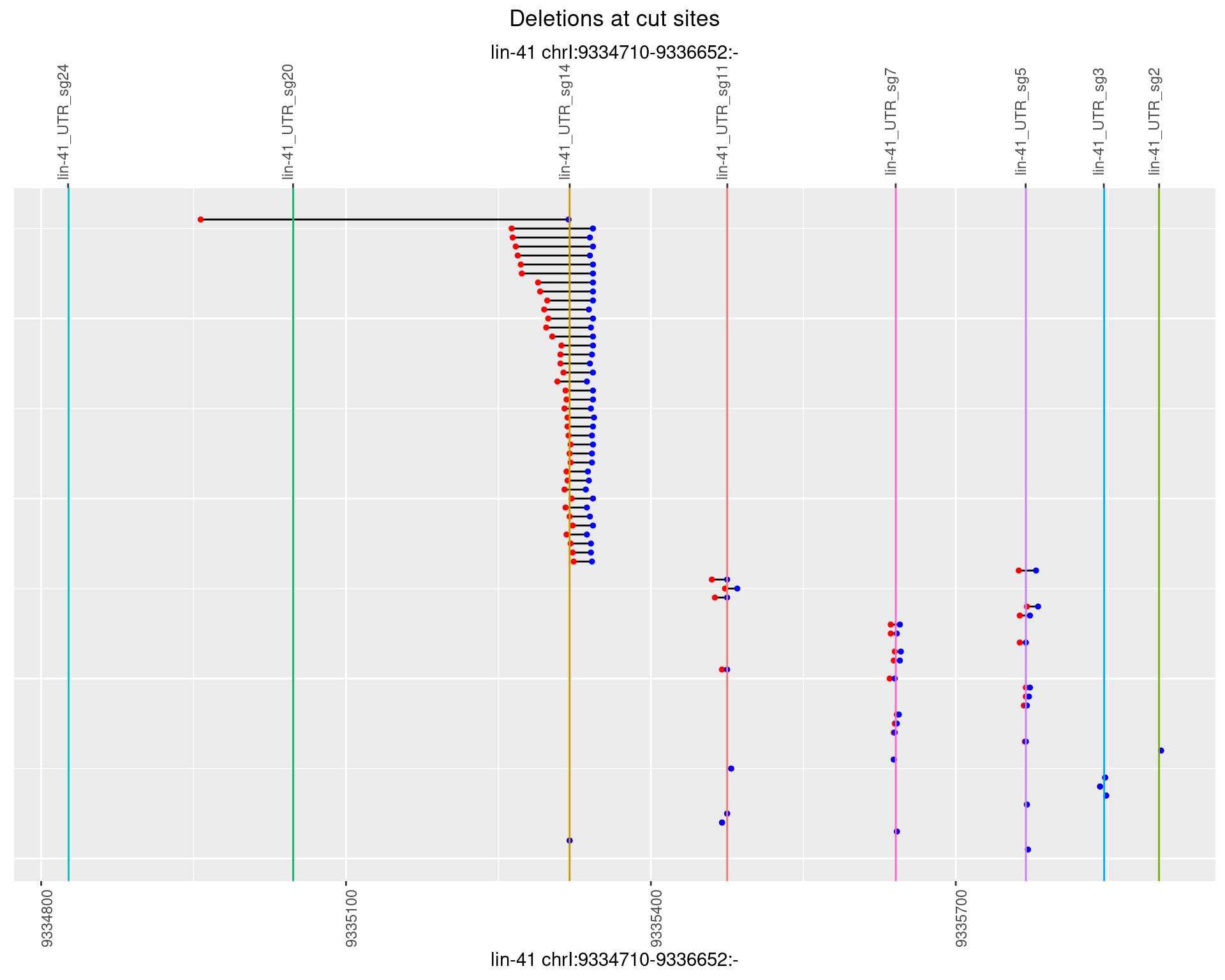
1.61.4 Freq: 0.001
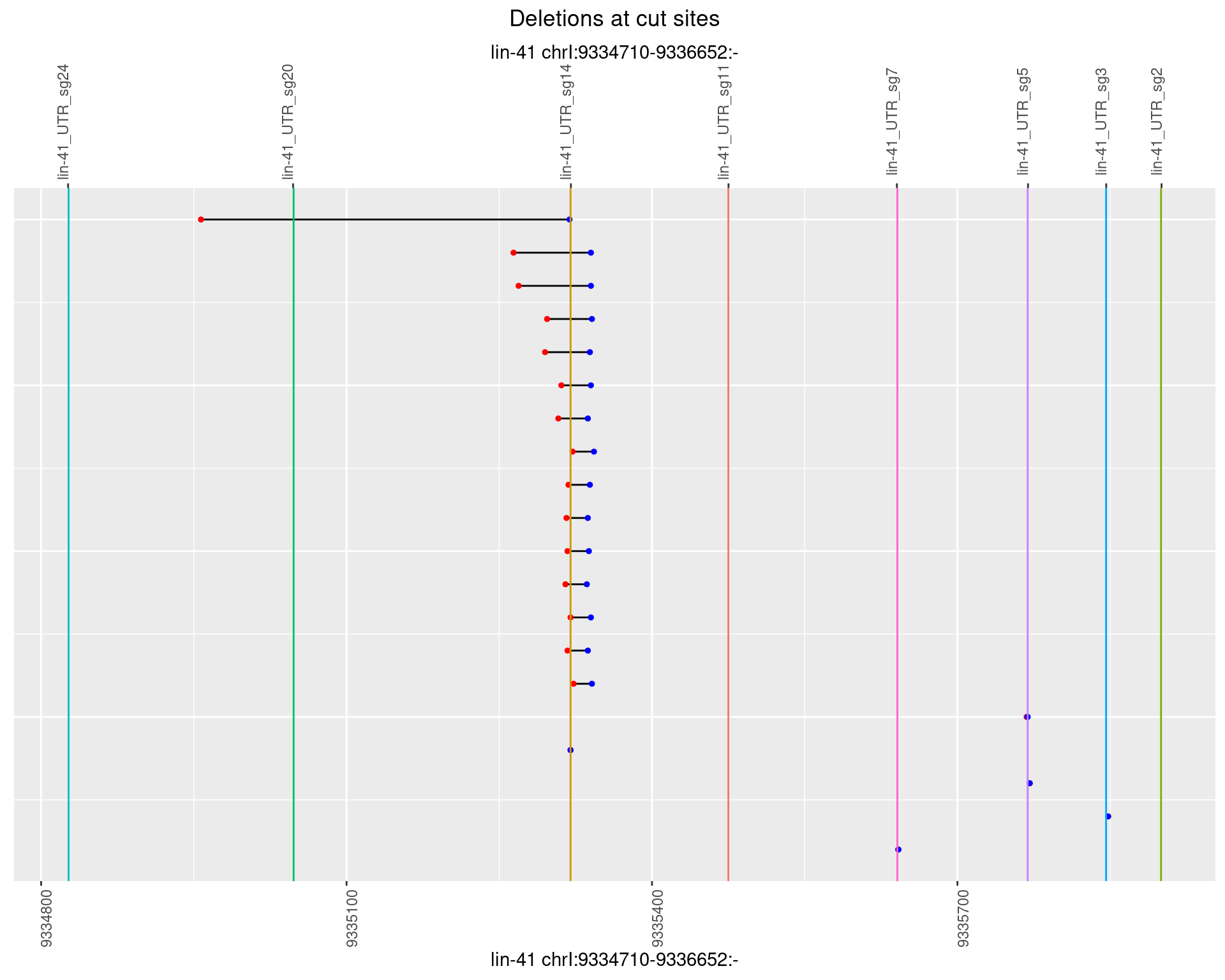
1.61.5 Freq: 0.01
No plot to show
1.61.6 Freq: 0.1
No plot to show
1.62 lin41_RNA_L1_N2_3end_UMI
1.62.1 Freq: 0
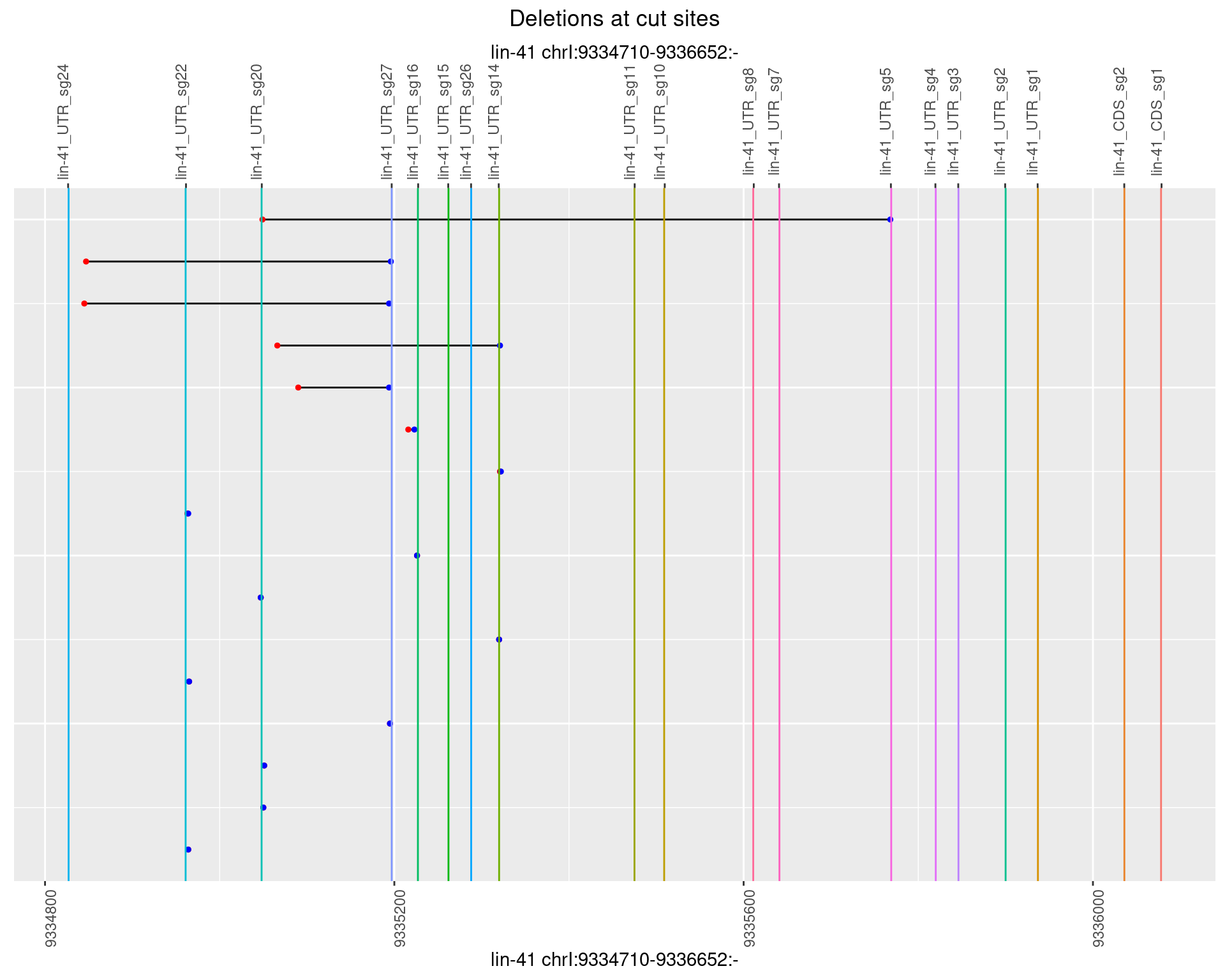
1.62.2 Freq: 1e-05
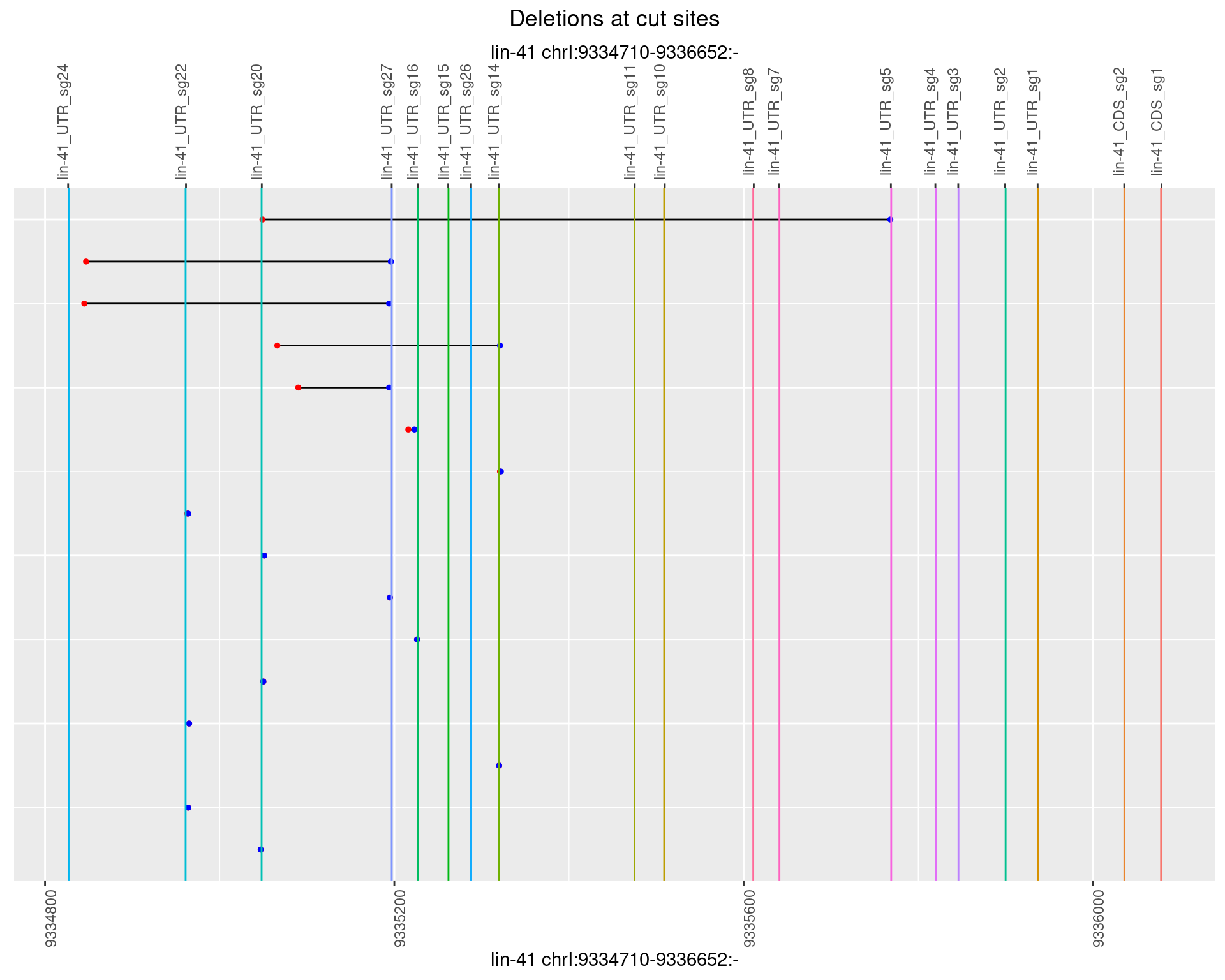
1.62.3 Freq: 1e-04
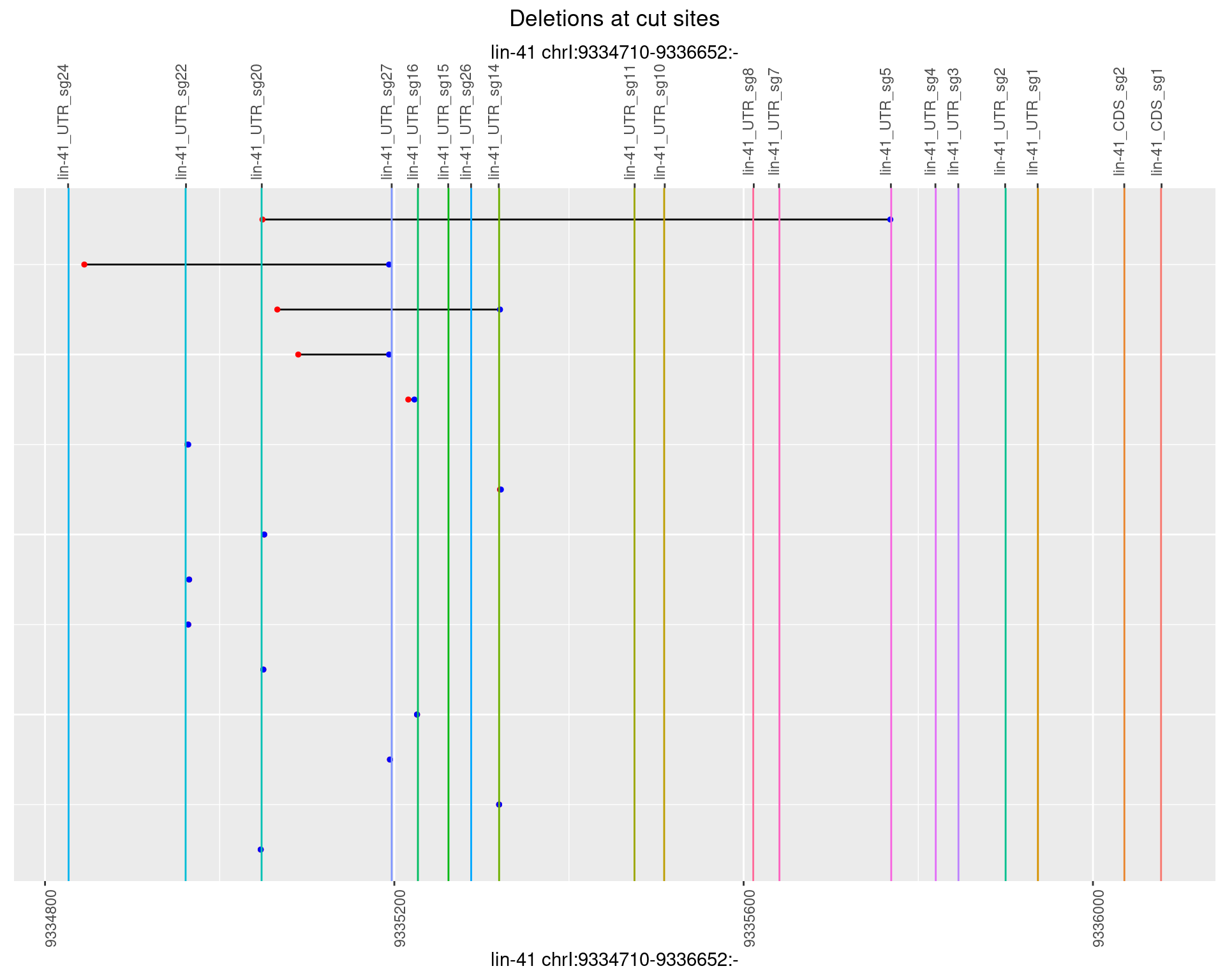
1.62.4 Freq: 0.001
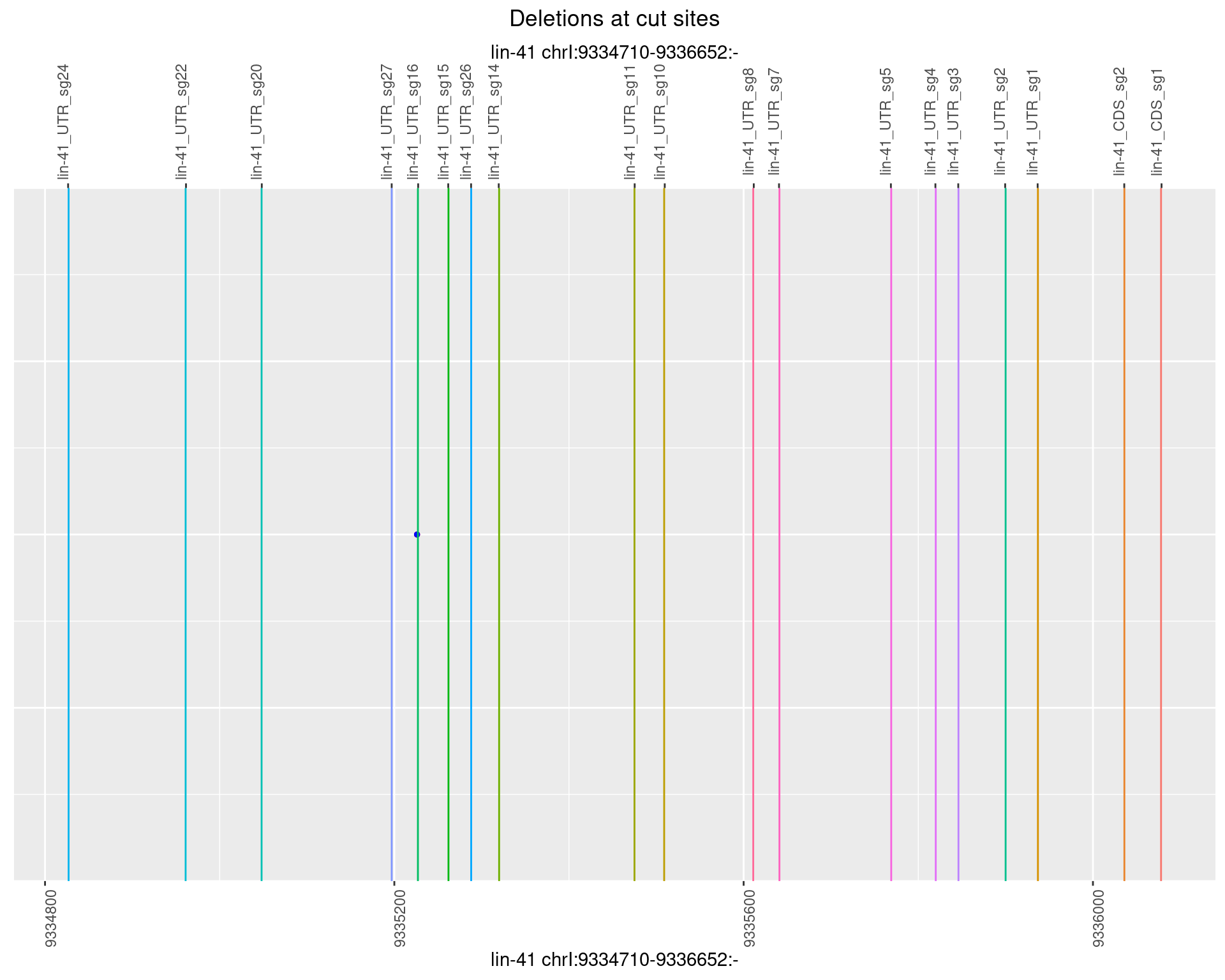
1.62.5 Freq: 0.01
No plot to show
1.62.6 Freq: 0.1
No plot to show
1.63 lin41_RNA_L1_1516_3end_UMI
1.63.1 Freq: 0
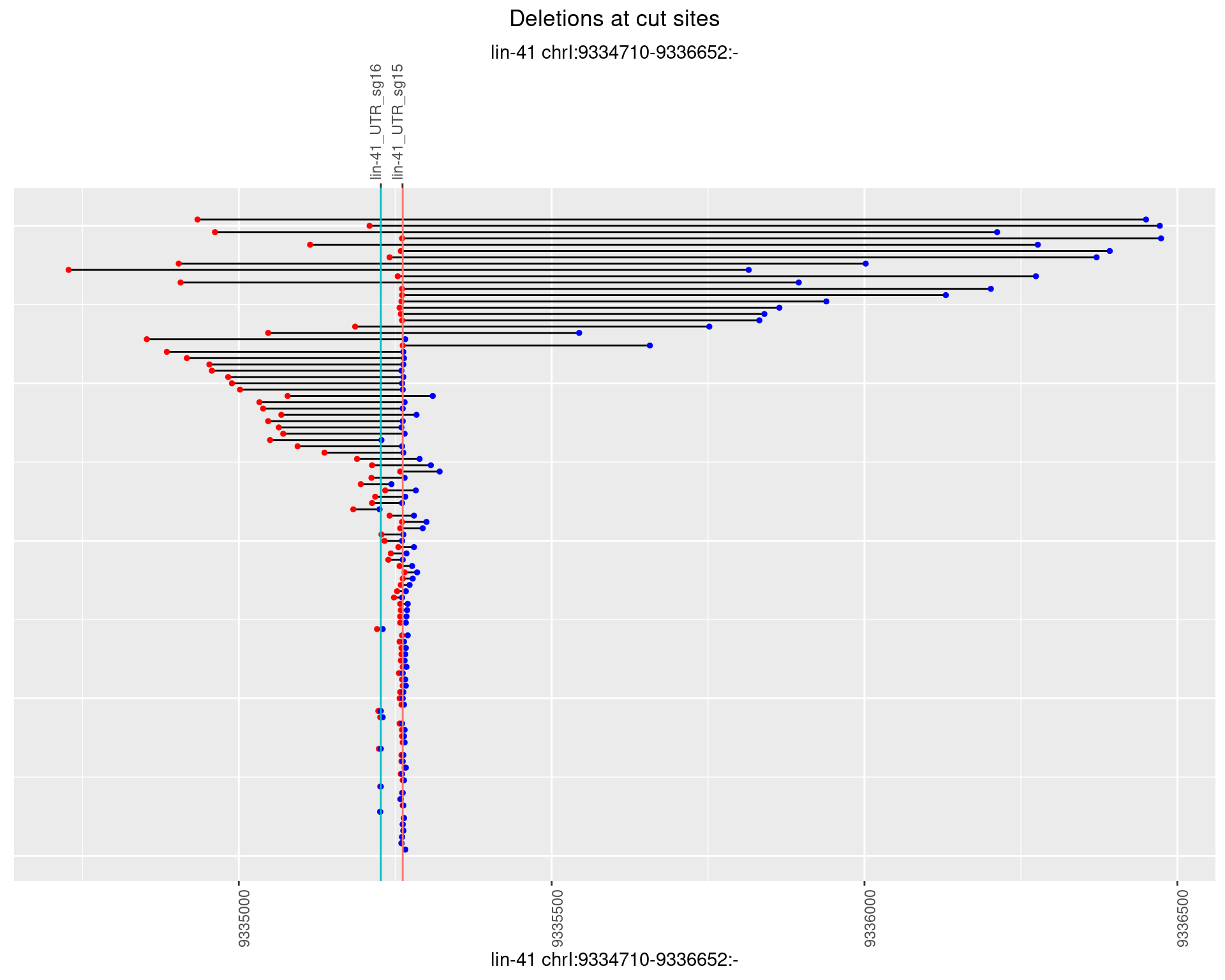
1.63.2 Freq: 1e-05
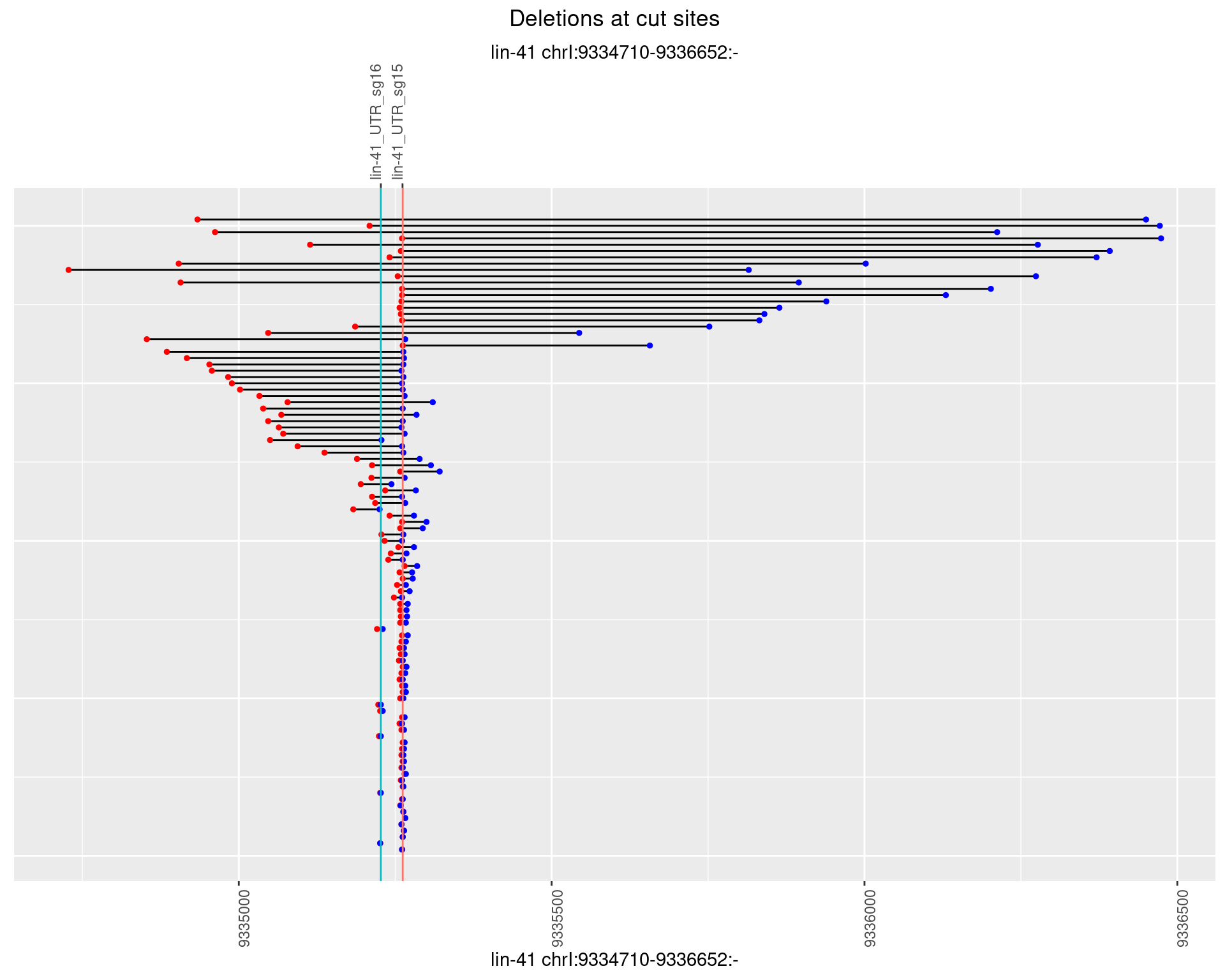
1.63.3 Freq: 1e-04
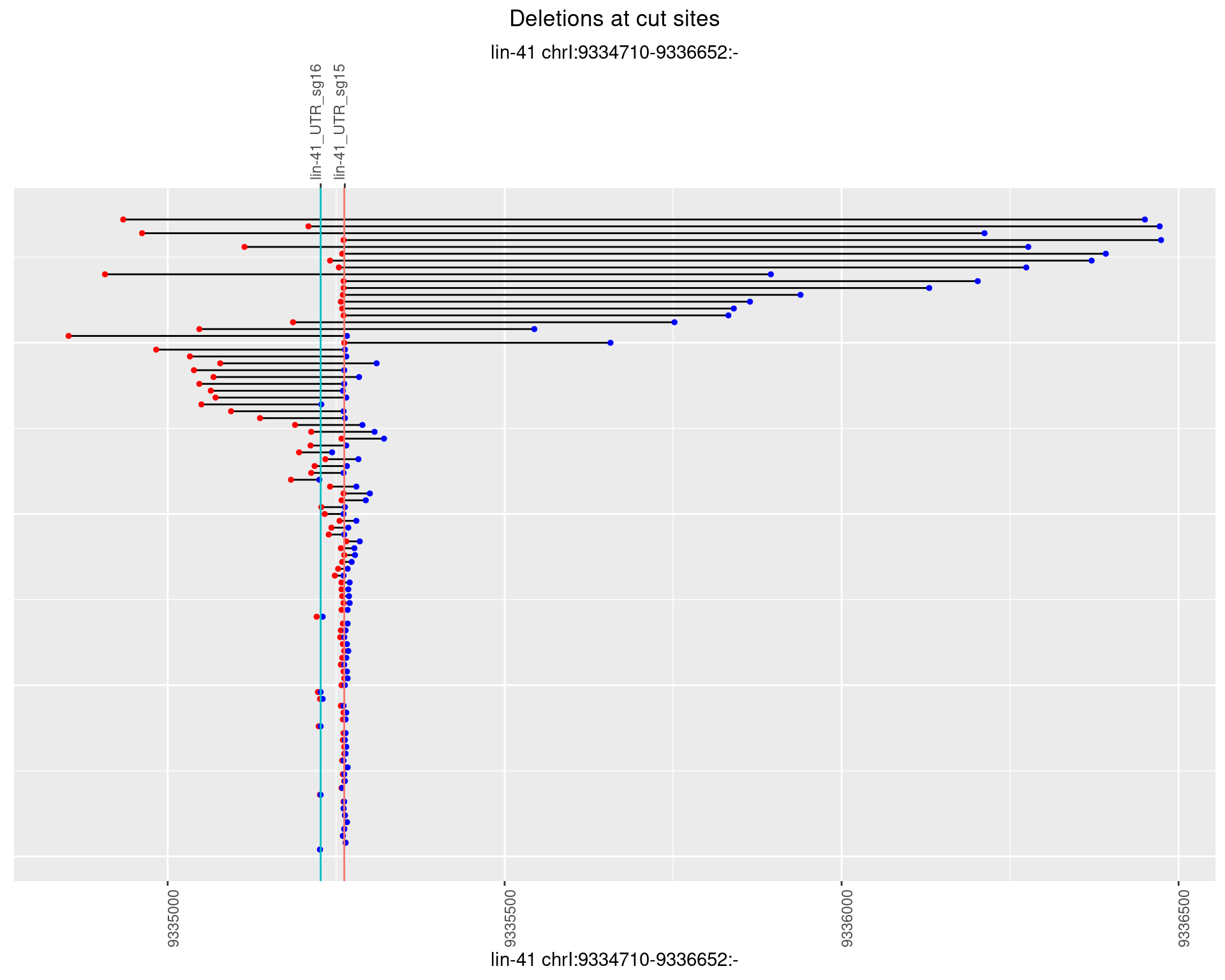
1.63.4 Freq: 0.001
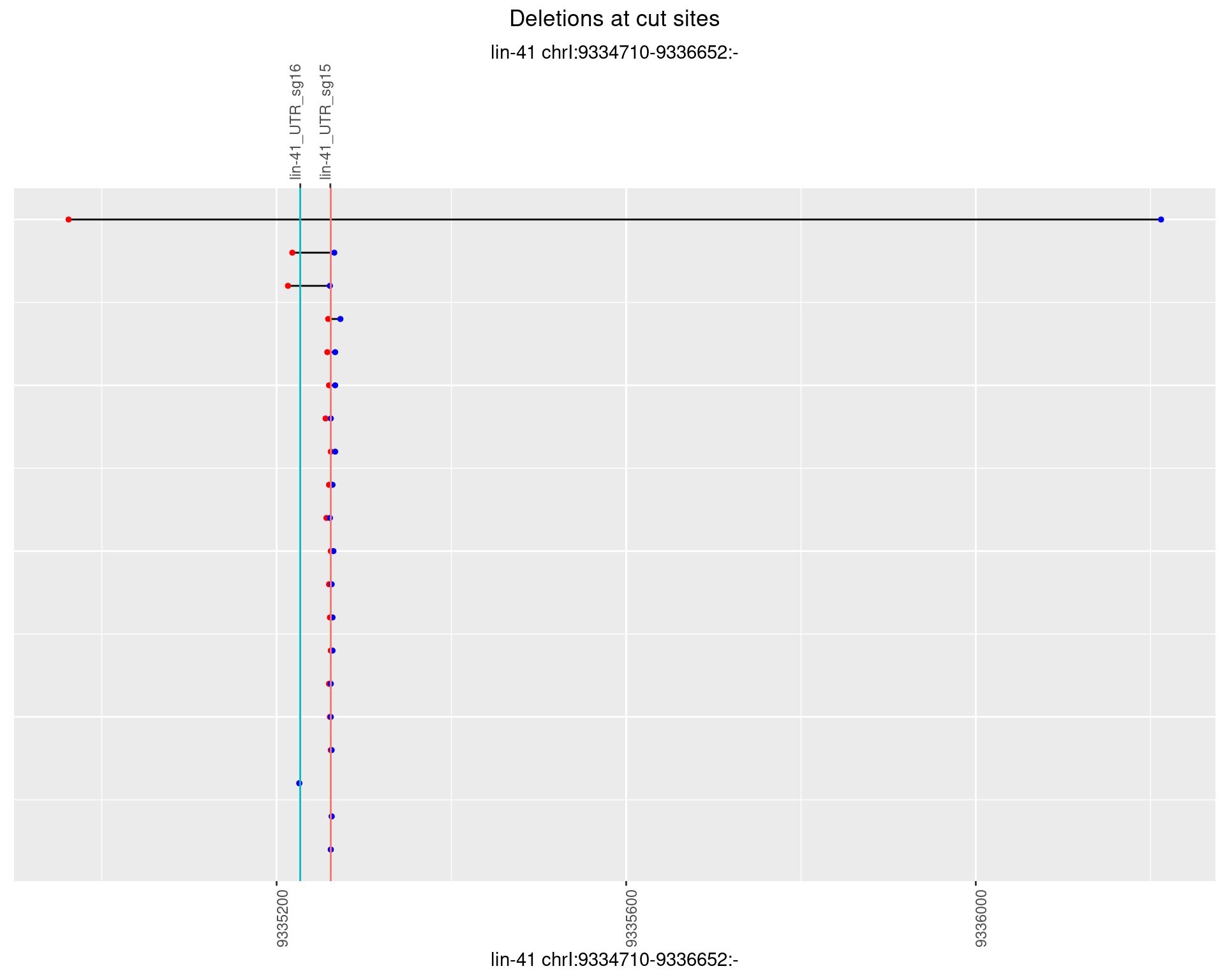
1.63.5 Freq: 0.01
1.63.6 Freq: 0.1
No plot to show
1.64 lin41_RNA_L1_2627_3end_UMI
1.64.1 Freq: 0
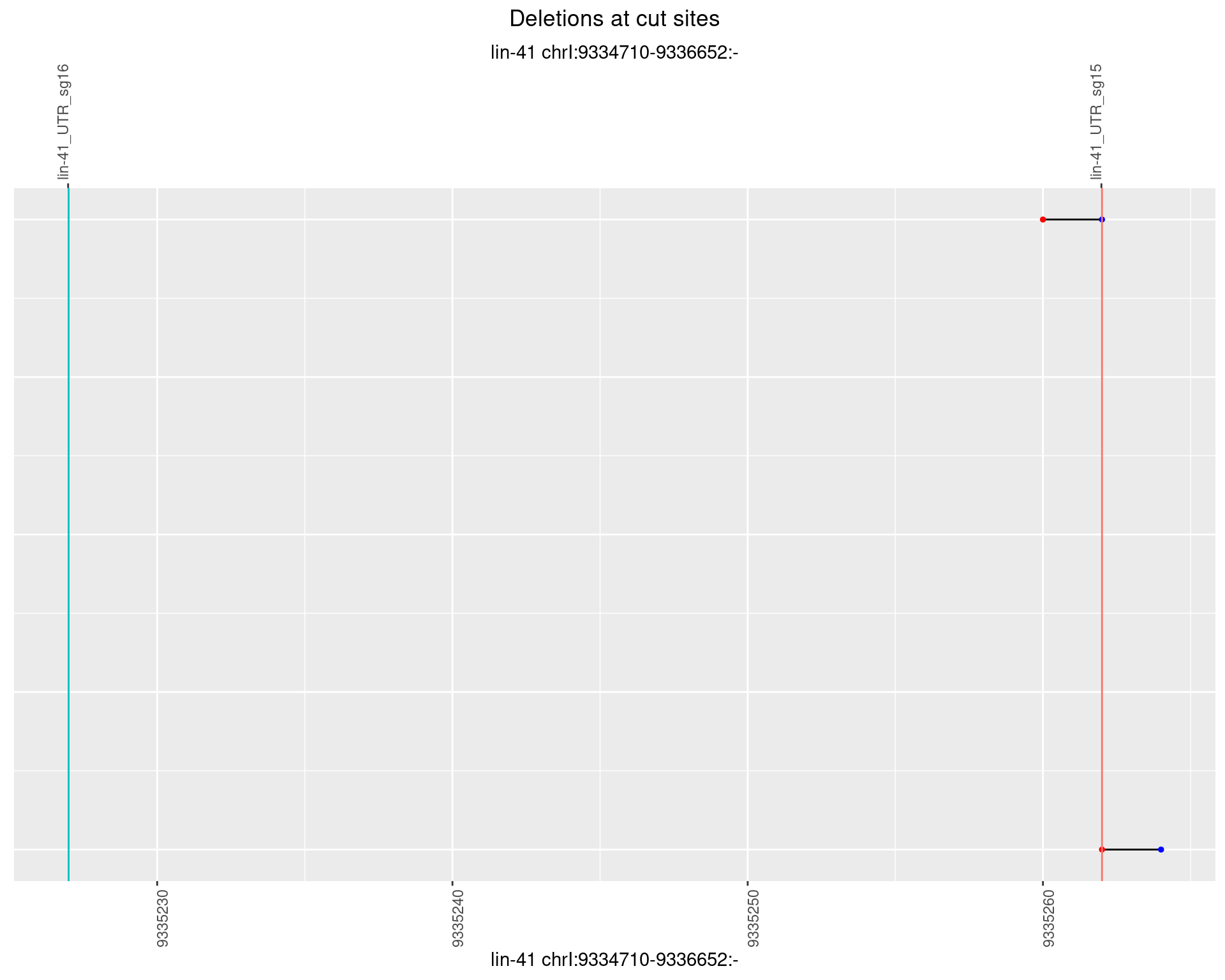
1.64.2 Freq: 1e-05
1.64.3 Freq: 1e-04
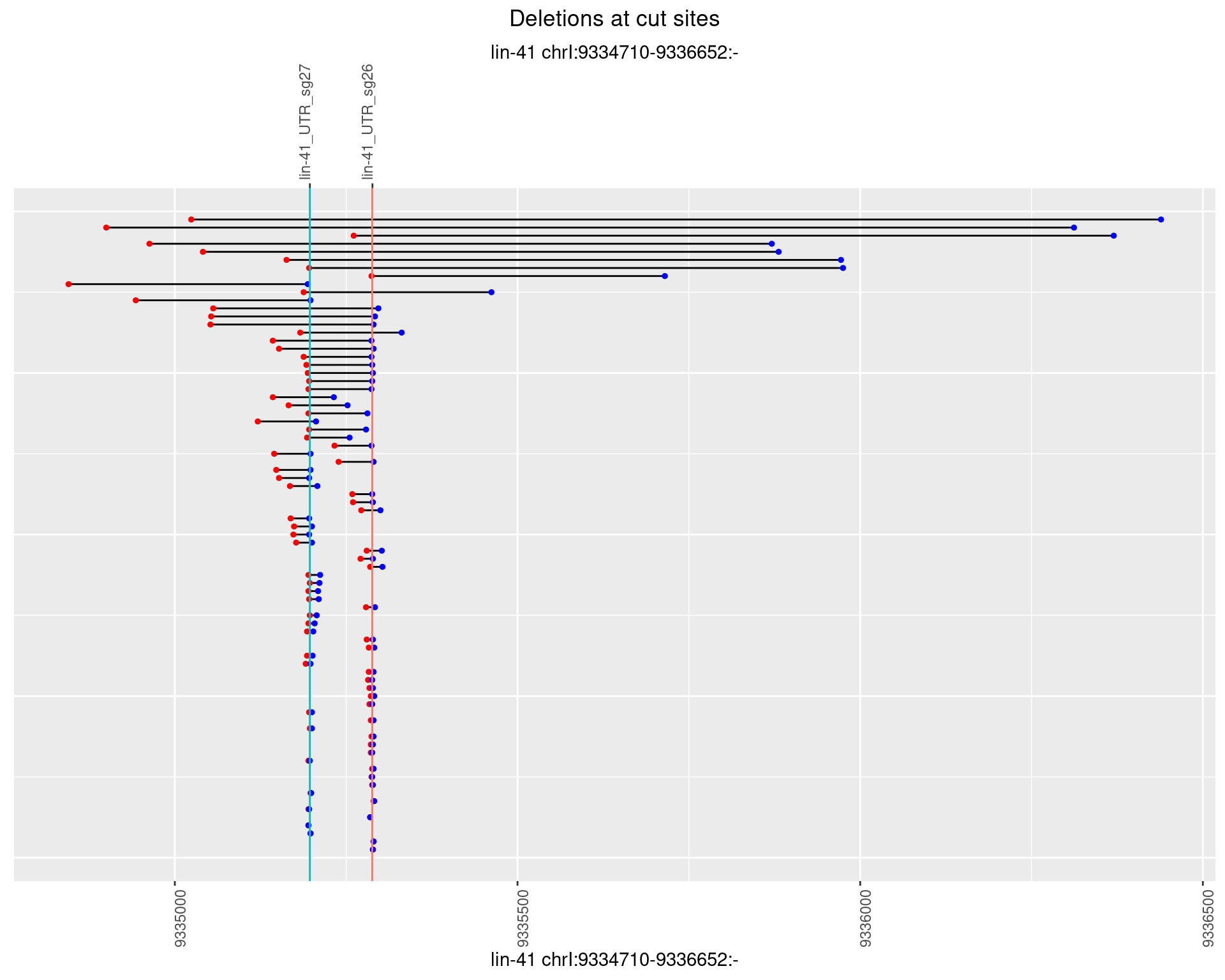
1.64.4 Freq: 0.001
1.64.5 Freq: 0.01
No plot to show
1.64.6 Freq: 0.1
No plot to show
1.65 lin41_RNA_L1_pool2_3end_UMI
1.65.1 Freq: 0
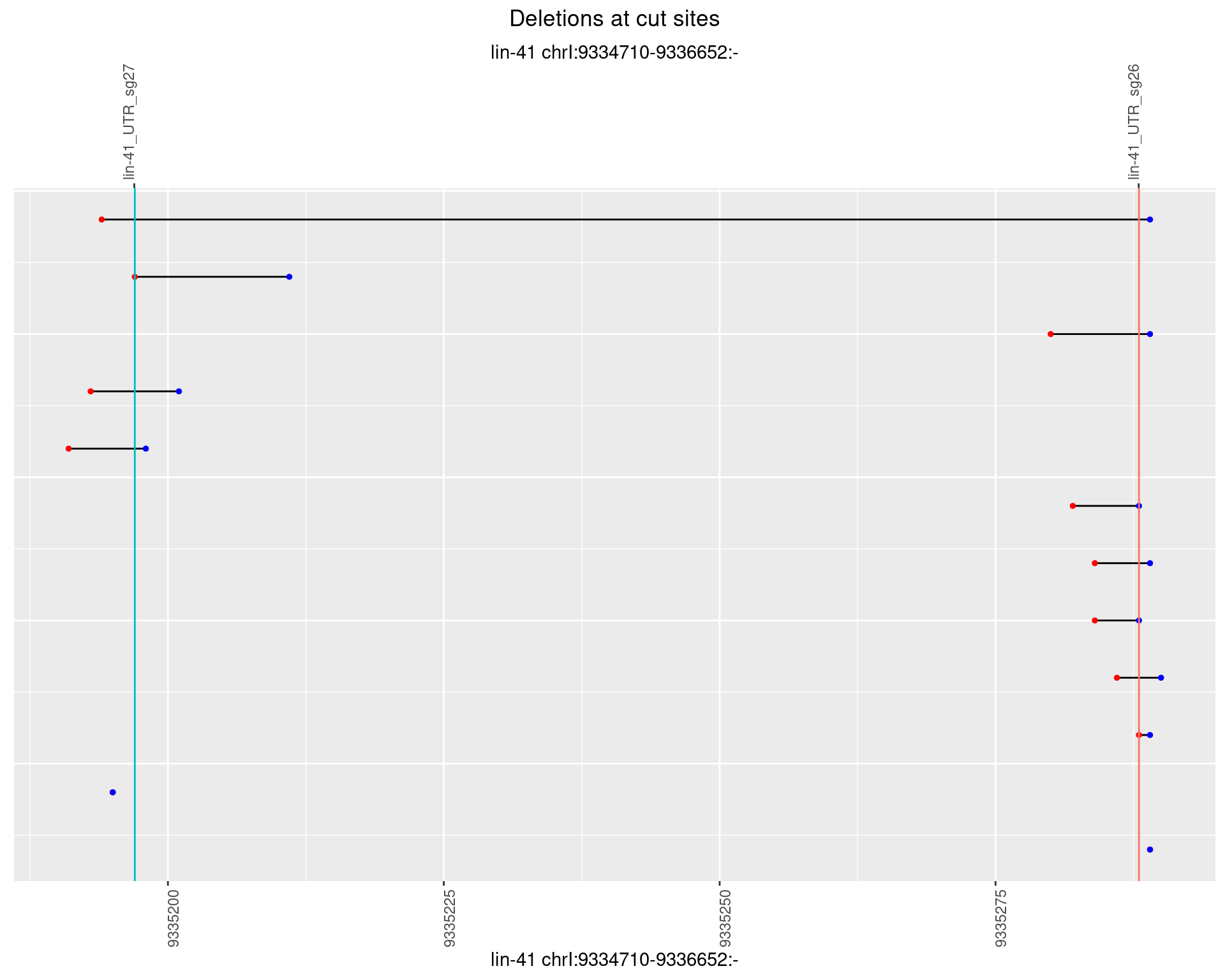
1.65.2 Freq: 1e-05
1.65.3 Freq: 1e-04
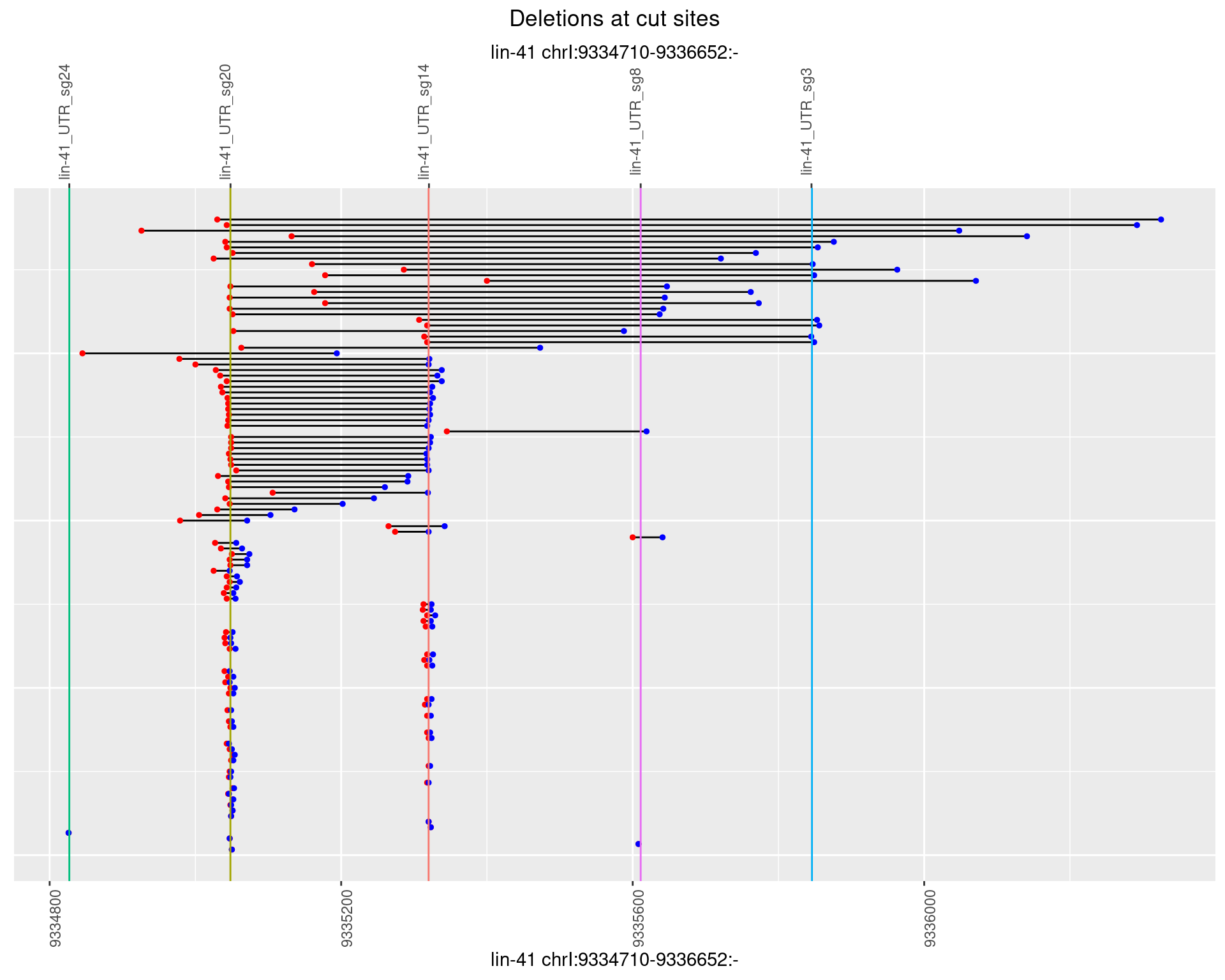
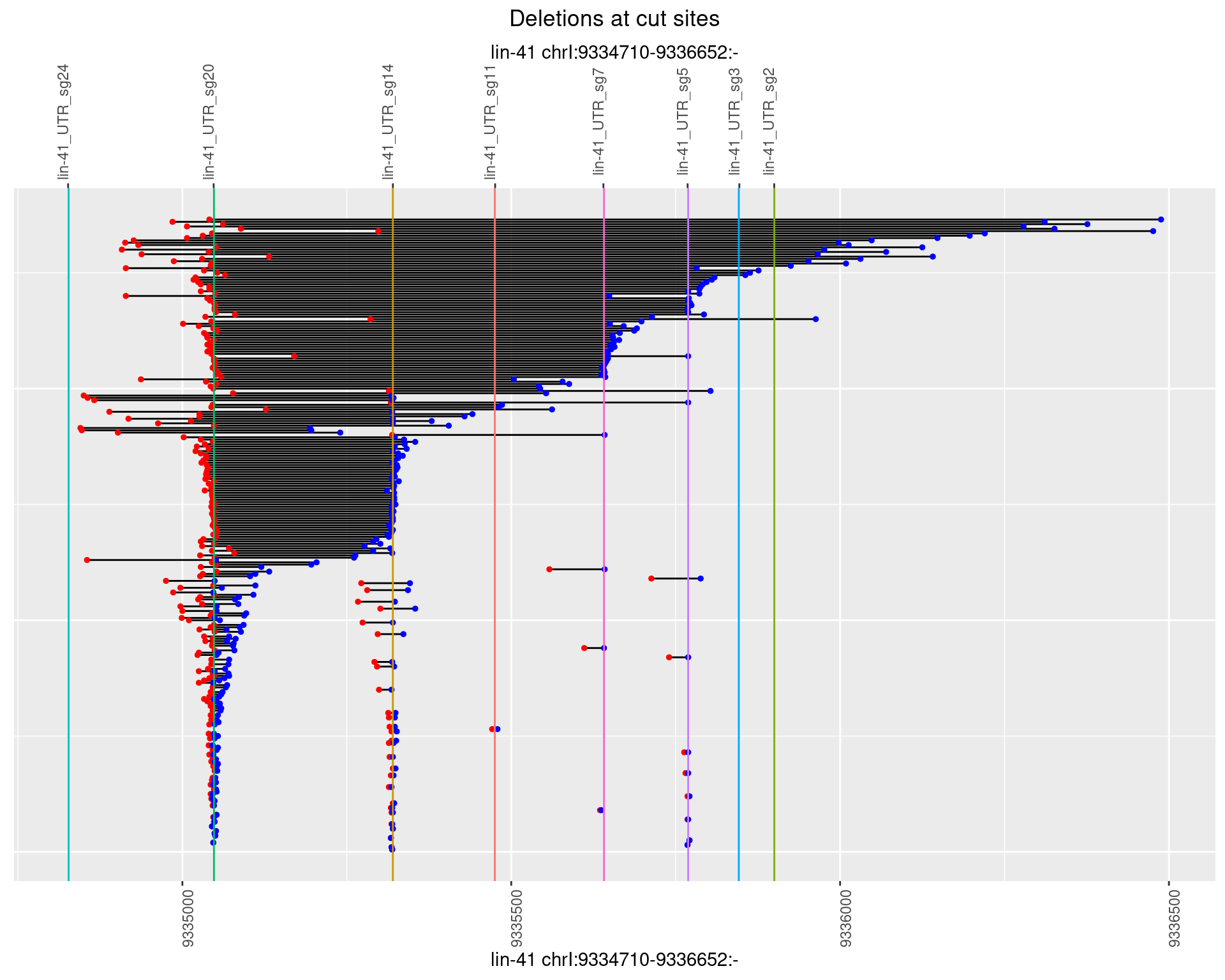
1.65.4 Freq: 0.001
1.65.5 Freq: 0.01
1.65.6 Freq: 0.1
No plot to show
1.66 lin41_RNA_L1_pool3_3end_UMI
1.66.1 Freq: 0
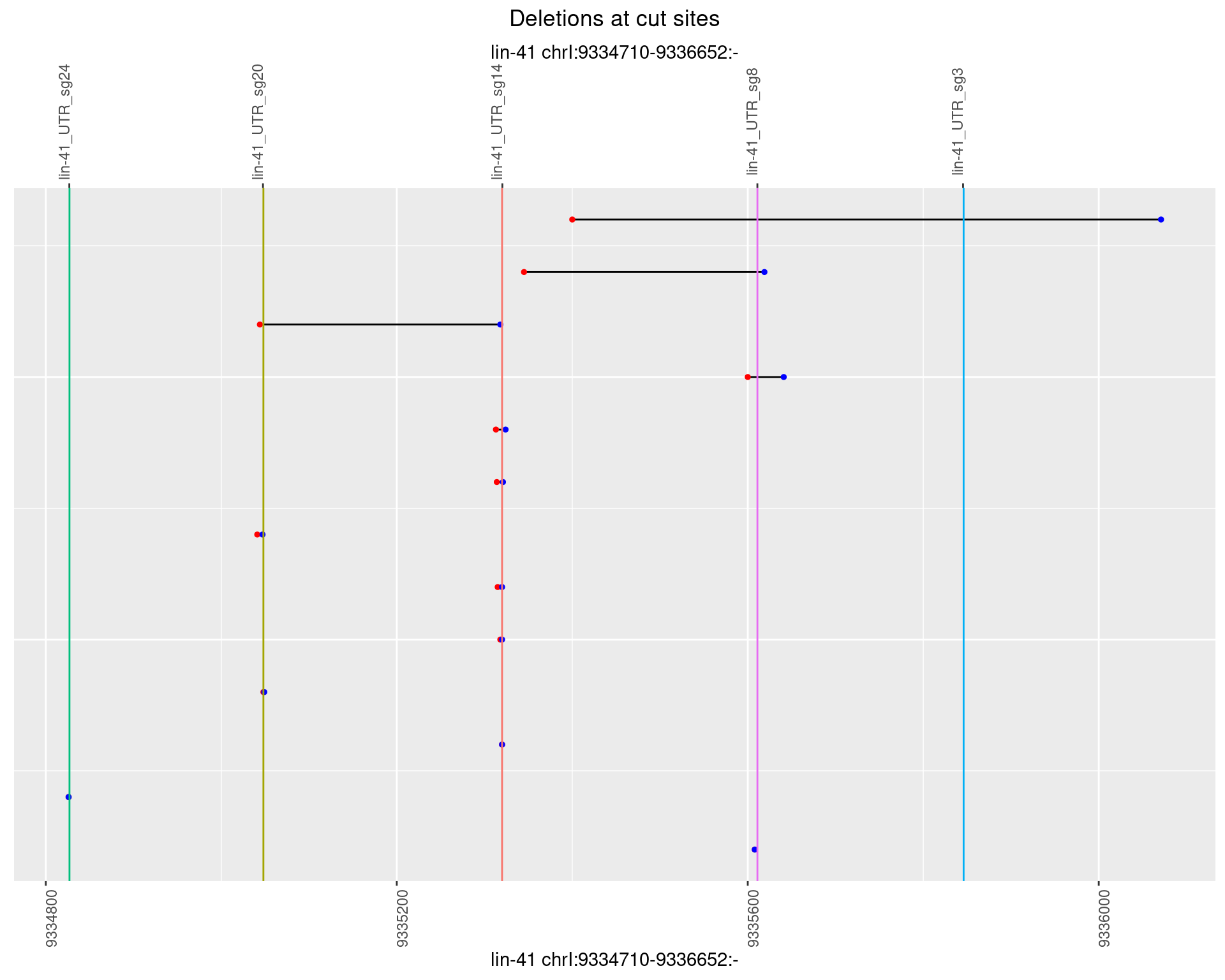
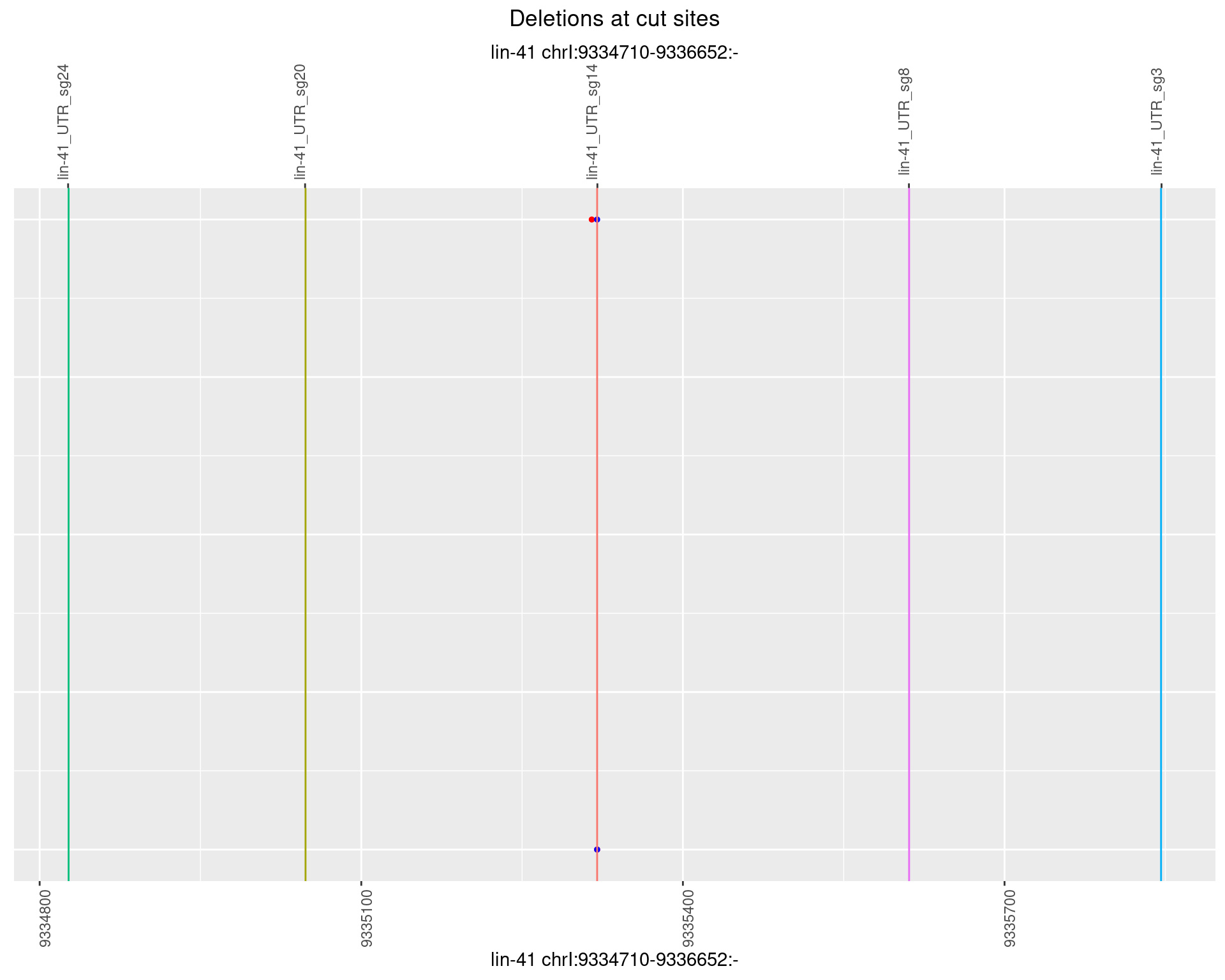
1.66.2 Freq: 1e-05
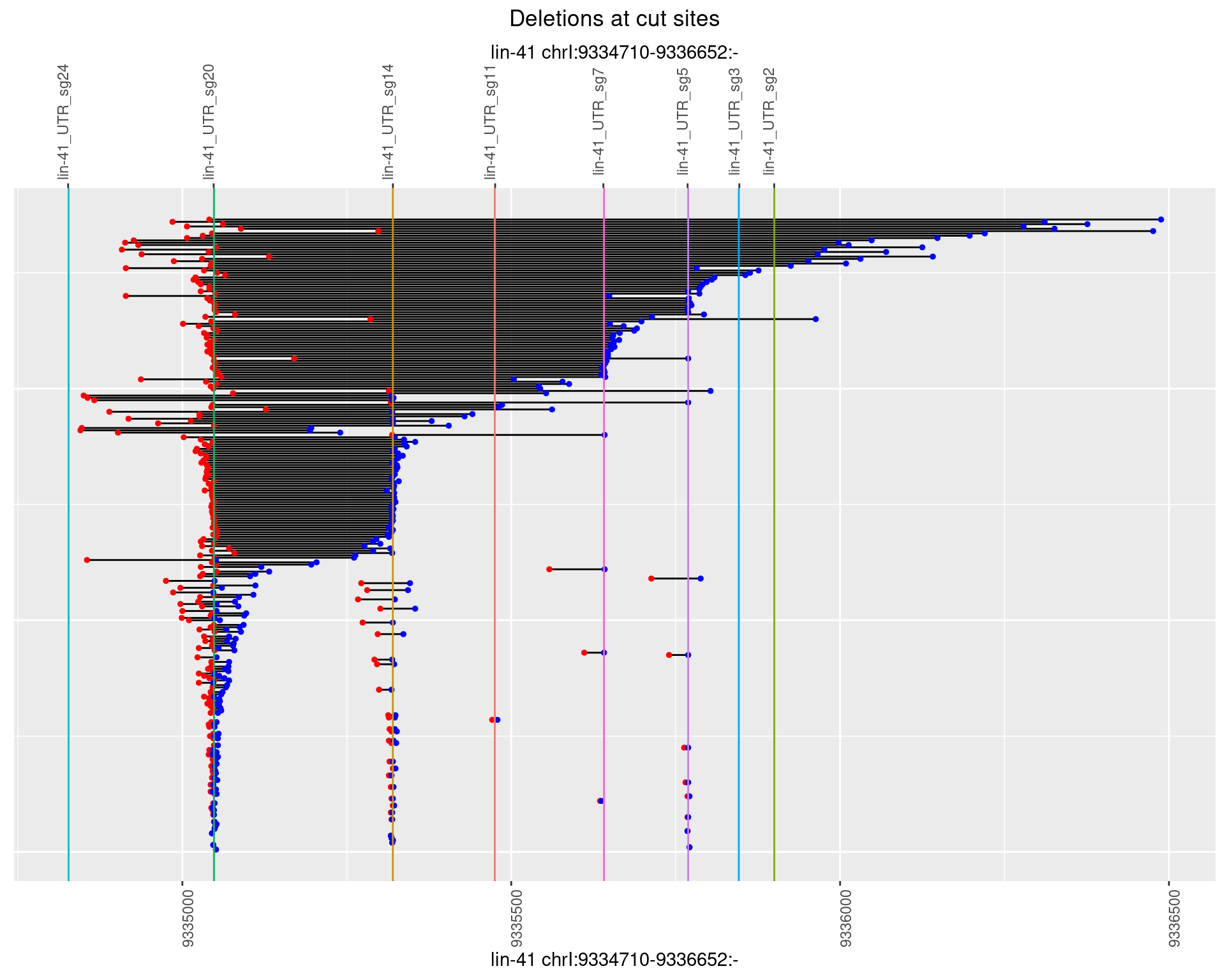
1.66.3 Freq: 1e-04
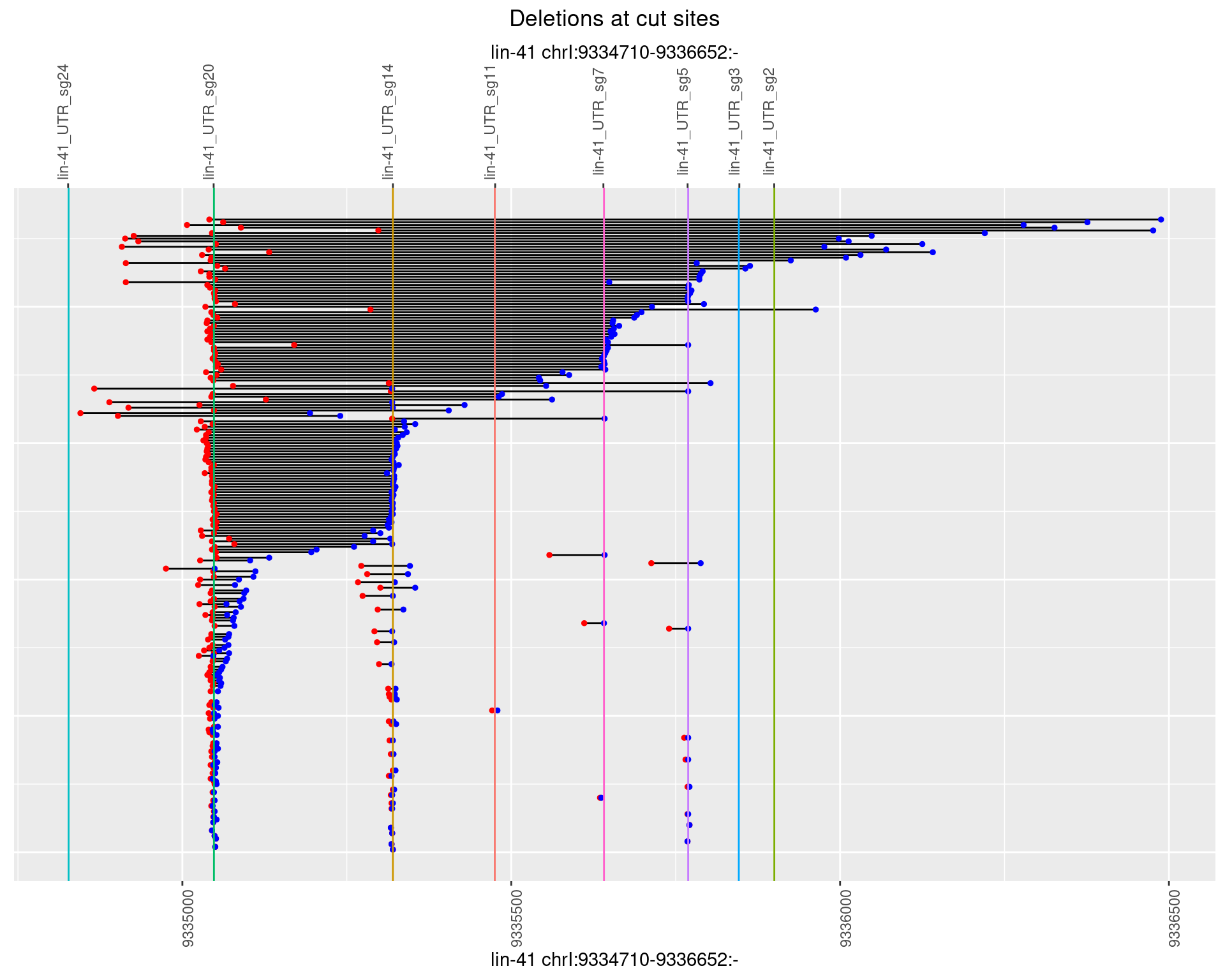
1.66.4 Freq: 0.001
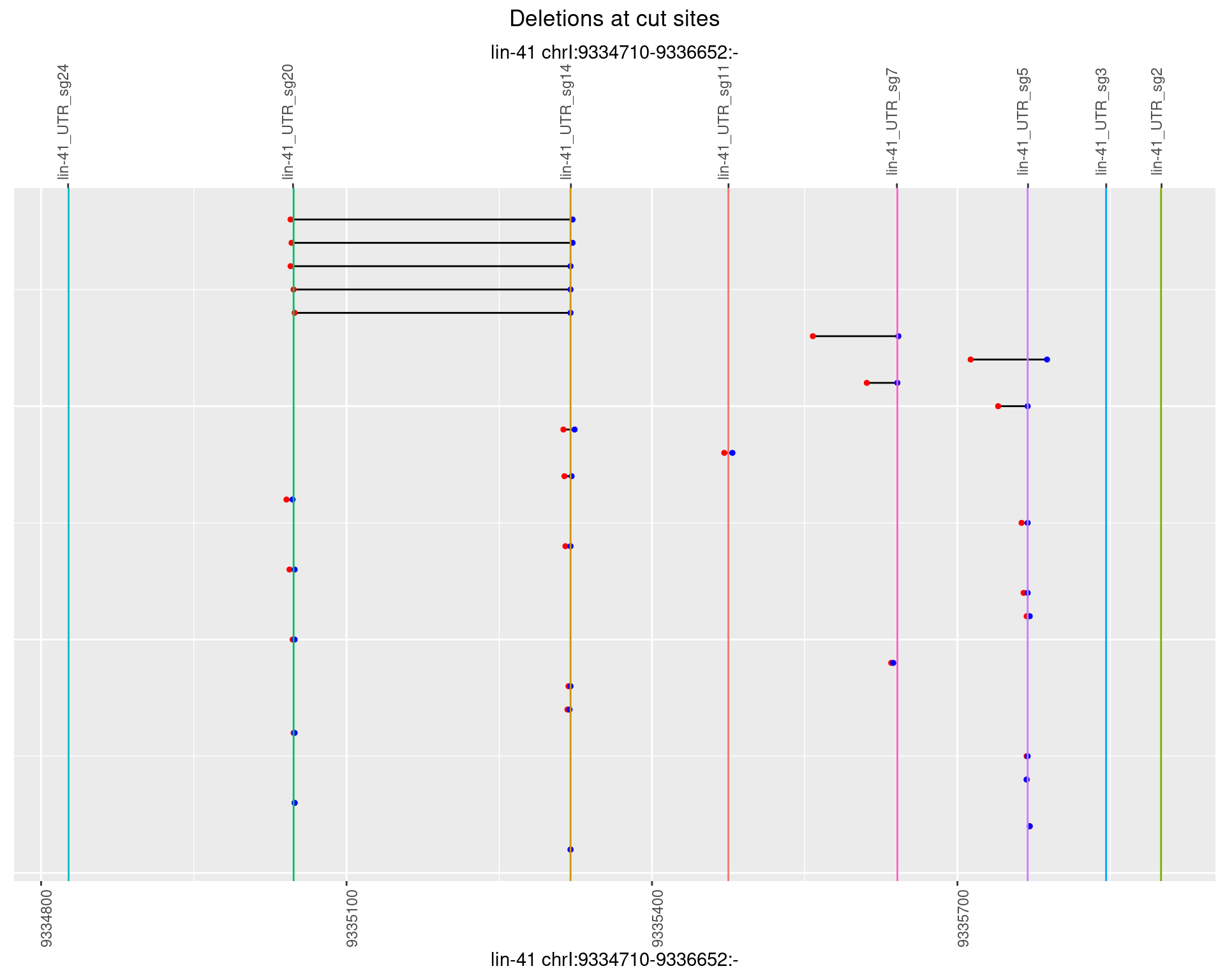
1.66.5 Freq: 0.01
1.66.6 Freq: 0.1
No plot to show
1.67 lin41_RNA_L4_N2_3end_UMI
1.67.1 Freq: 0
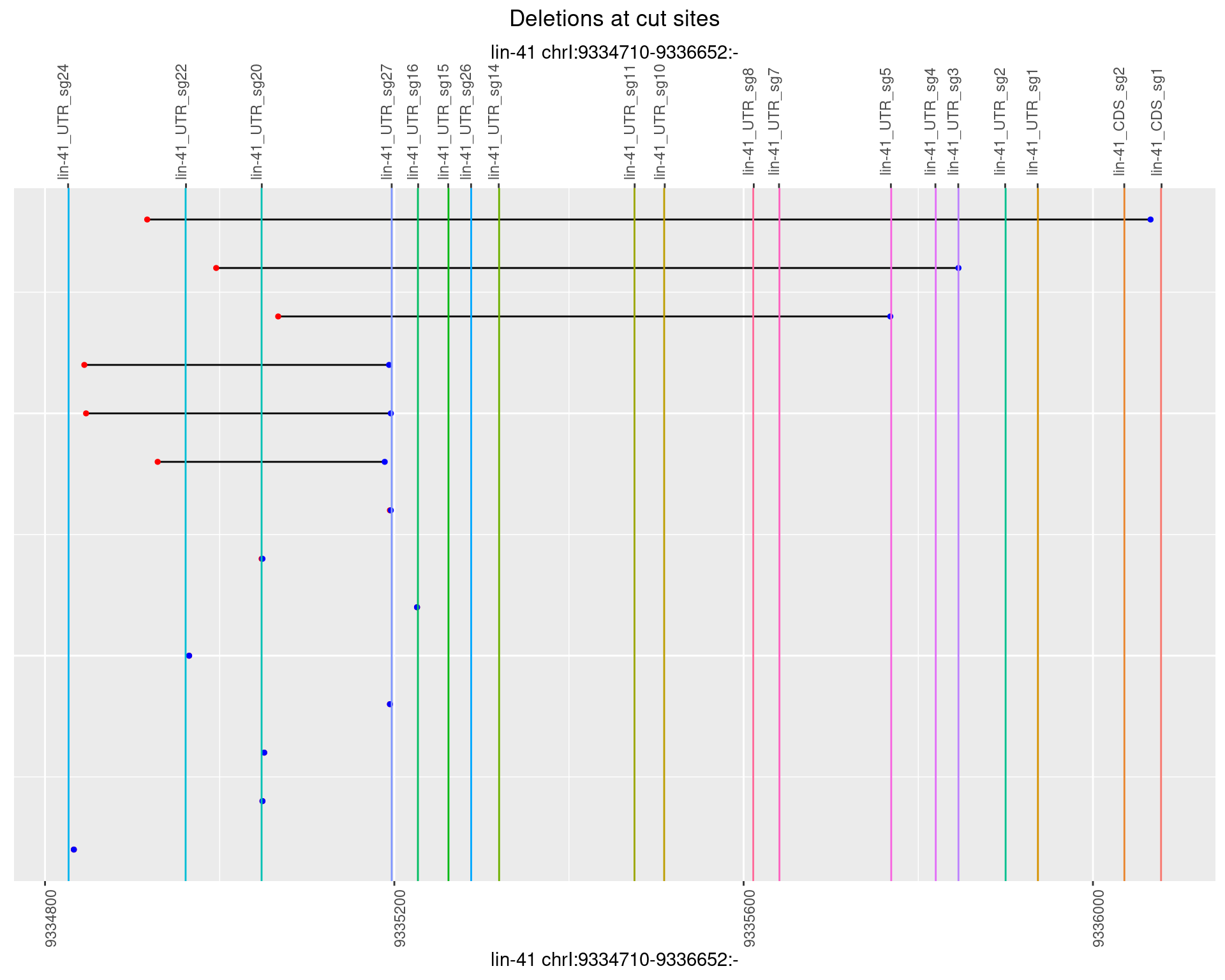
1.67.2 Freq: 1e-05
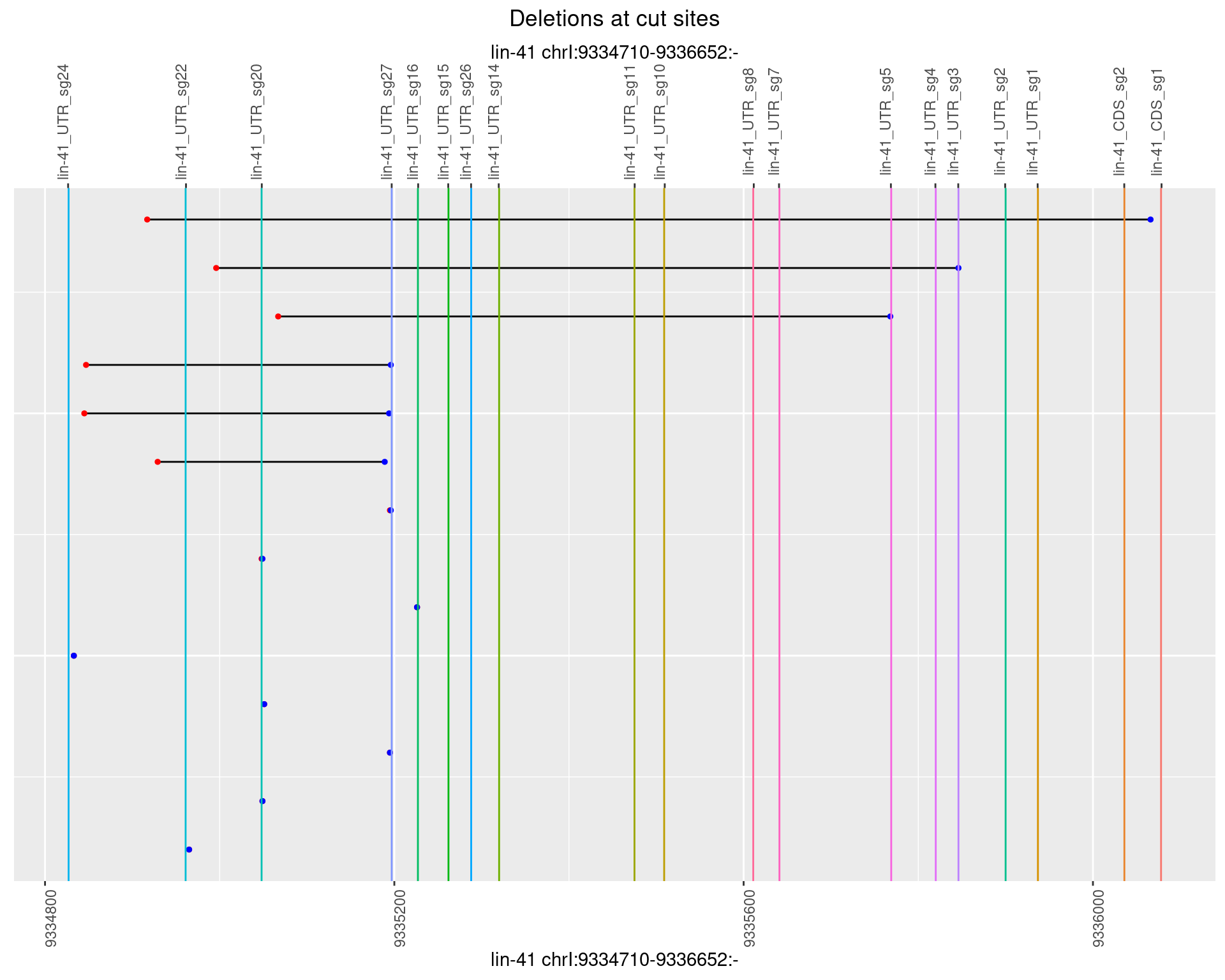
1.67.3 Freq: 1e-04
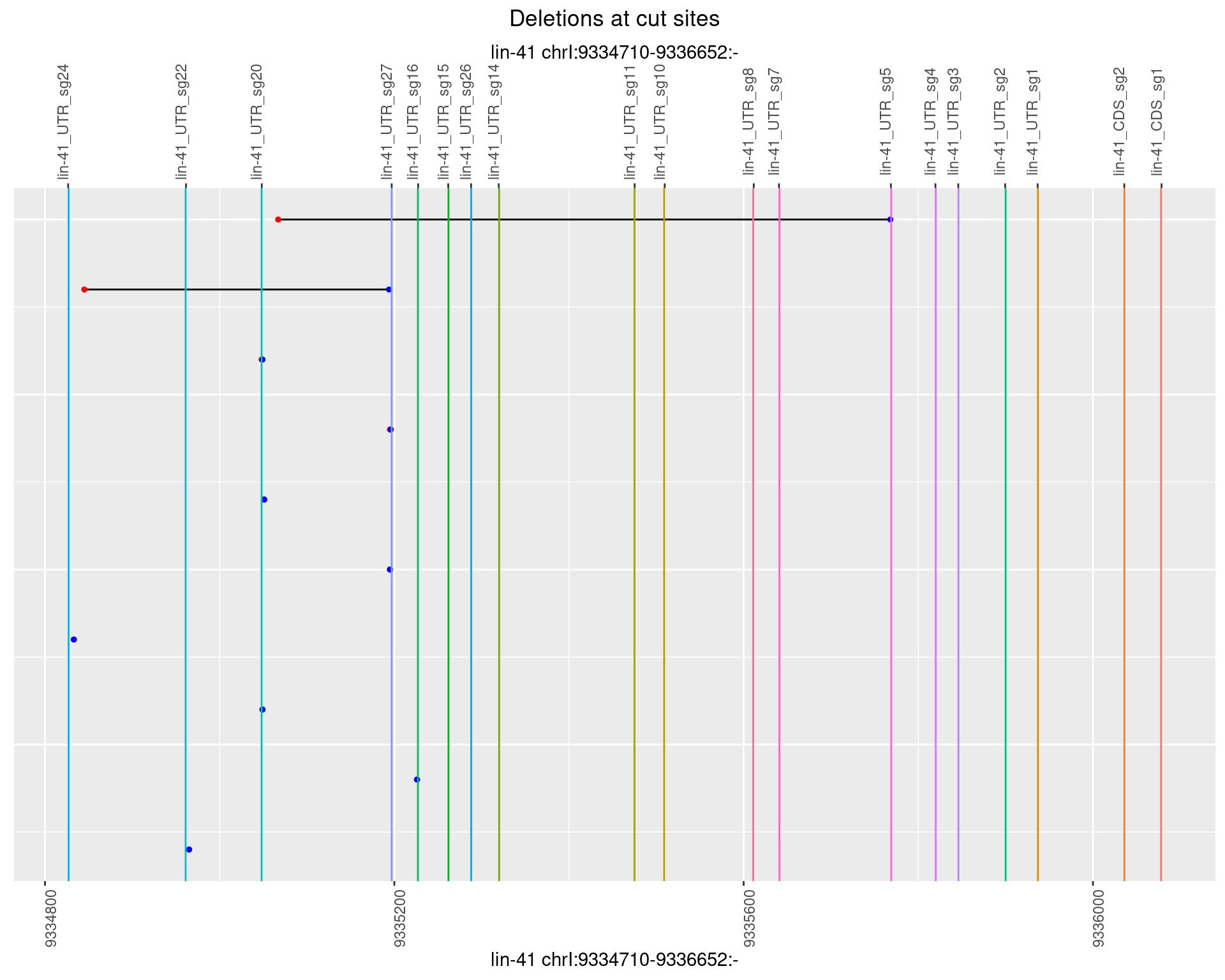
1.67.4 Freq: 0.001
1.67.5 Freq: 0.01
No plot to show
1.67.6 Freq: 0.1
No plot to show
1.68 lin41_RNA_L4_1516_3end_UMI
1.68.1 Freq: 0
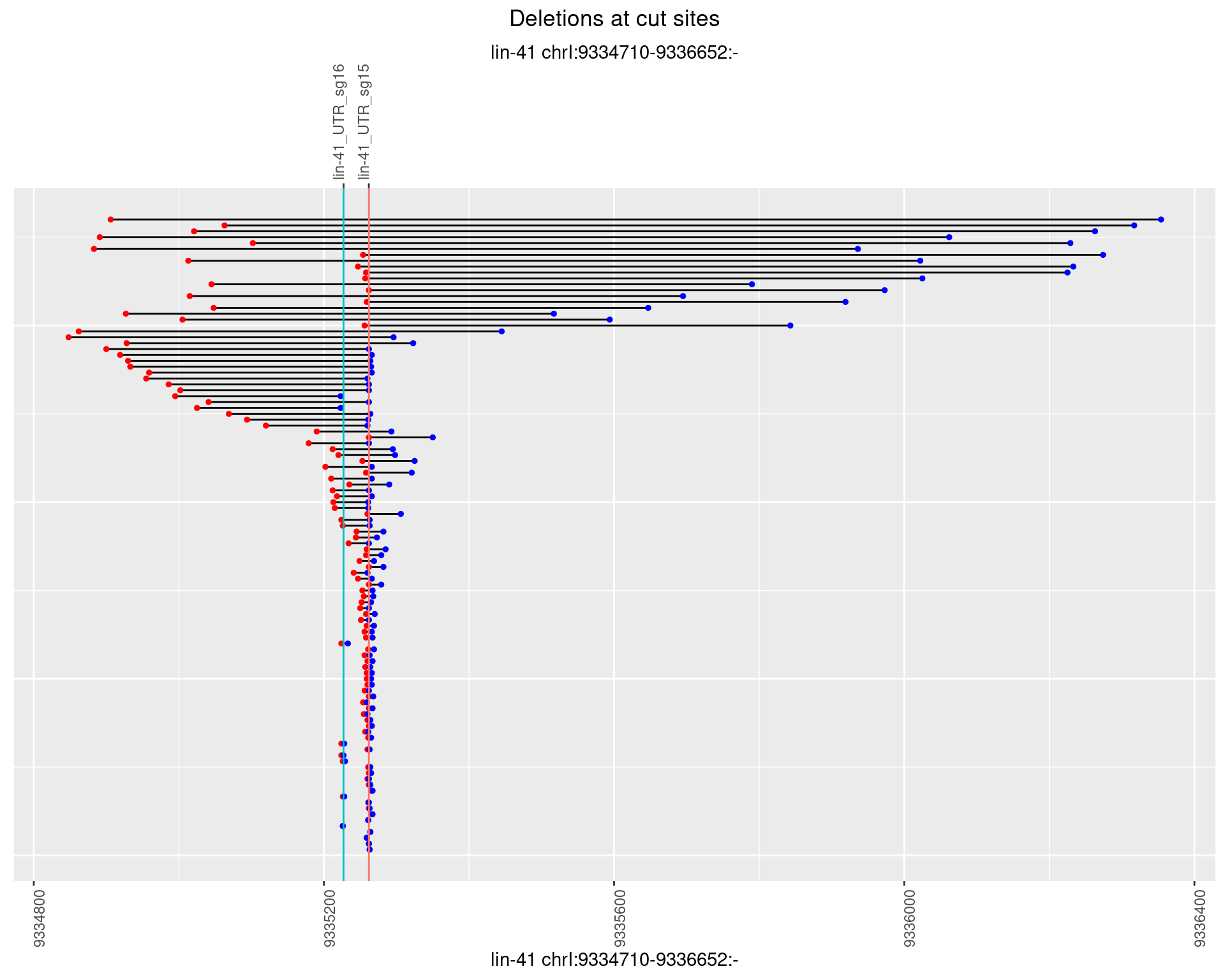
1.68.2 Freq: 1e-05
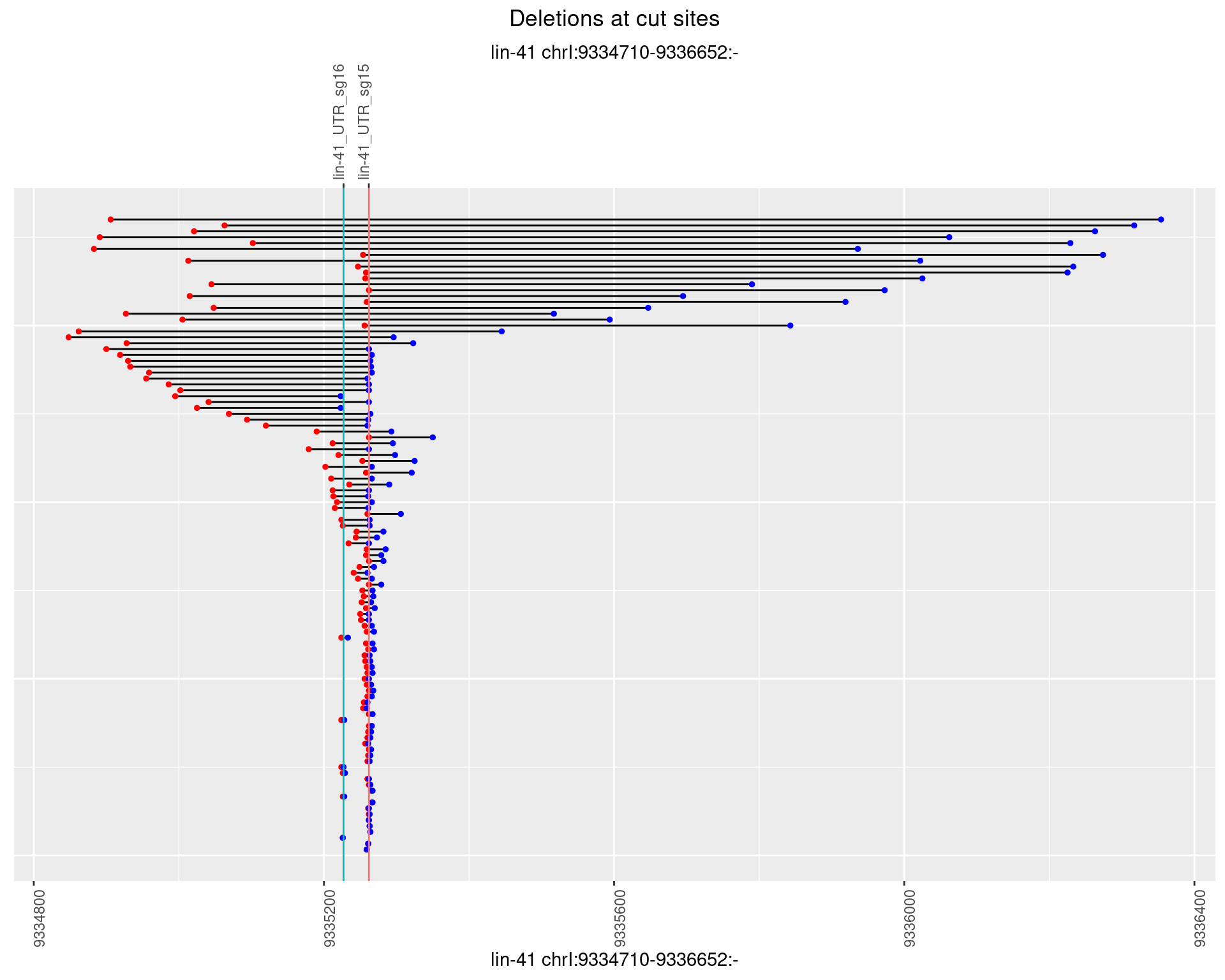
1.68.3 Freq: 1e-04
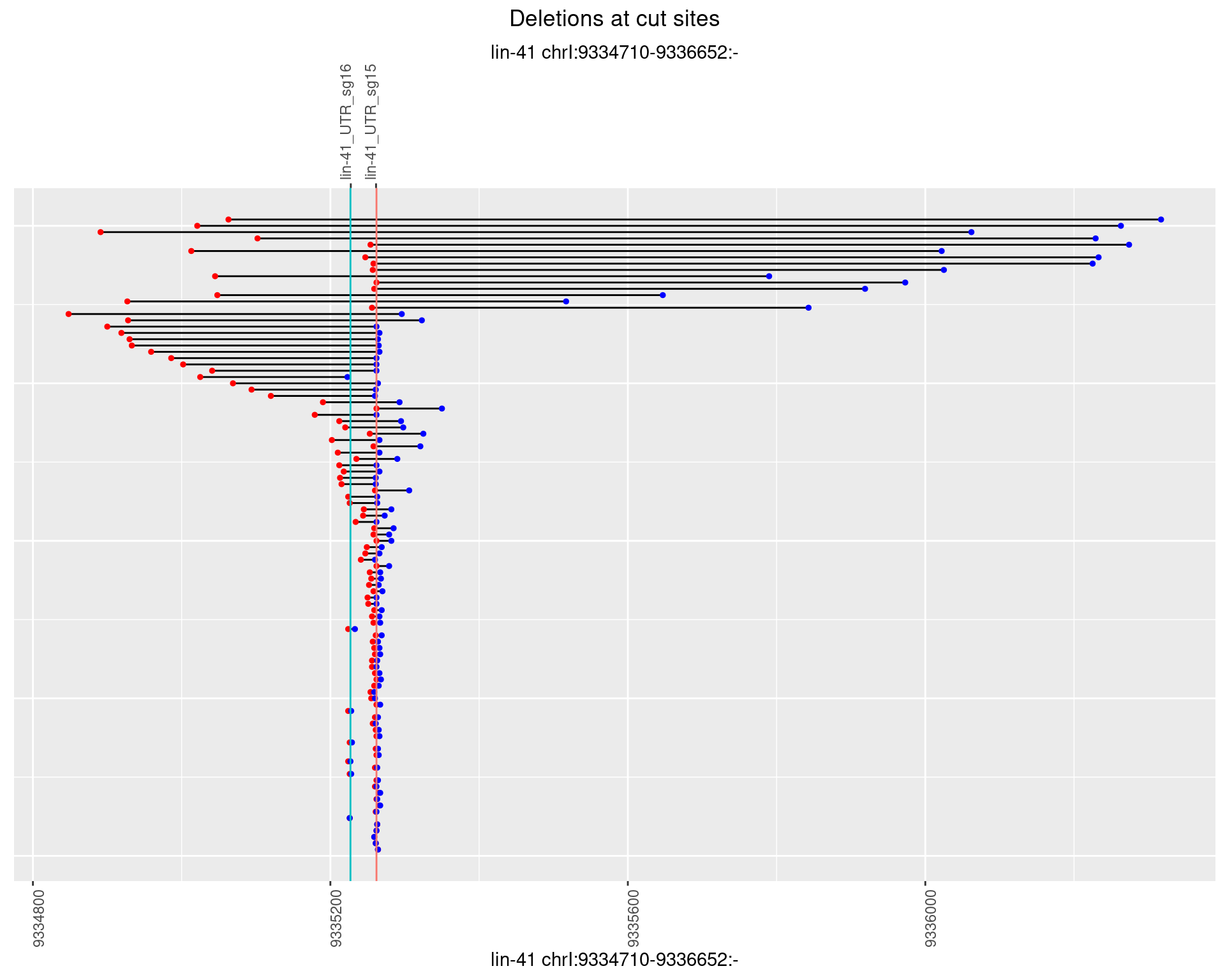
1.68.4 Freq: 0.001
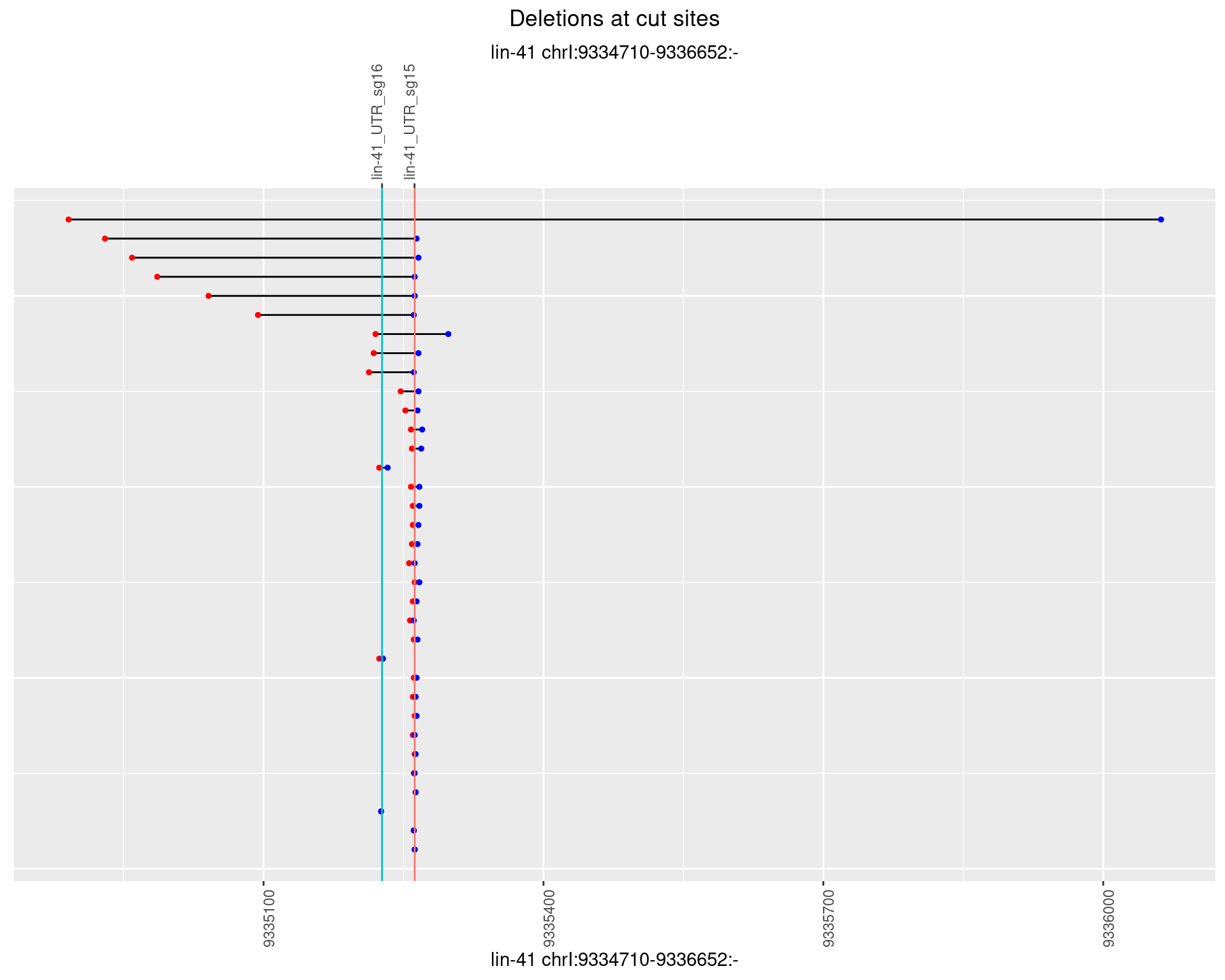
1.68.5 Freq: 0.01
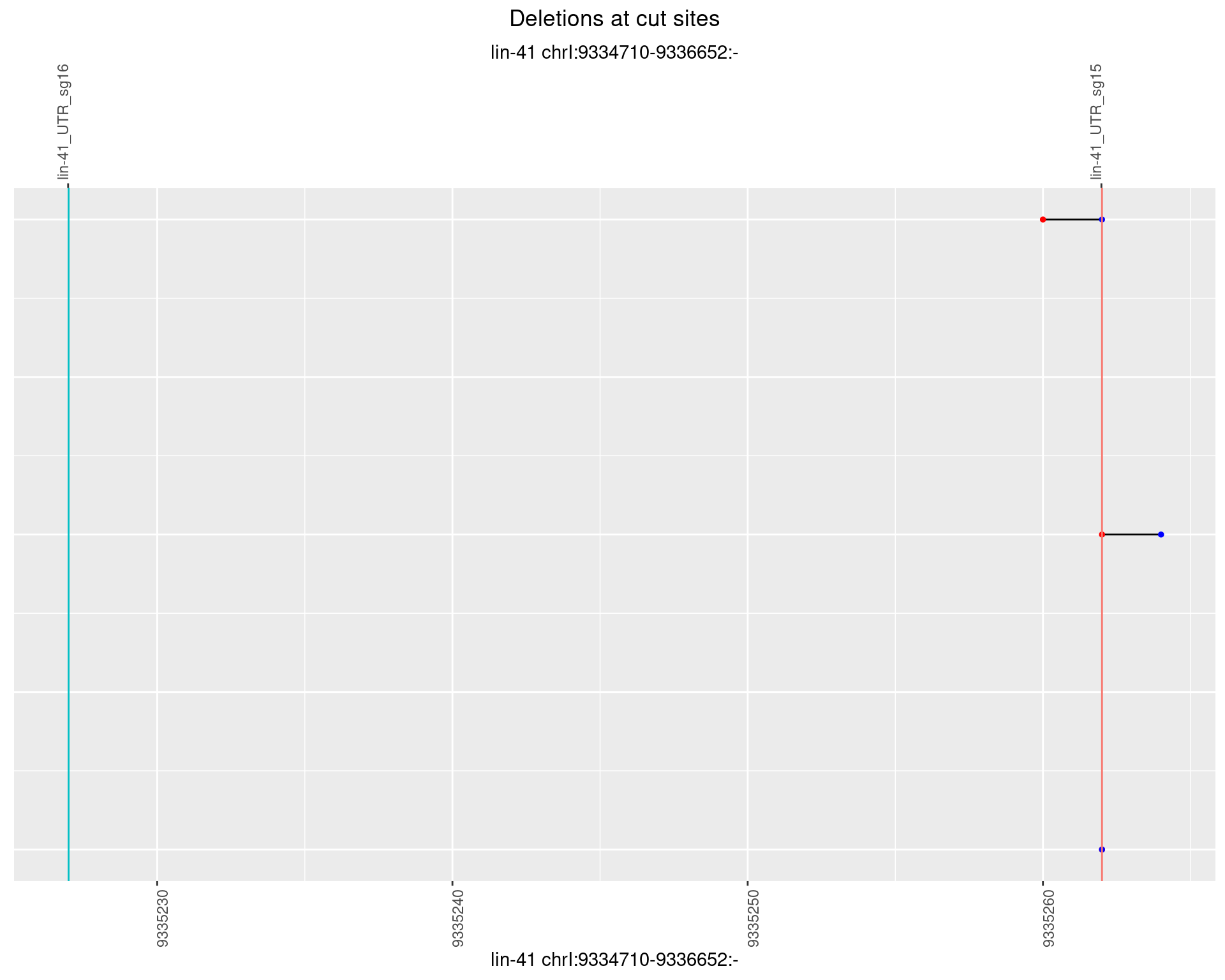
1.68.6 Freq: 0.1
No plot to show
1.69 lin41_RNA_L4_2627_3end_UMI
1.69.1 Freq: 0
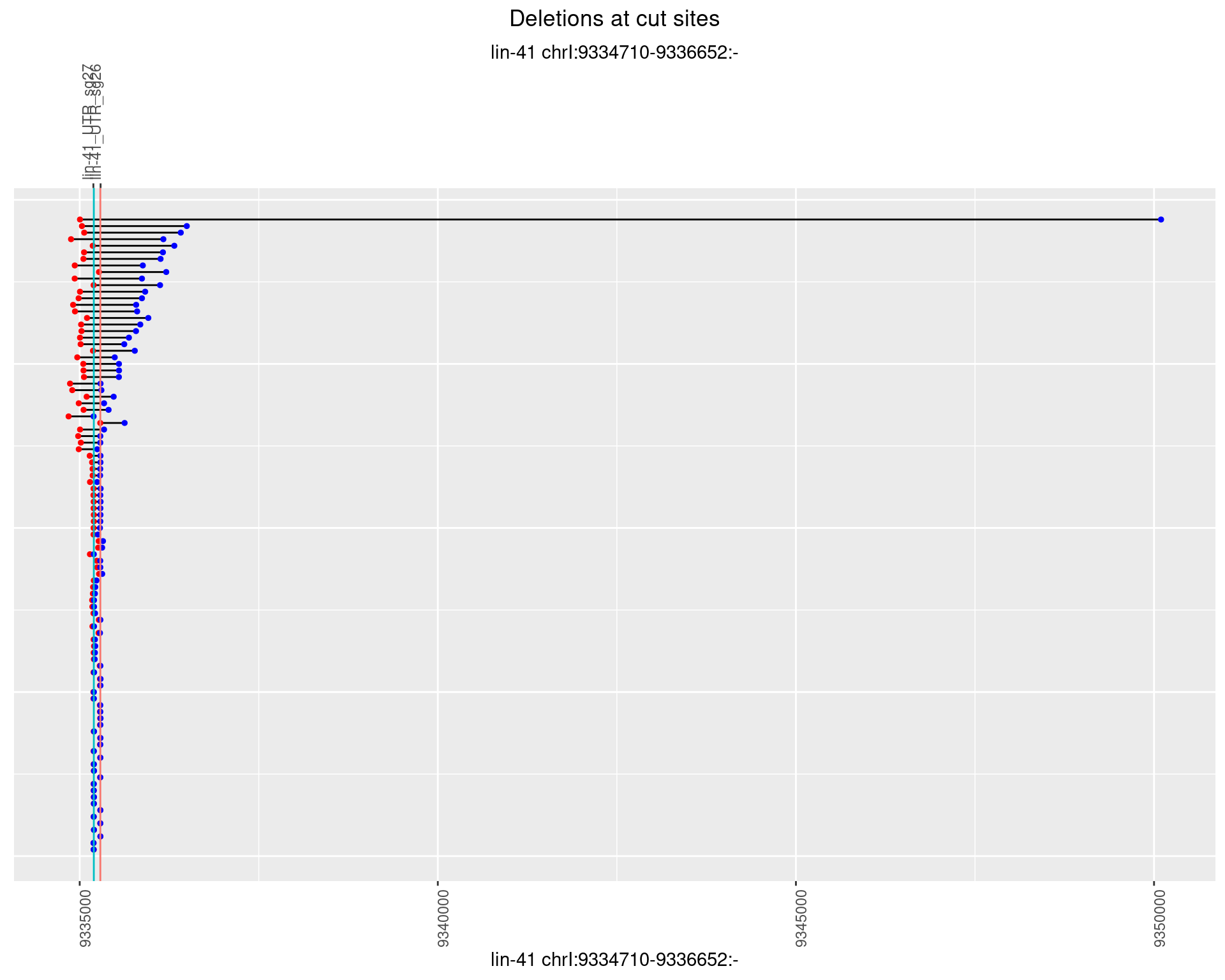
1.69.2 Freq: 1e-05
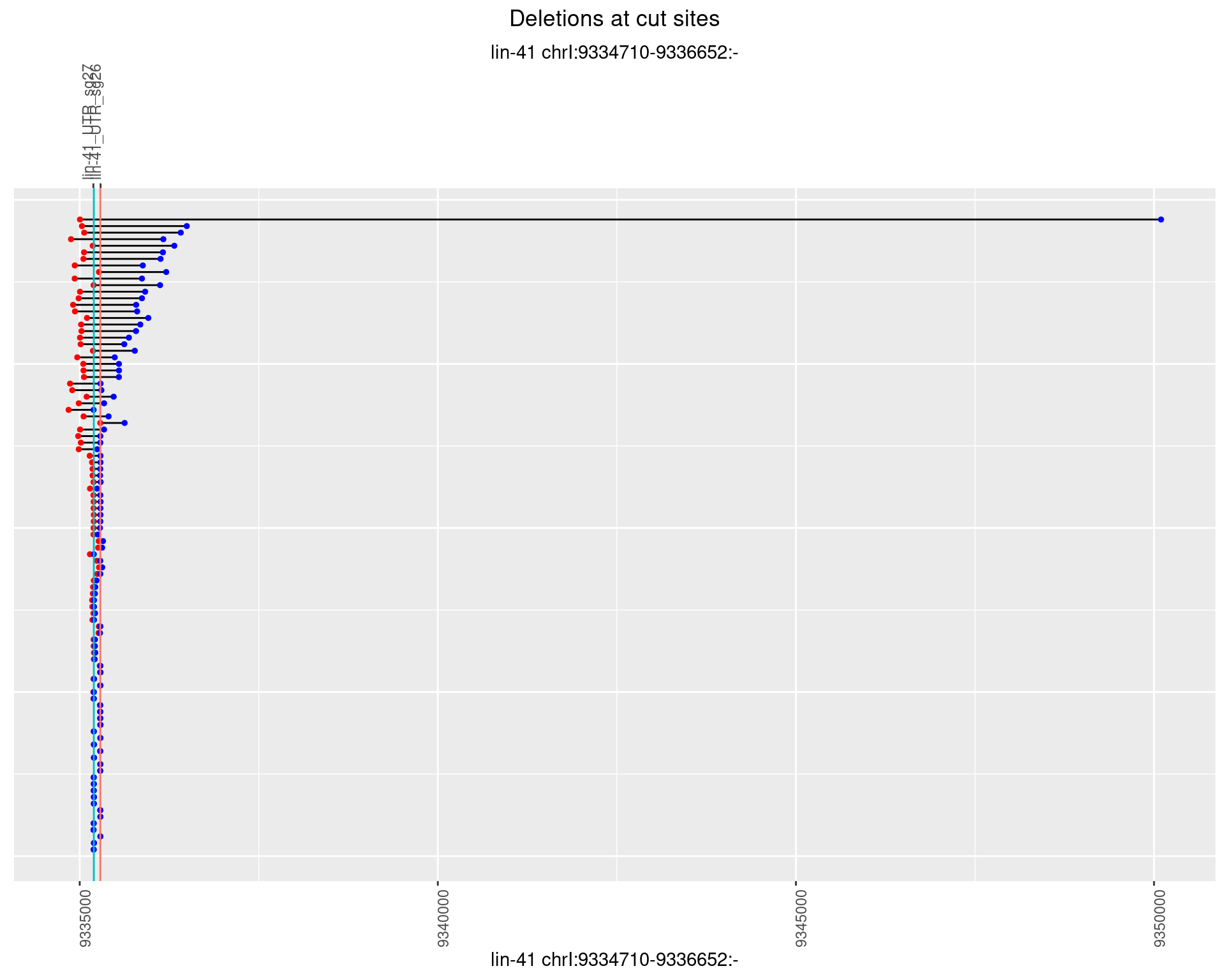
1.69.3 Freq: 1e-04
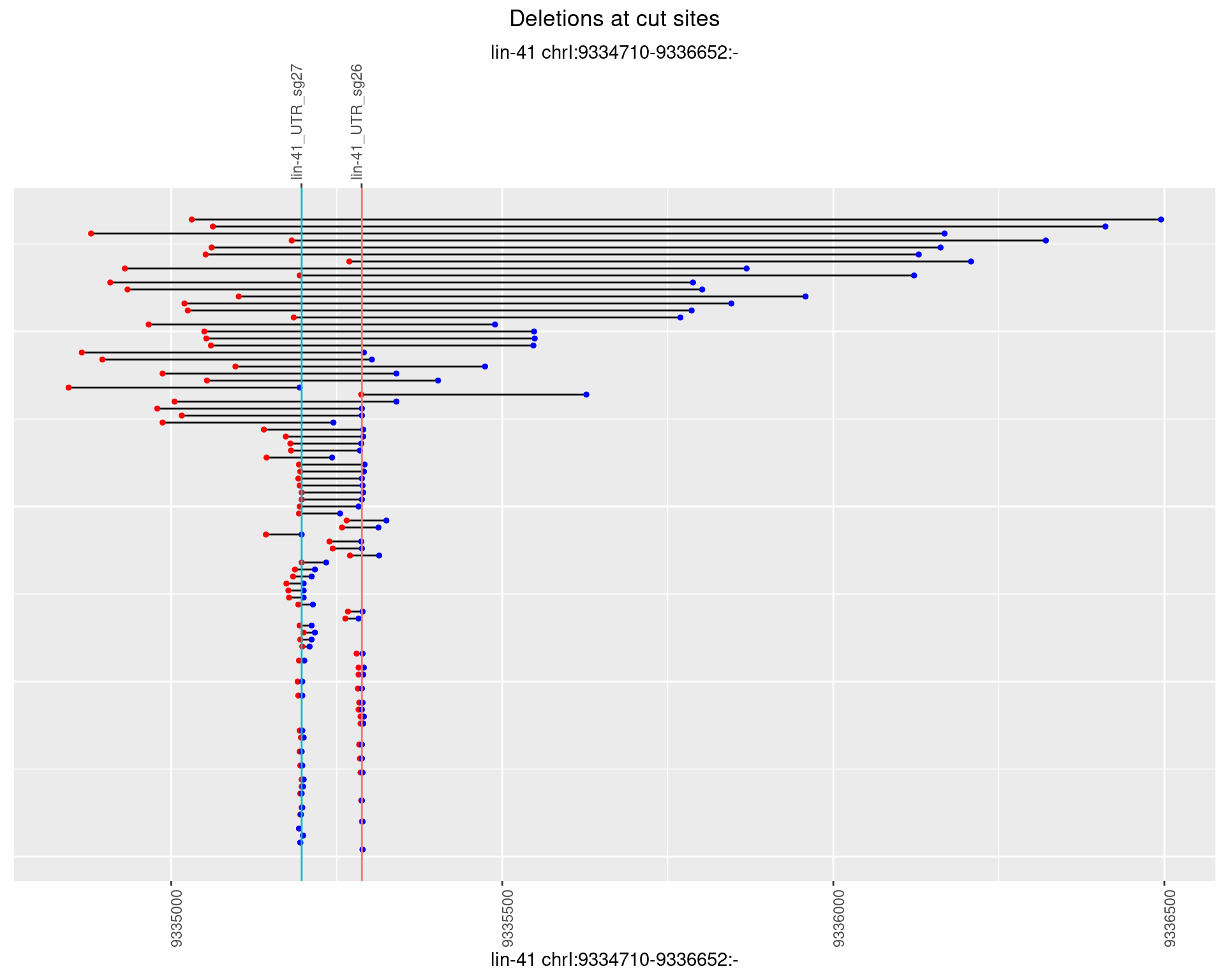
1.69.4 Freq: 0.001
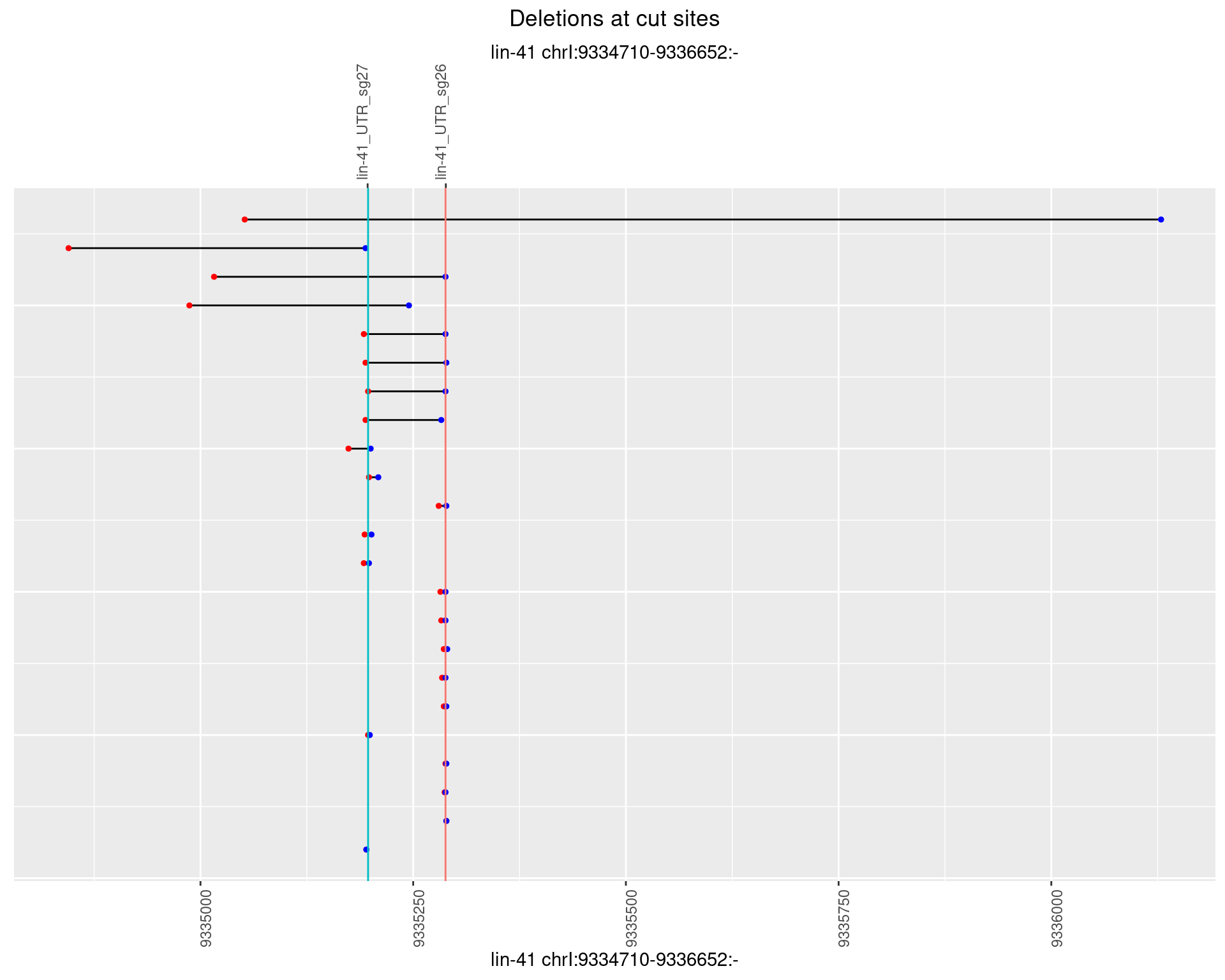
1.69.5 Freq: 0.01
No plot to show
1.69.6 Freq: 0.1
No plot to show
1.70 lin41_RNA_L4_pool2_3end_UMI
1.70.1 Freq: 0
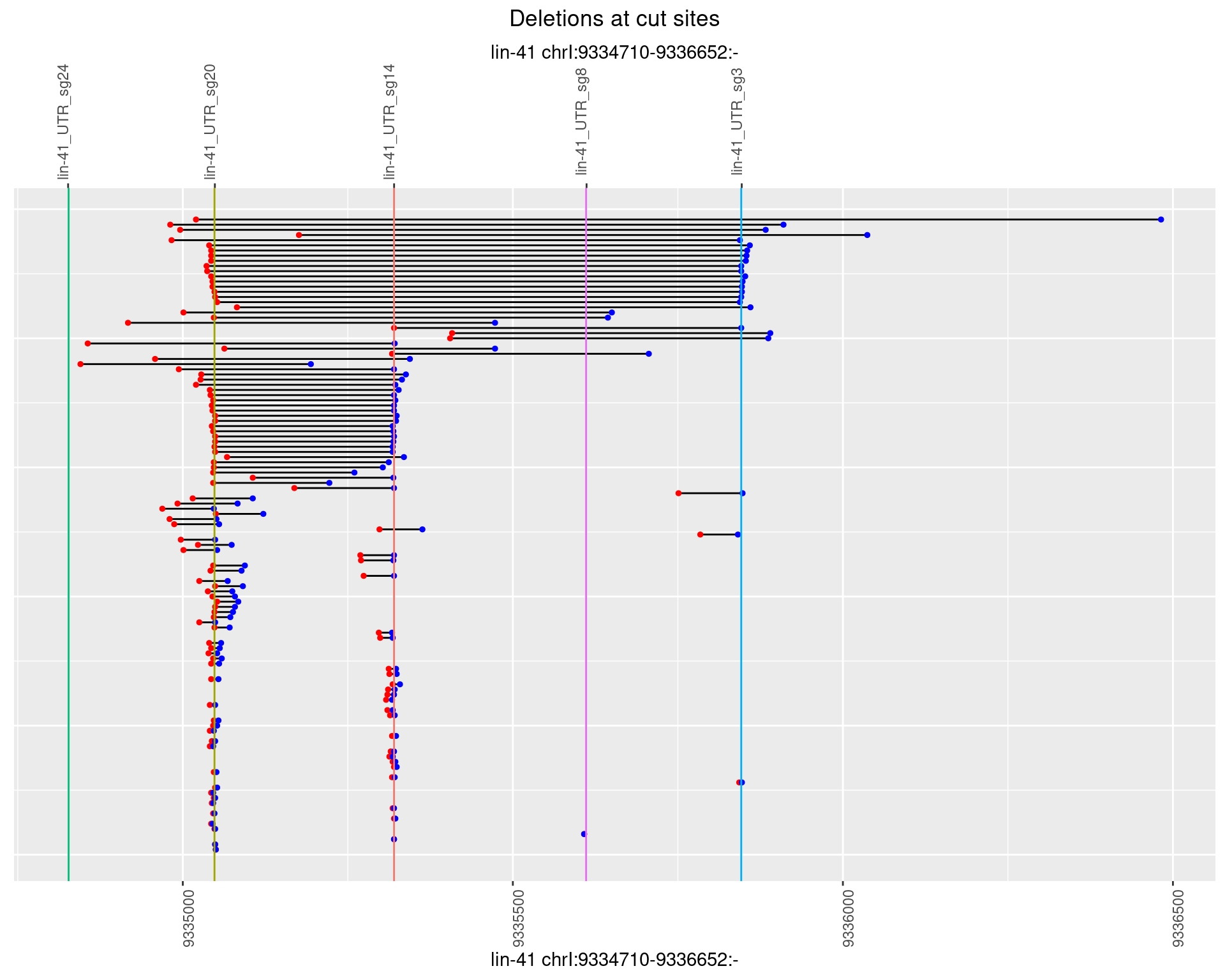
1.70.2 Freq: 1e-05
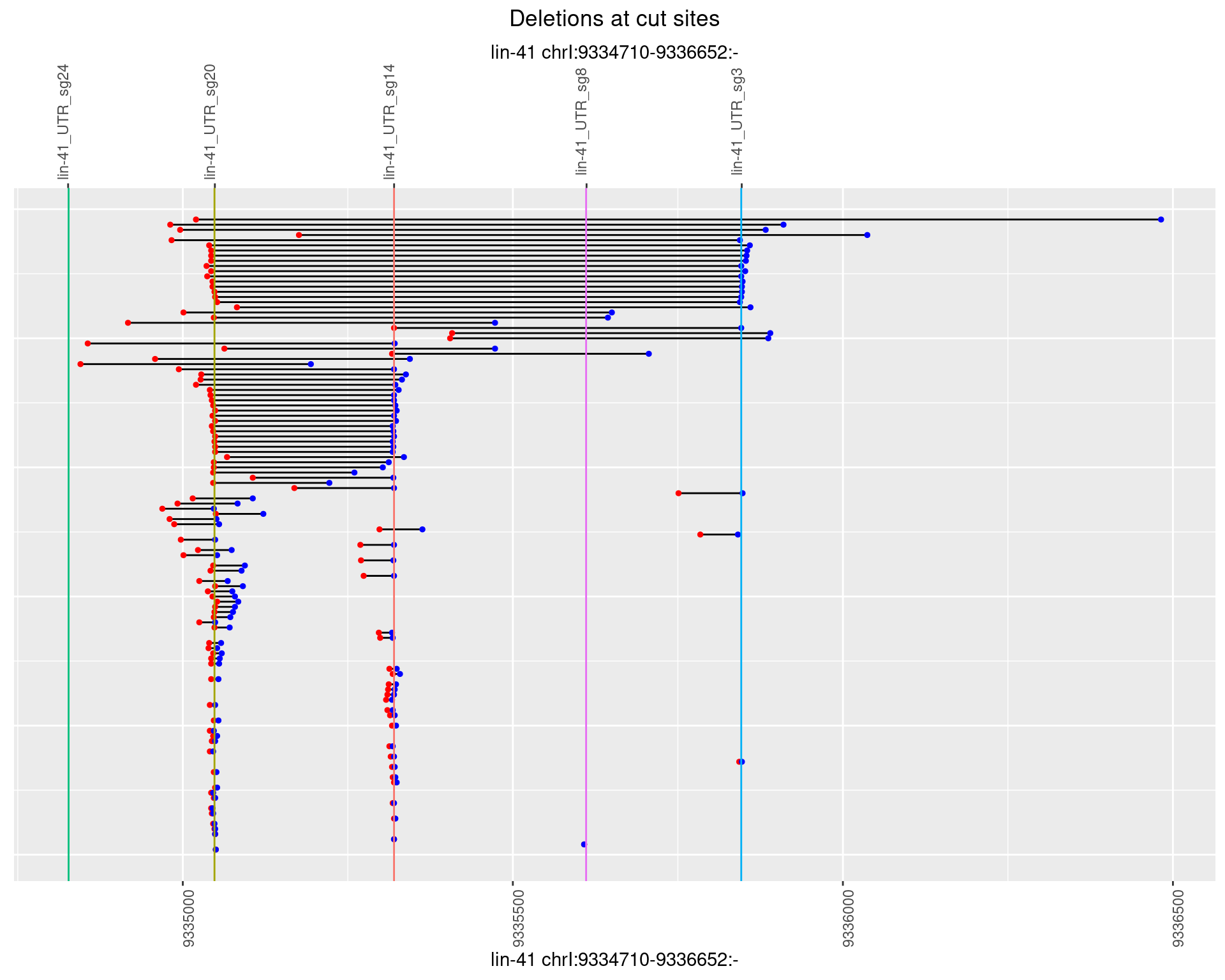
1.70.3 Freq: 1e-04
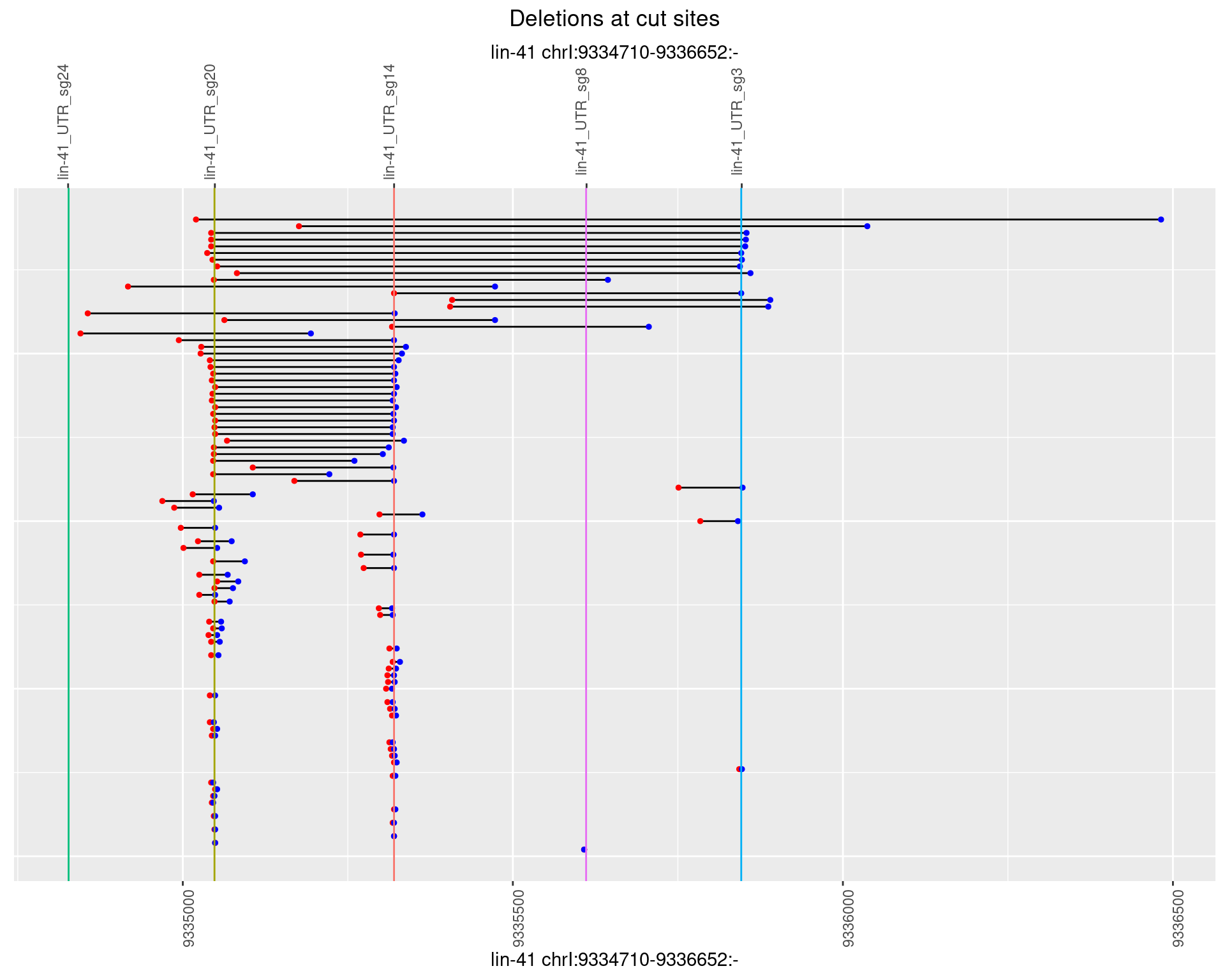
1.70.4 Freq: 0.001
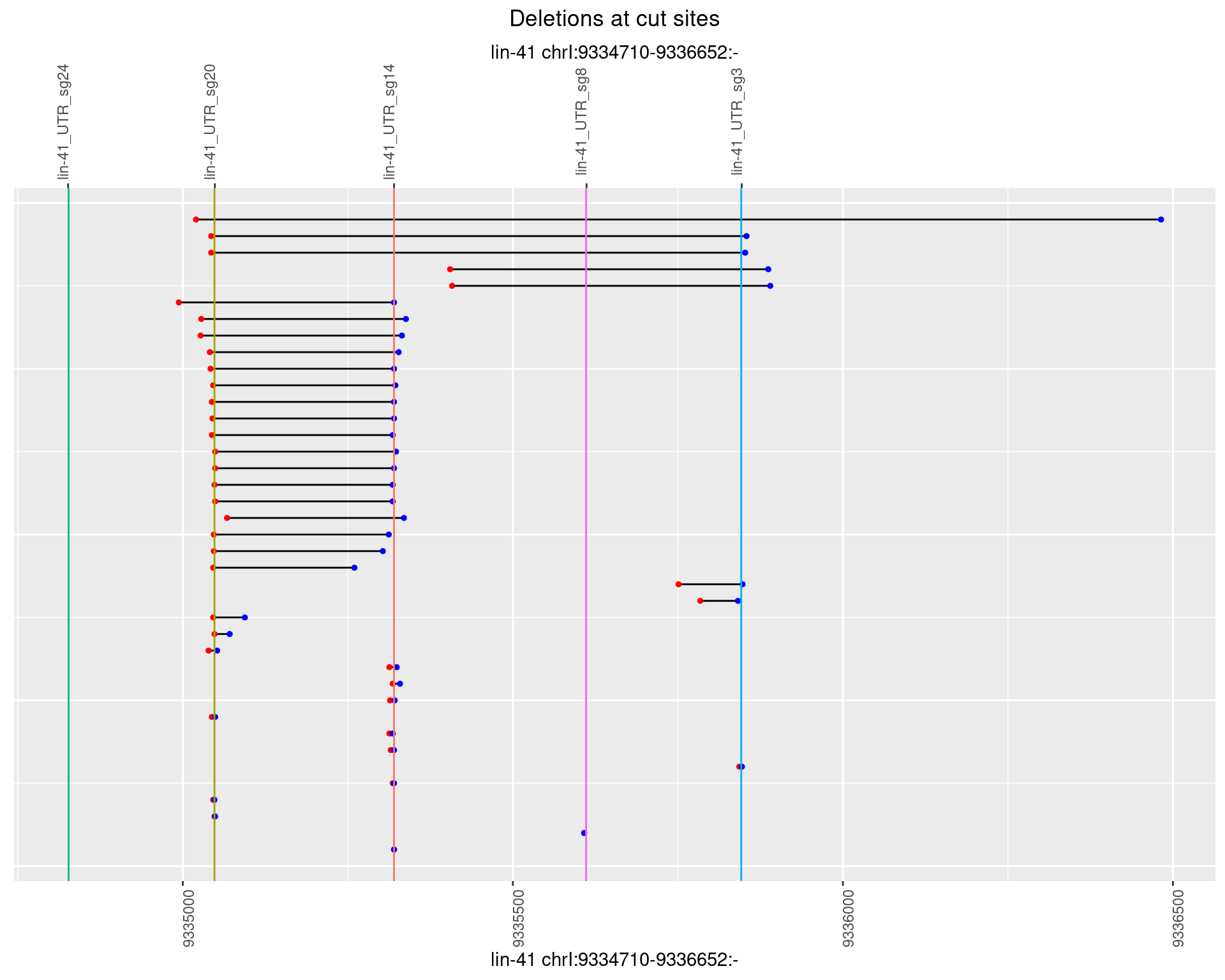
1.70.5 Freq: 0.01
1.70.6 Freq: 0.1
No plot to show
1.71 lin41_RNA_L4_pool3_3end_UMI
1.71.1 Freq: 0
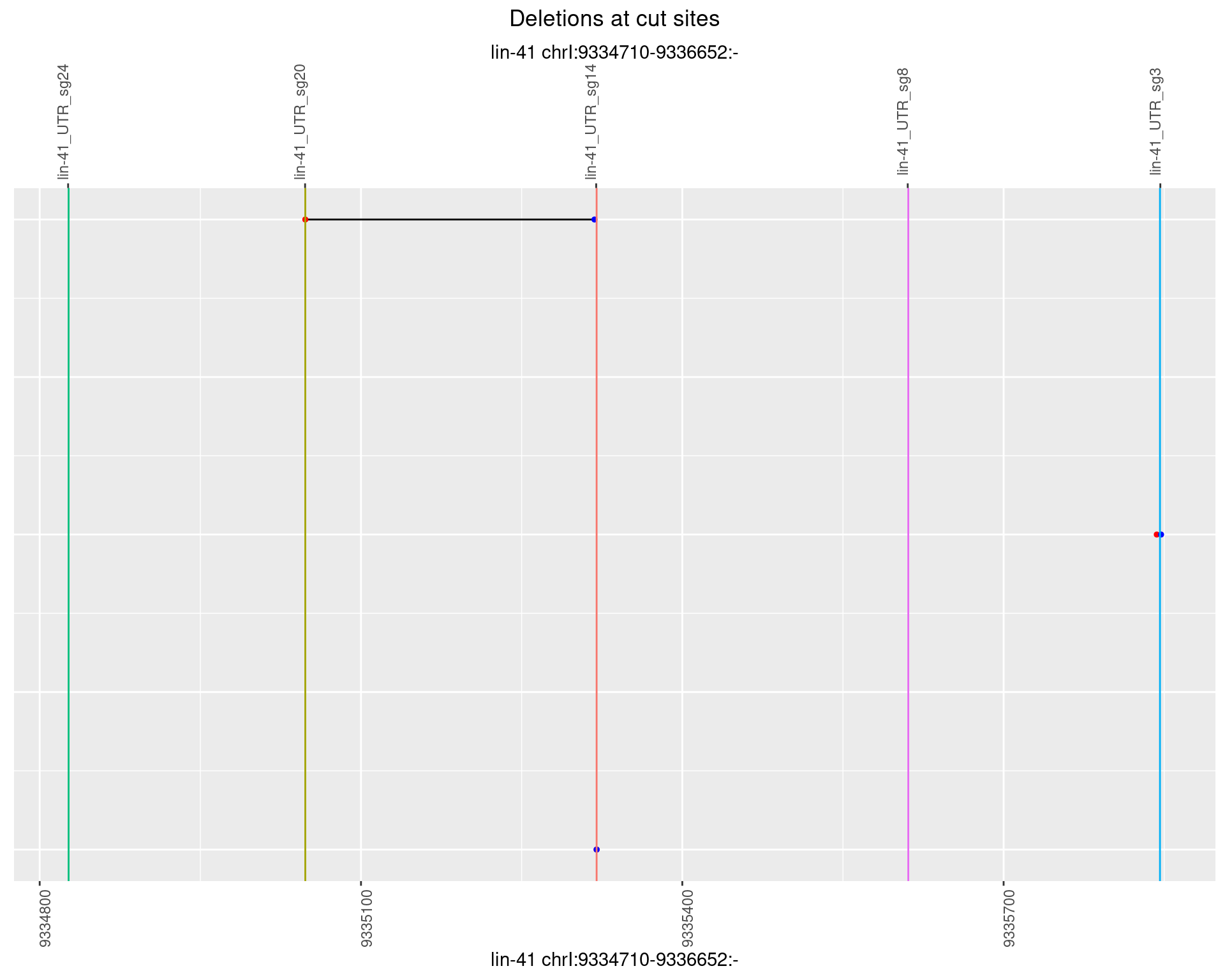
1.71.2 Freq: 1e-05
1.71.3 Freq: 1e-04
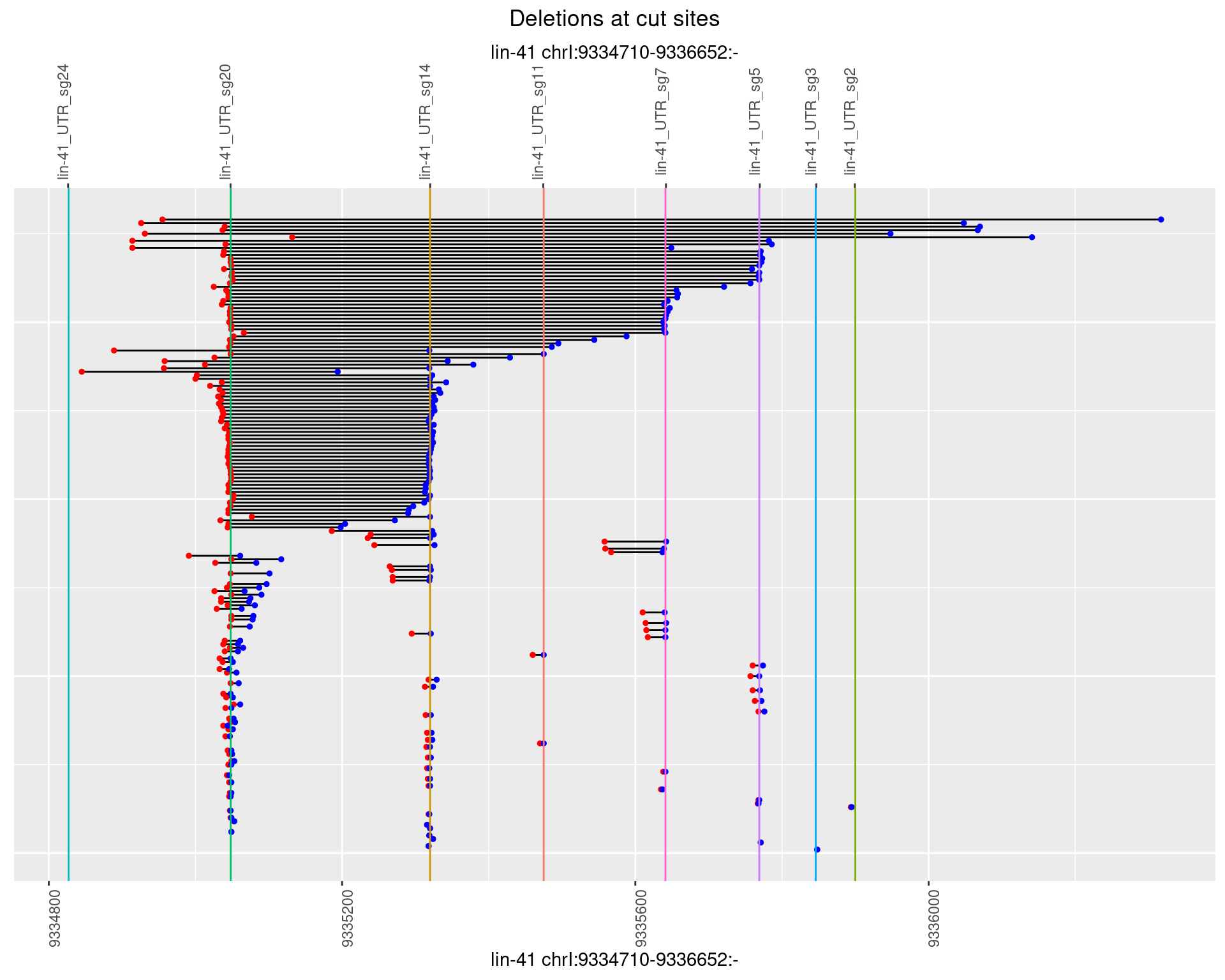
1.71.4 Freq: 0.001
1.71.5 Freq: 0.01
1.71.6 Freq: 0.1
No plot to show
1.72 lin41_RNA_L1_N2_middle_UMI
1.72.1 Freq: 0
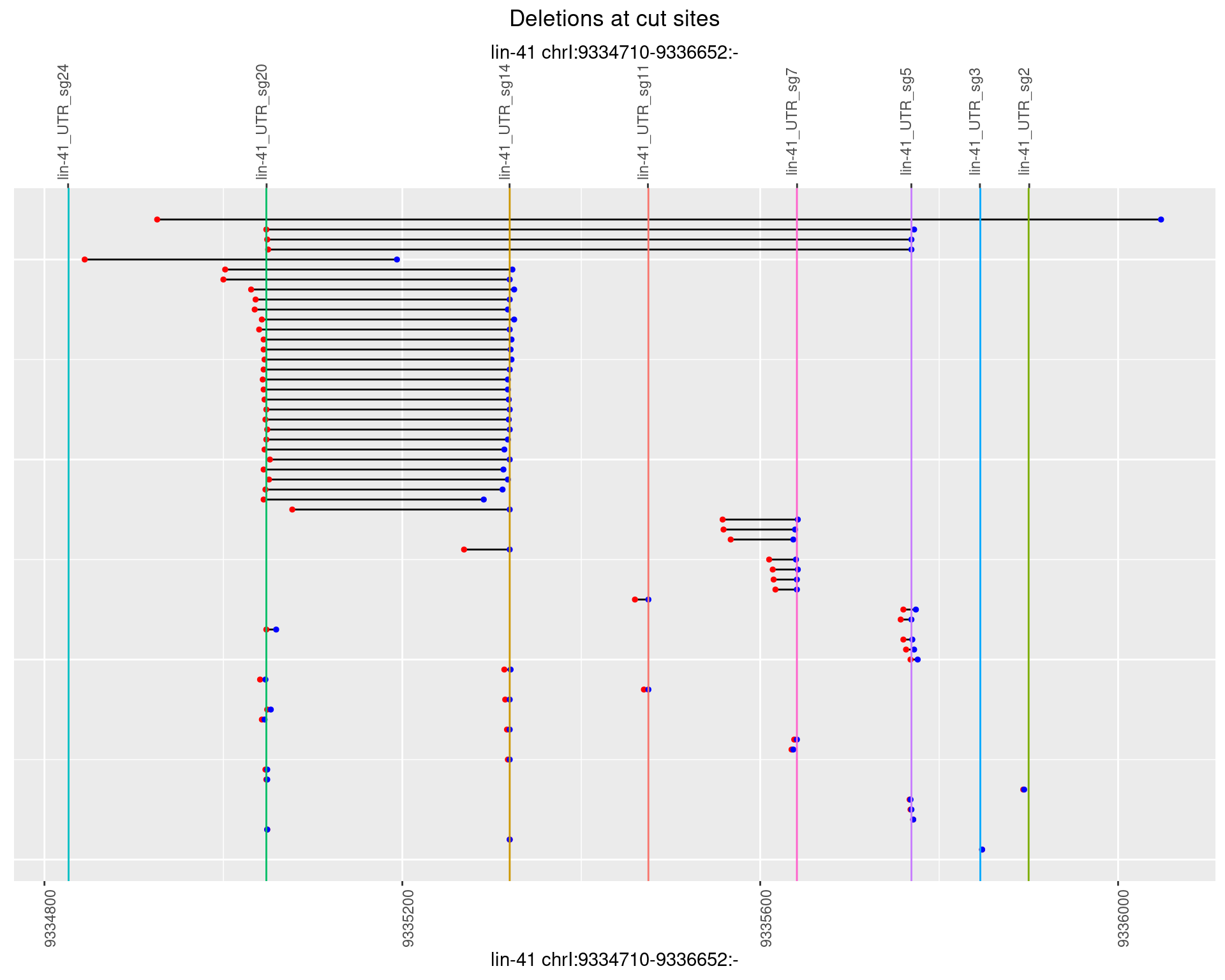
1.72.2 Freq: 1e-05
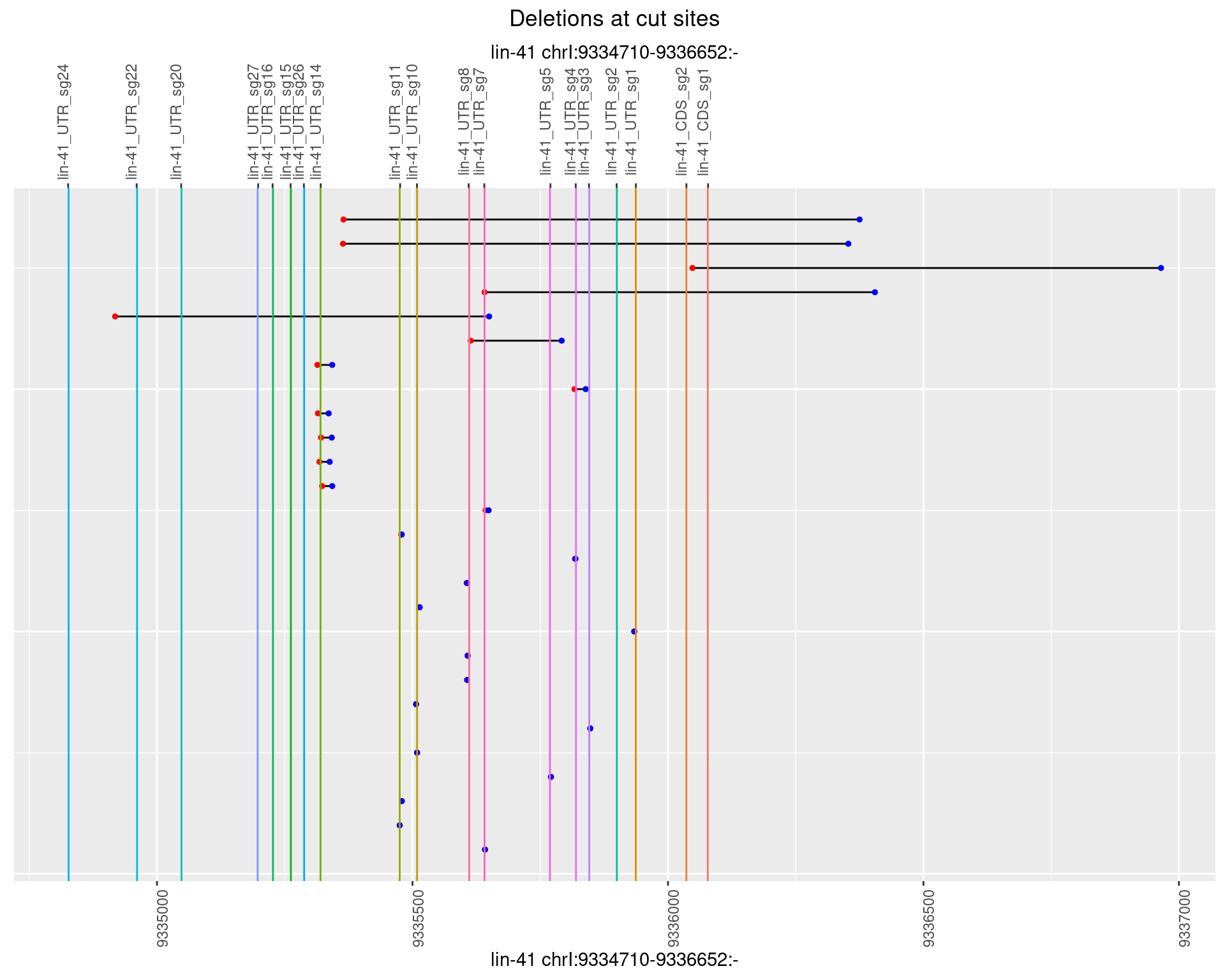
1.72.3 Freq: 1e-04
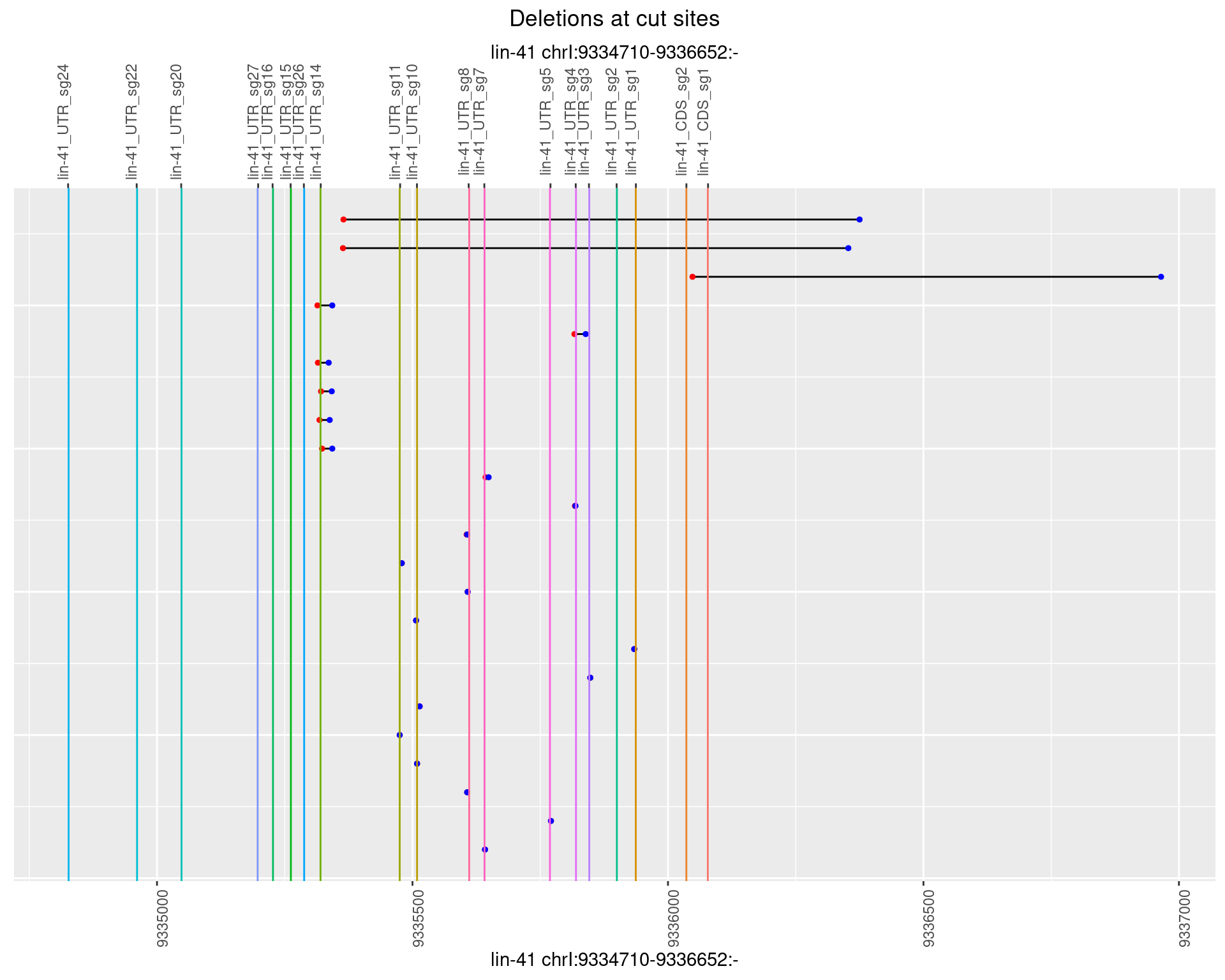
1.72.4 Freq: 0.001
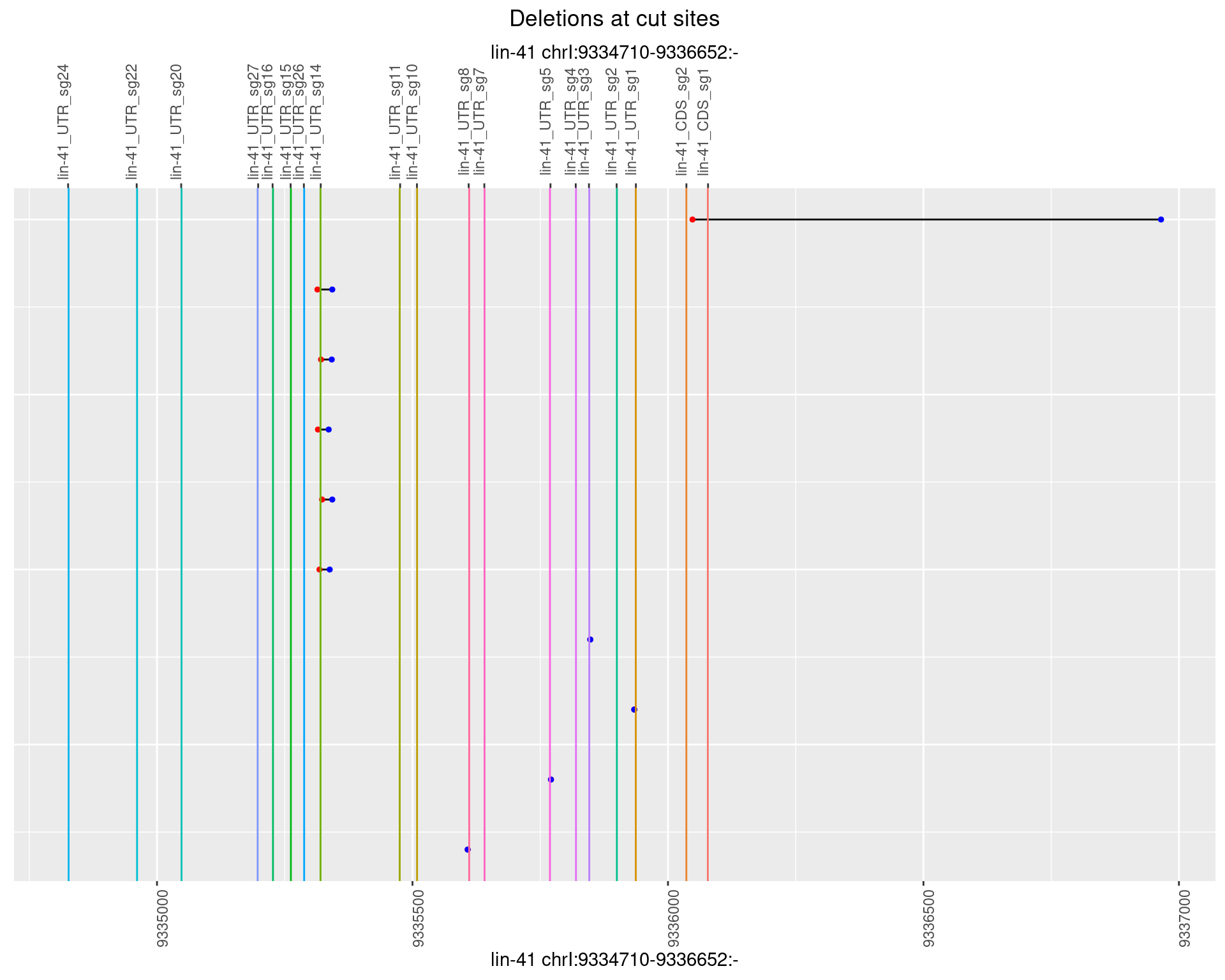
1.72.5 Freq: 0.01
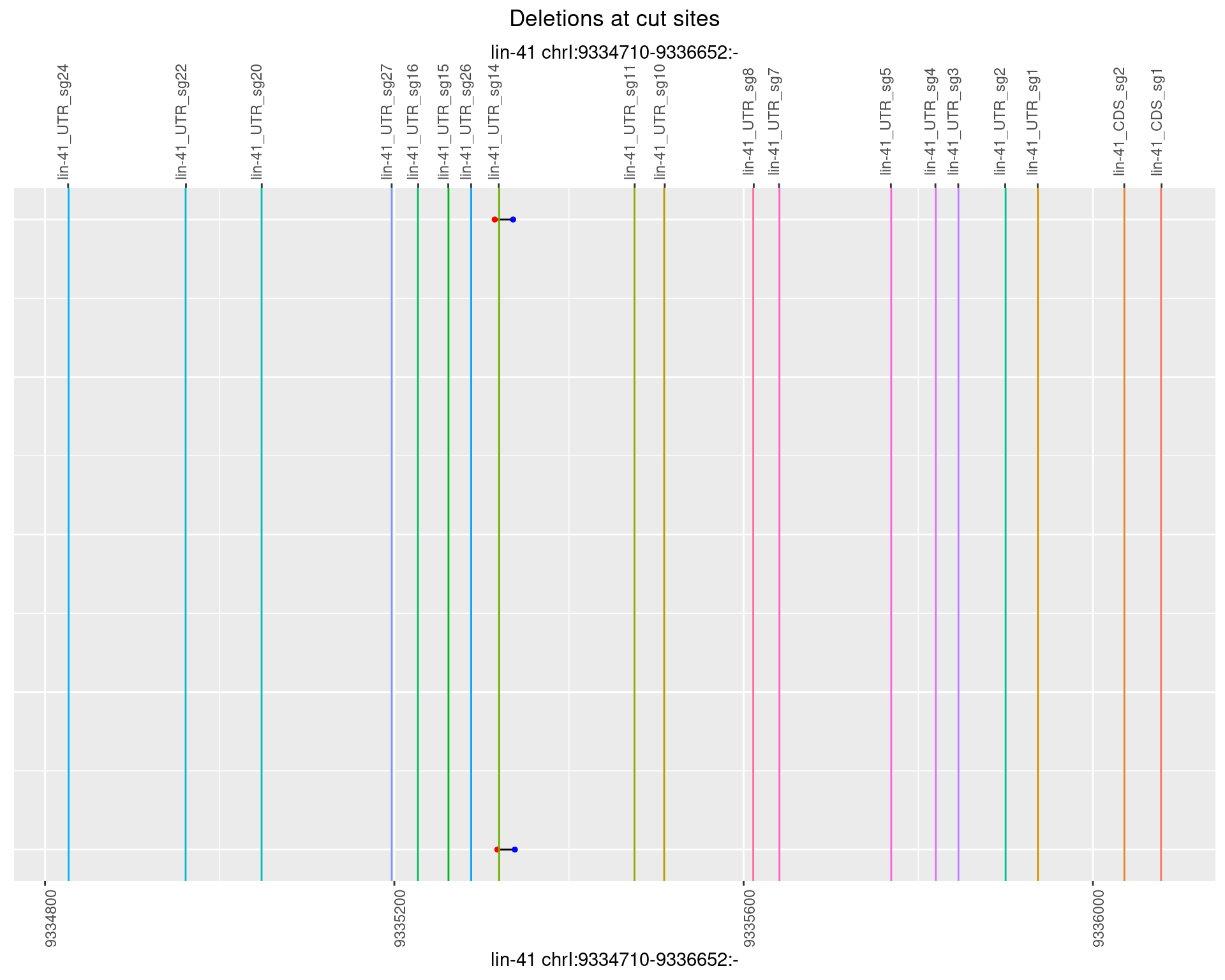
1.72.6 Freq: 0.1
No plot to show
1.73 lin41_RNA_L1_1516_middle_UMI
1.73.1 Freq: 0
No plot to show
1.73.2 Freq: 1e-05
No plot to show
1.73.3 Freq: 1e-04
No plot to show
1.73.4 Freq: 0.001
No plot to show
1.73.5 Freq: 0.01
No plot to show
1.73.6 Freq: 0.1
No plot to show
1.74 lin41_RNA_L1_2627_middle_UMI
1.74.1 Freq: 0
No plot to show
1.74.2 Freq: 1e-05
No plot to show
1.74.3 Freq: 1e-04
No plot to show
1.74.4 Freq: 0.001
No plot to show
1.74.5 Freq: 0.01
No plot to show
1.74.6 Freq: 0.1
No plot to show
1.75 lin41_RNA_L1_pool2_middle_UMI
1.75.1 Freq: 0
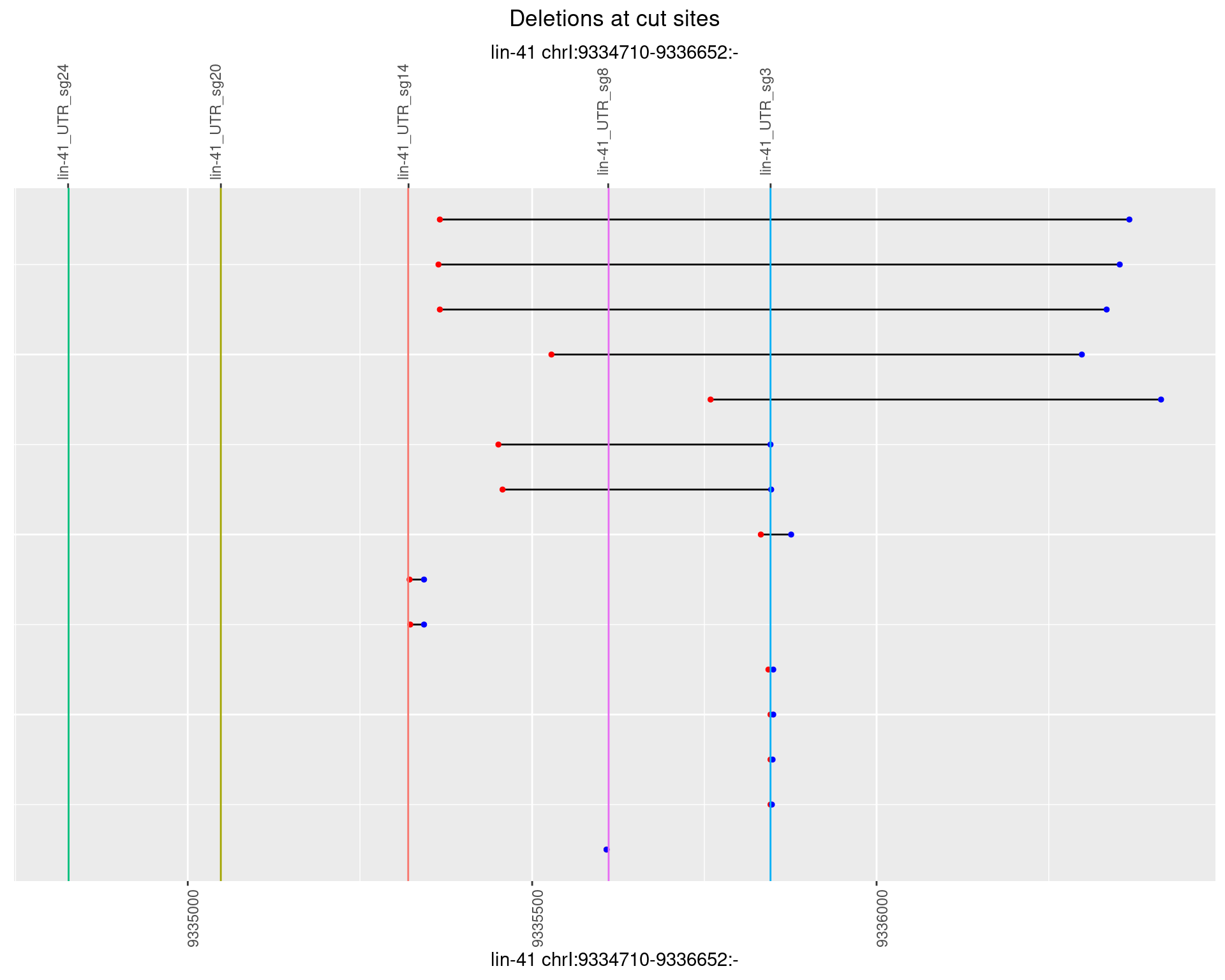
1.75.2 Freq: 1e-05
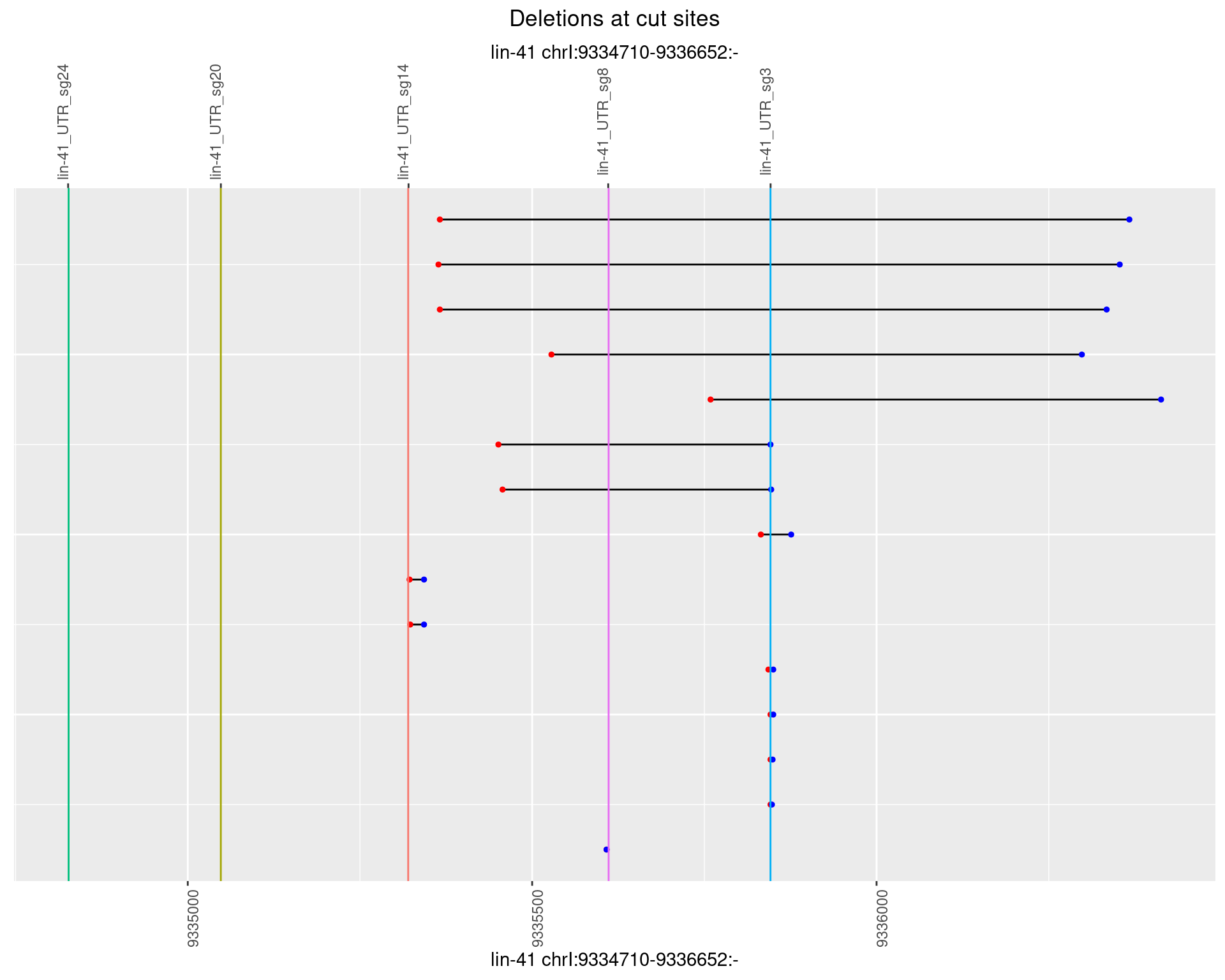
1.75.3 Freq: 1e-04
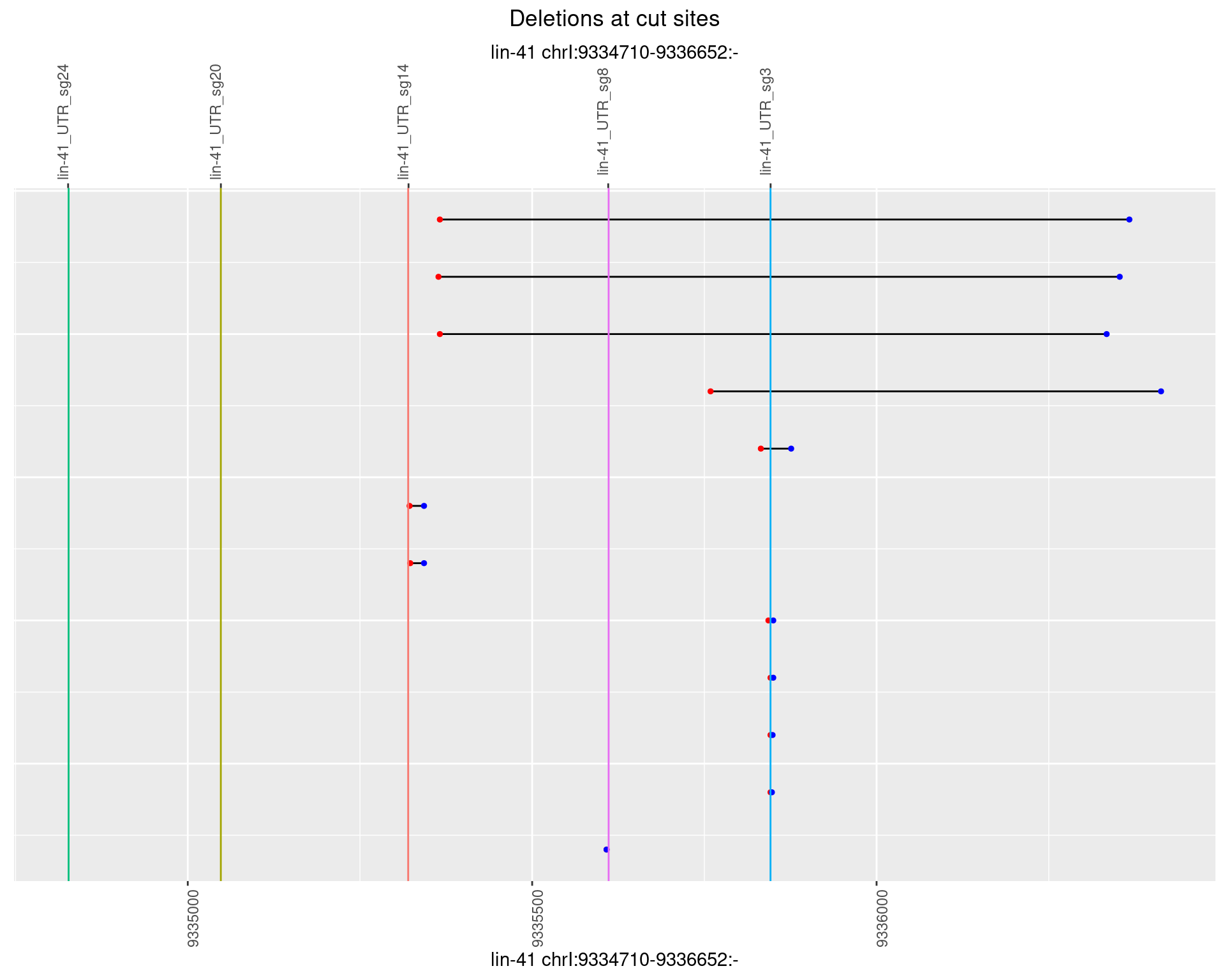
1.75.4 Freq: 0.001
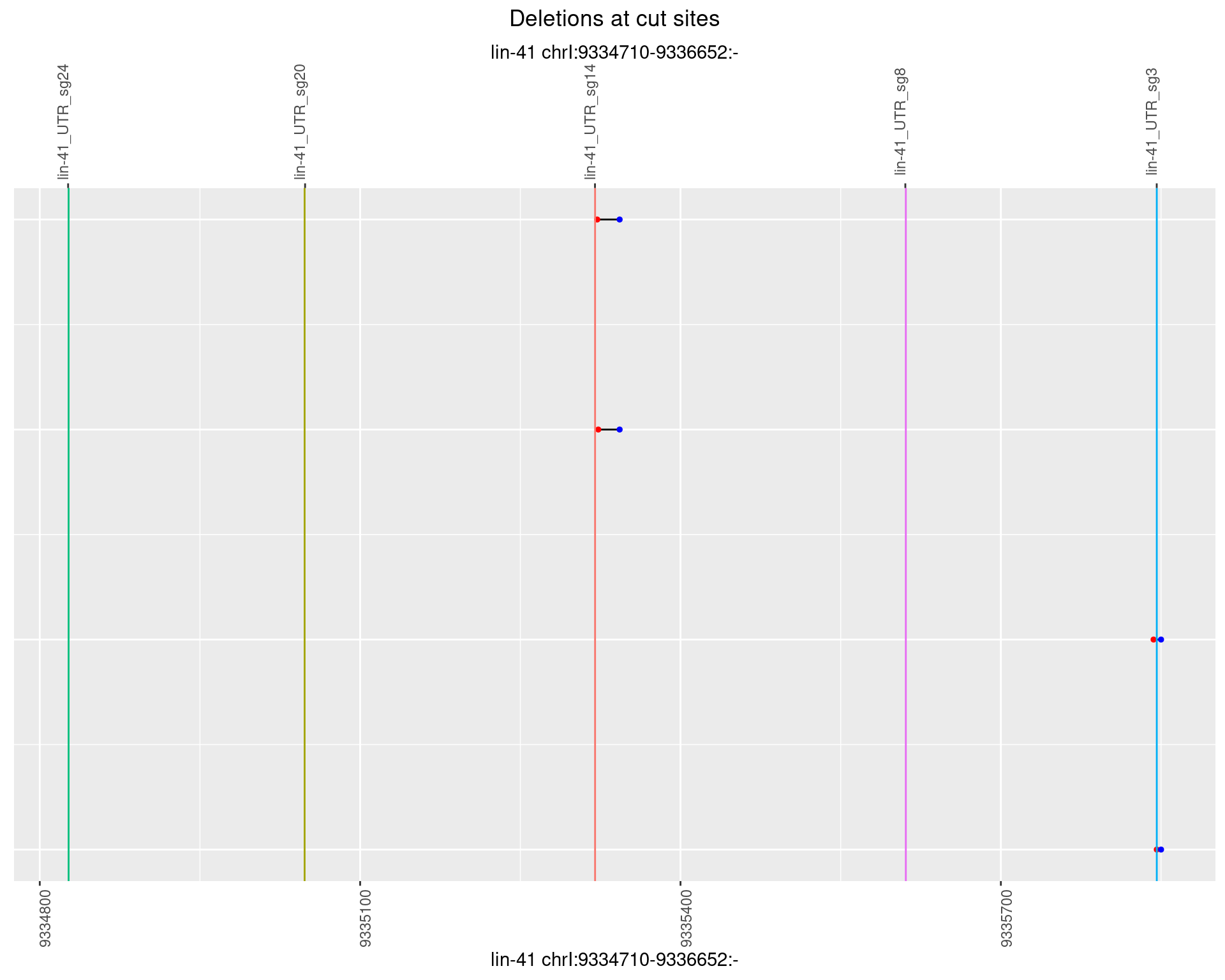
1.75.5 Freq: 0.01
No plot to show
1.75.6 Freq: 0.1
No plot to show
1.76 lin41_RNA_L1_pool3_middle_UMI
1.76.1 Freq: 0
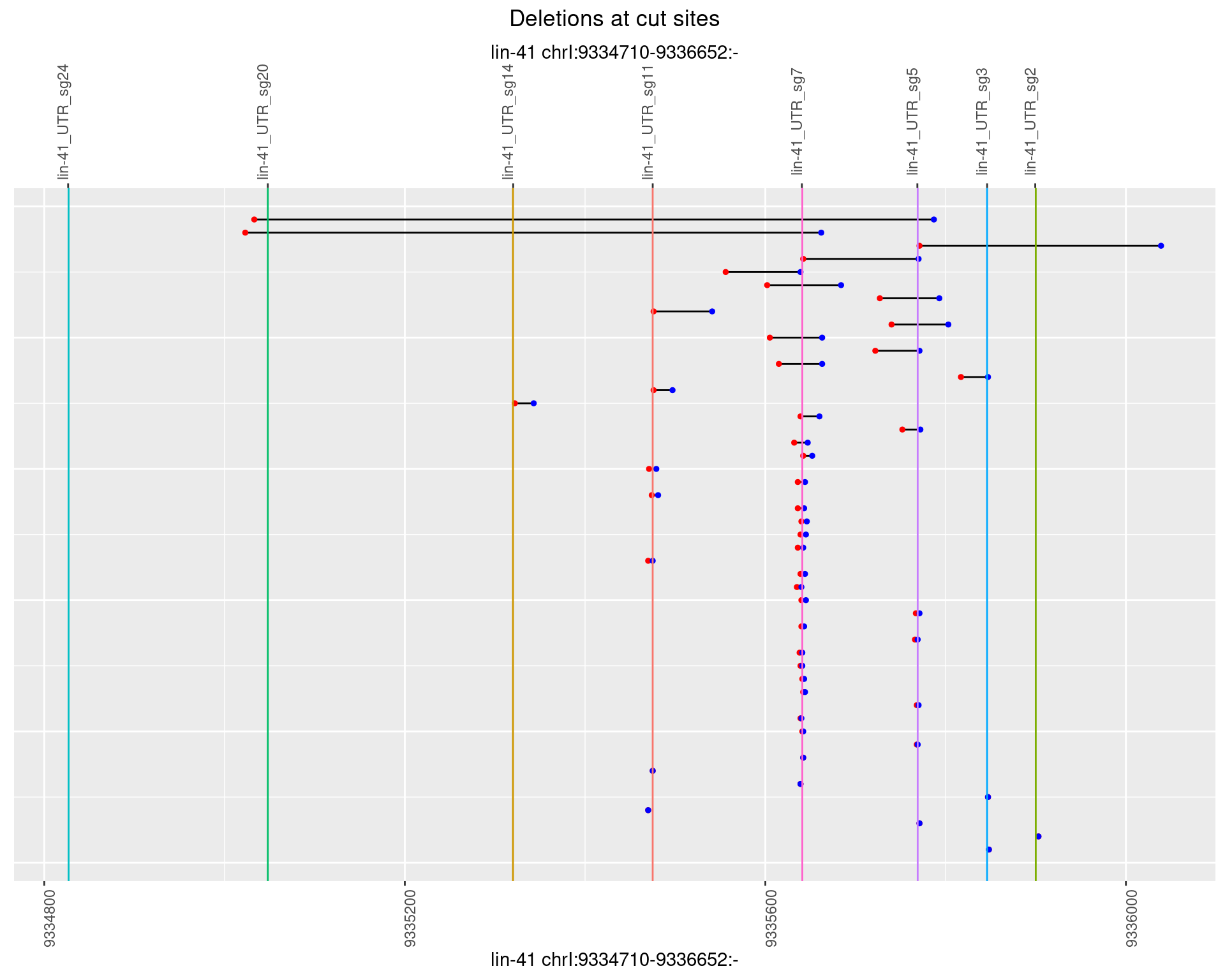
1.76.2 Freq: 1e-05
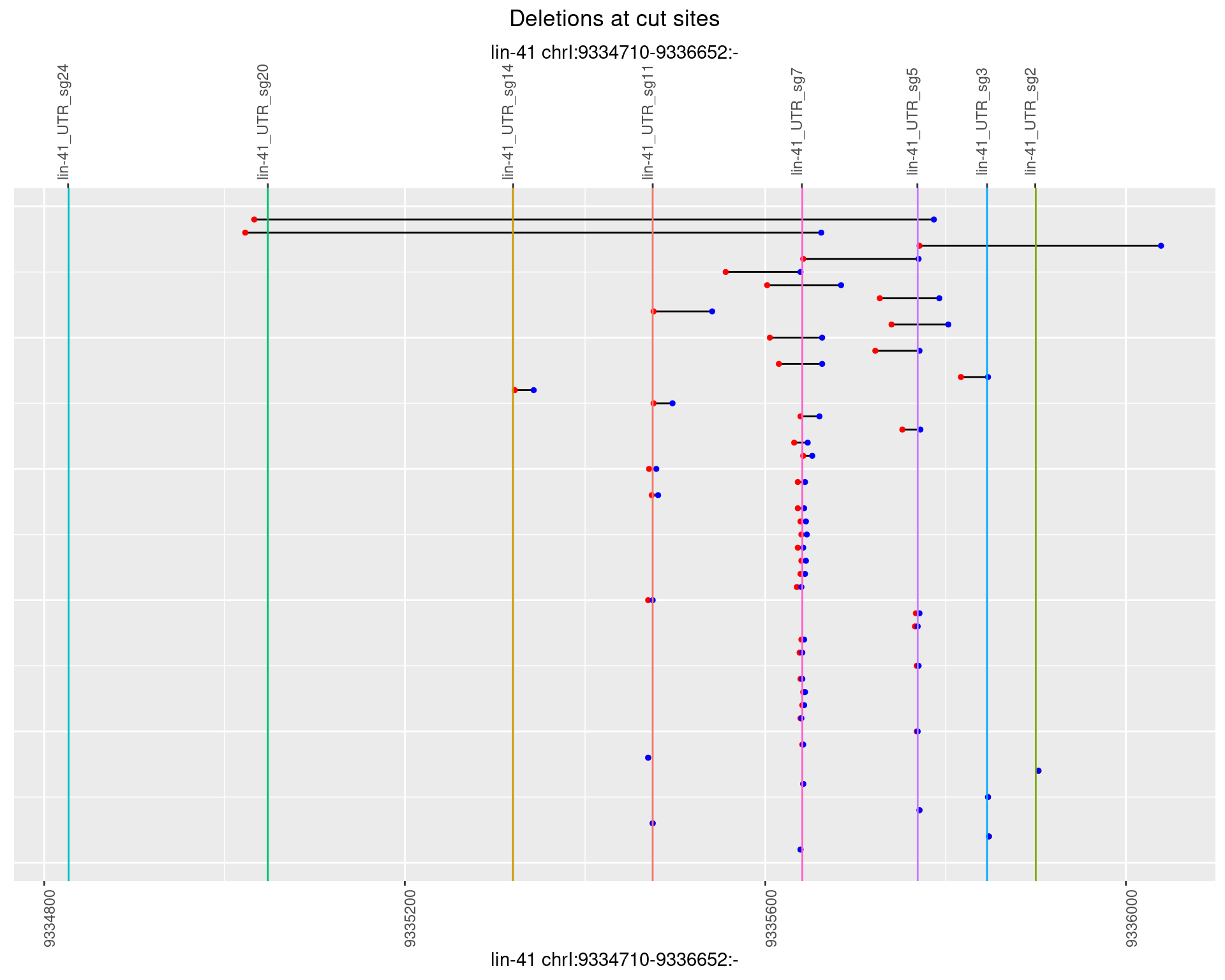
1.76.3 Freq: 1e-04
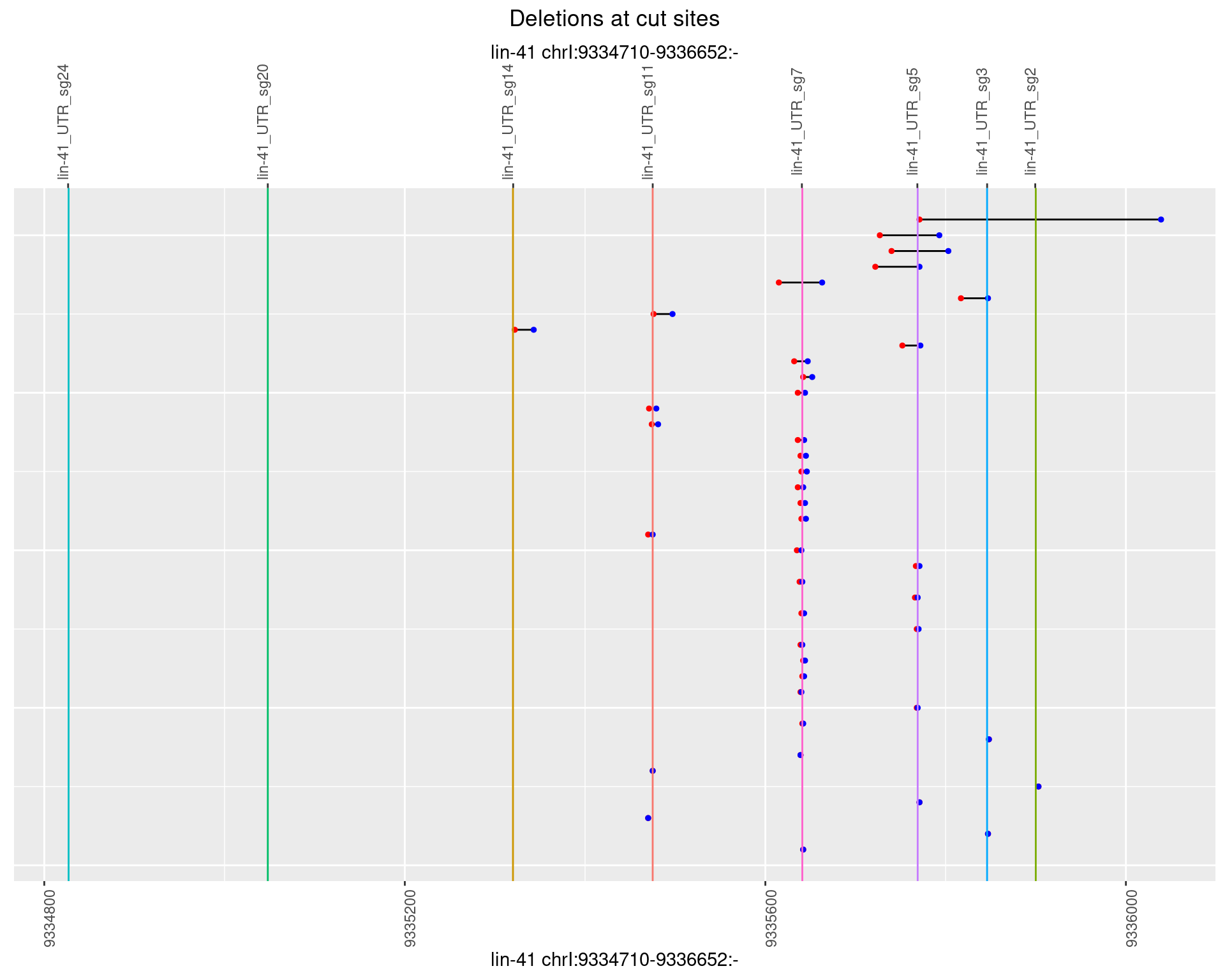
1.76.4 Freq: 0.001
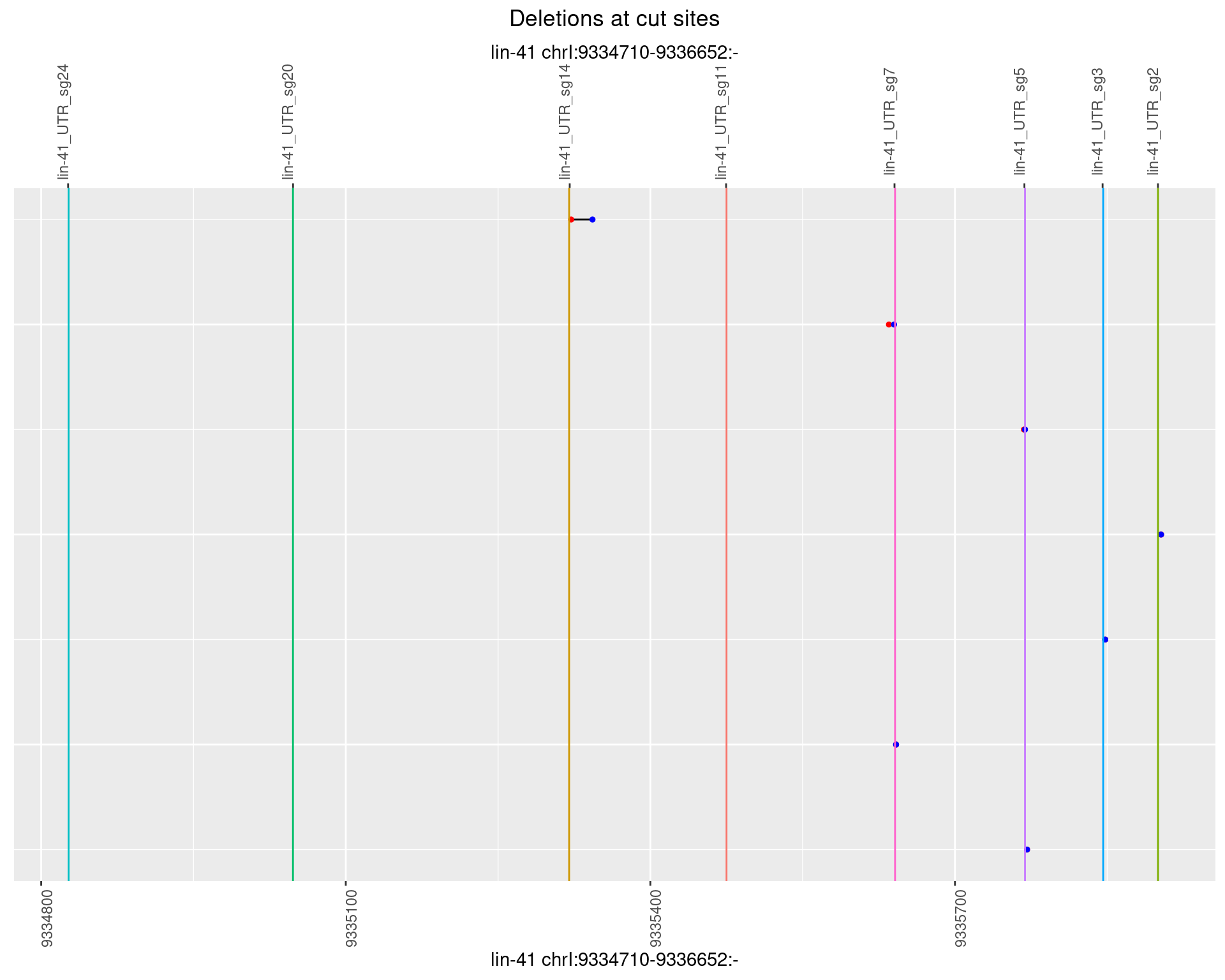
1.76.5 Freq: 0.01
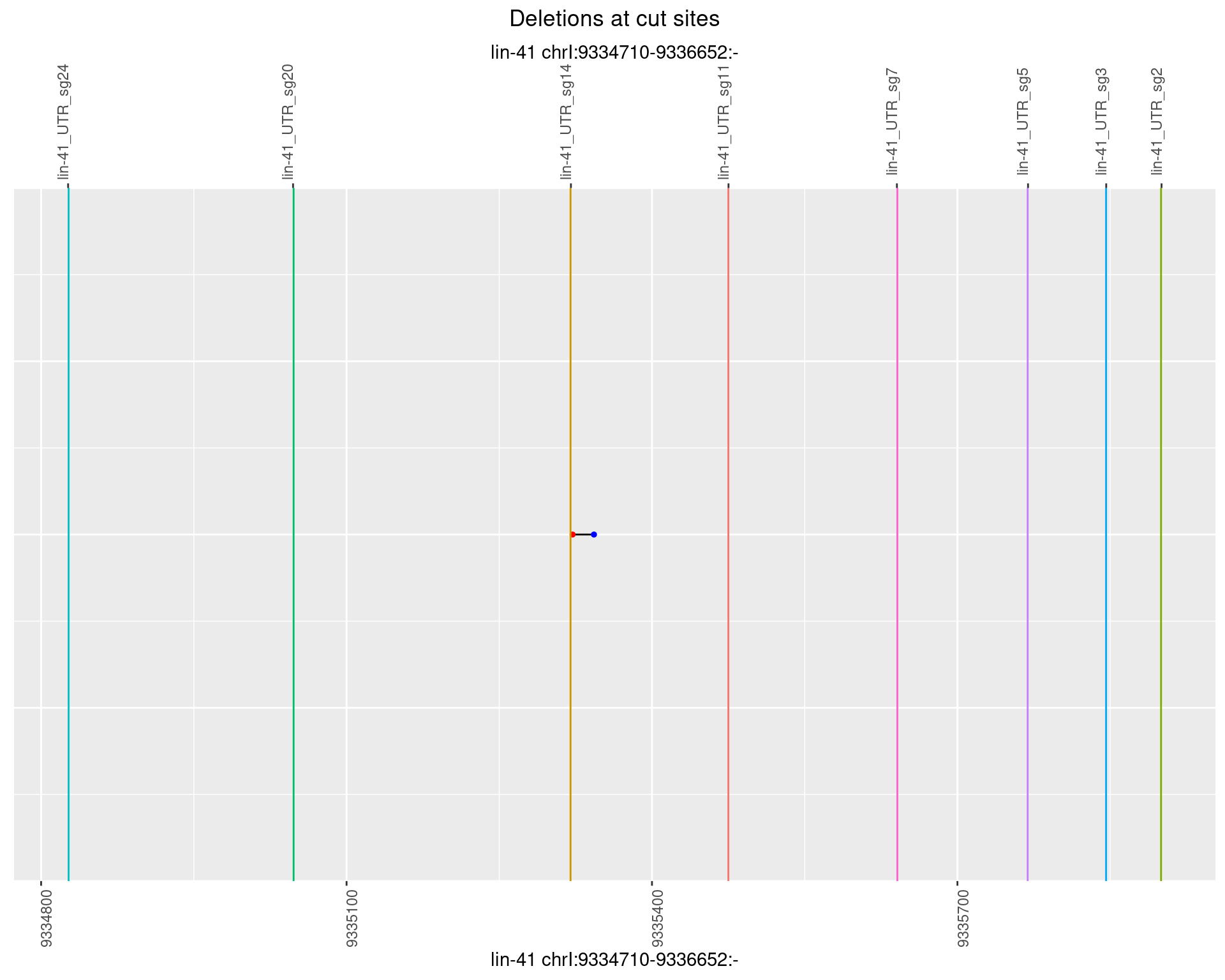
1.76.6 Freq: 0.1
No plot to show
1.77 lin41_RNA_L4_N2_middle_UMI
1.77.1 Freq: 0
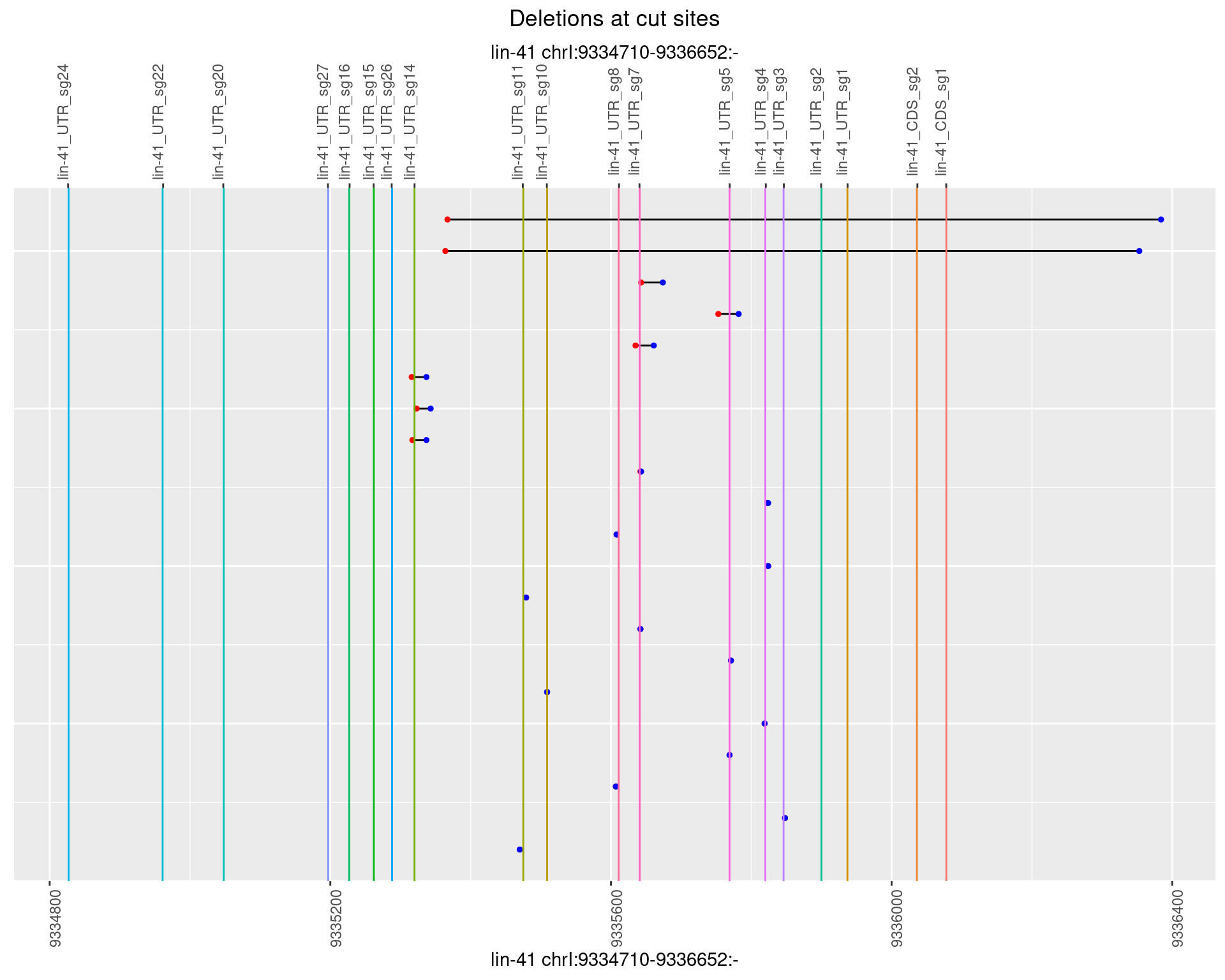
1.77.2 Freq: 1e-05
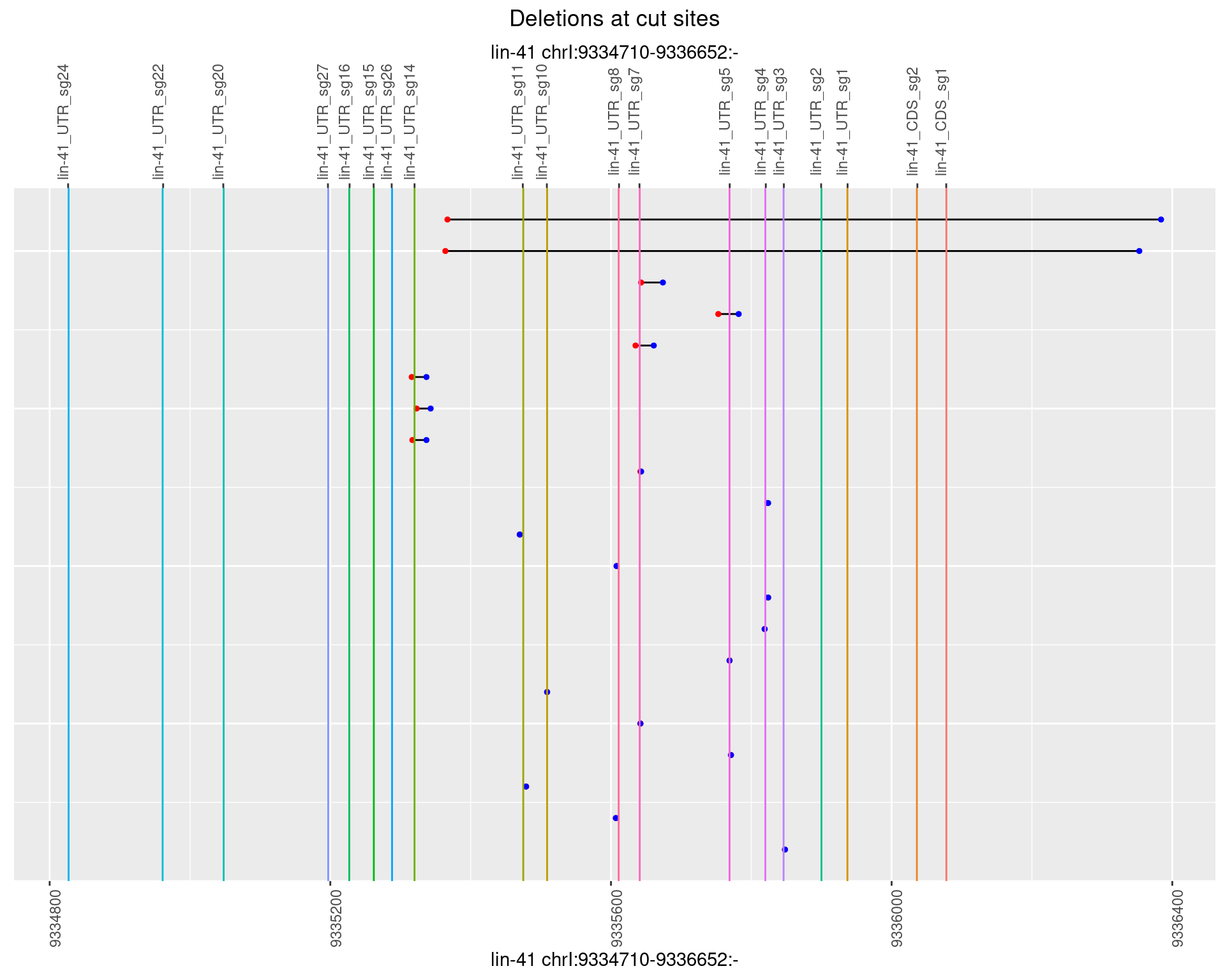
1.77.3 Freq: 1e-04
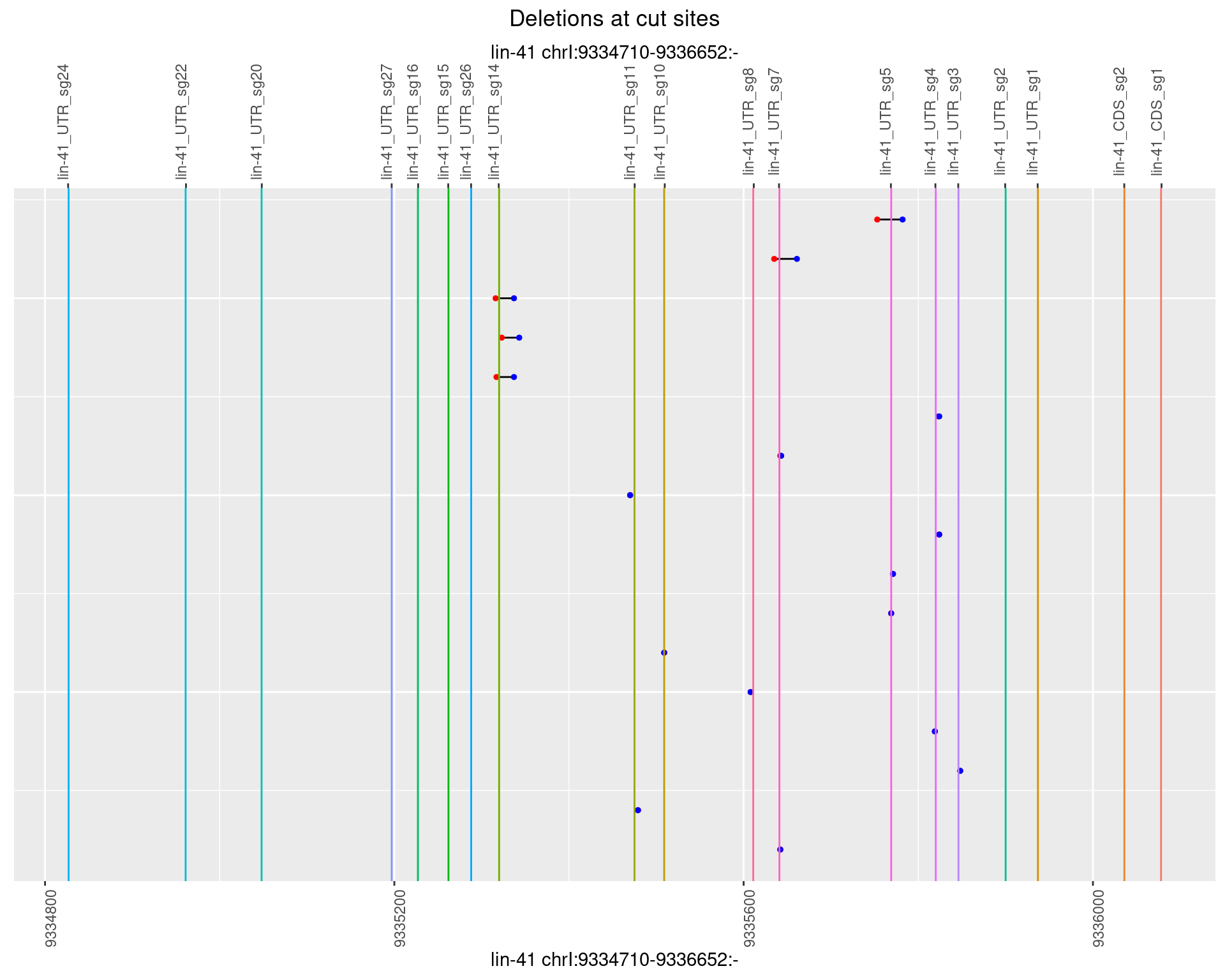
1.77.4 Freq: 0.001
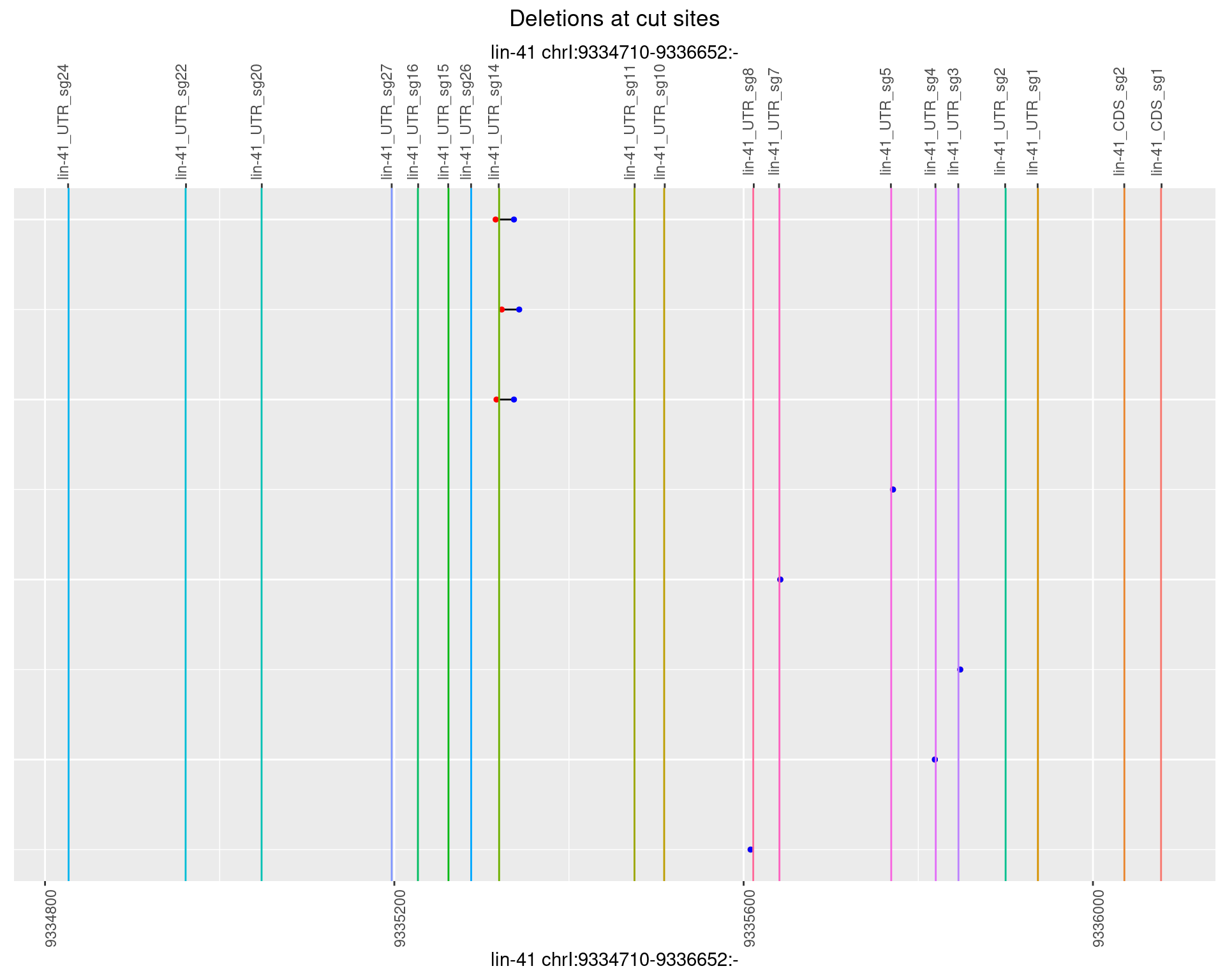
1.77.5 Freq: 0.01
1.77.6 Freq: 0.1
No plot to show
1.78 lin41_RNA_L4_1516_middle_UMI
1.78.1 Freq: 0
No plot to show
1.78.2 Freq: 1e-05
No plot to show
1.78.3 Freq: 1e-04
No plot to show
1.78.4 Freq: 0.001
No plot to show
1.78.5 Freq: 0.01
No plot to show
1.78.6 Freq: 0.1
No plot to show
1.79 lin41_RNA_L4_2627_middle_UMI
1.79.1 Freq: 0
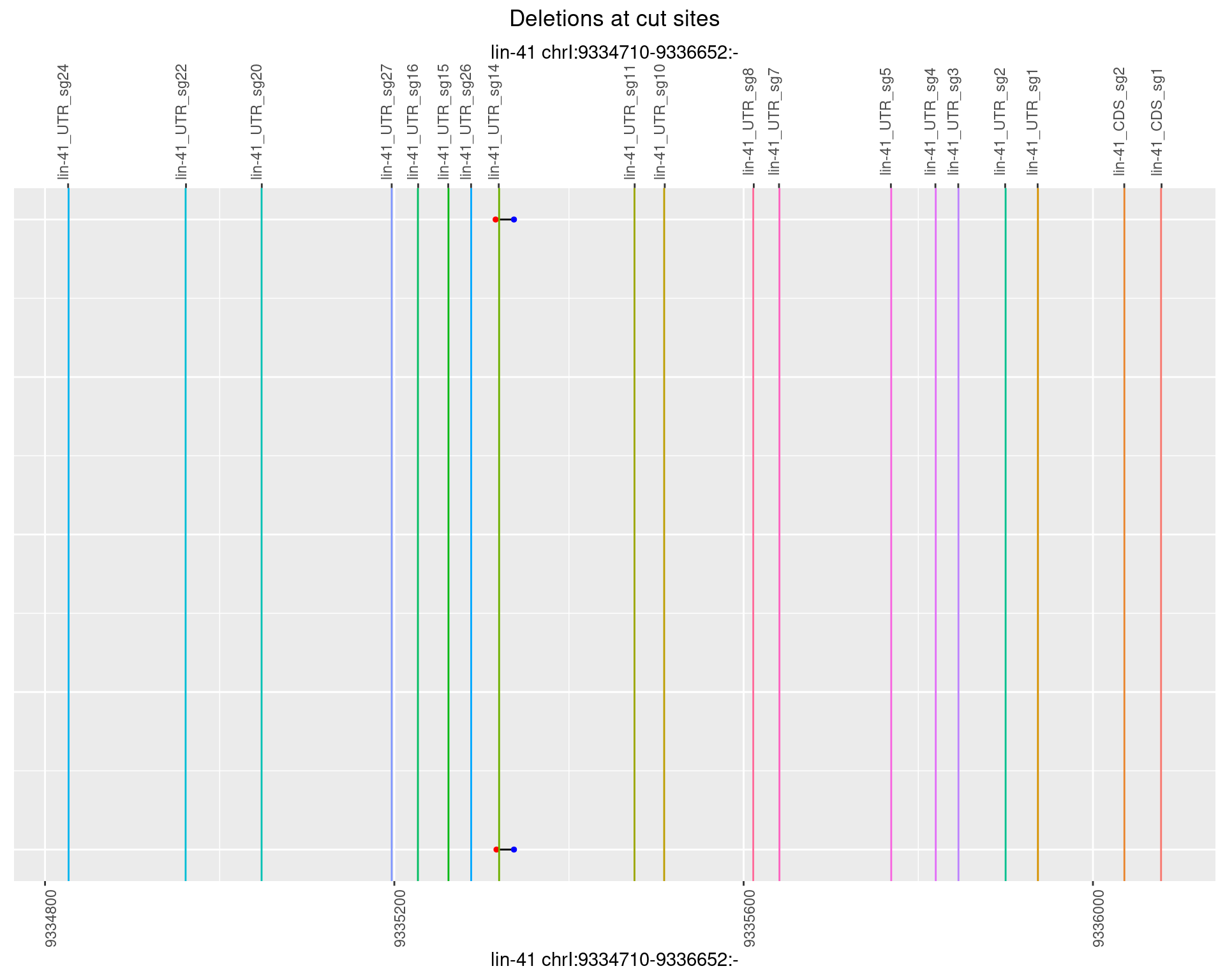
1.79.2 Freq: 1e-05
1.79.3 Freq: 1e-04
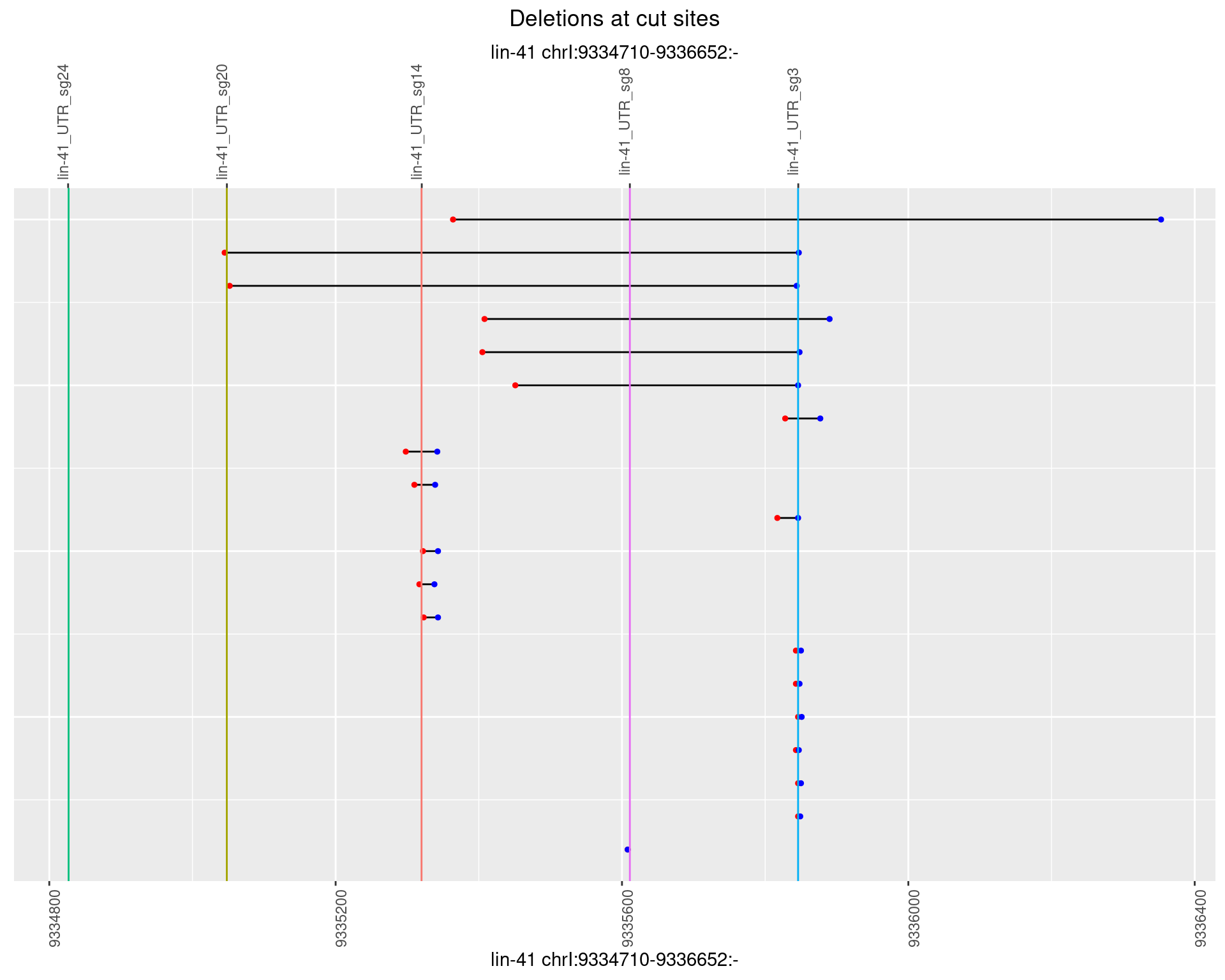
1.79.4 Freq: 0.001
1.79.5 Freq: 0.01
No plot to show
1.79.6 Freq: 0.1
No plot to show
1.80 lin41_RNA_L4_pool2_middle_UMI
1.80.1 Freq: 0
1.80.2 Freq: 1e-05
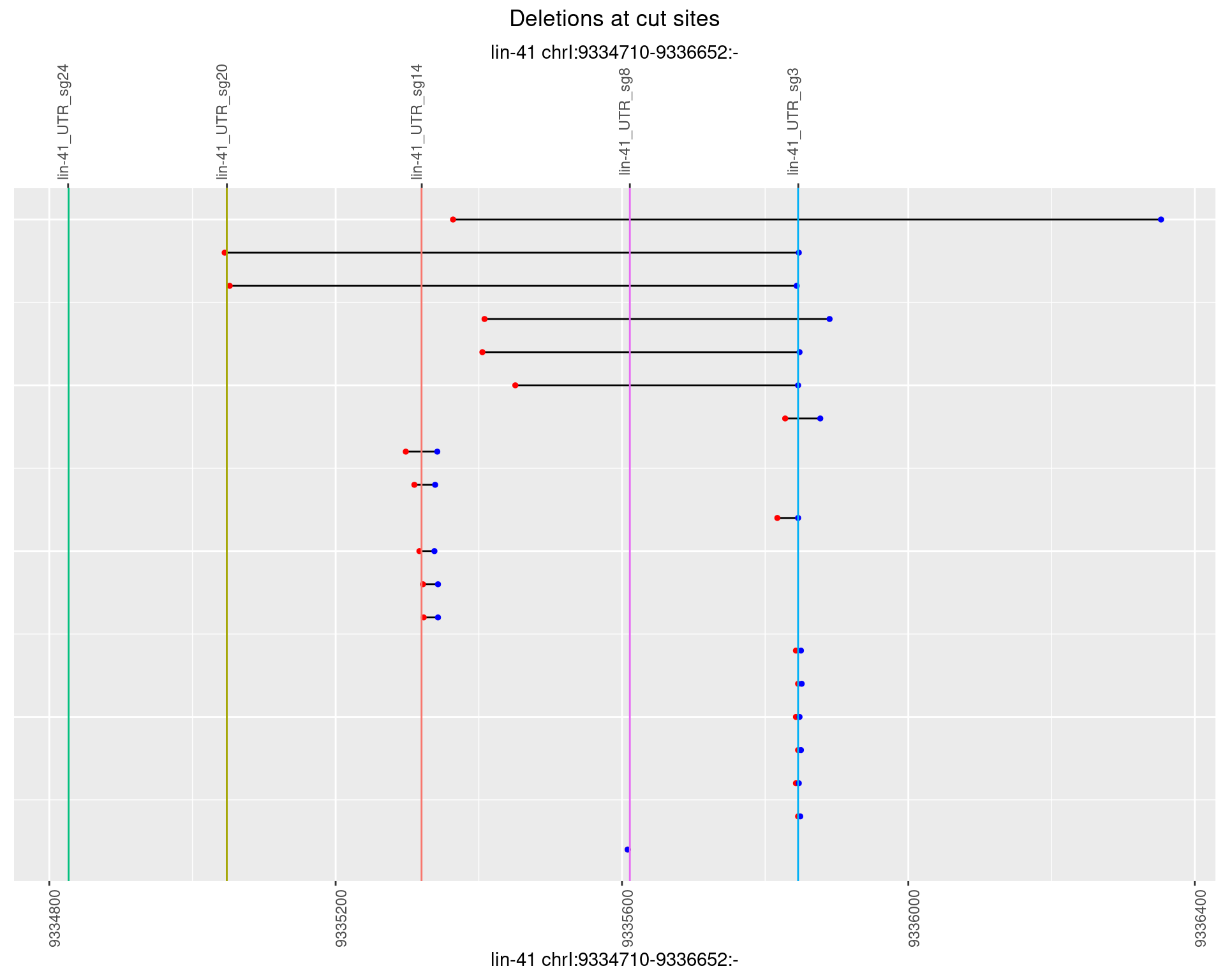
1.80.3 Freq: 1e-04
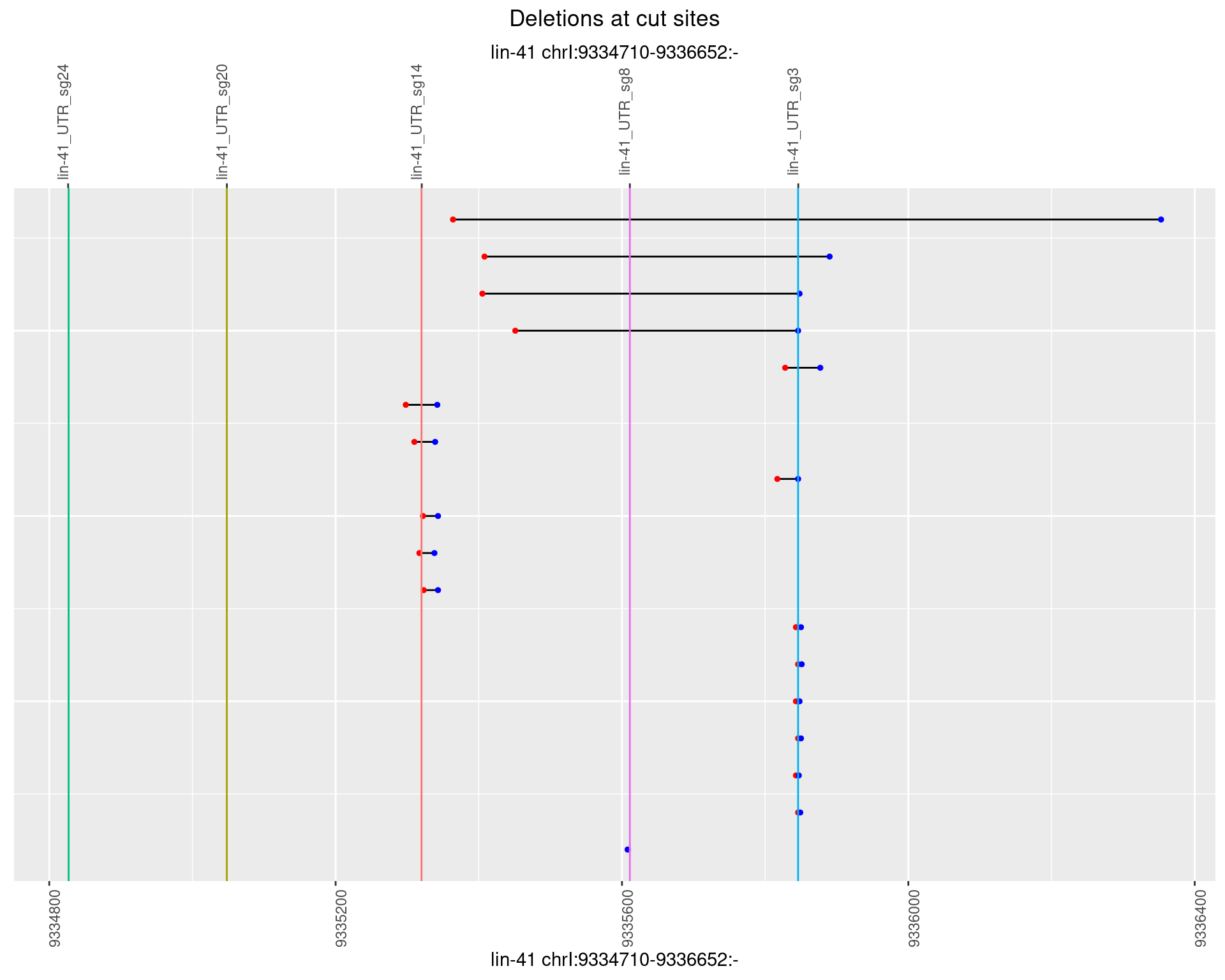
1.80.4 Freq: 0.001
1.80.5 Freq: 0.01
1.80.6 Freq: 0.1
No plot to show
1.81 lin41_RNA_L4_pool3_middle_UMI
1.81.1 Freq: 0
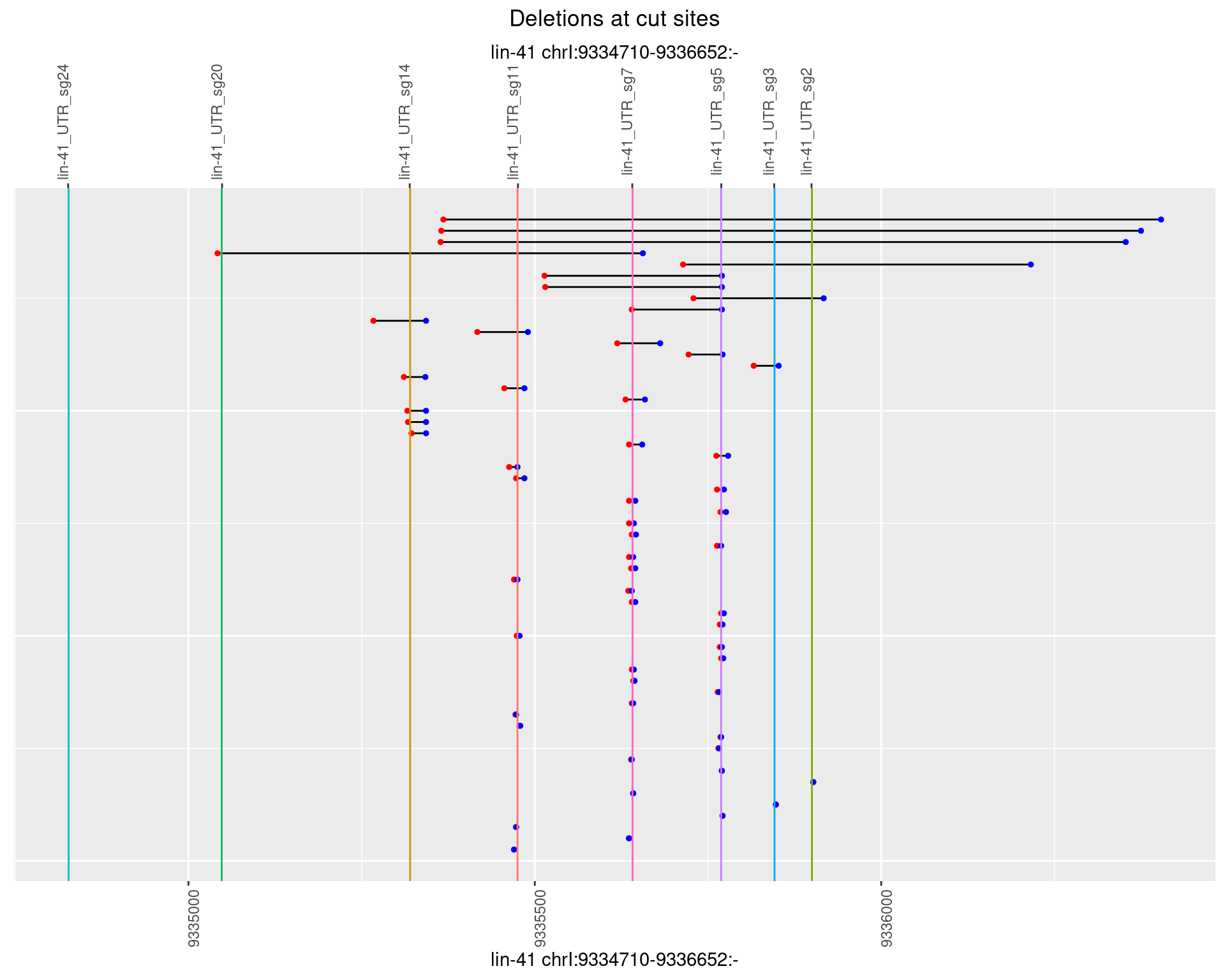
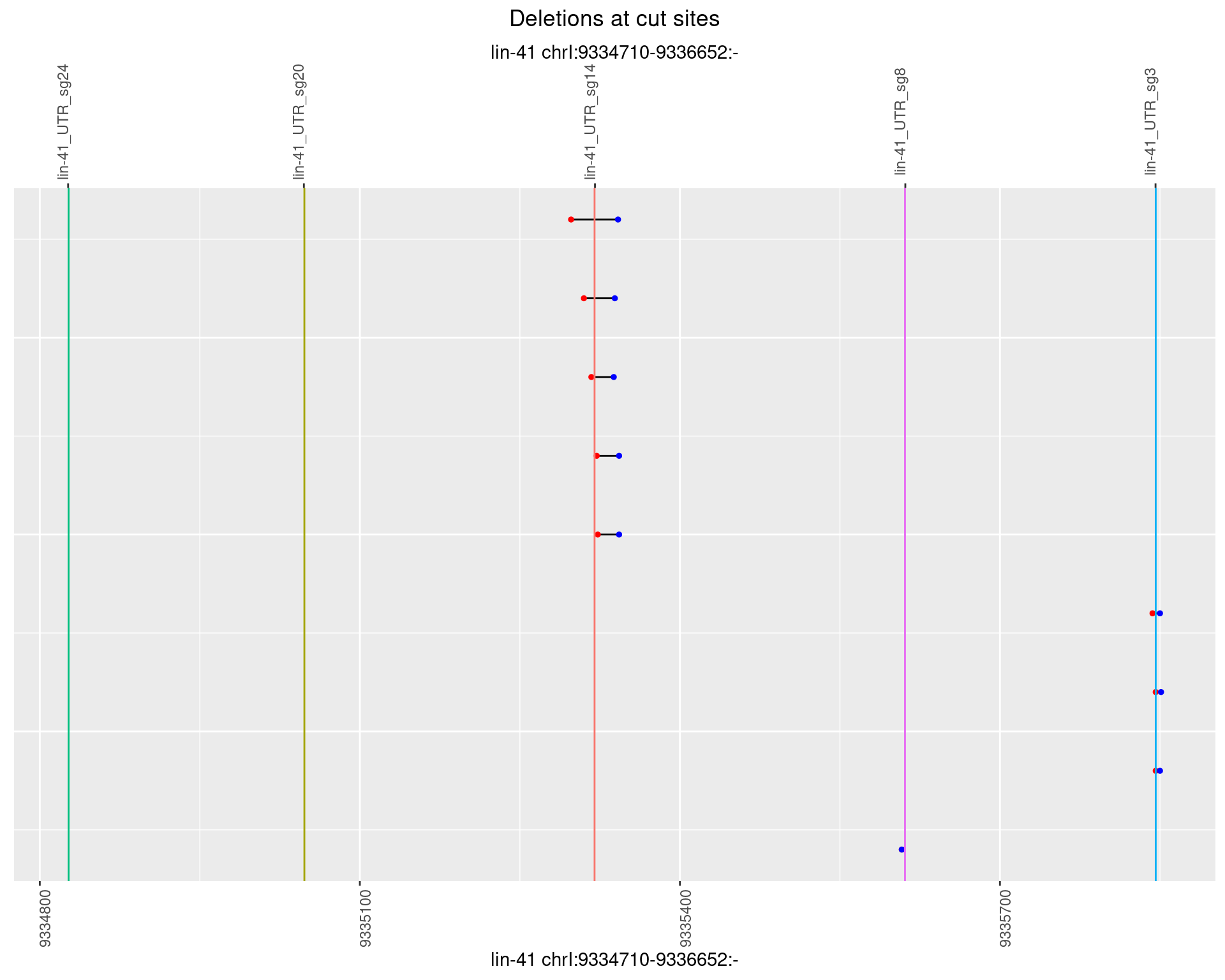
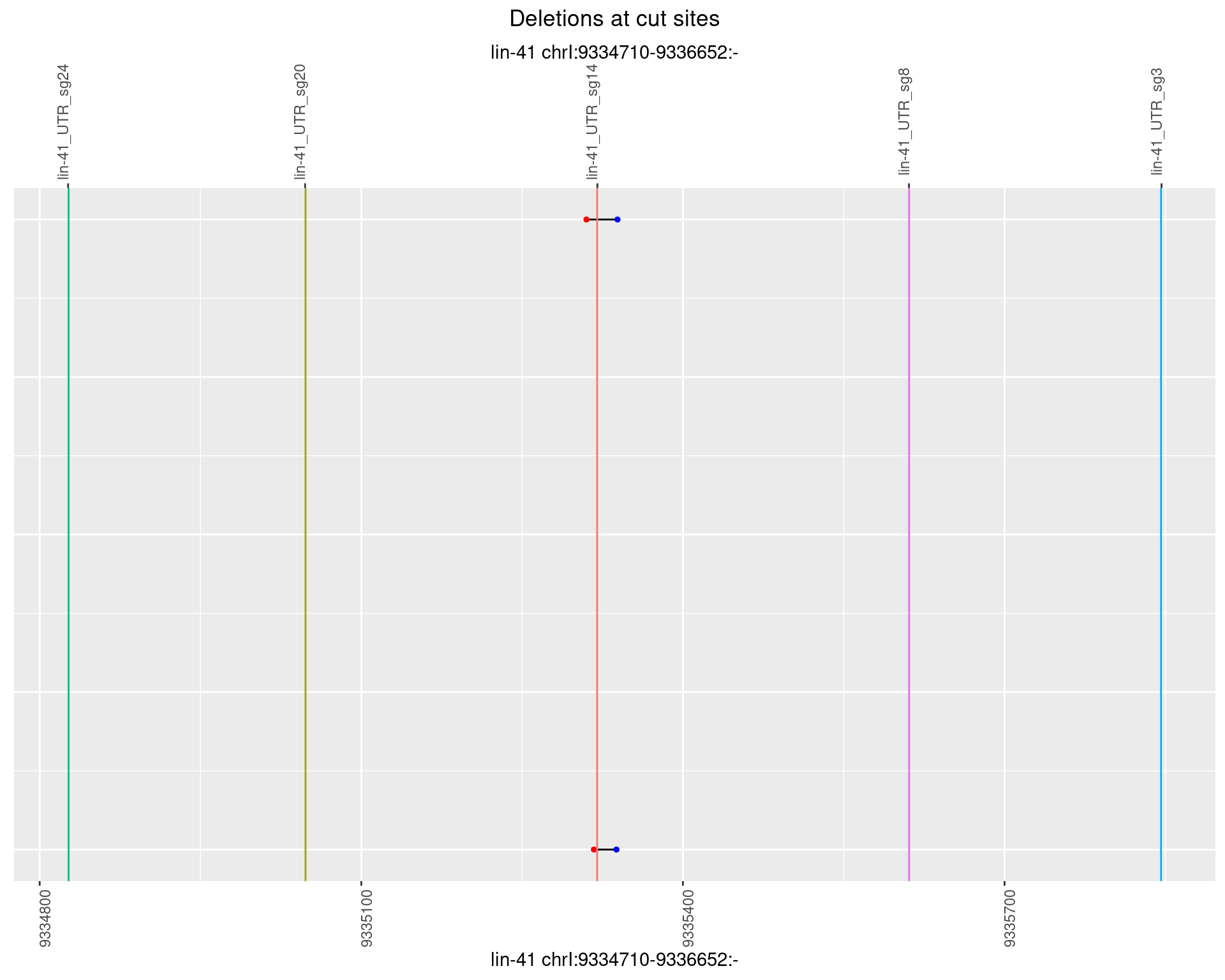
1.81.2 Freq: 1e-05
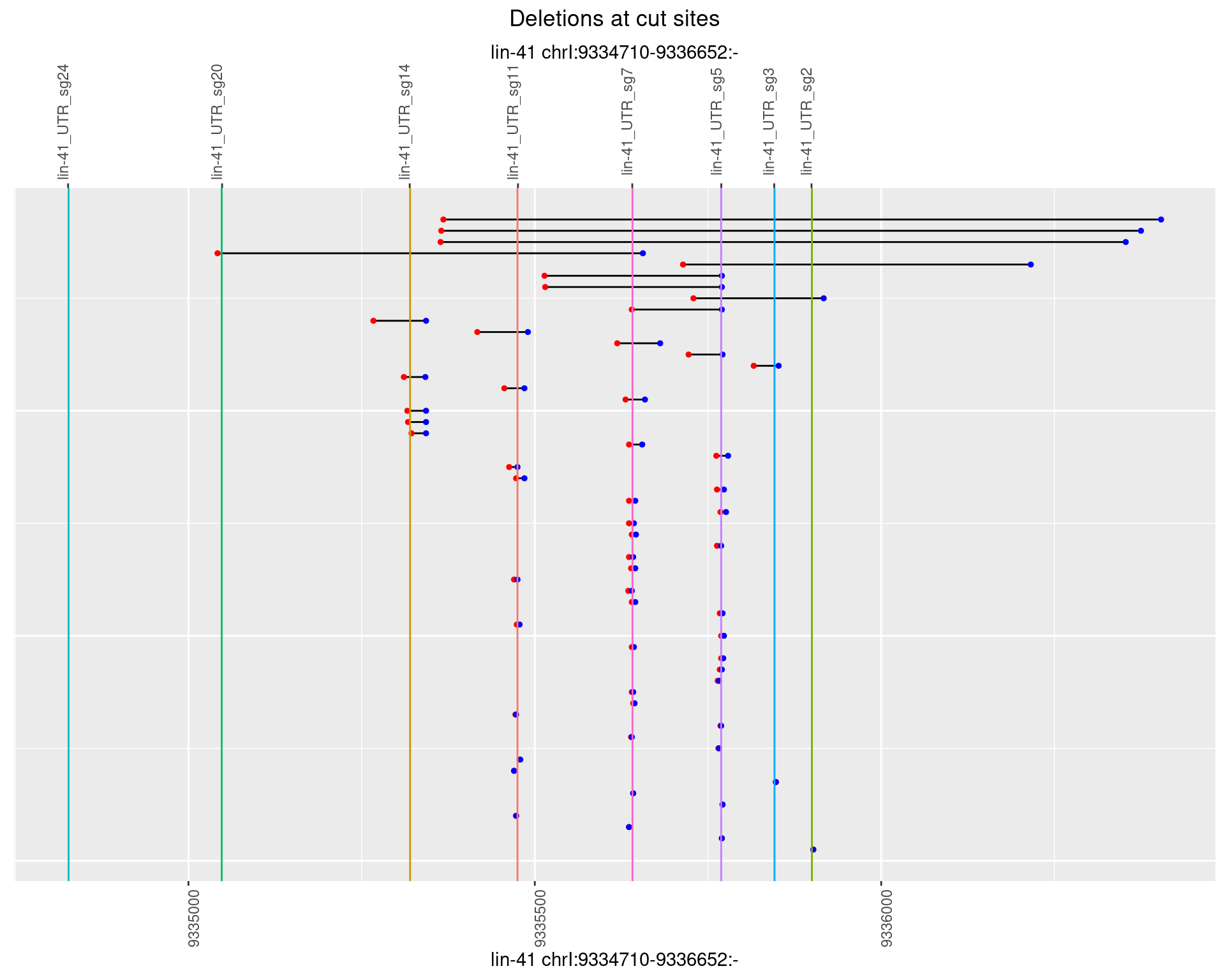
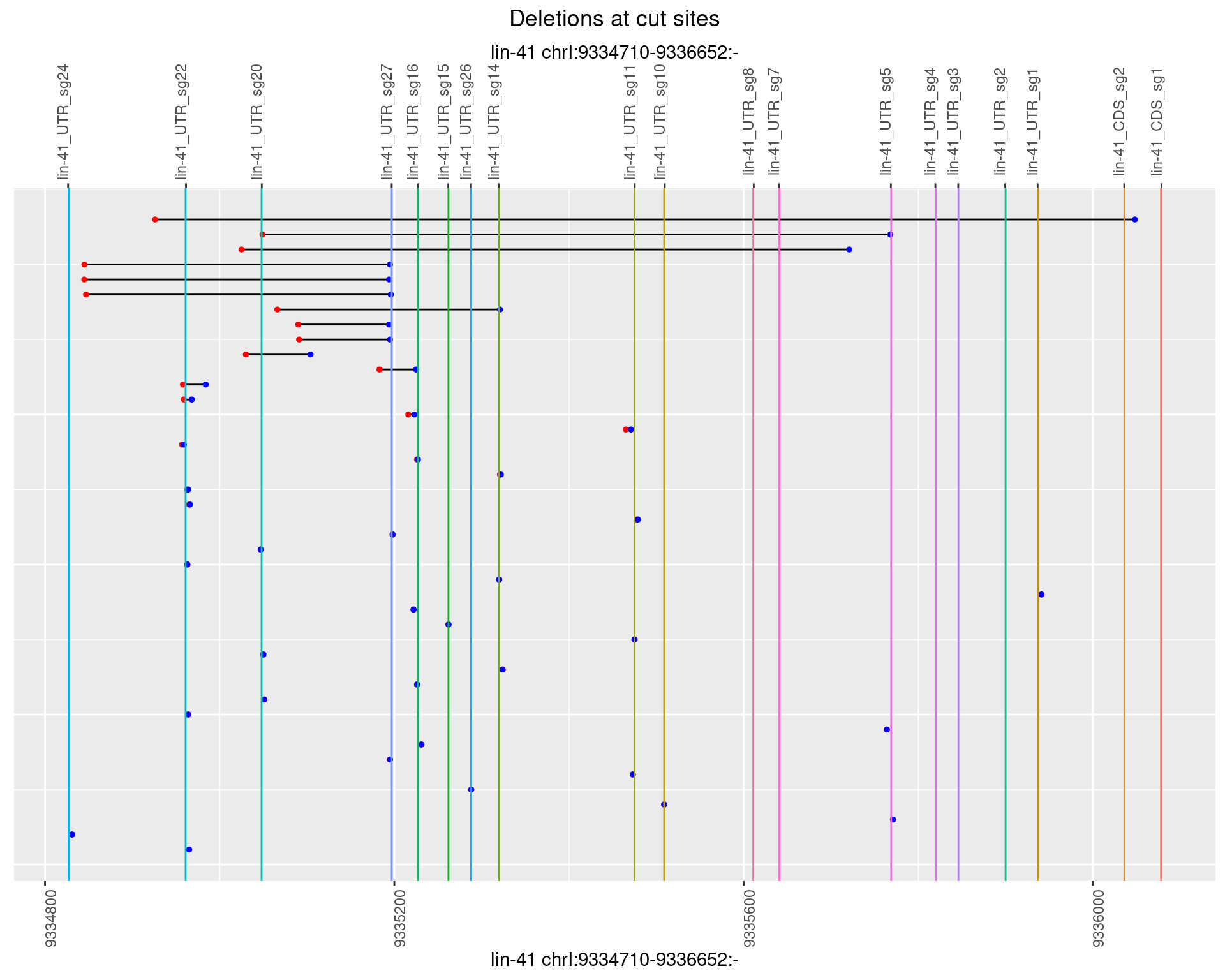
1.81.3 Freq: 1e-04
1.81.4 Freq: 0.001
1.81.5 Freq: 0.01
No plot to show
1.81.6 Freq: 0.1
No plot to show
1.82 lin41_RNA_L1_N2_3end_UMI_R2
1.82.1 Freq: 0
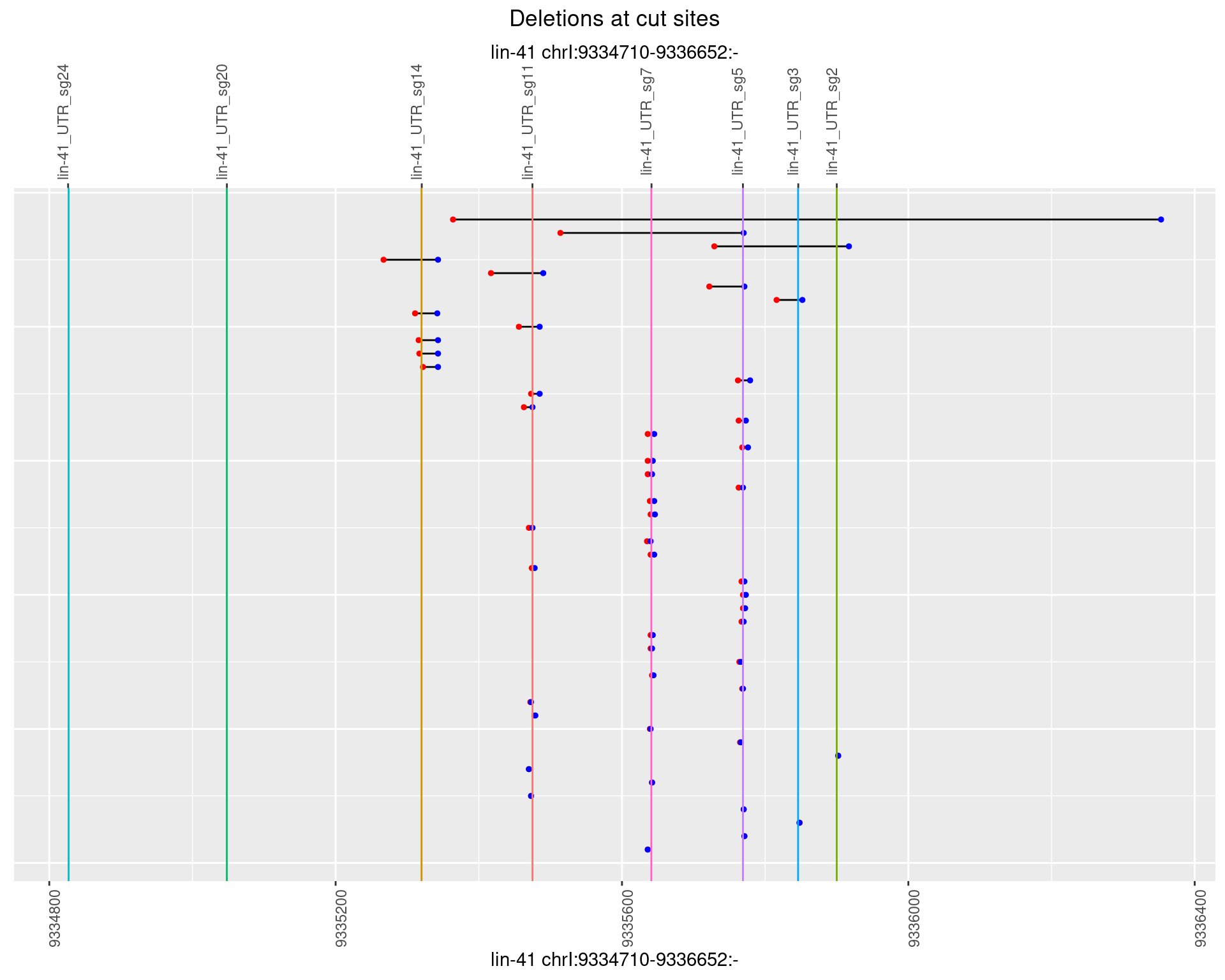
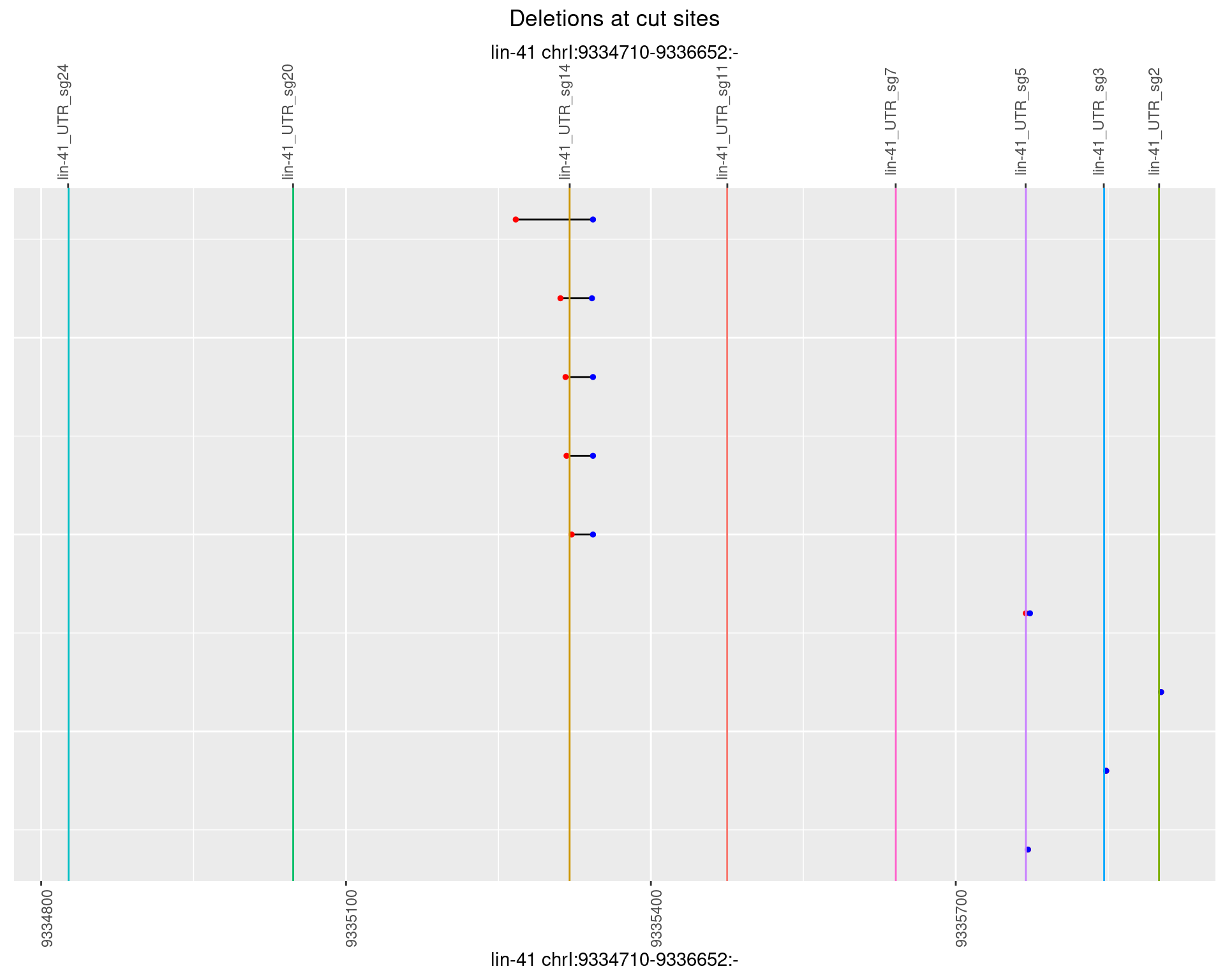
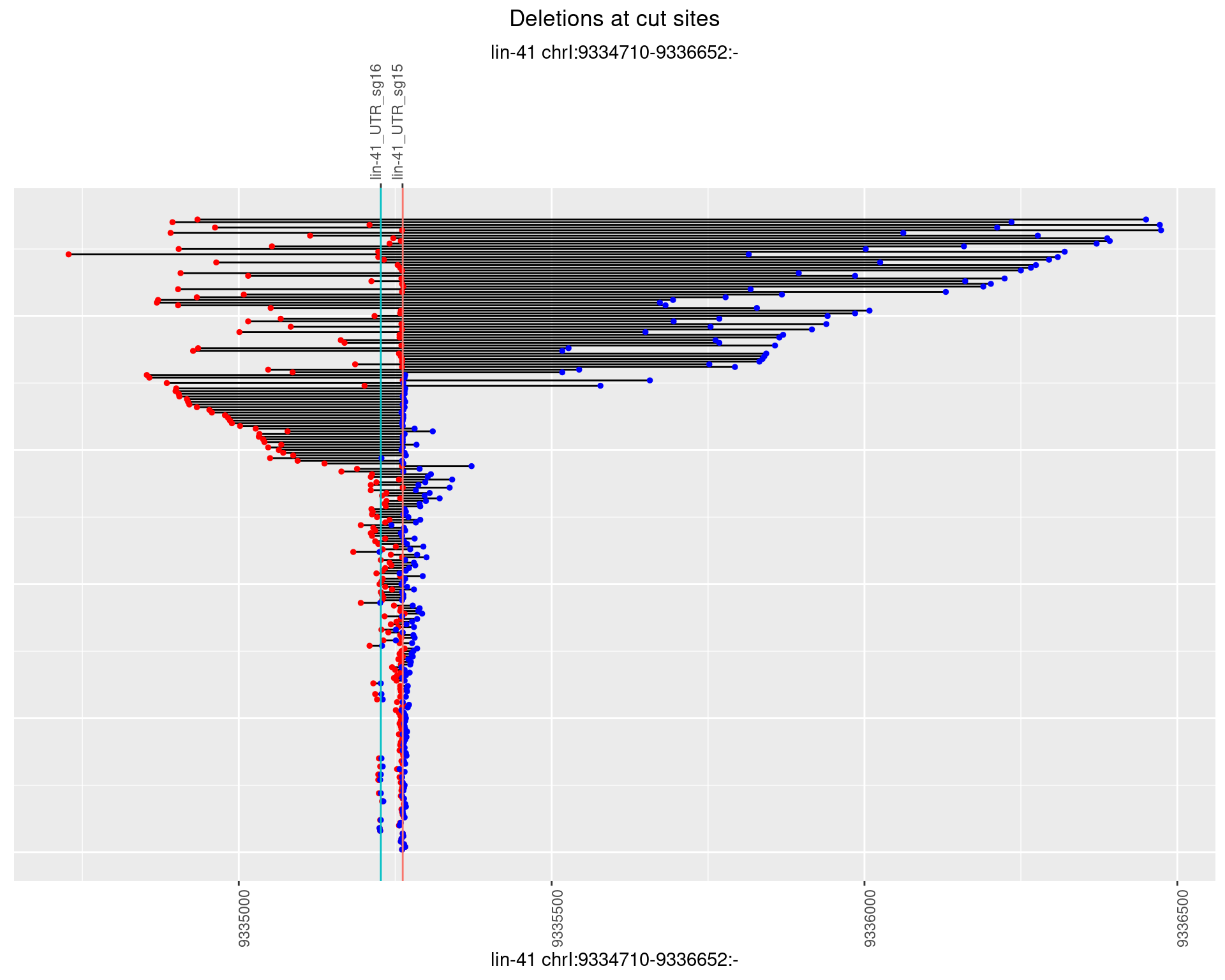
1.82.2 Freq: 1e-05
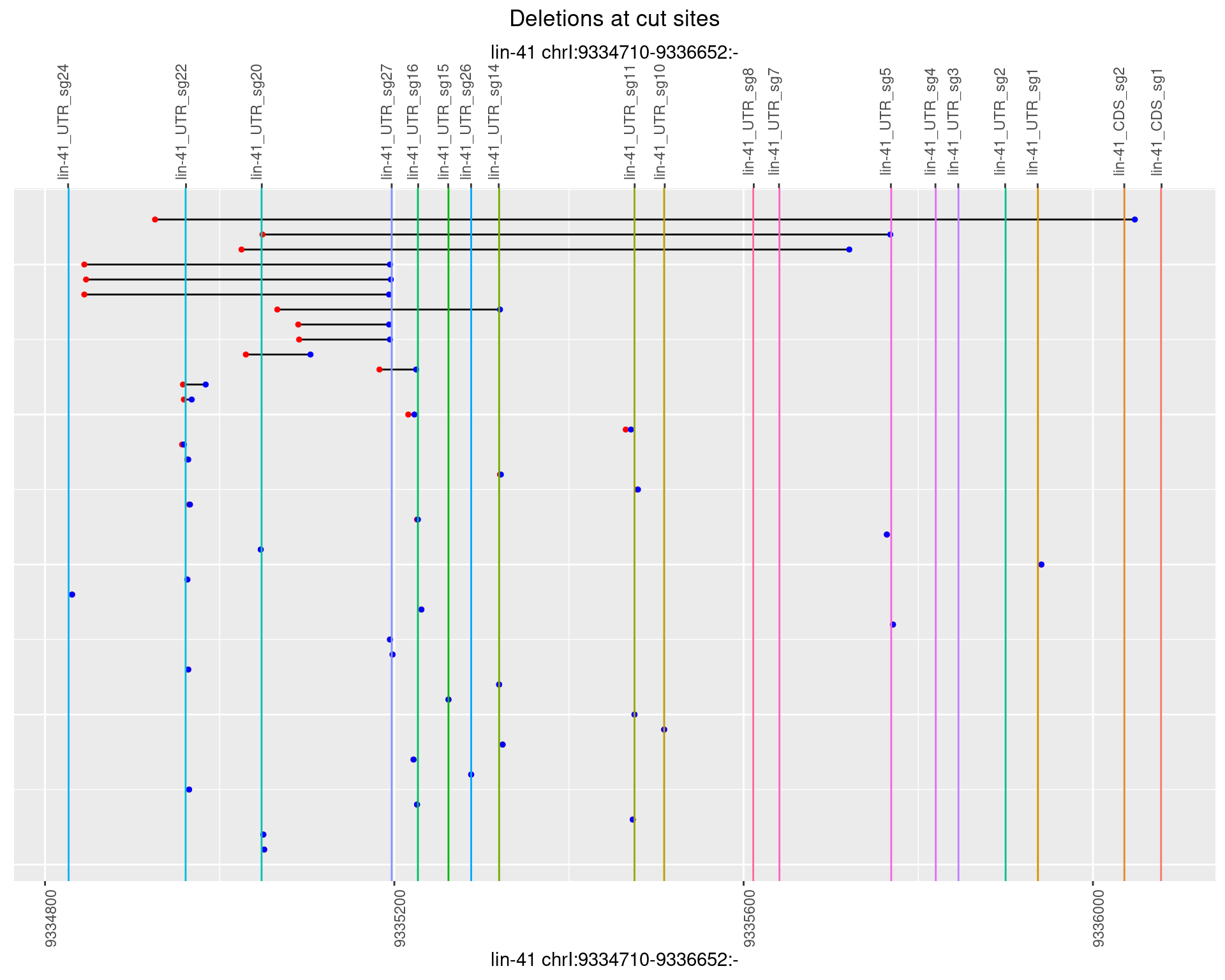
1.82.3 Freq: 1e-04
1.82.4 Freq: 0.001
1.82.5 Freq: 0.01
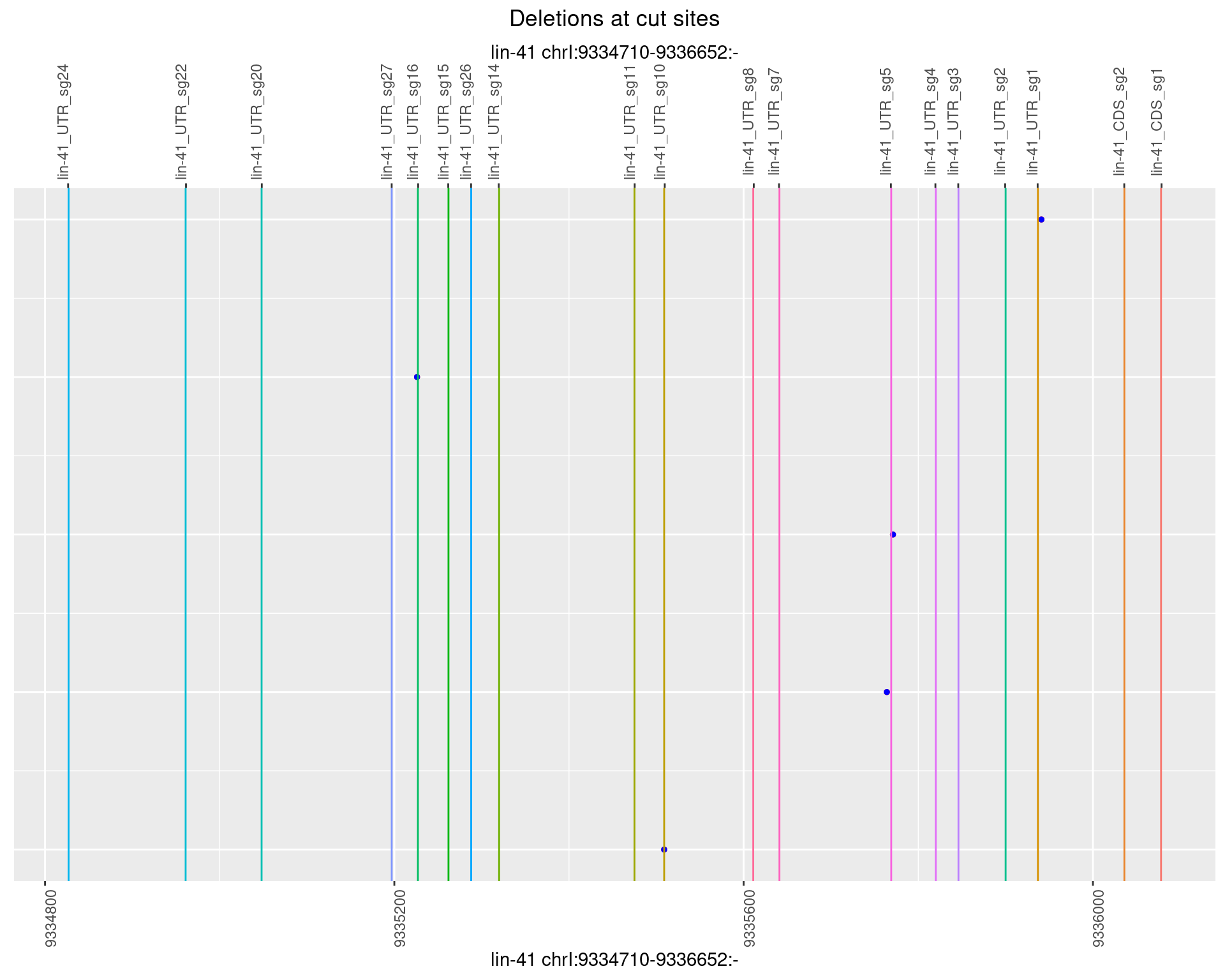
No plot to show
1.82.6 Freq: 0.1
No plot to show
1.83 lin41_RNA_L1_1516_3end_UMI_R2
1.83.1 Freq: 0
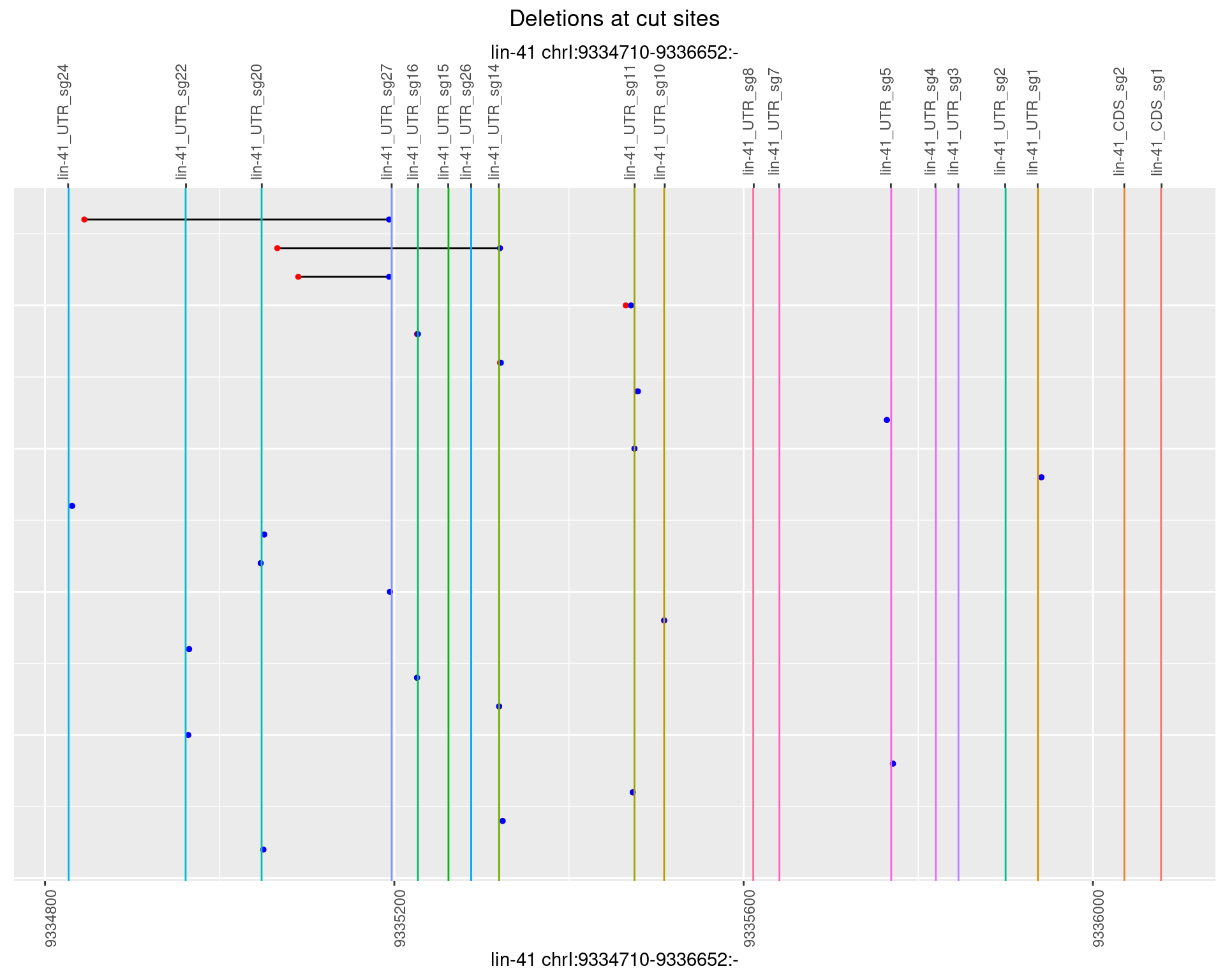
1.83.2 Freq: 1e-05
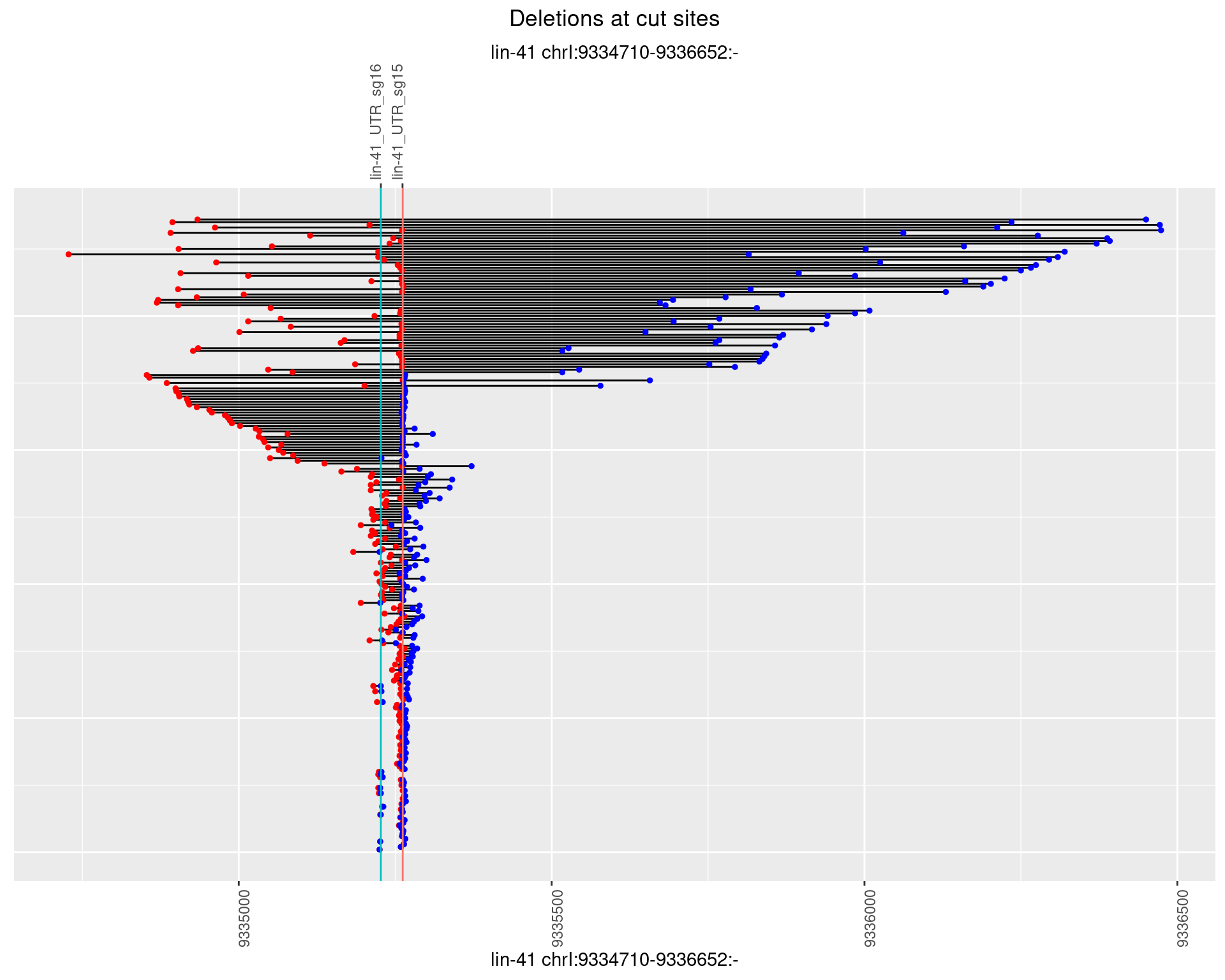
1.83.3 Freq: 1e-04
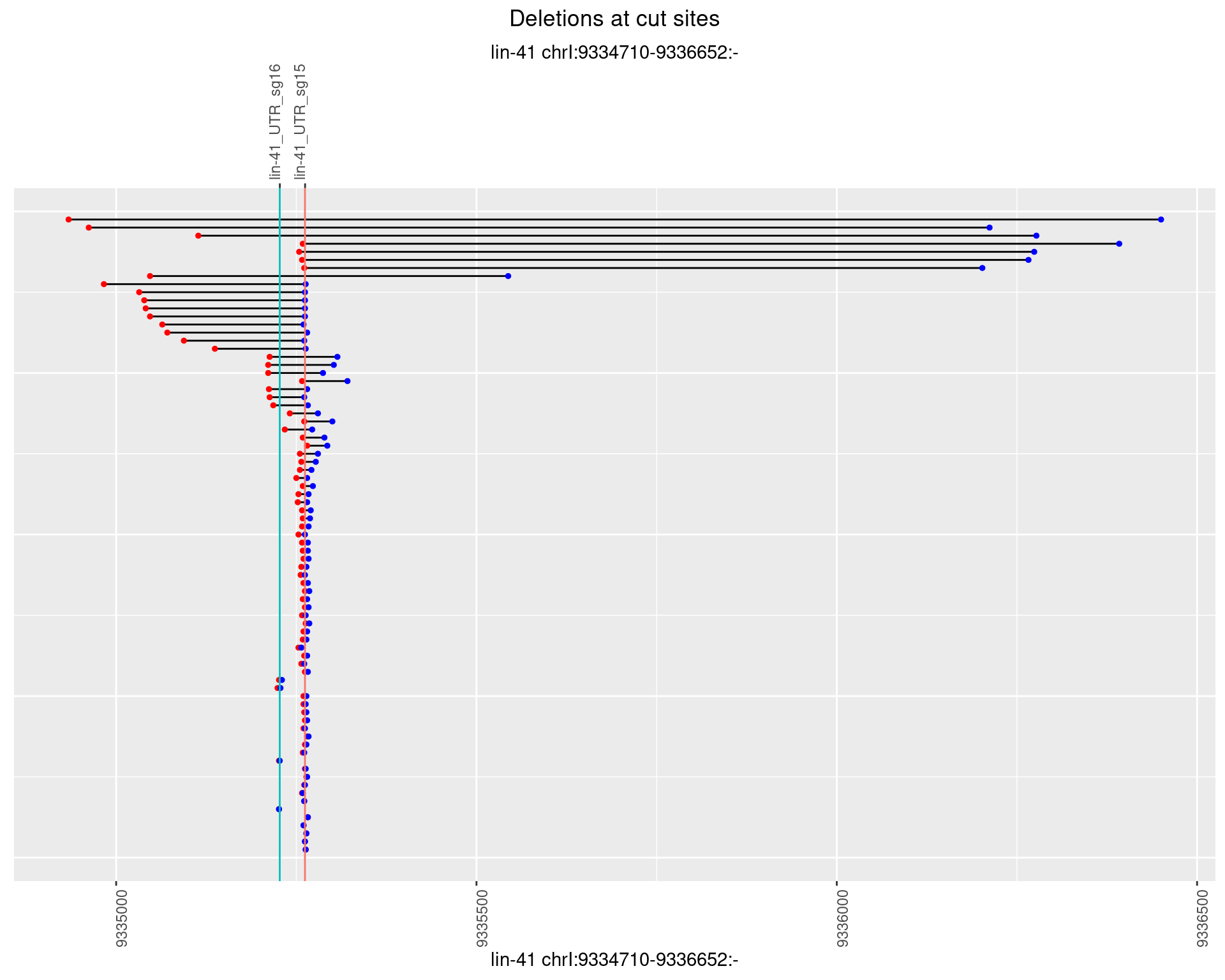
1.83.4 Freq: 0.001
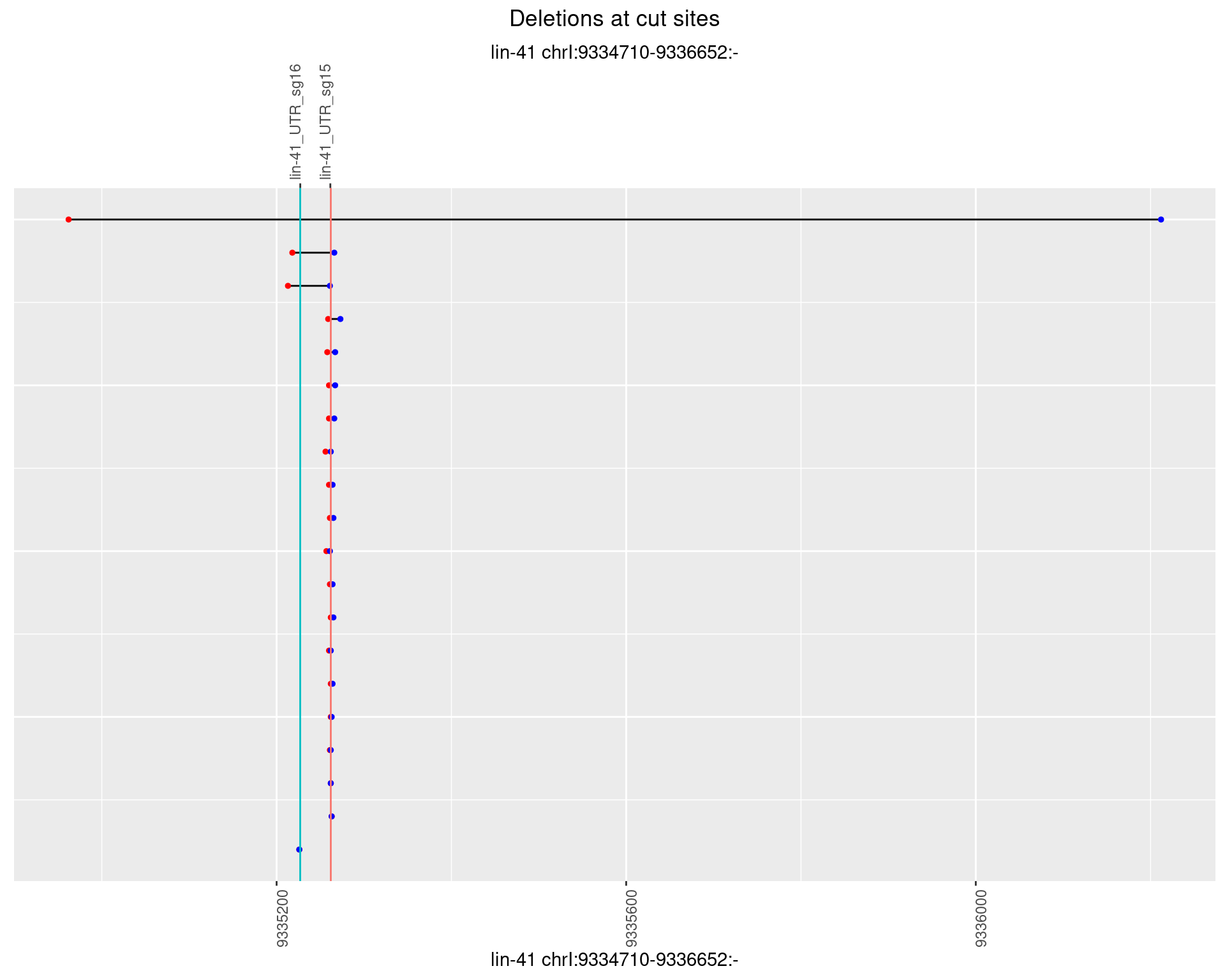
1.83.5 Freq: 0.01
1.83.6 Freq: 0.1
No plot to show
1.84 lin41_RNA_L1_2627_3end_UMI_R2
1.84.1 Freq: 0
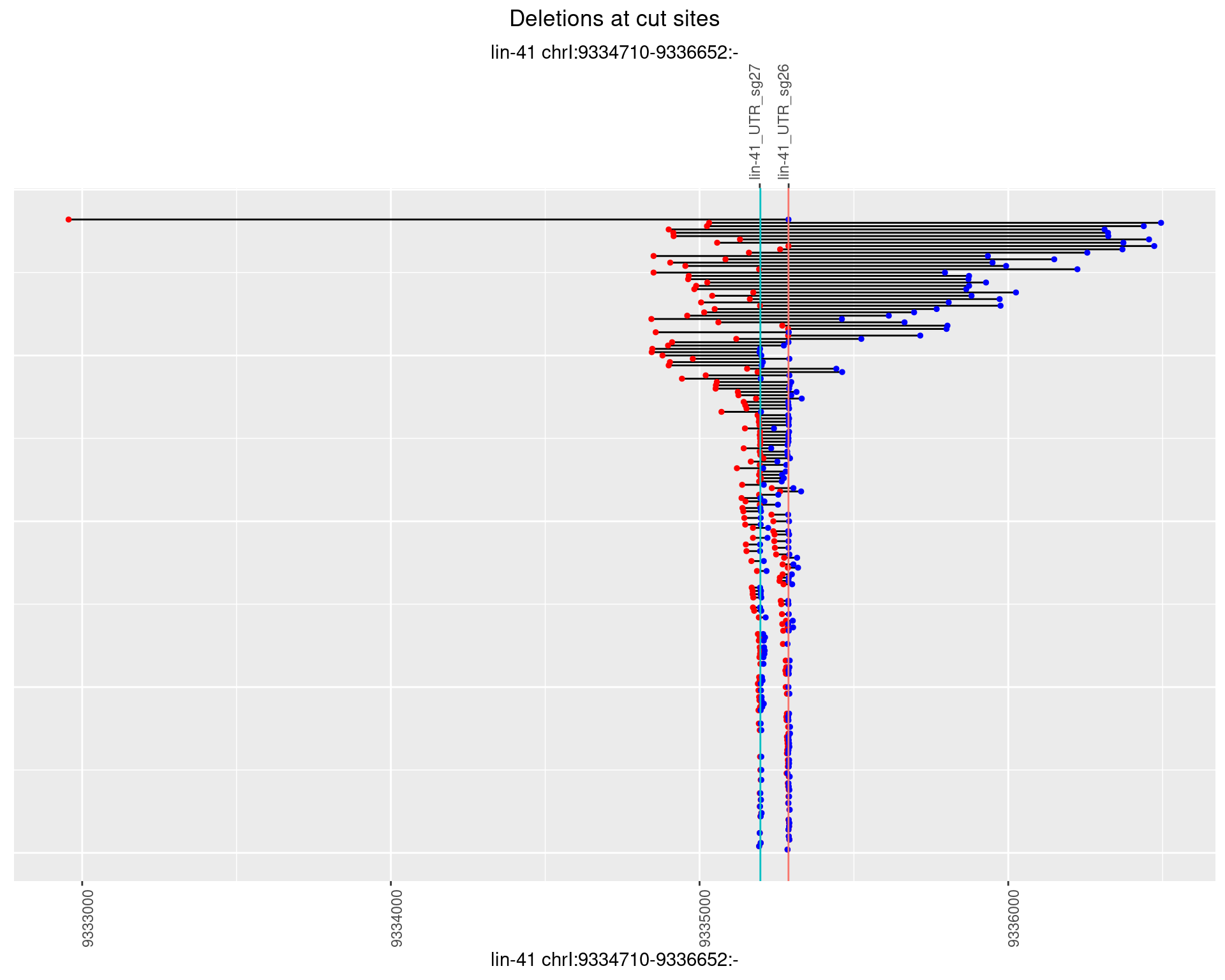
1.84.2 Freq: 1e-05
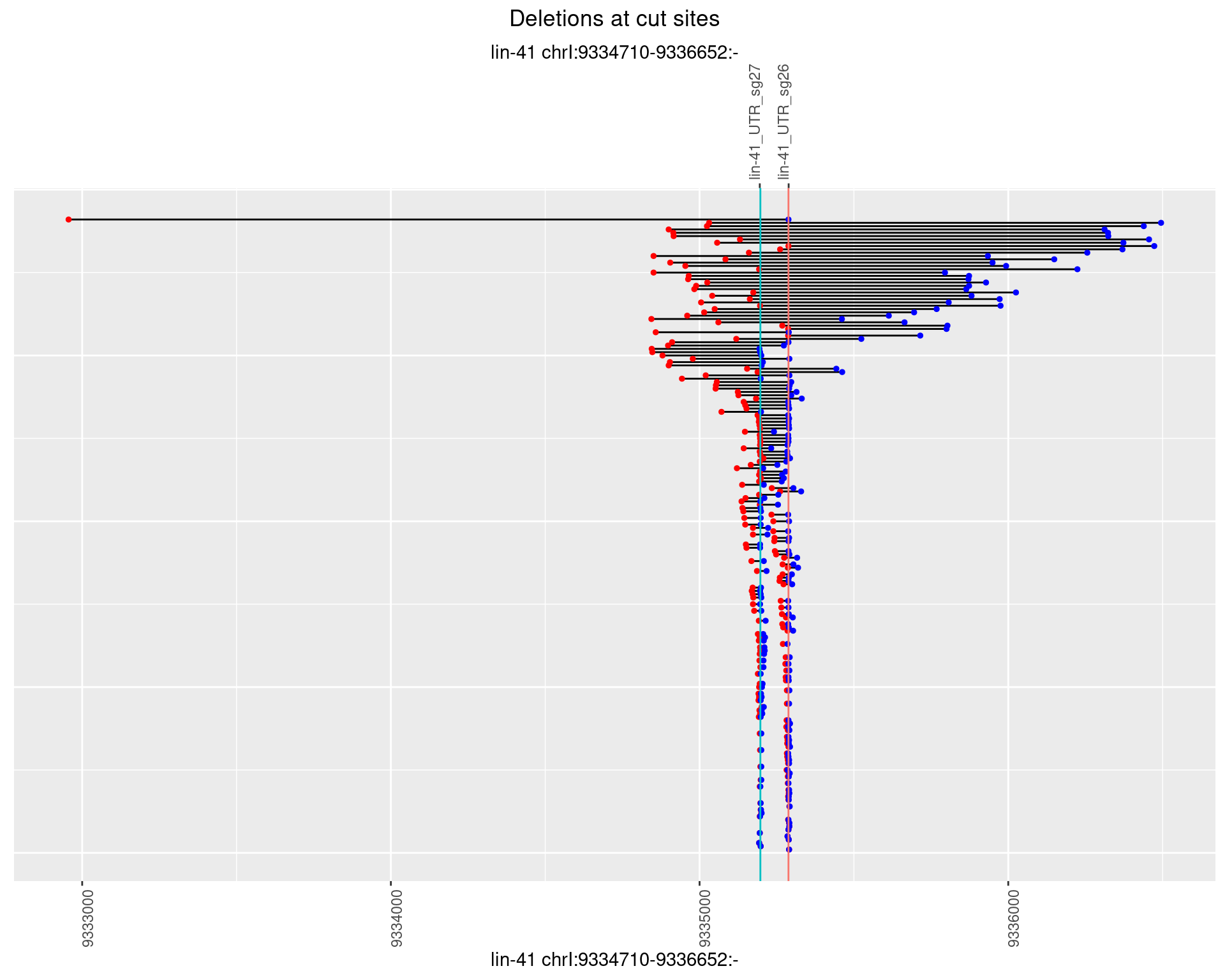
1.84.3 Freq: 1e-04
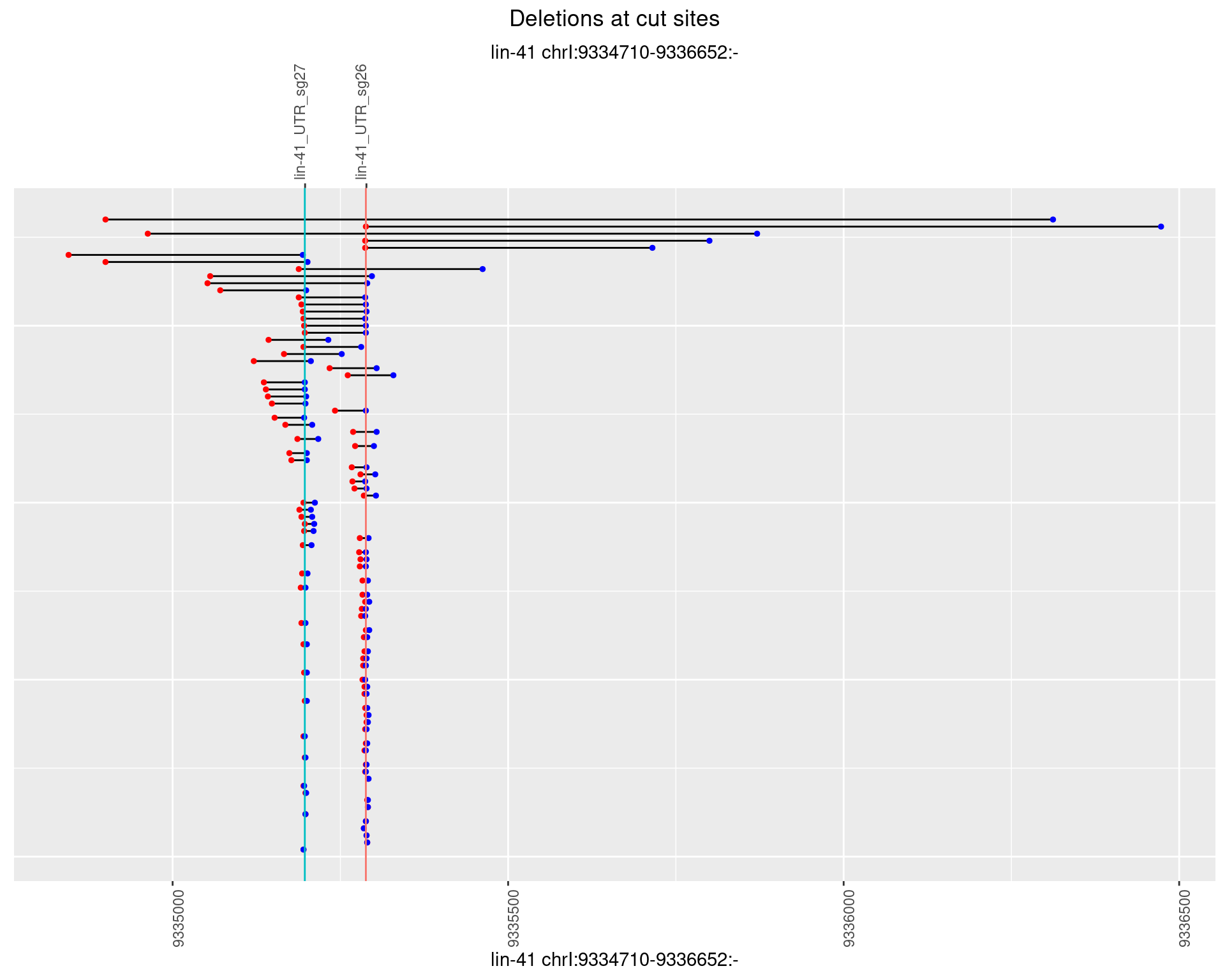
1.84.4 Freq: 0.001
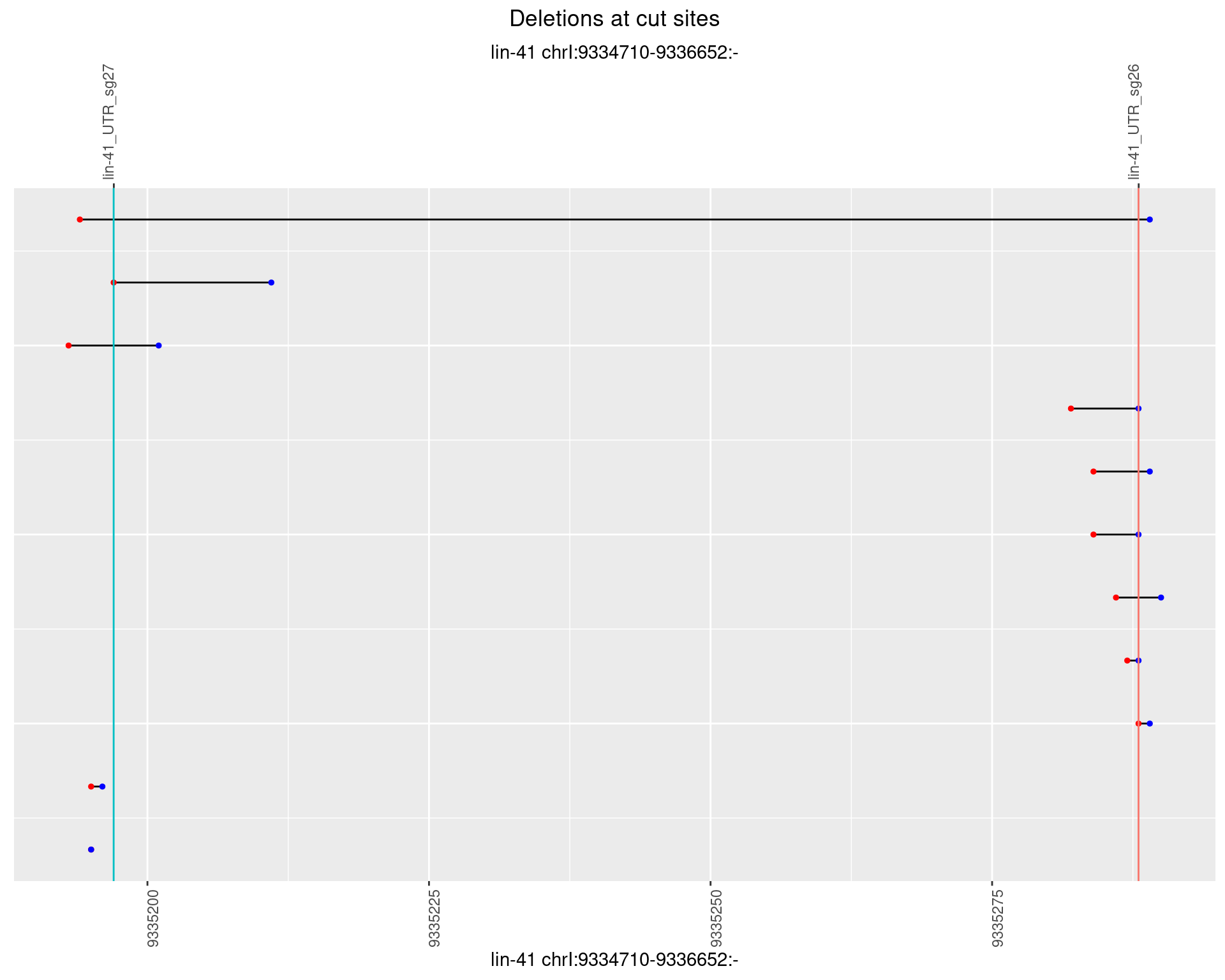
1.84.5 Freq: 0.01
No plot to show
1.84.6 Freq: 0.1
No plot to show
1.85 lin41_RNA_L1_pool2_3end_UMI_R2
1.85.1 Freq: 0
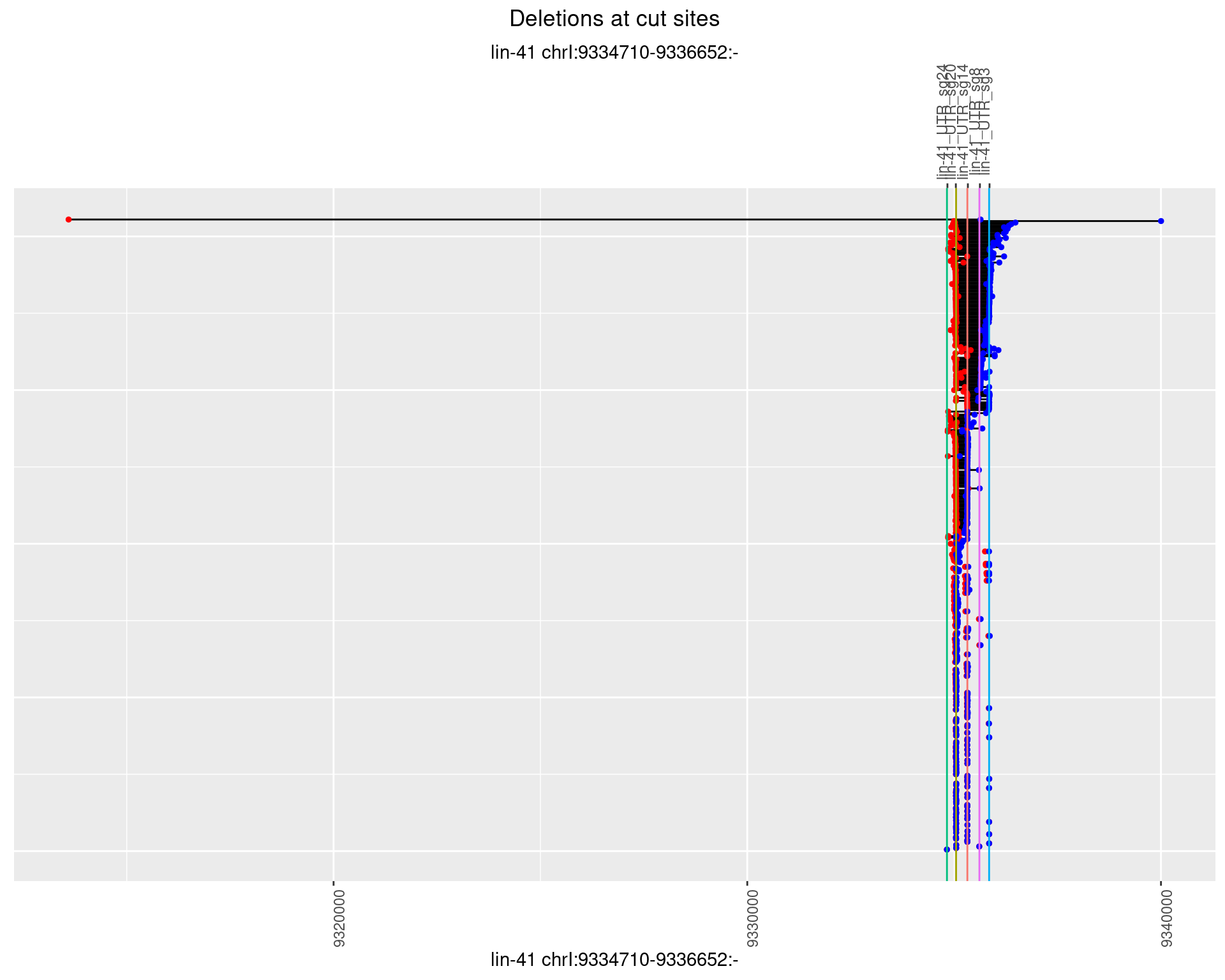
1.85.2 Freq: 1e-05
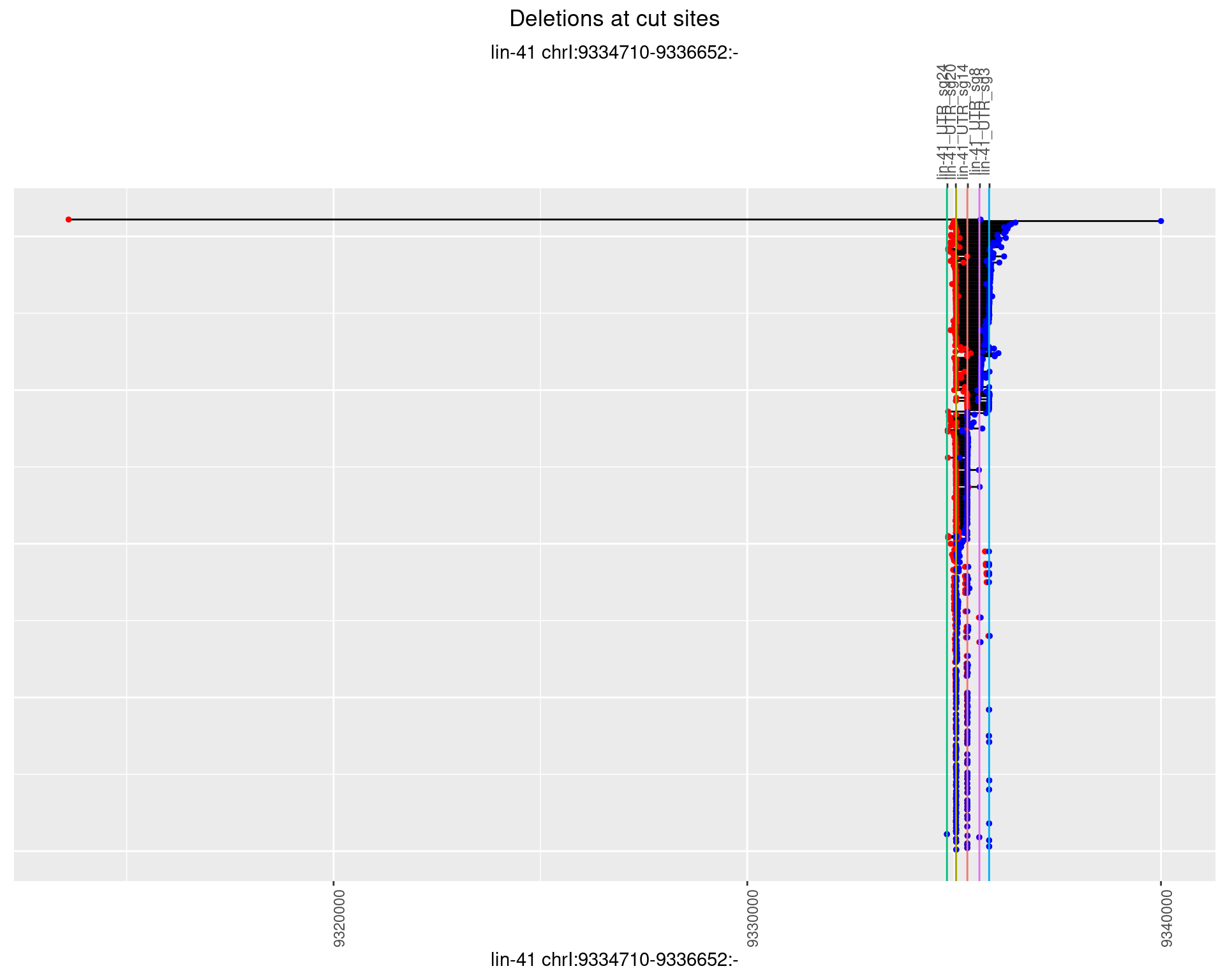
1.85.3 Freq: 1e-04
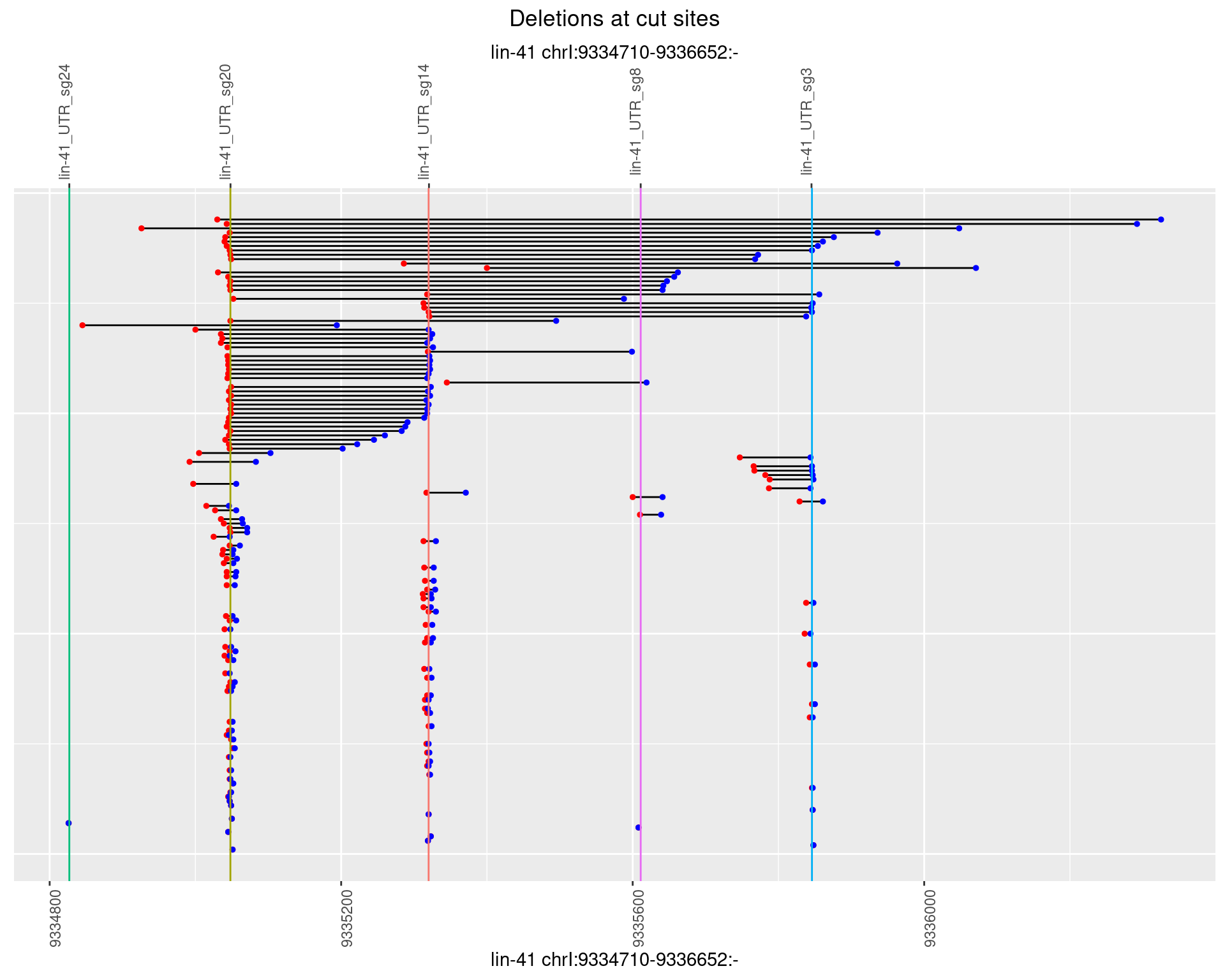
1.85.4 Freq: 0.001
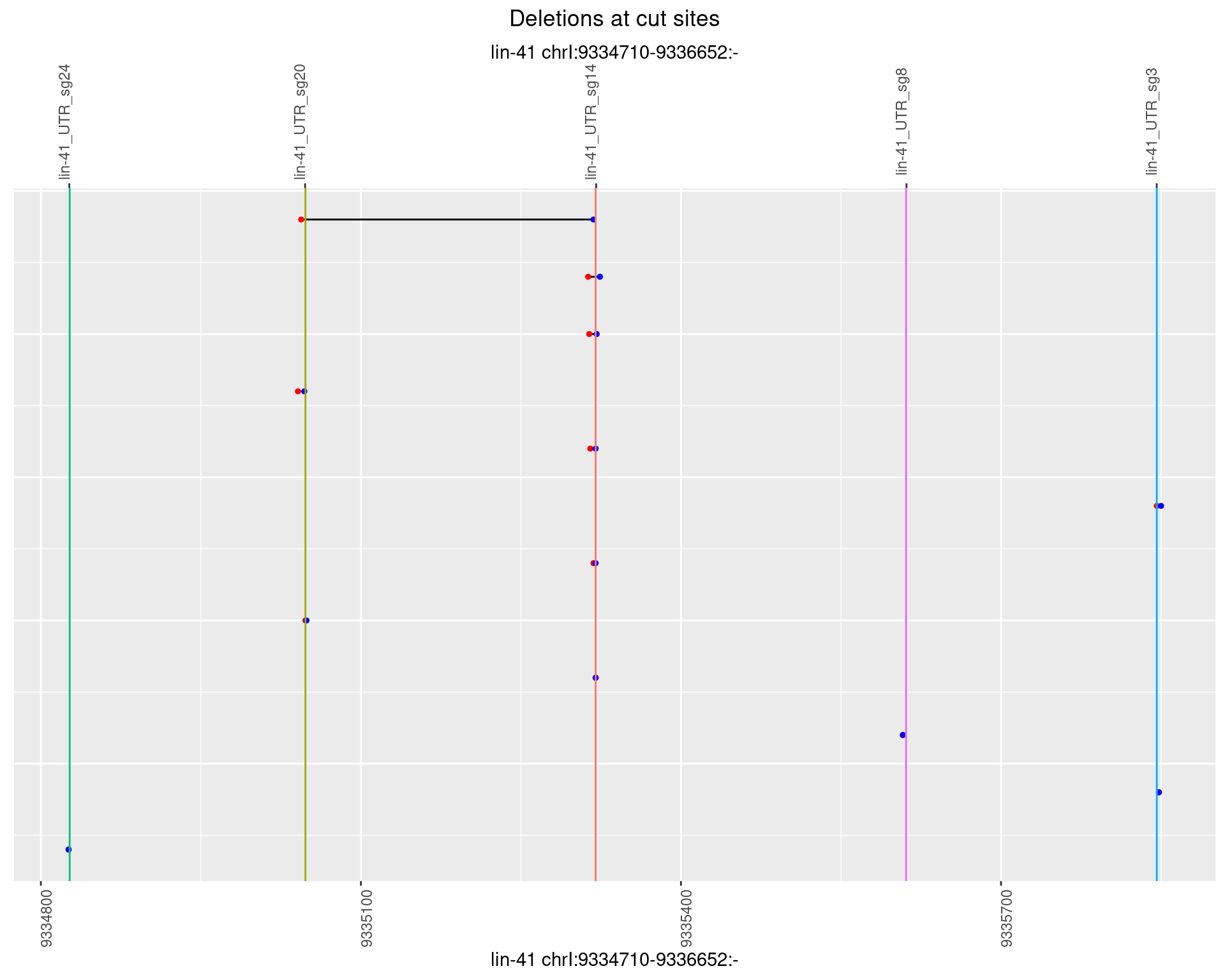
1.85.5 Freq: 0.01
1.85.6 Freq: 0.1
No plot to show
1.86 lin41_RNA_L1_pool3_3end_UMI_R2
1.86.1 Freq: 0
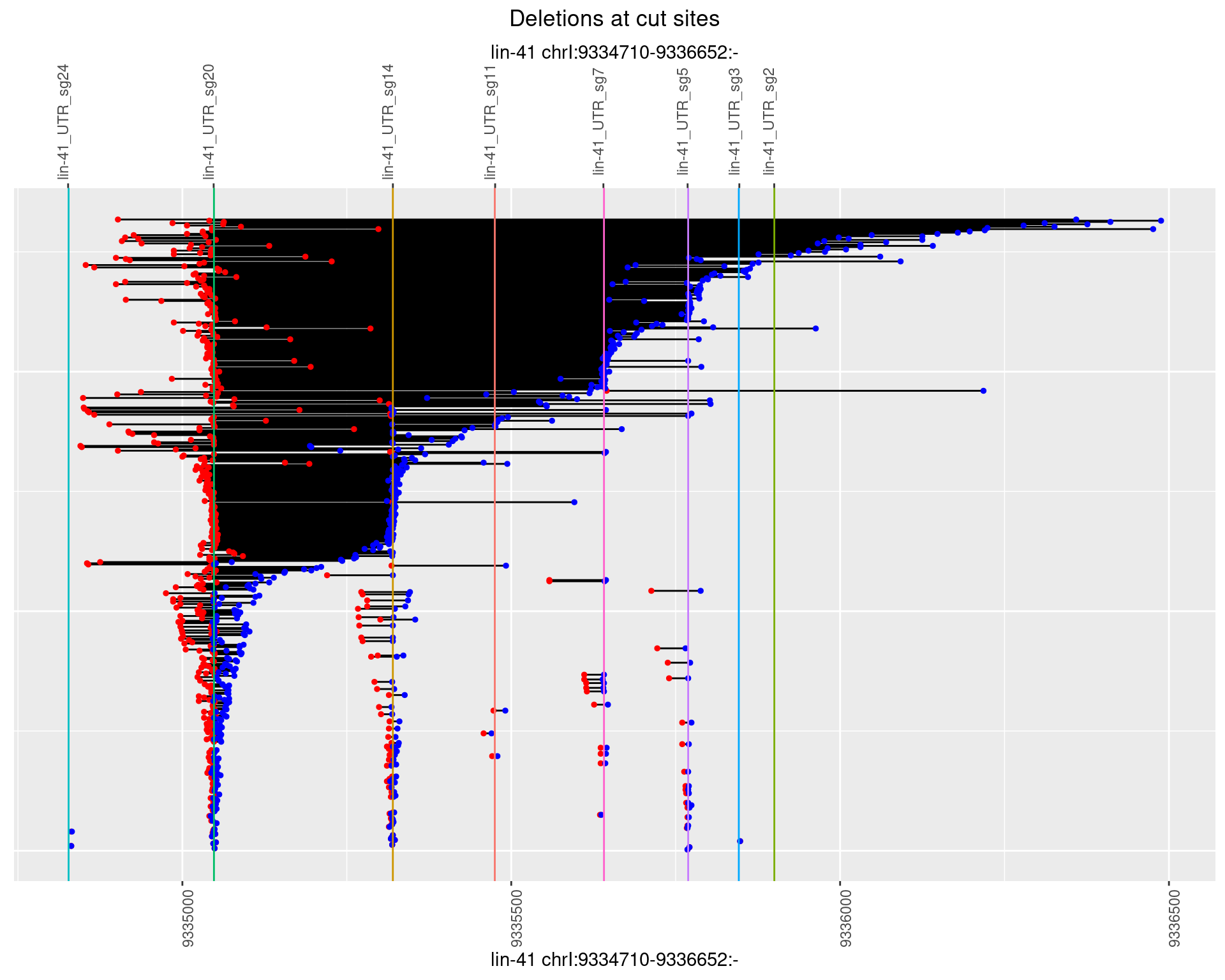
1.86.2 Freq: 1e-05
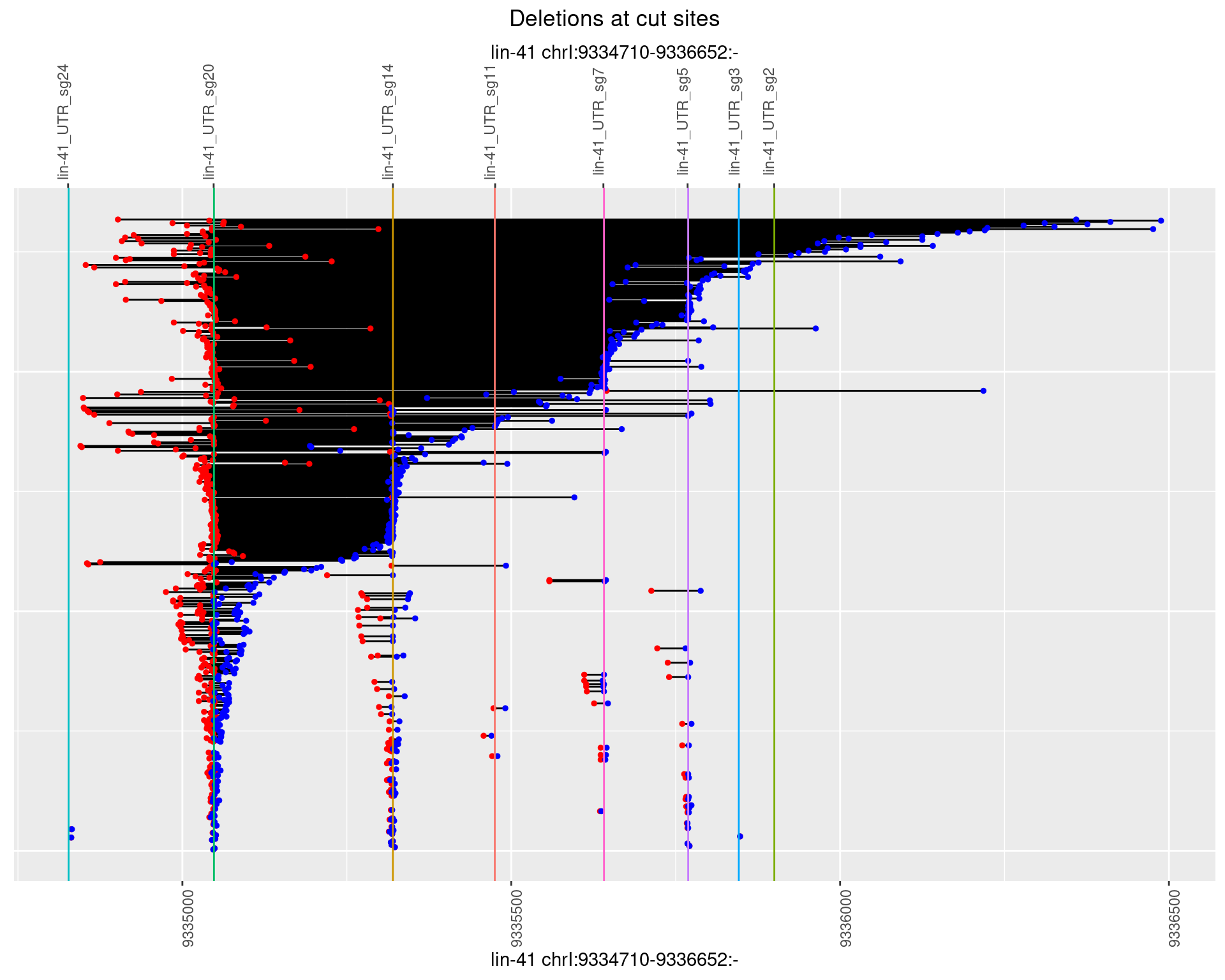
1.86.3 Freq: 1e-04
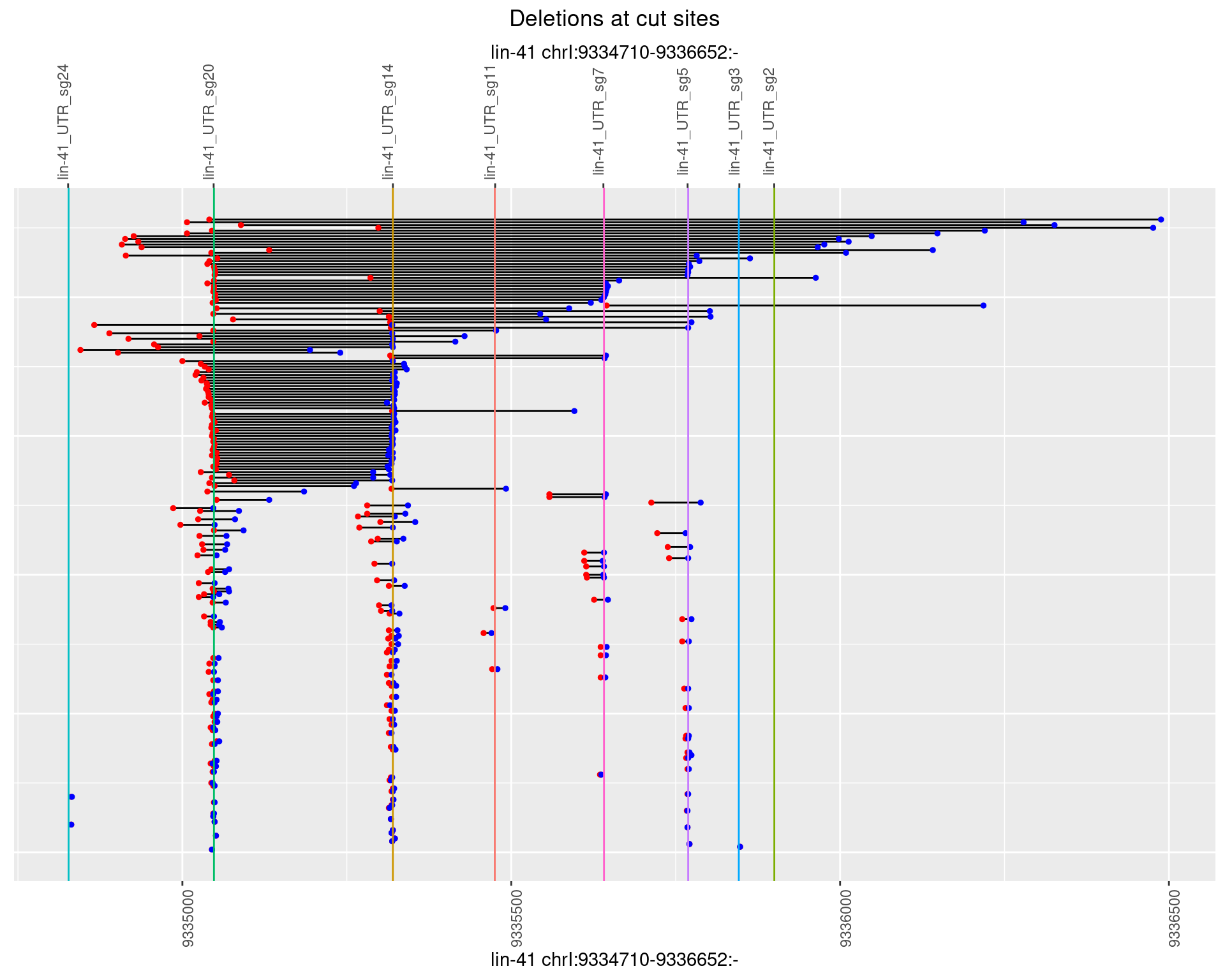
1.86.4 Freq: 0.001
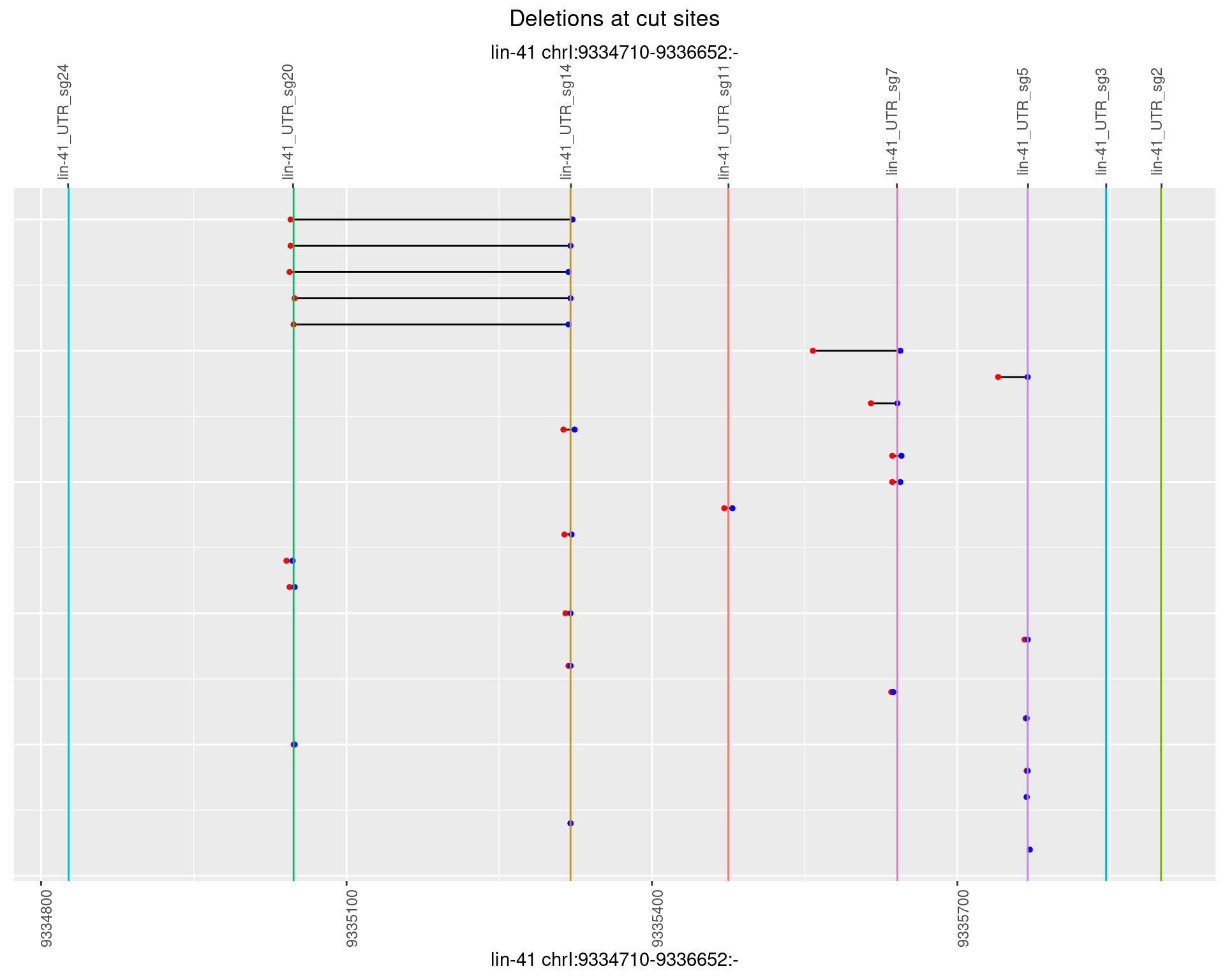
1.86.5 Freq: 0.01
1.86.6 Freq: 0.1
No plot to show
1.87 lin41_RNA_L4_N2_3end_UMI_R2
1.87.1 Freq: 0
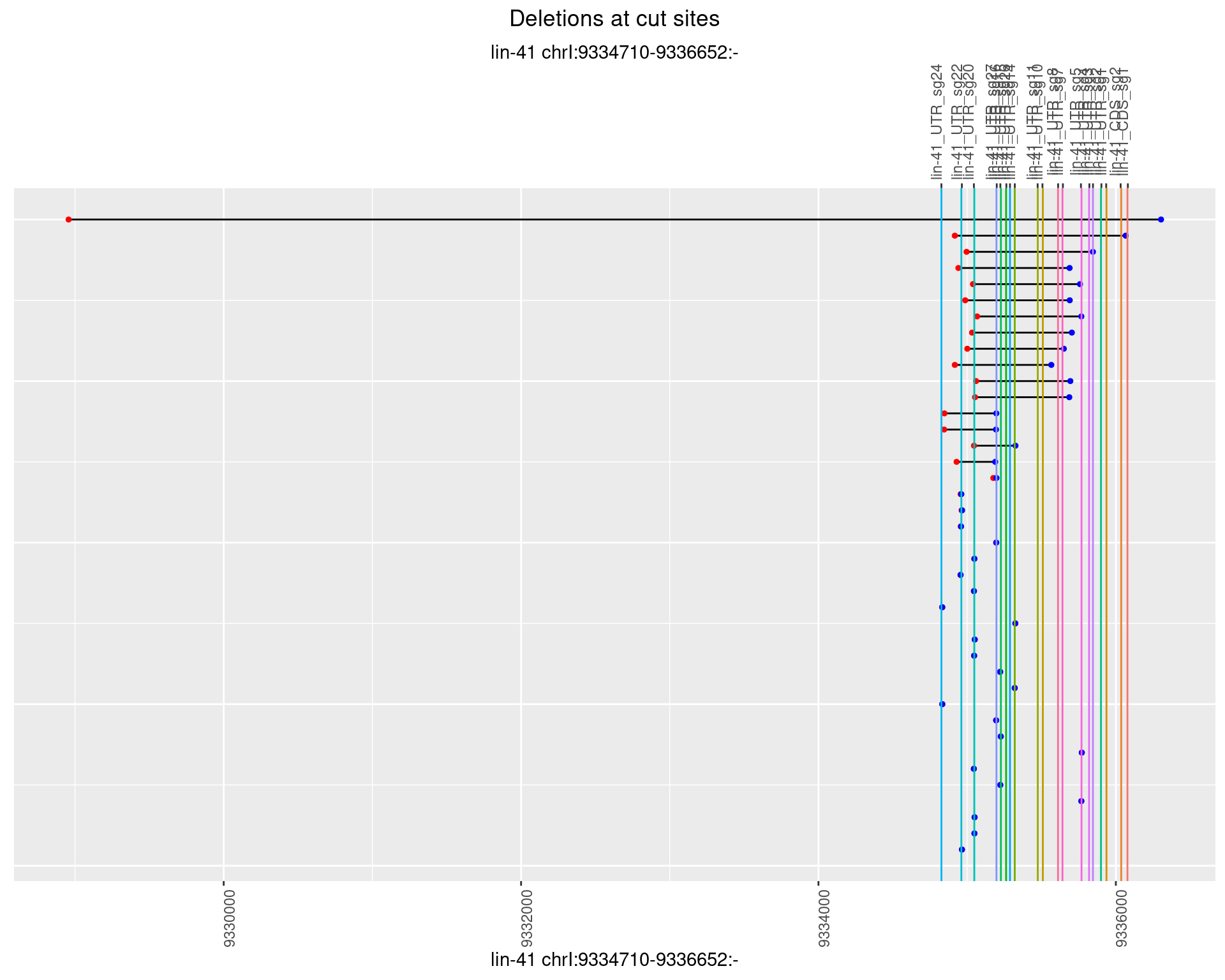
1.87.2 Freq: 1e-05
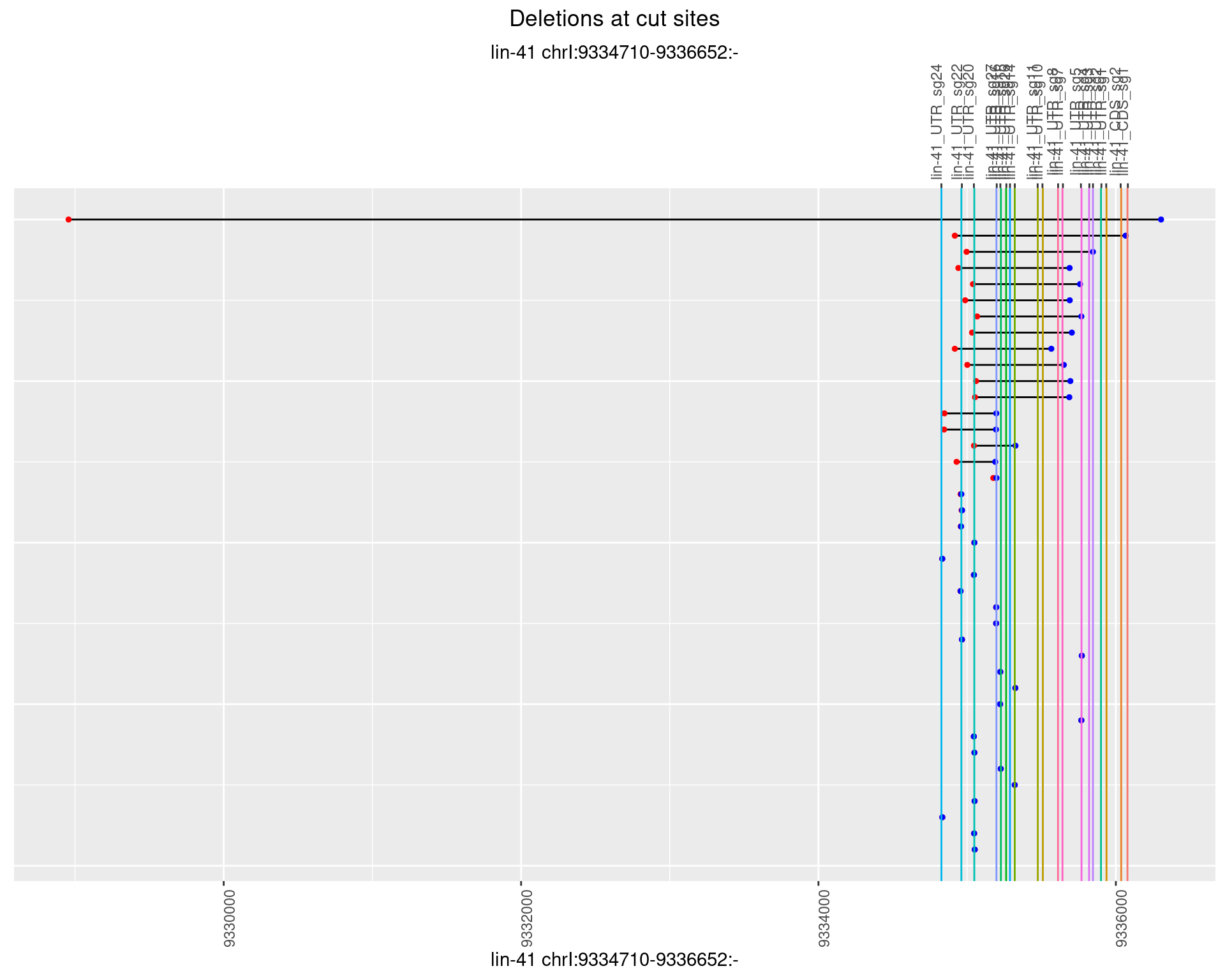
1.87.3 Freq: 1e-04
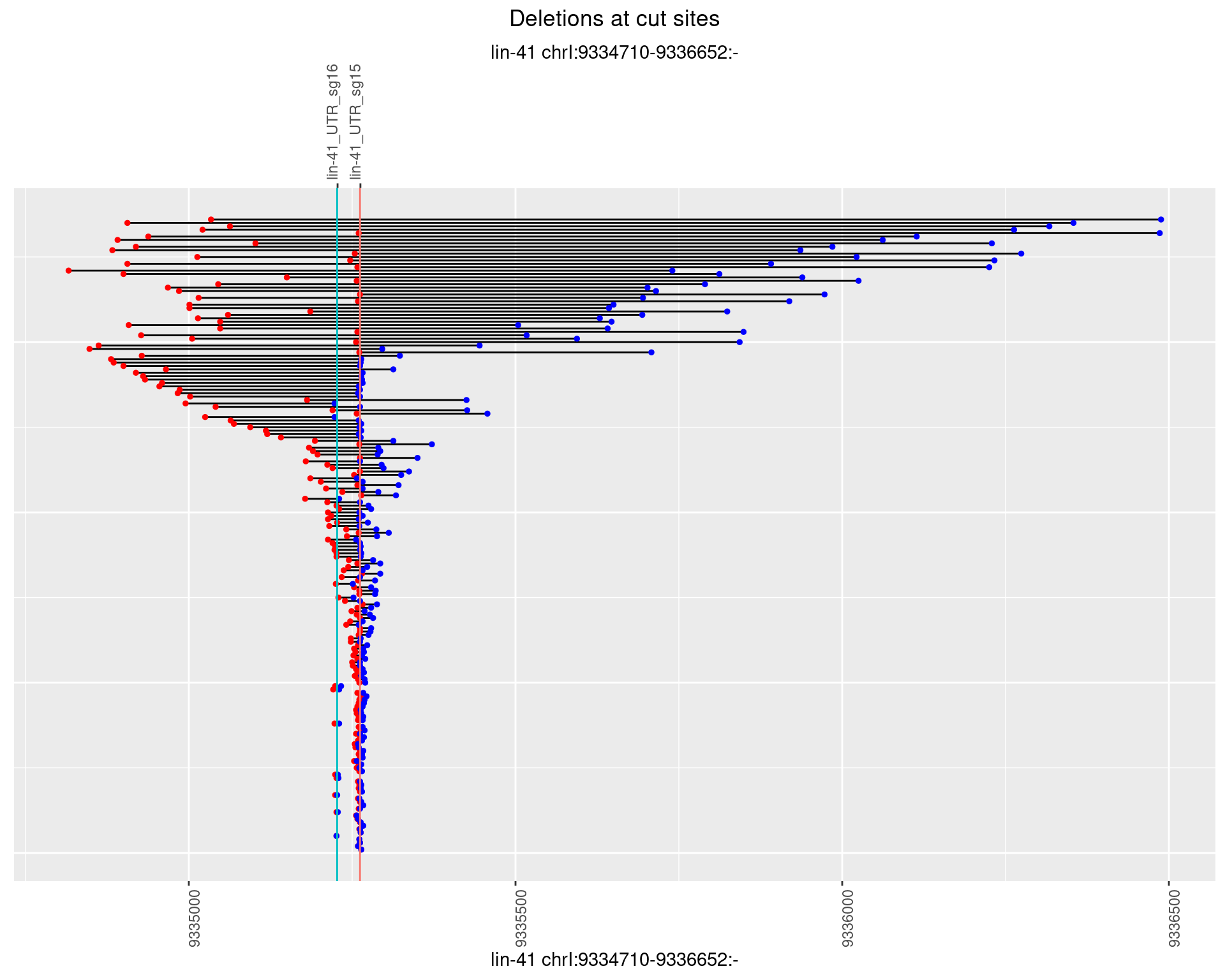
1.87.4 Freq: 0.001
1.87.5 Freq: 0.01
No plot to show
1.87.6 Freq: 0.1
No plot to show
1.88 lin41_RNA_L4_1516_3end_UMI_R2
1.88.1 Freq: 0
1.88.2 Freq: 1e-05
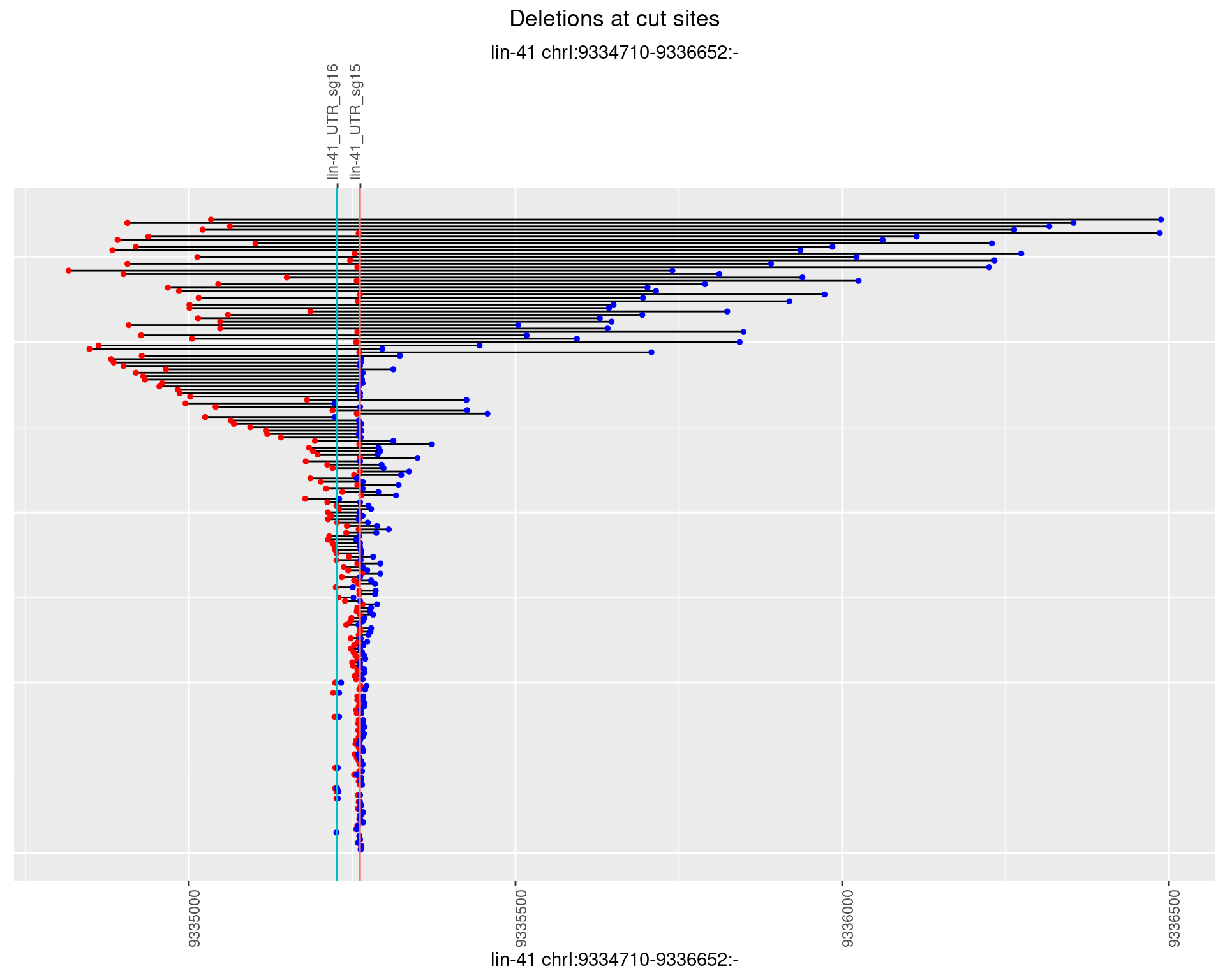
1.88.3 Freq: 1e-04
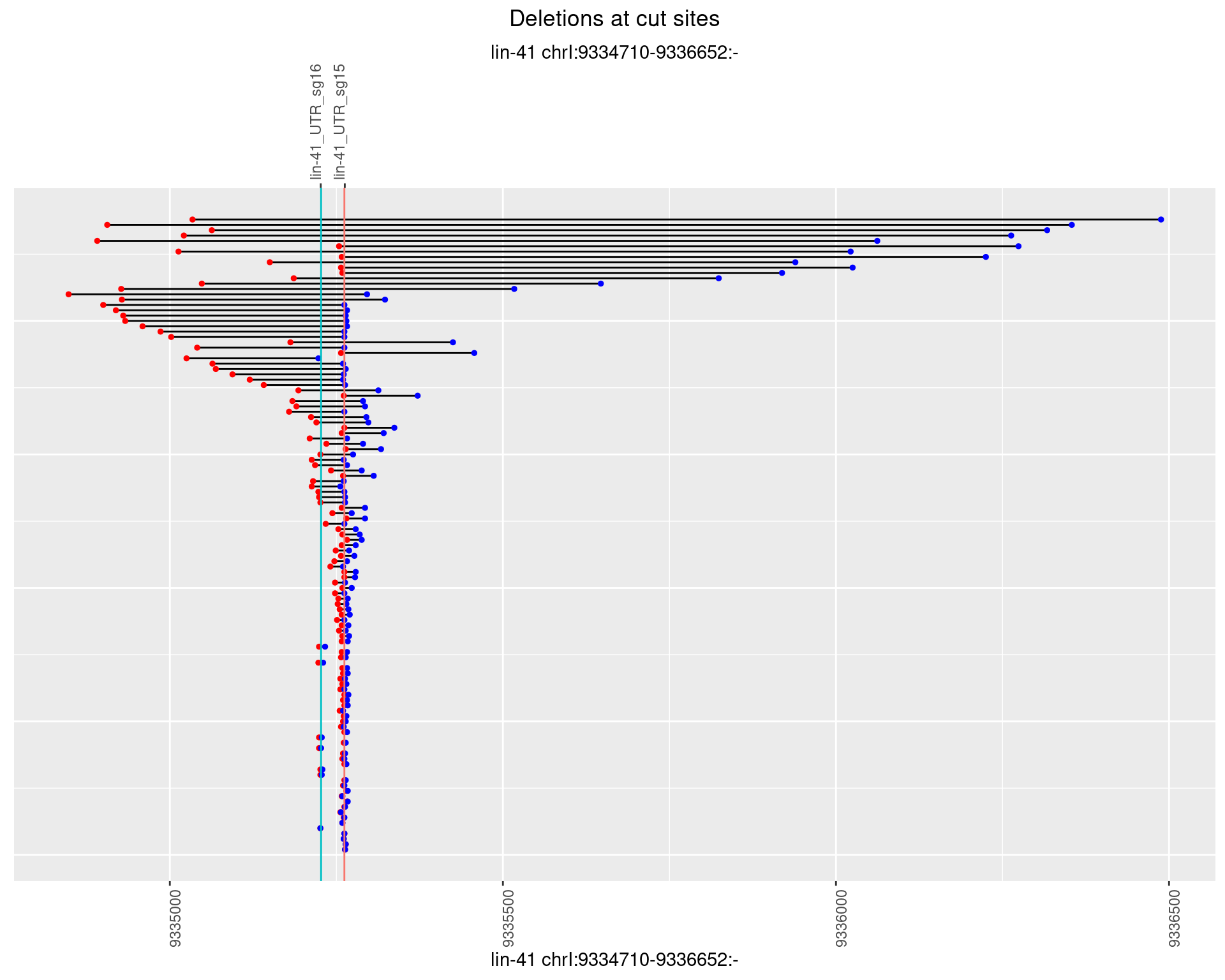
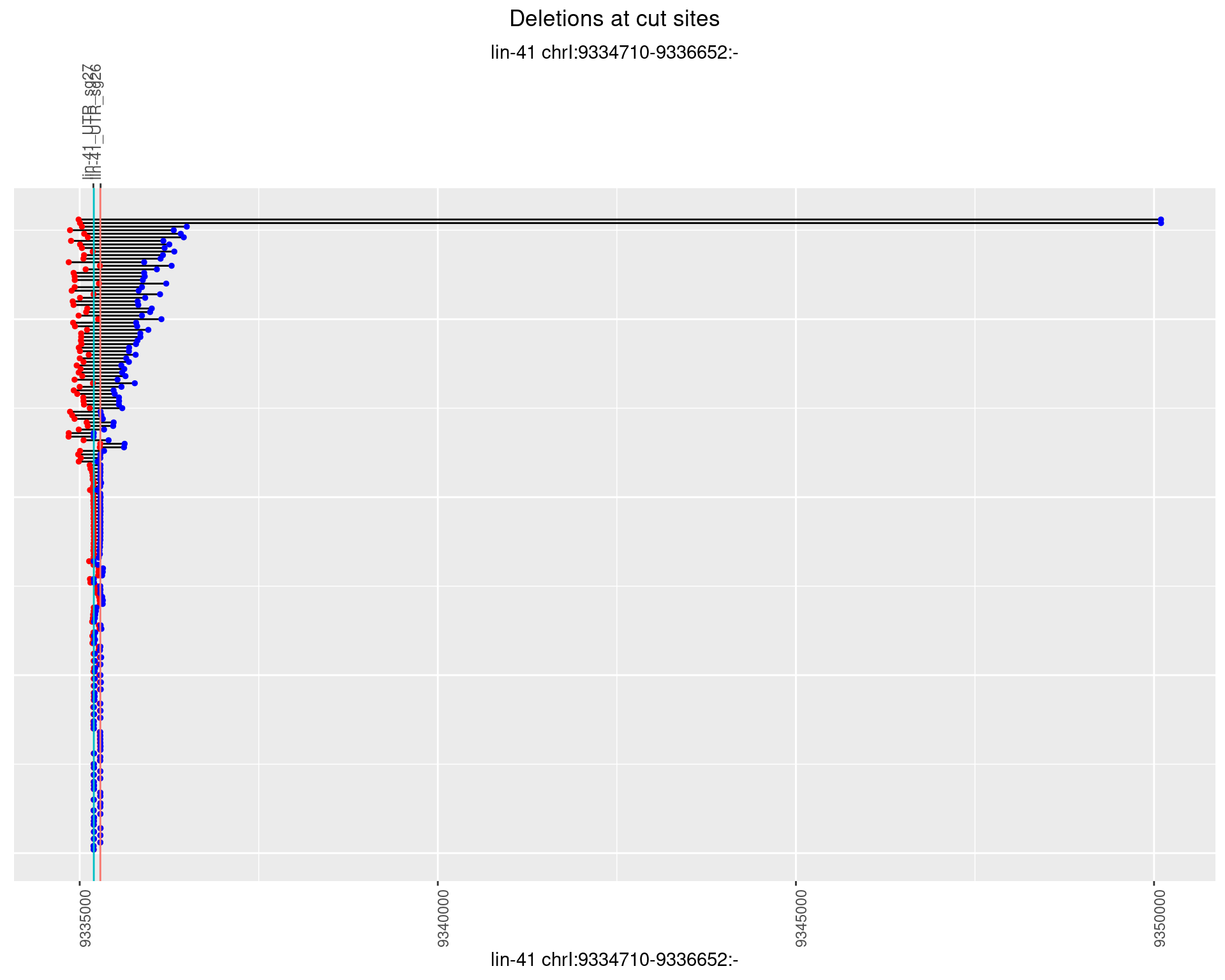
1.88.4 Freq: 0.001
1.88.5 Freq: 0.01
1.88.6 Freq: 0.1
No plot to show
1.89 lin41_RNA_L4_2627_3end_UMI_R2
1.89.1 Freq: 0
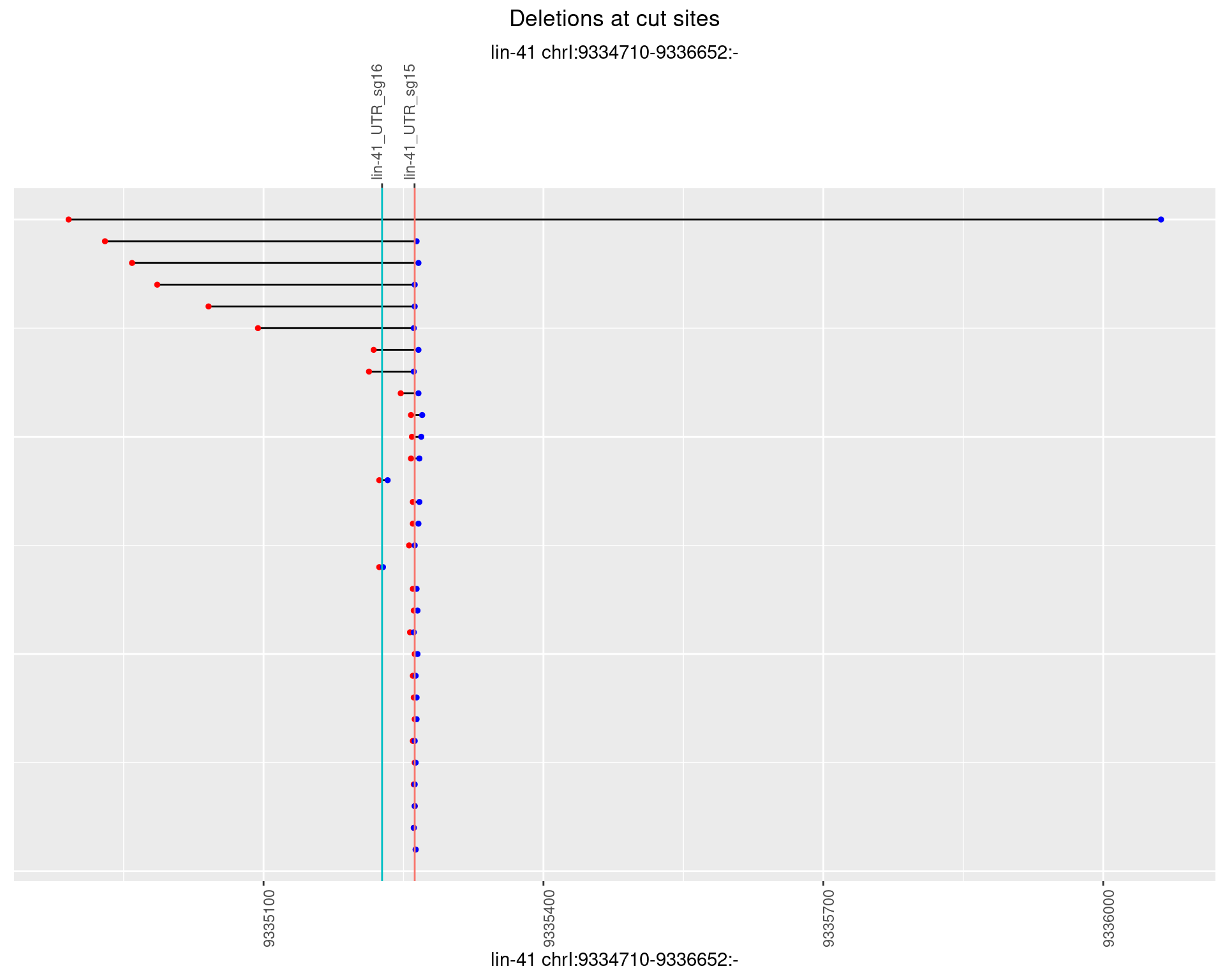
1.89.2 Freq: 1e-05
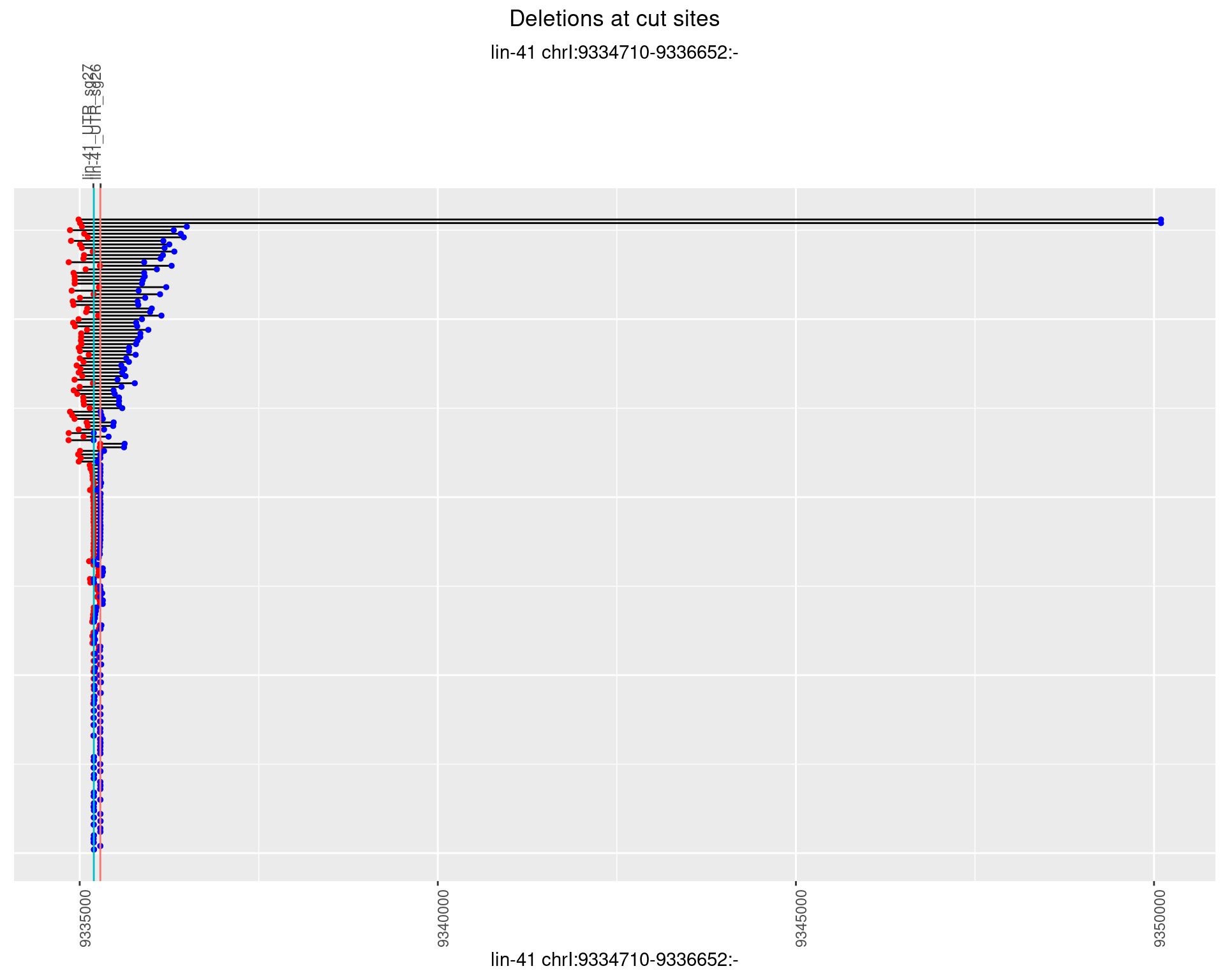
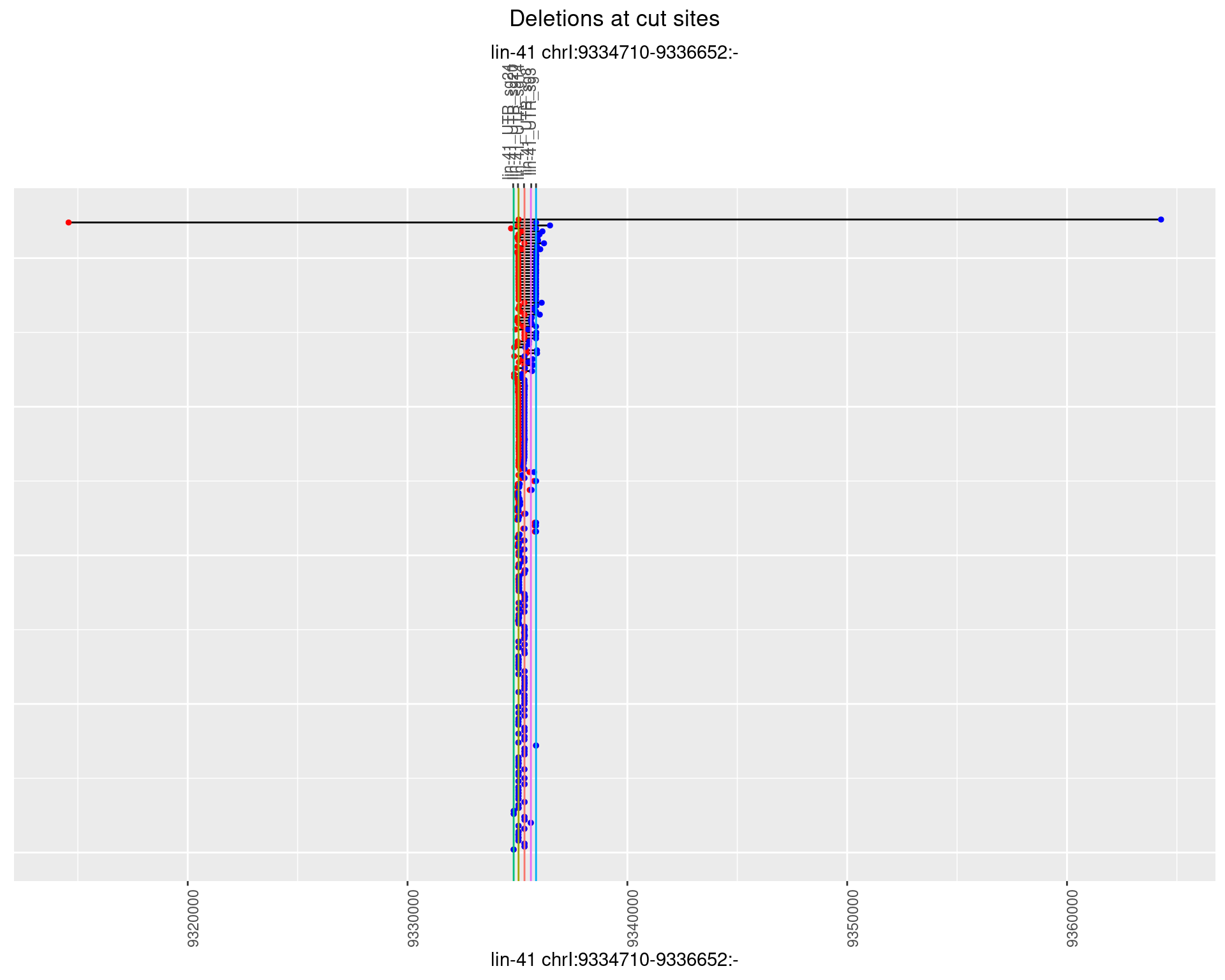
1.89.3 Freq: 1e-04
1.89.4 Freq: 0.001
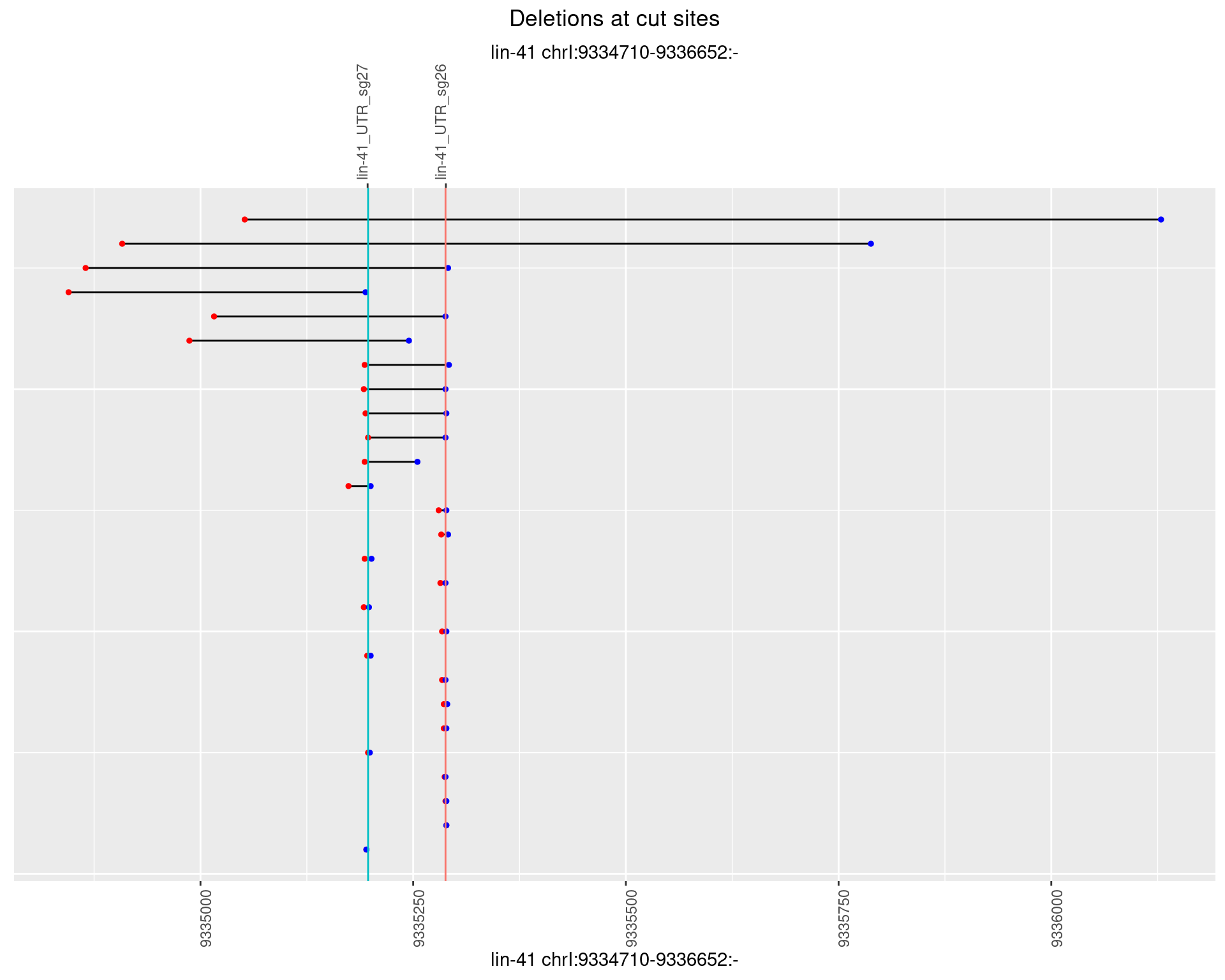
1.89.5 Freq: 0.01
No plot to show
1.89.6 Freq: 0.1
No plot to show
1.90 lin41_RNA_L4_pool2_3end_UMI_R2
1.90.1 Freq: 0
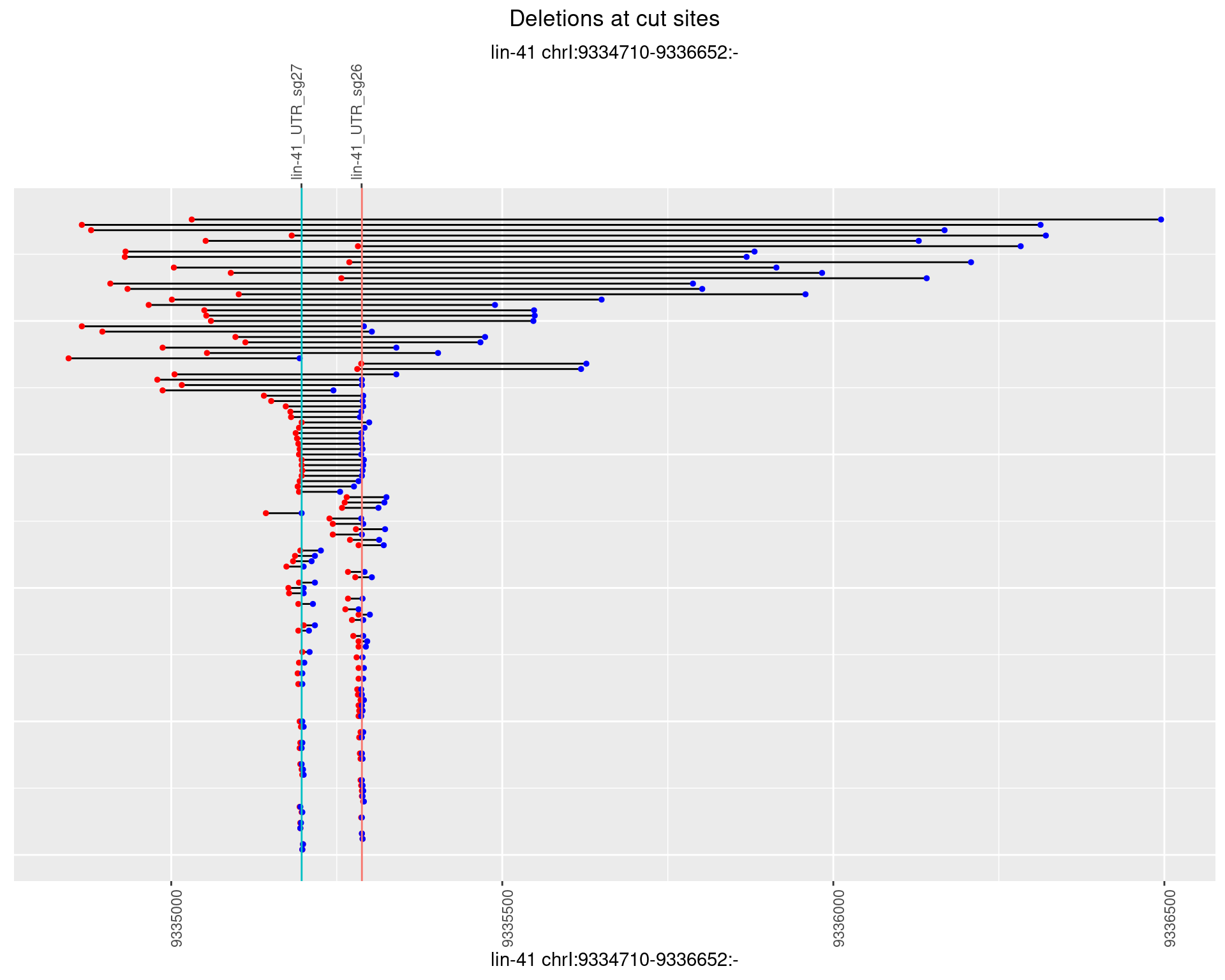
1.90.2 Freq: 1e-05
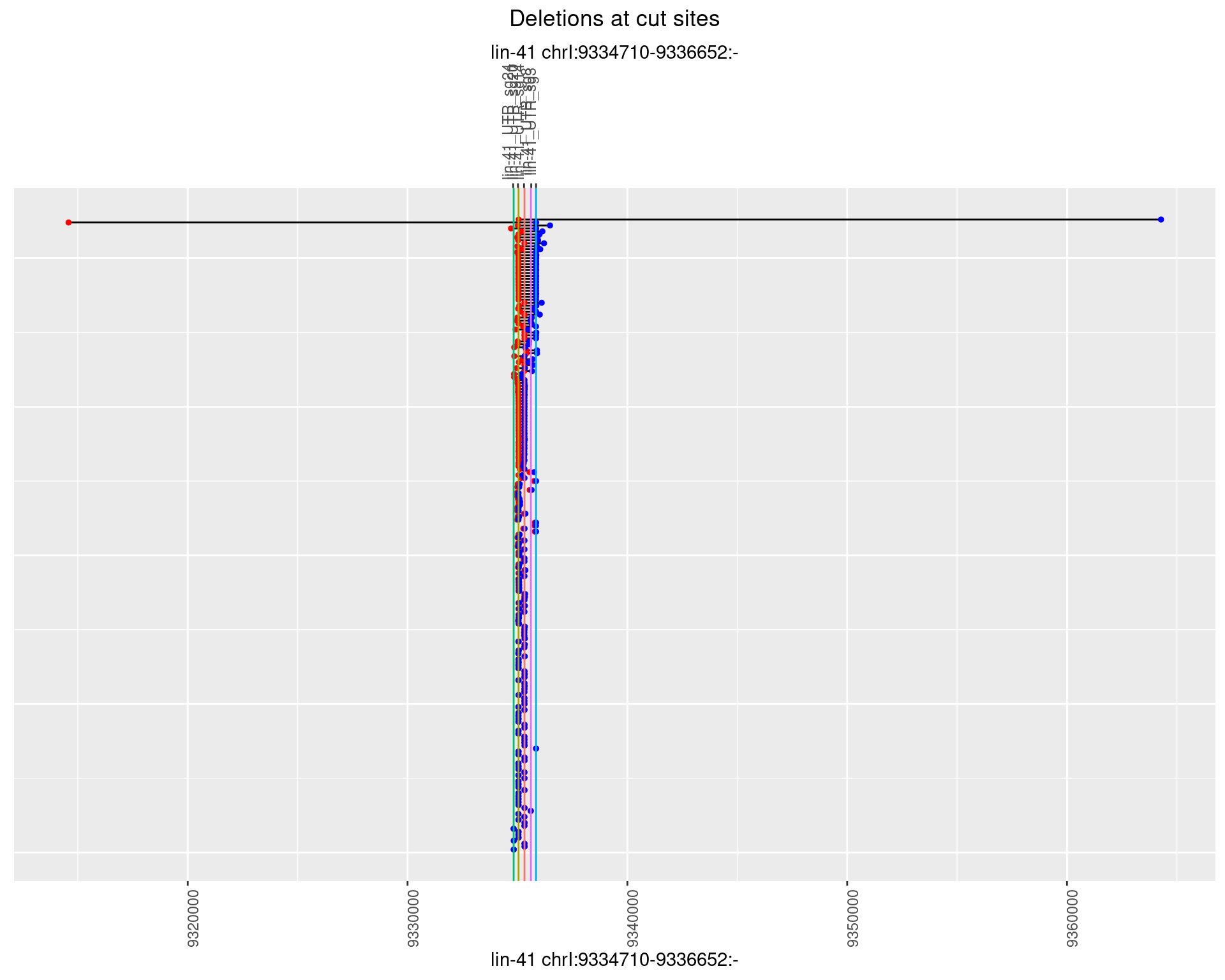
1.90.3 Freq: 1e-04
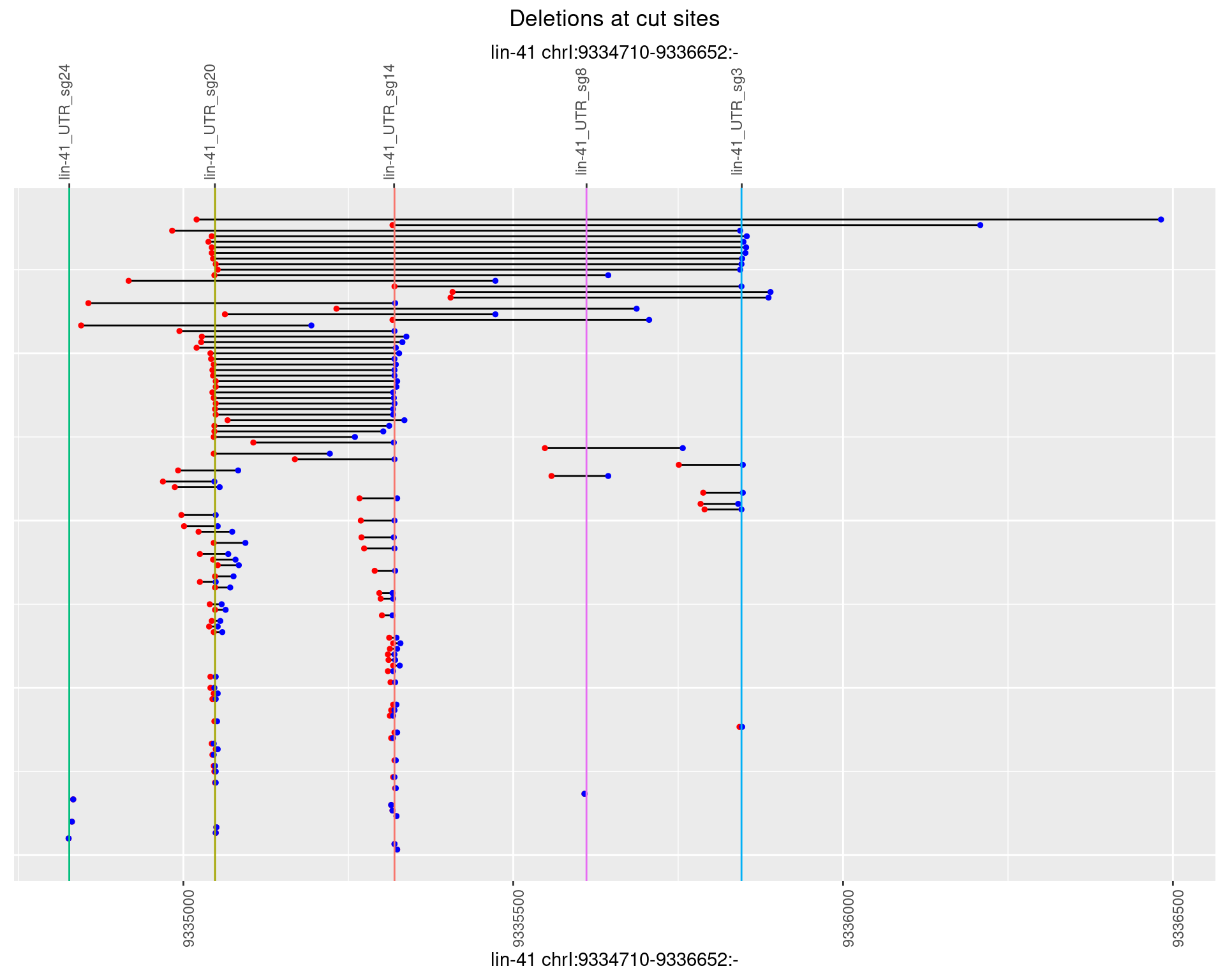
1.90.4 Freq: 0.001
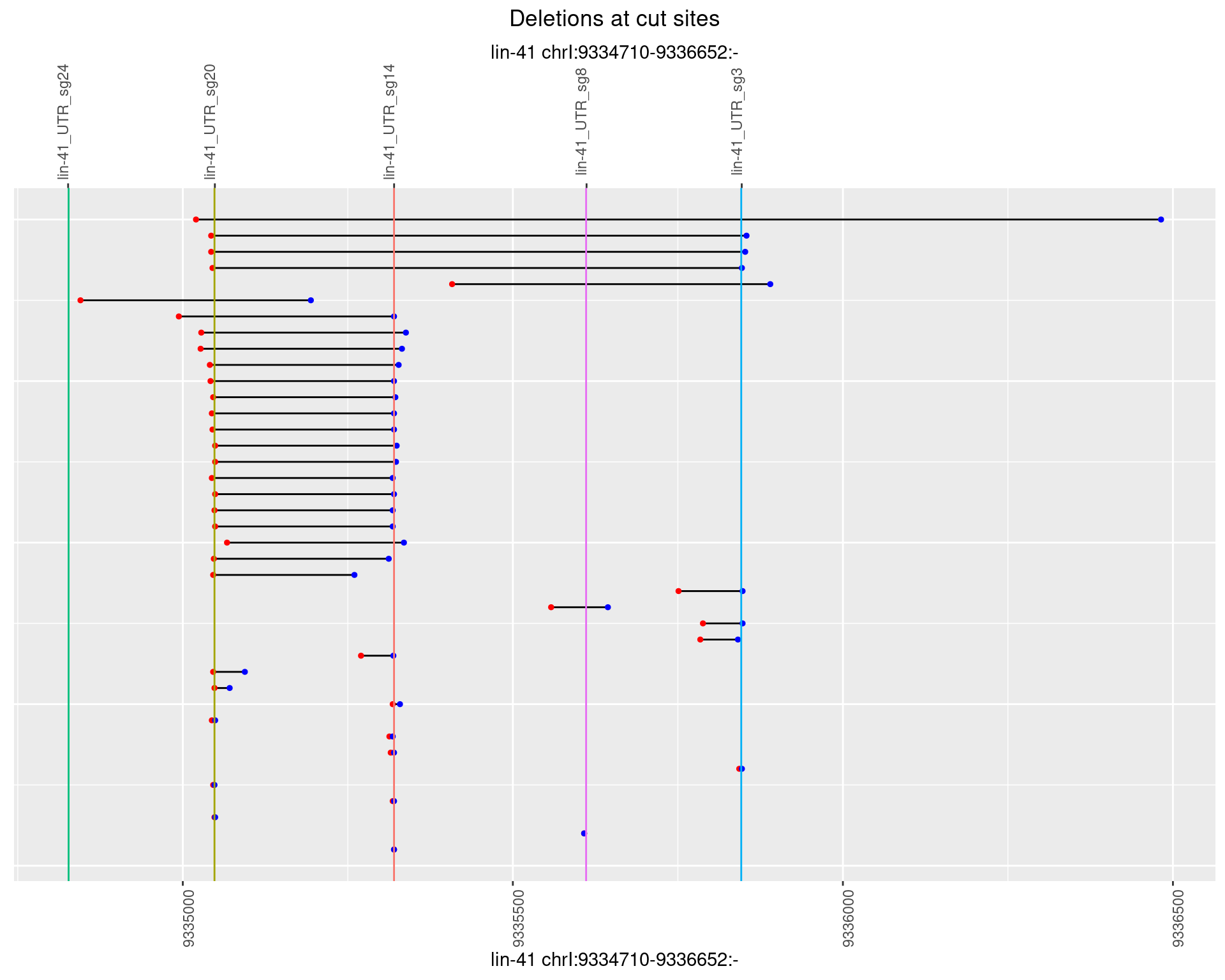
1.90.5 Freq: 0.01
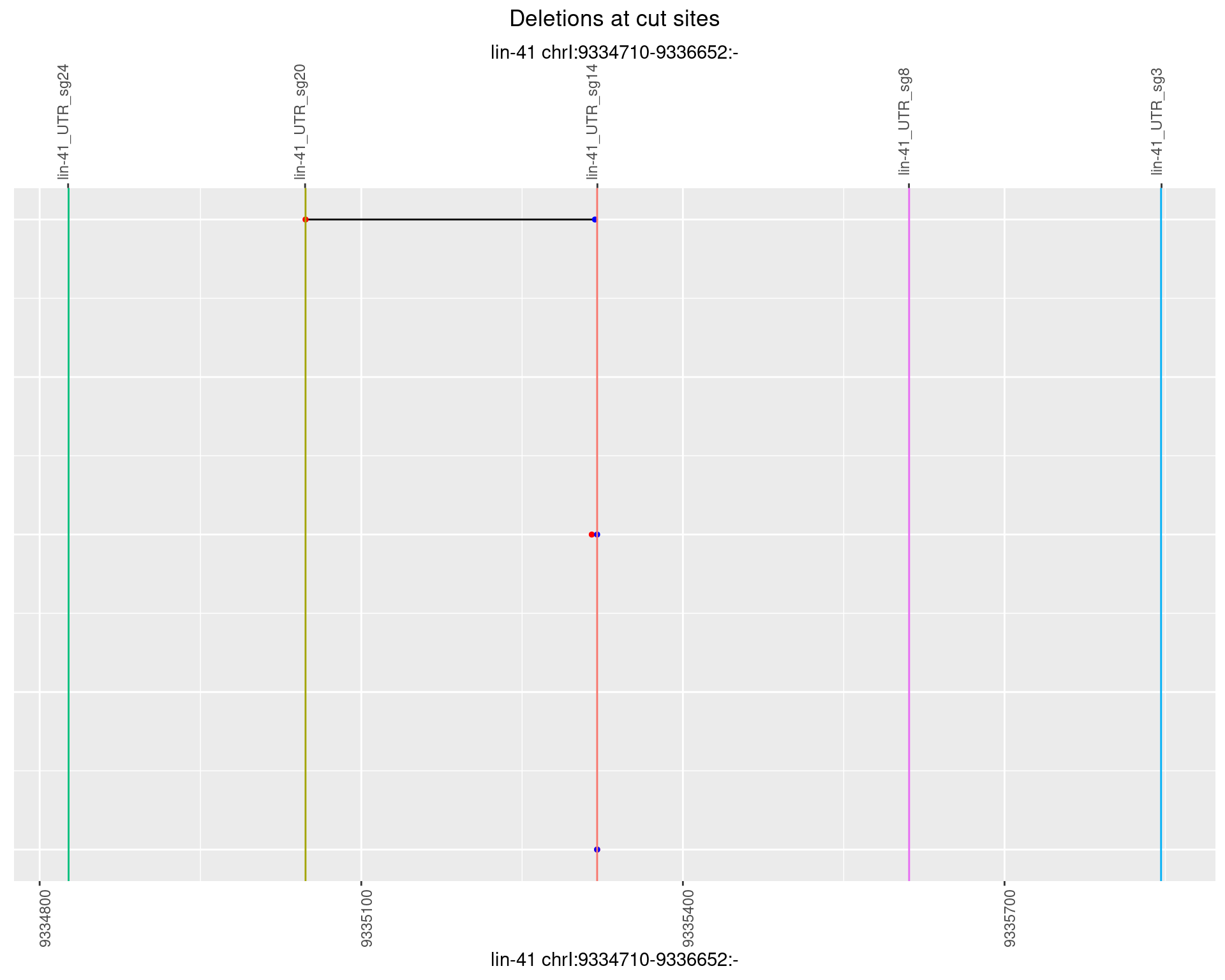
1.90.6 Freq: 0.1
No plot to show
1.91 lin41_RNA_L4_pool3_3end_UMI_R2
1.91.1 Freq: 0
1.91.2 Freq: 1e-05
1.91.3 Freq: 1e-04
1.91.4 Freq: 0.001
1.91.5 Freq: 0.01
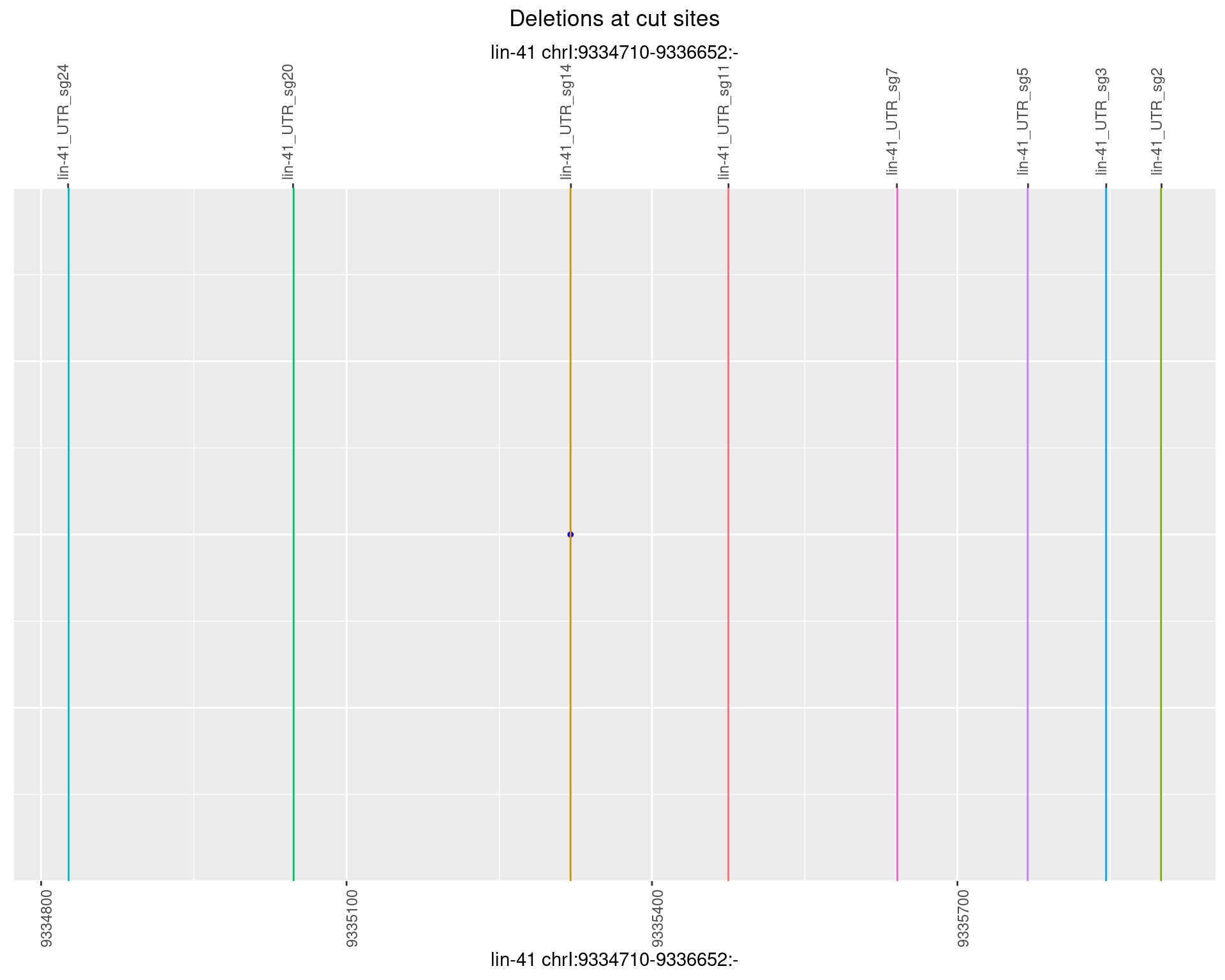
1.91.6 Freq: 0.1
No plot to show
1.92 lin41_RNA_L1_N2_middle_UMI_R2
1.92.1 Freq: 0
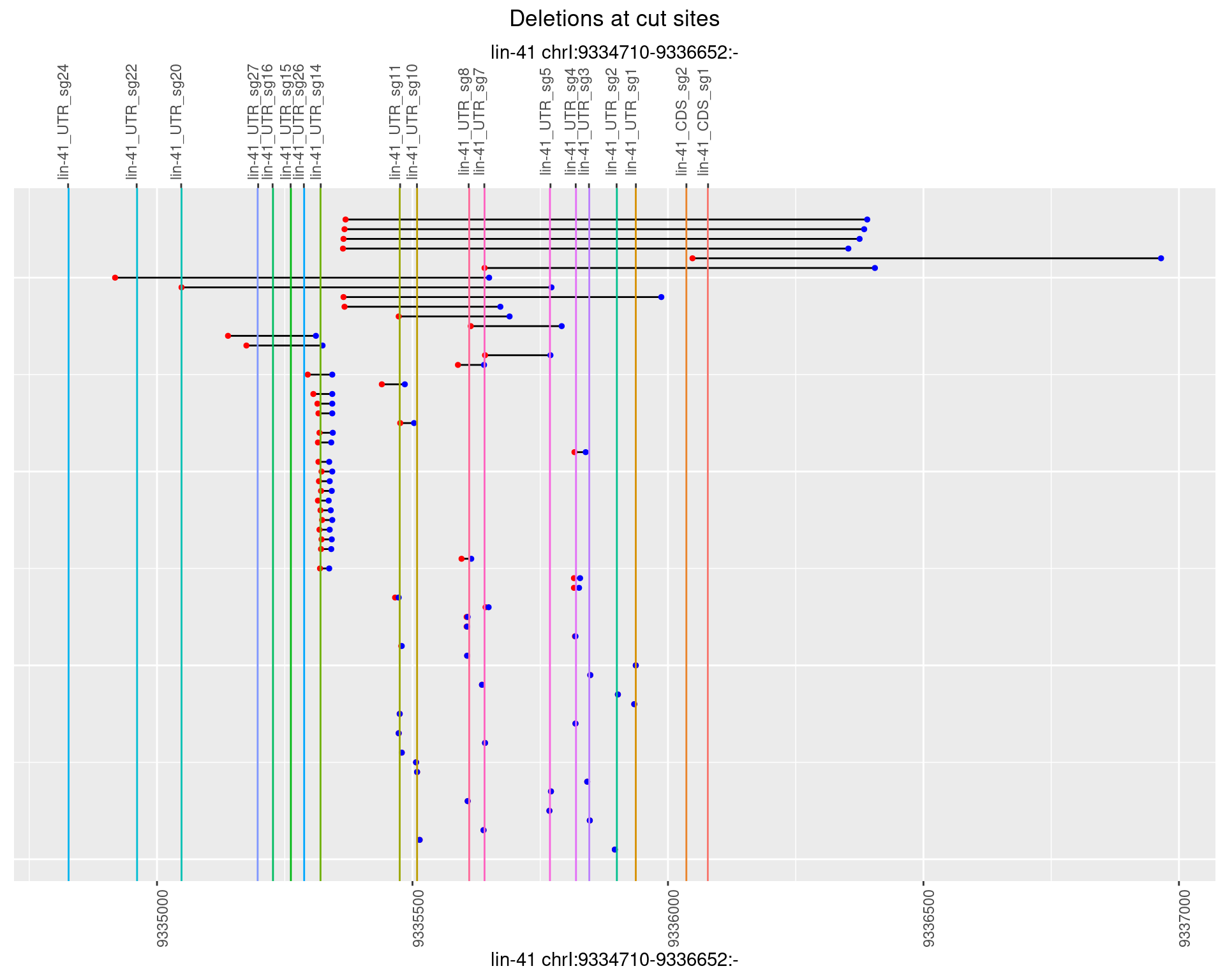
1.92.2 Freq: 1e-05
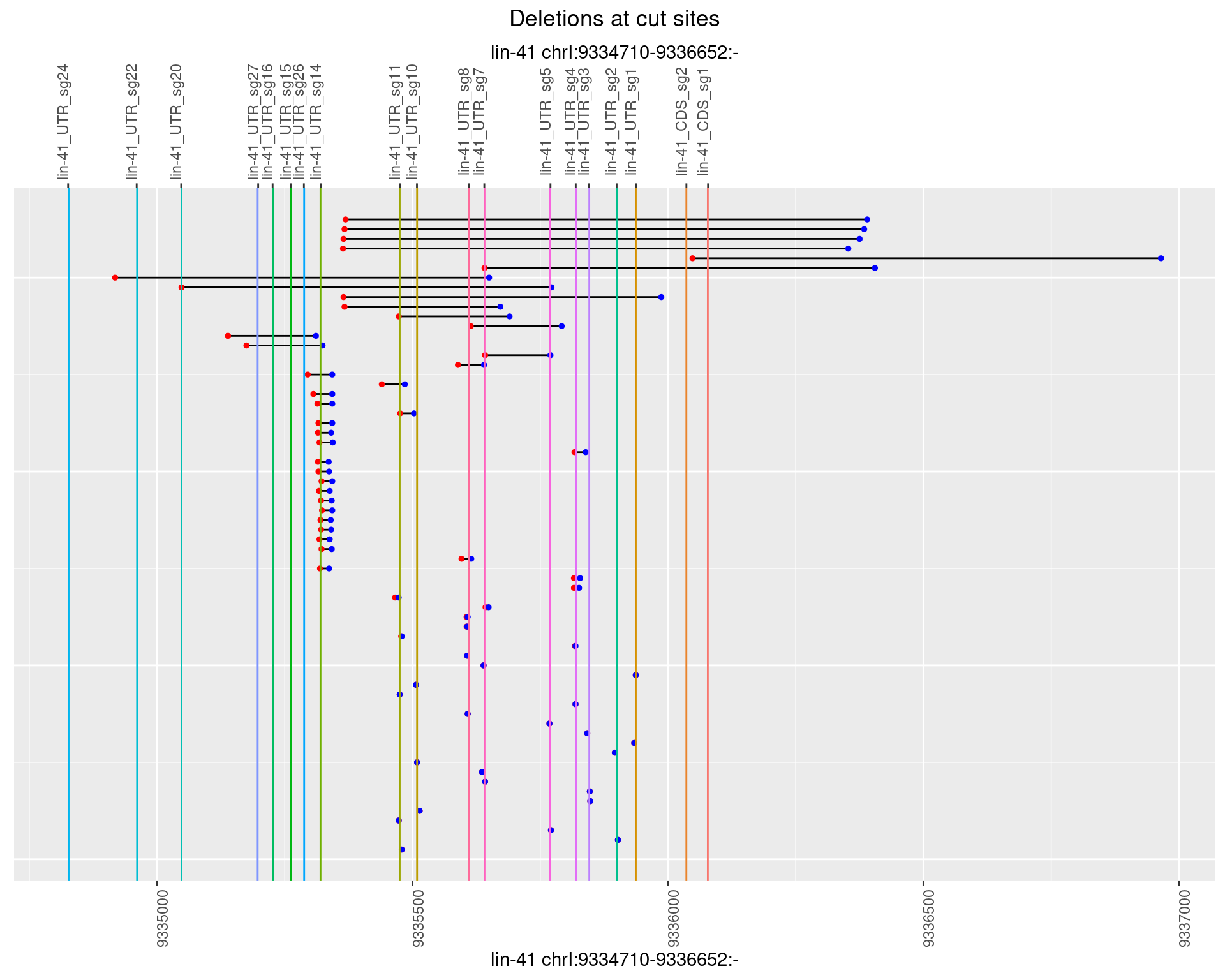
1.92.3 Freq: 1e-04
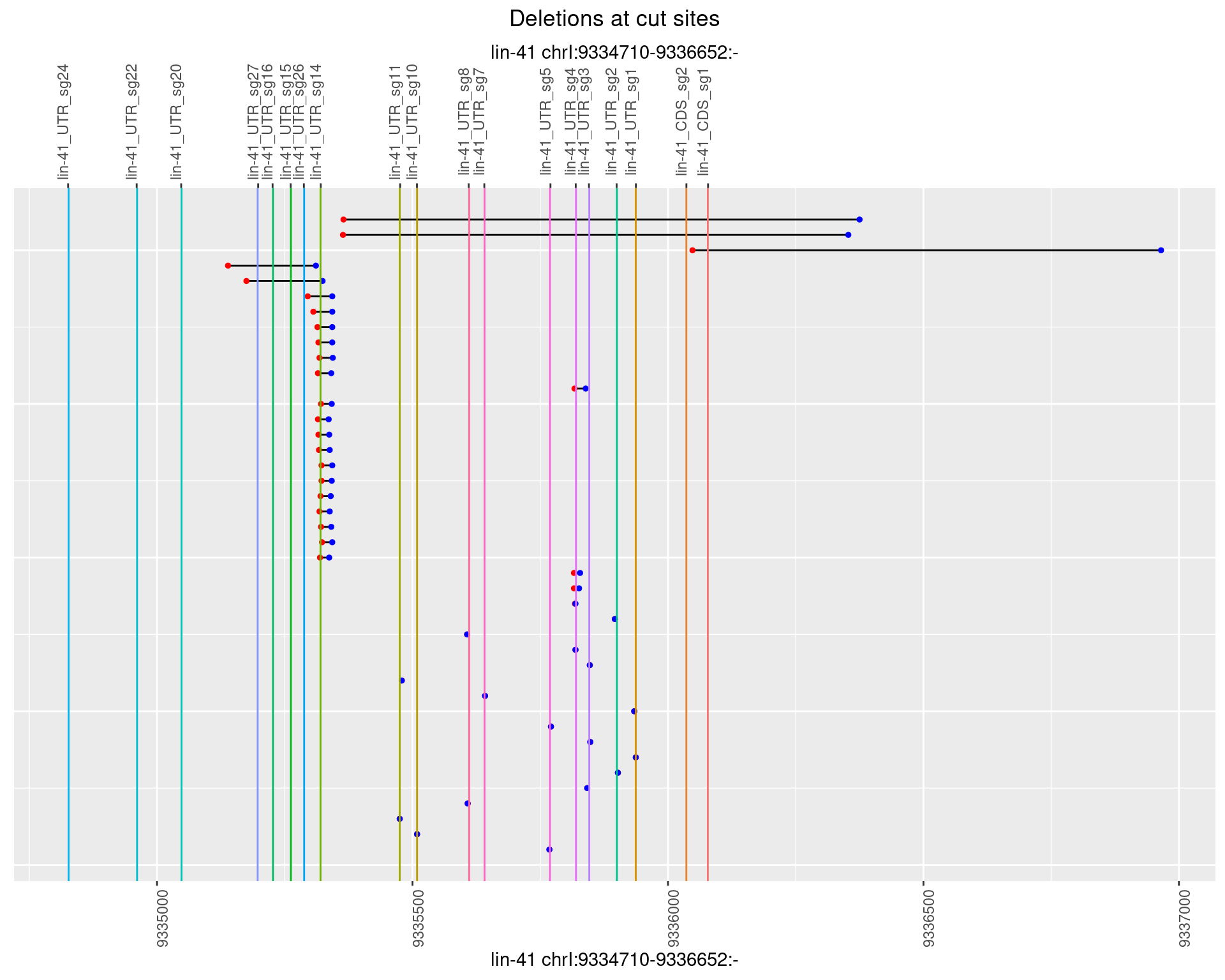
1.92.4 Freq: 0.001
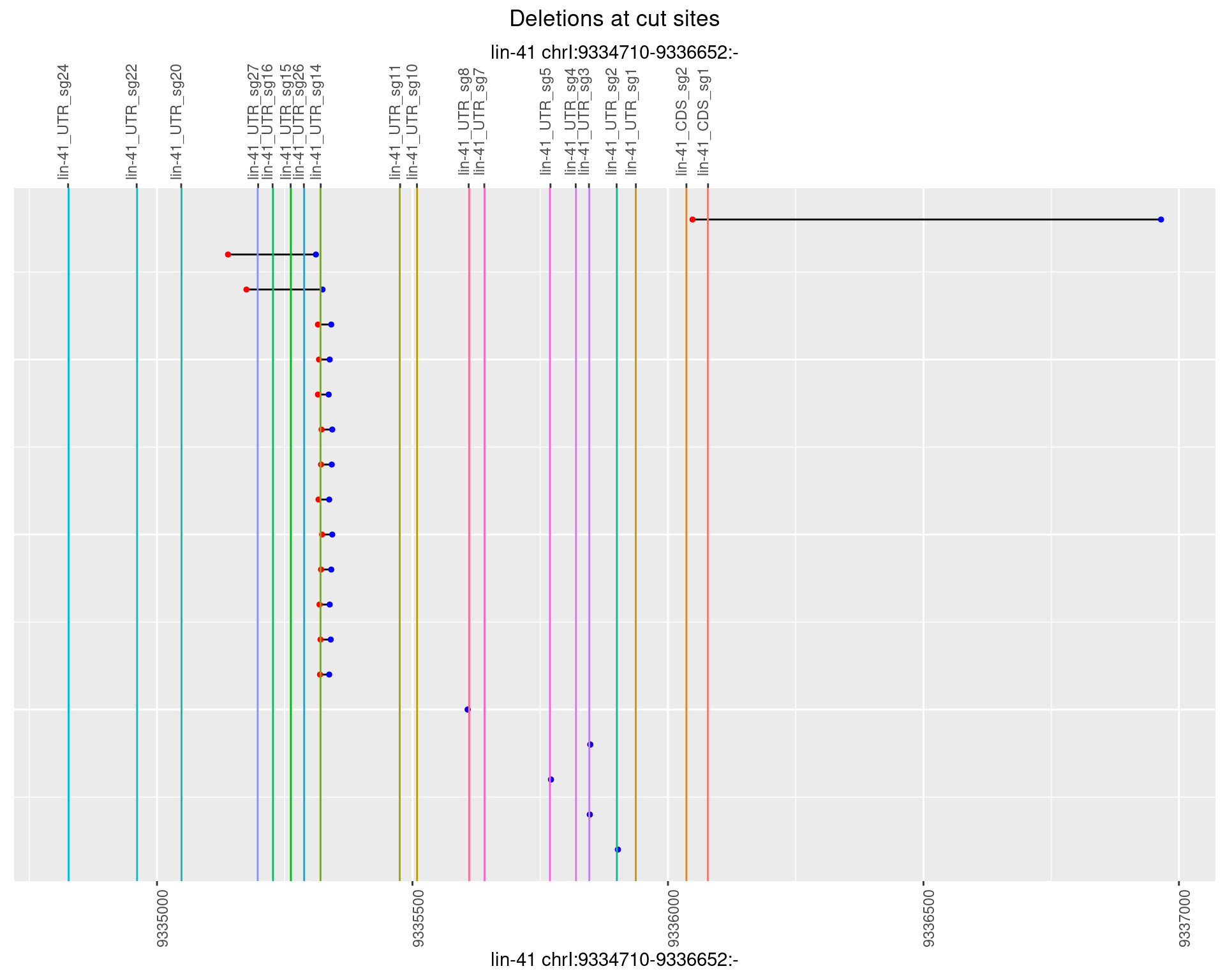
1.92.5 Freq: 0.01
1.92.6 Freq: 0.1
No plot to show
1.93 lin41_RNA_L1_1516_middle_UMI_R2
1.93.1 Freq: 0
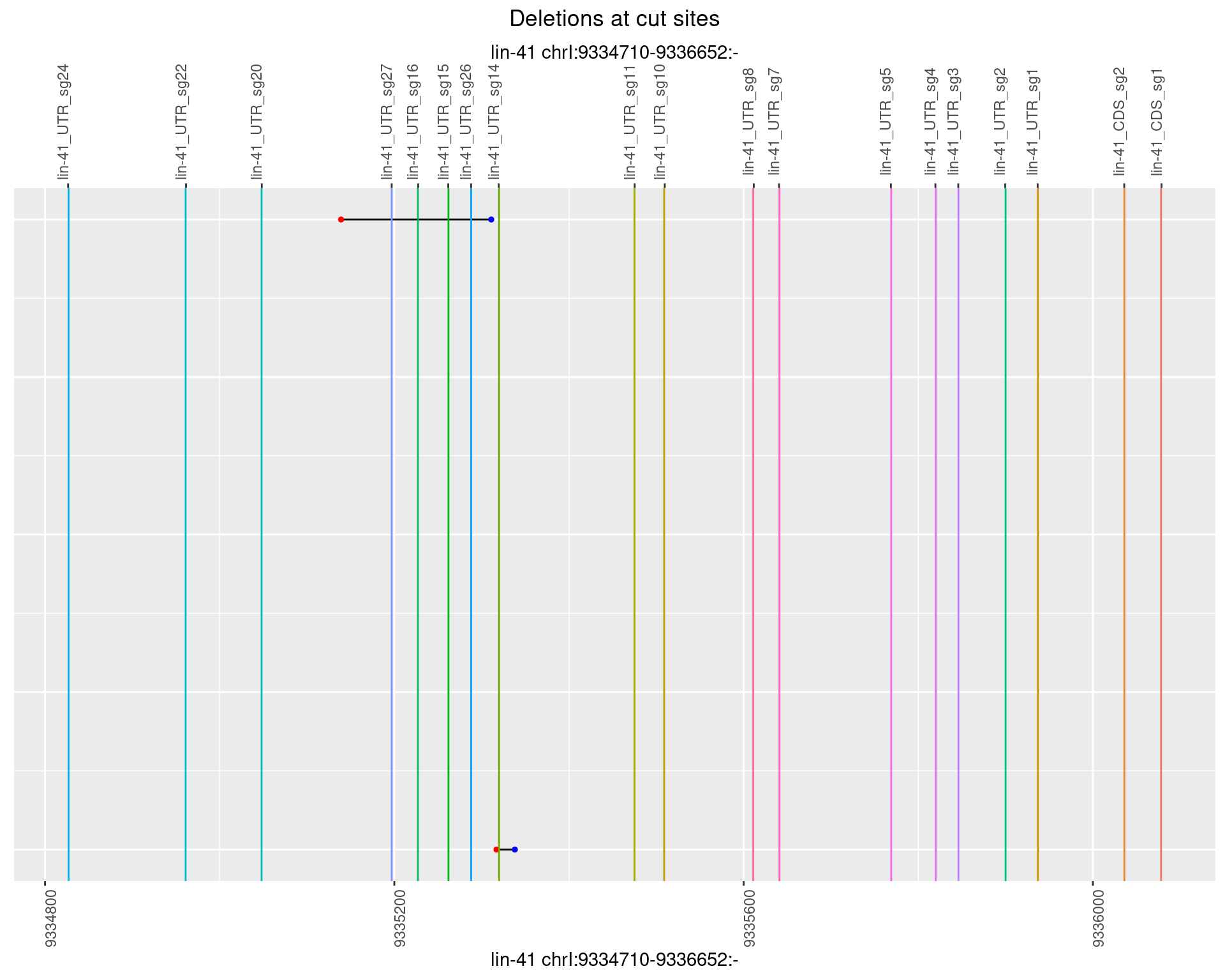
1.93.2 Freq: 1e-05
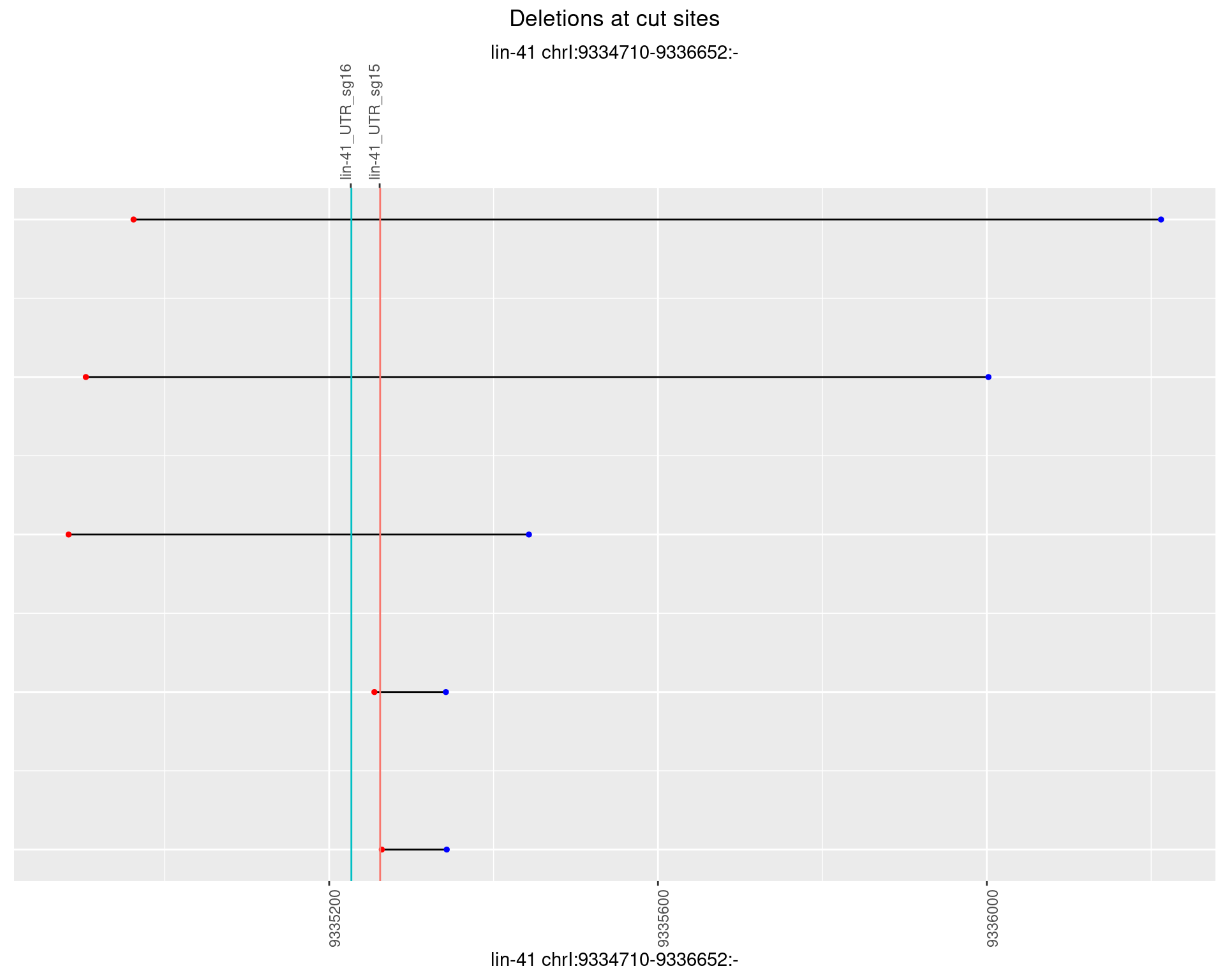
1.93.3 Freq: 1e-04
1.93.4 Freq: 0.001
No plot to show
1.93.5 Freq: 0.01
No plot to show
1.93.6 Freq: 0.1
No plot to show
1.94 lin41_RNA_L1_2627_middle_UMI_R2
1.94.1 Freq: 0
1.94.2 Freq: 1e-05
1.94.3 Freq: 1e-04
1.94.4 Freq: 0.001
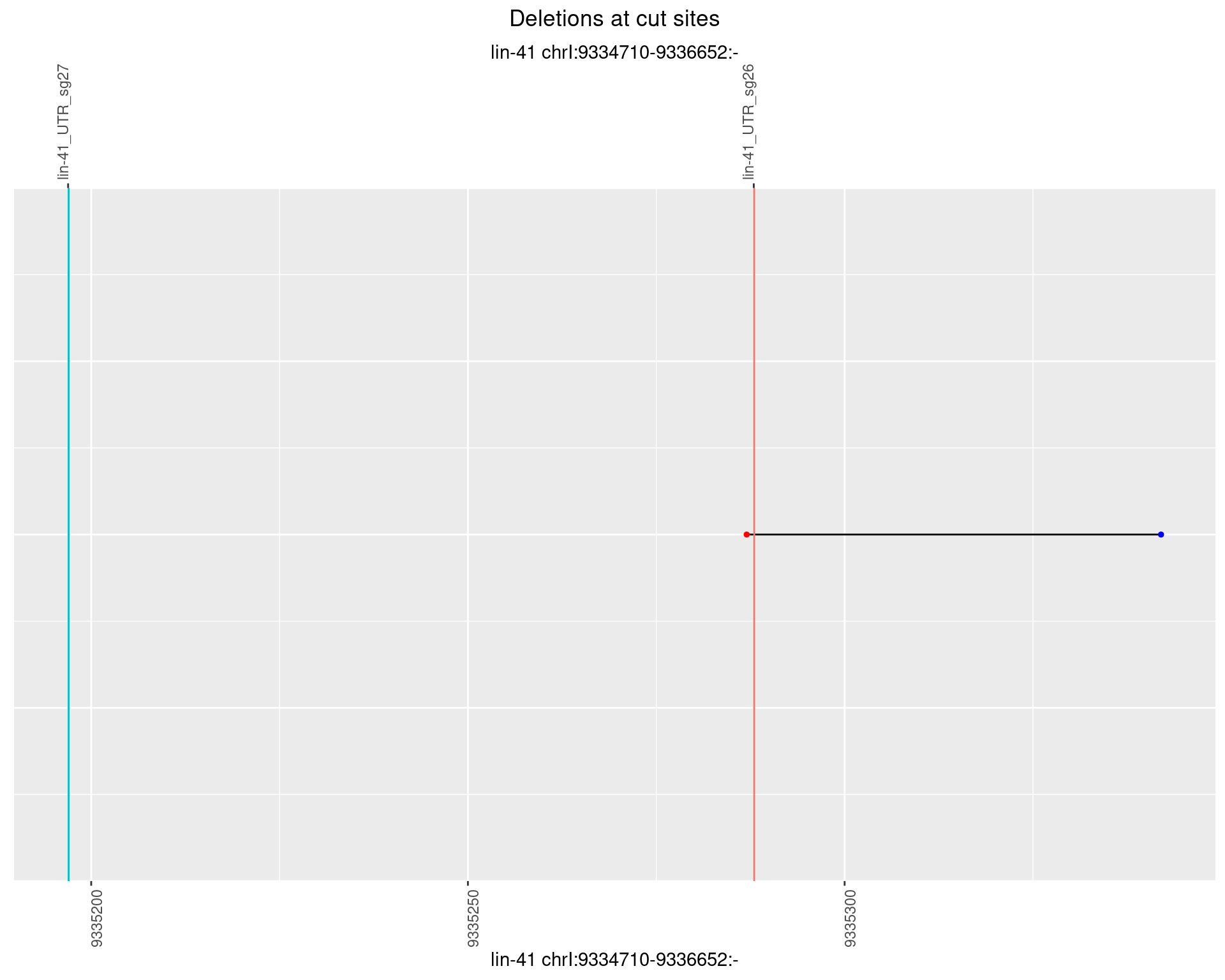
1.94.5 Freq: 0.01
No plot to show
1.94.6 Freq: 0.1
No plot to show
1.95 lin41_RNA_L1_pool2_middle_UMI_R2
1.95.1 Freq: 0
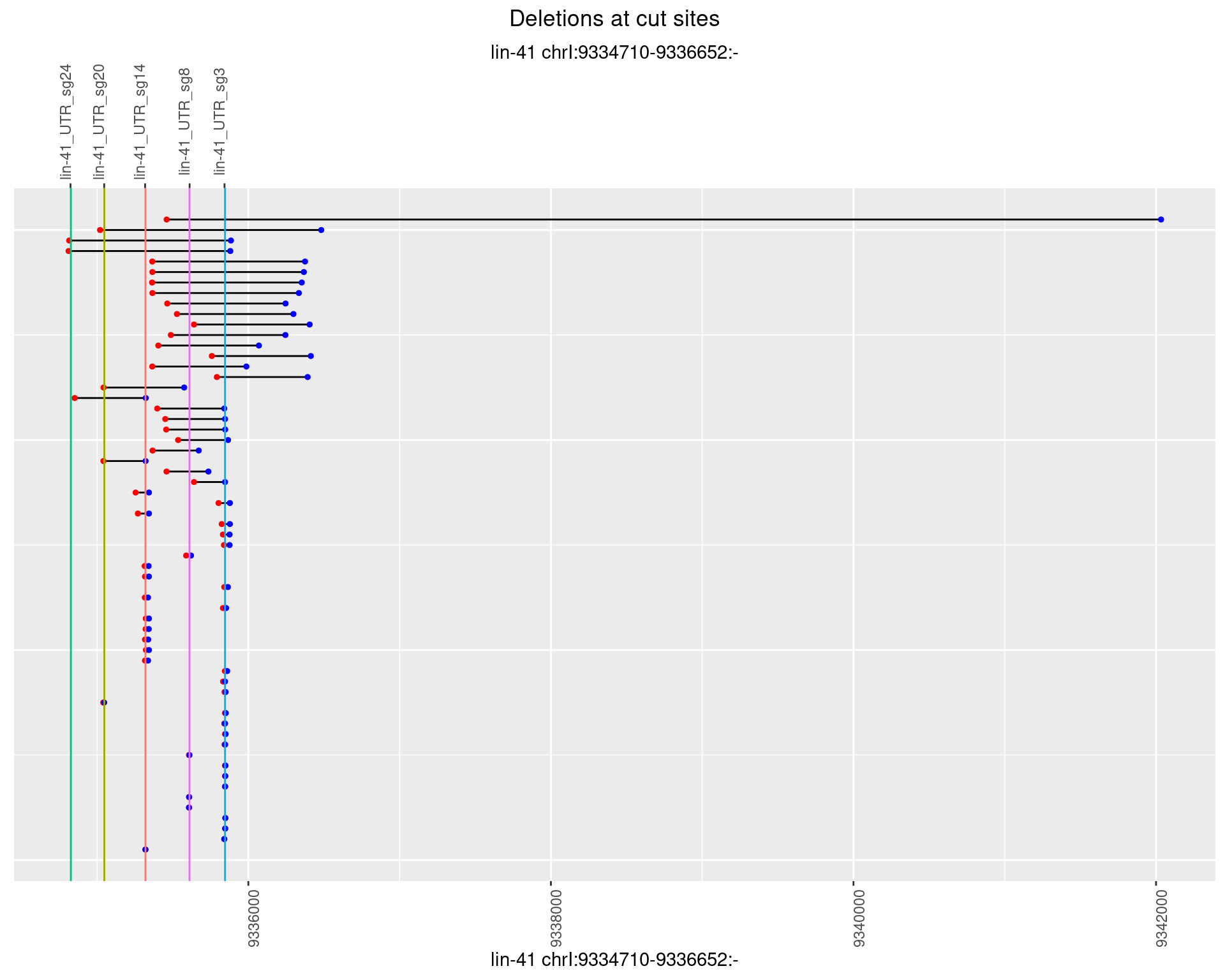
1.95.2 Freq: 1e-05
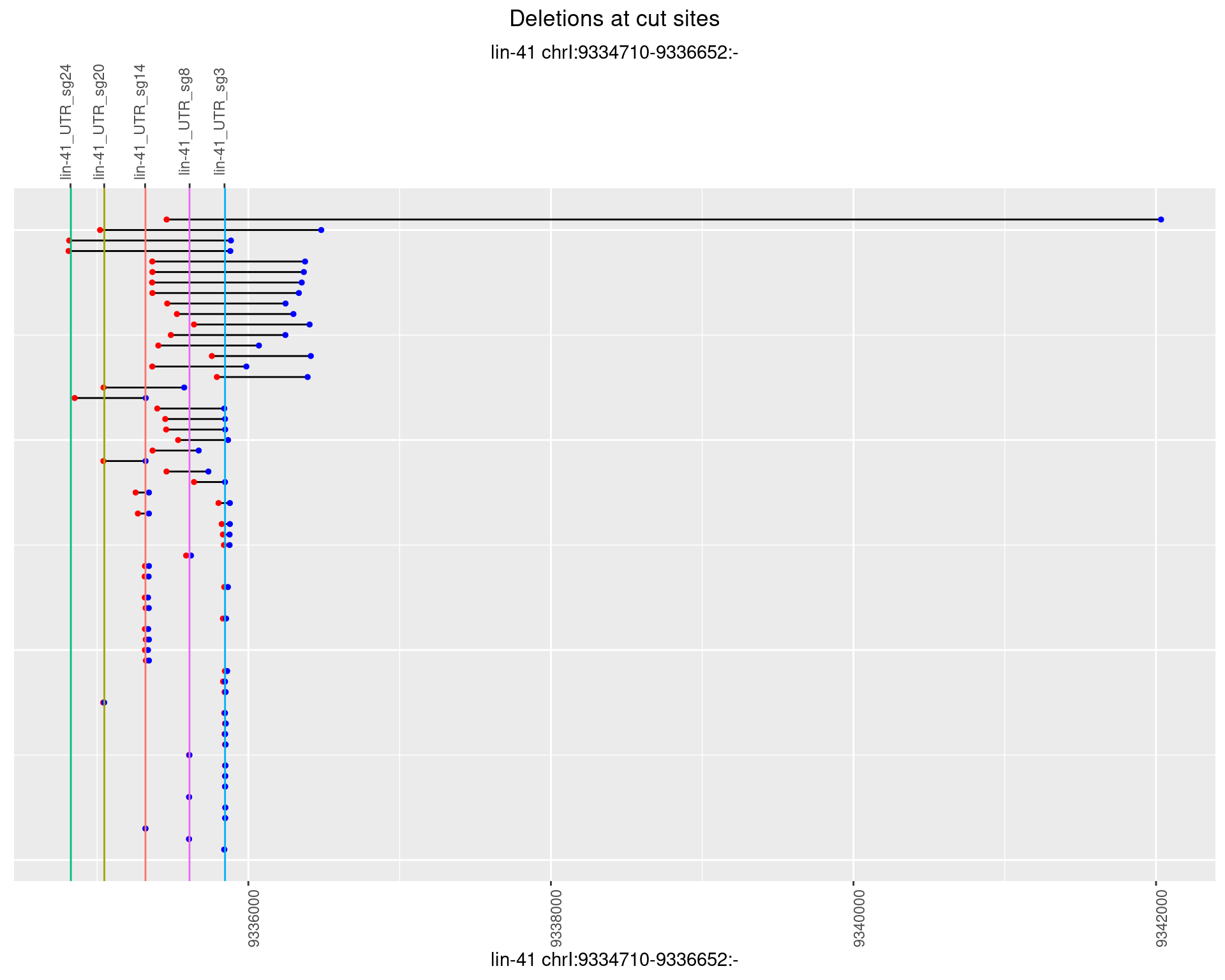
1.95.3 Freq: 1e-04
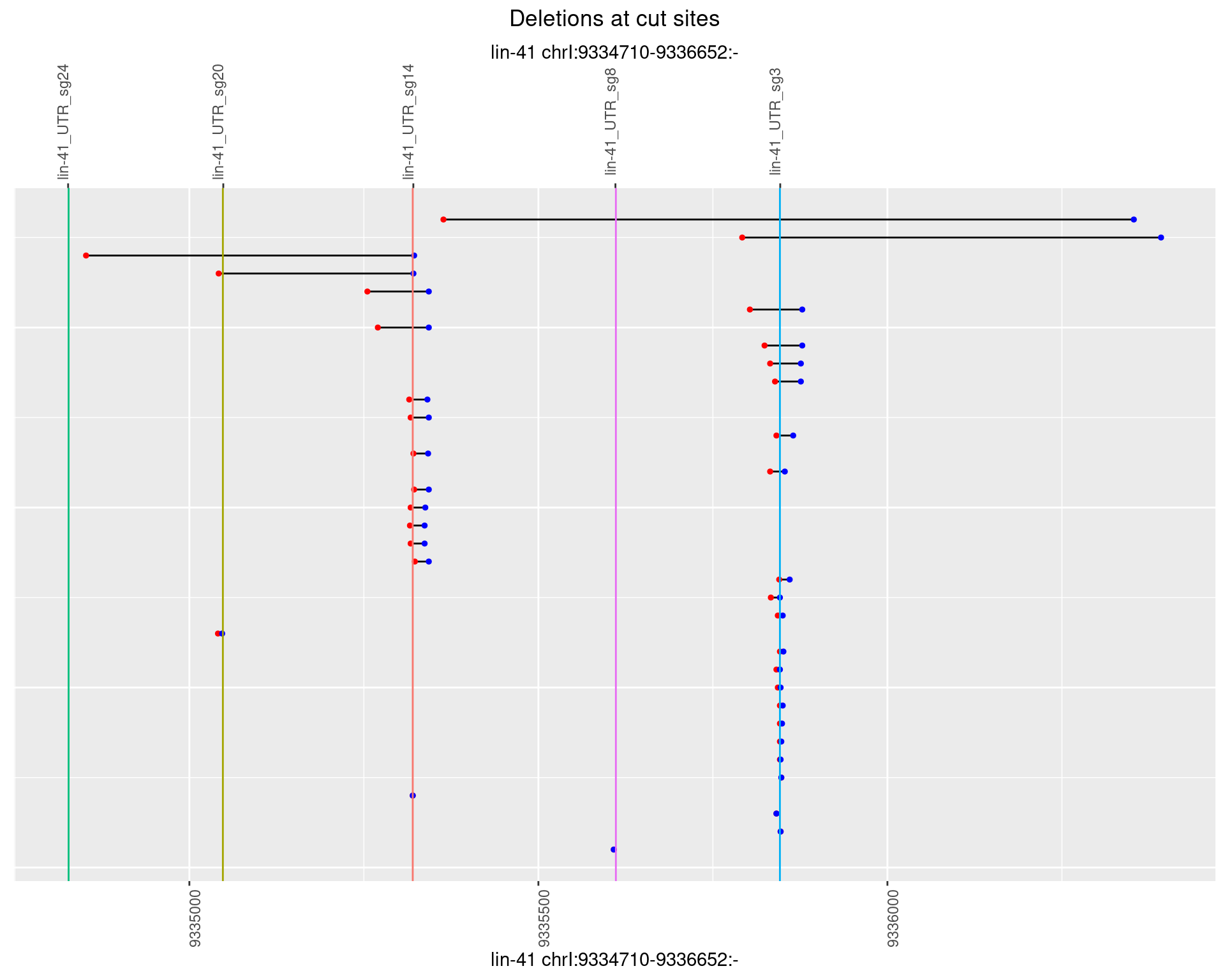
1.95.4 Freq: 0.001
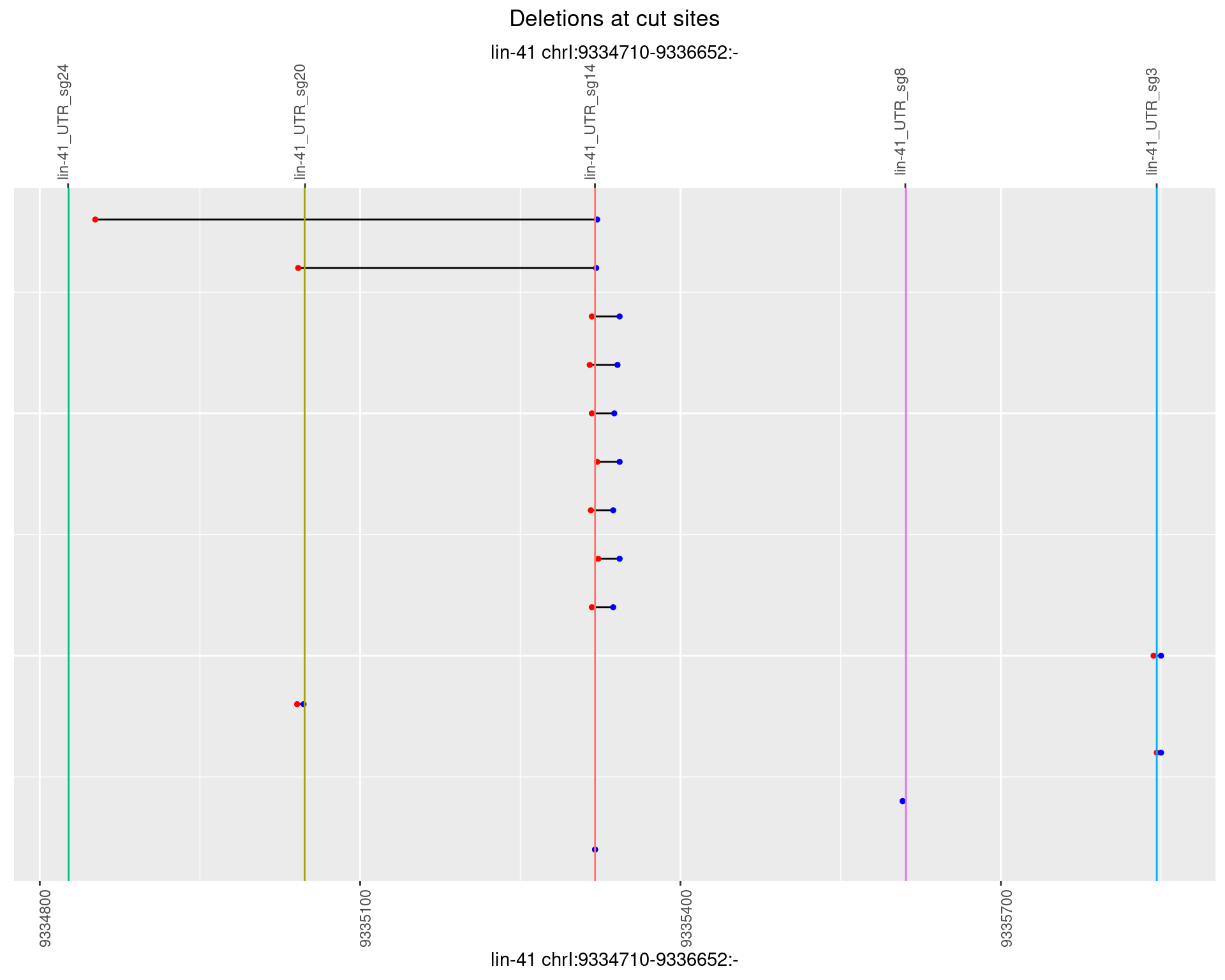
1.95.5 Freq: 0.01
1.95.6 Freq: 0.1
No plot to show
1.96 lin41_RNA_L1_pool3_middle_UMI_R2
1.96.1 Freq: 0
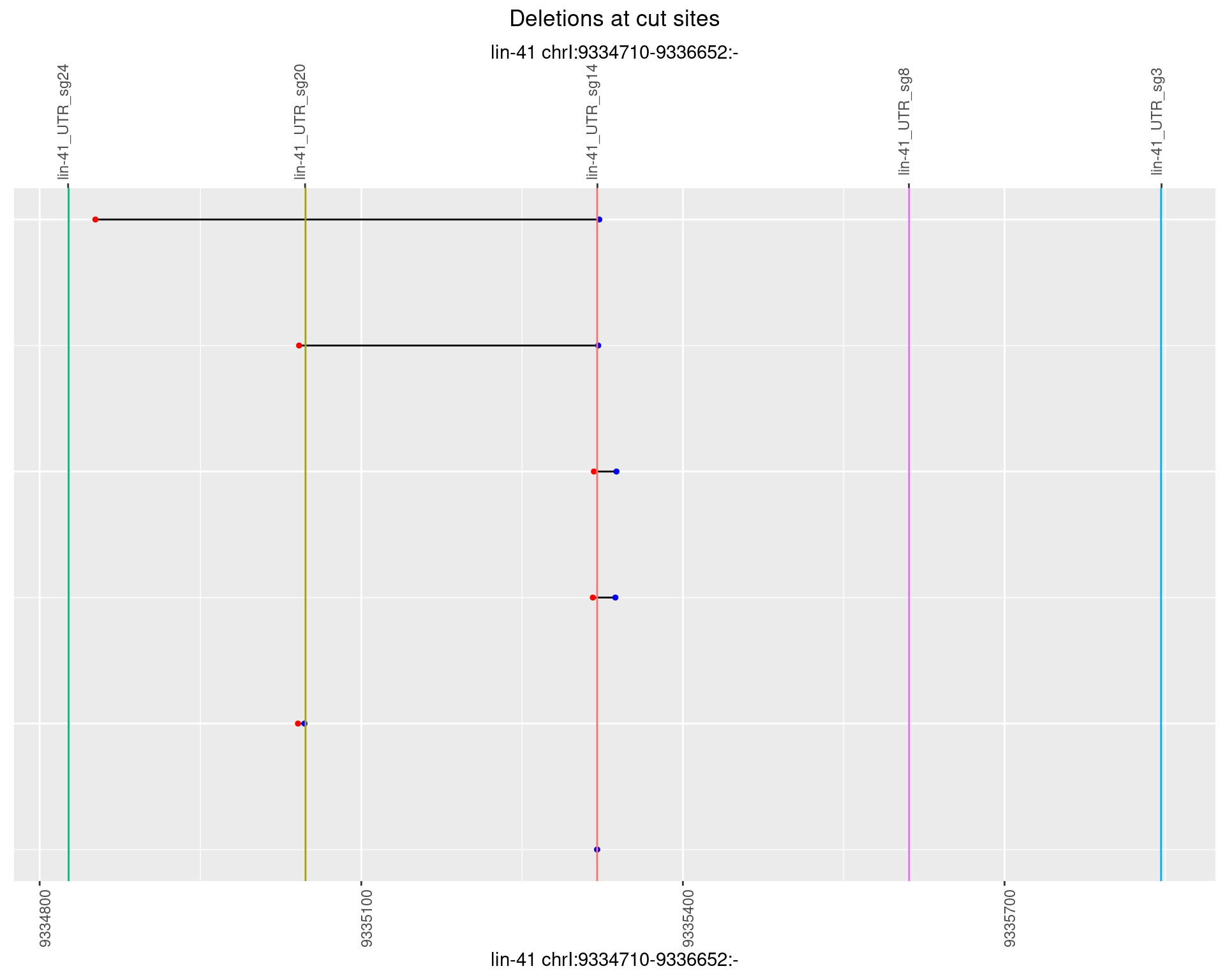
1.96.2 Freq: 1e-05
1.96.3 Freq: 1e-04
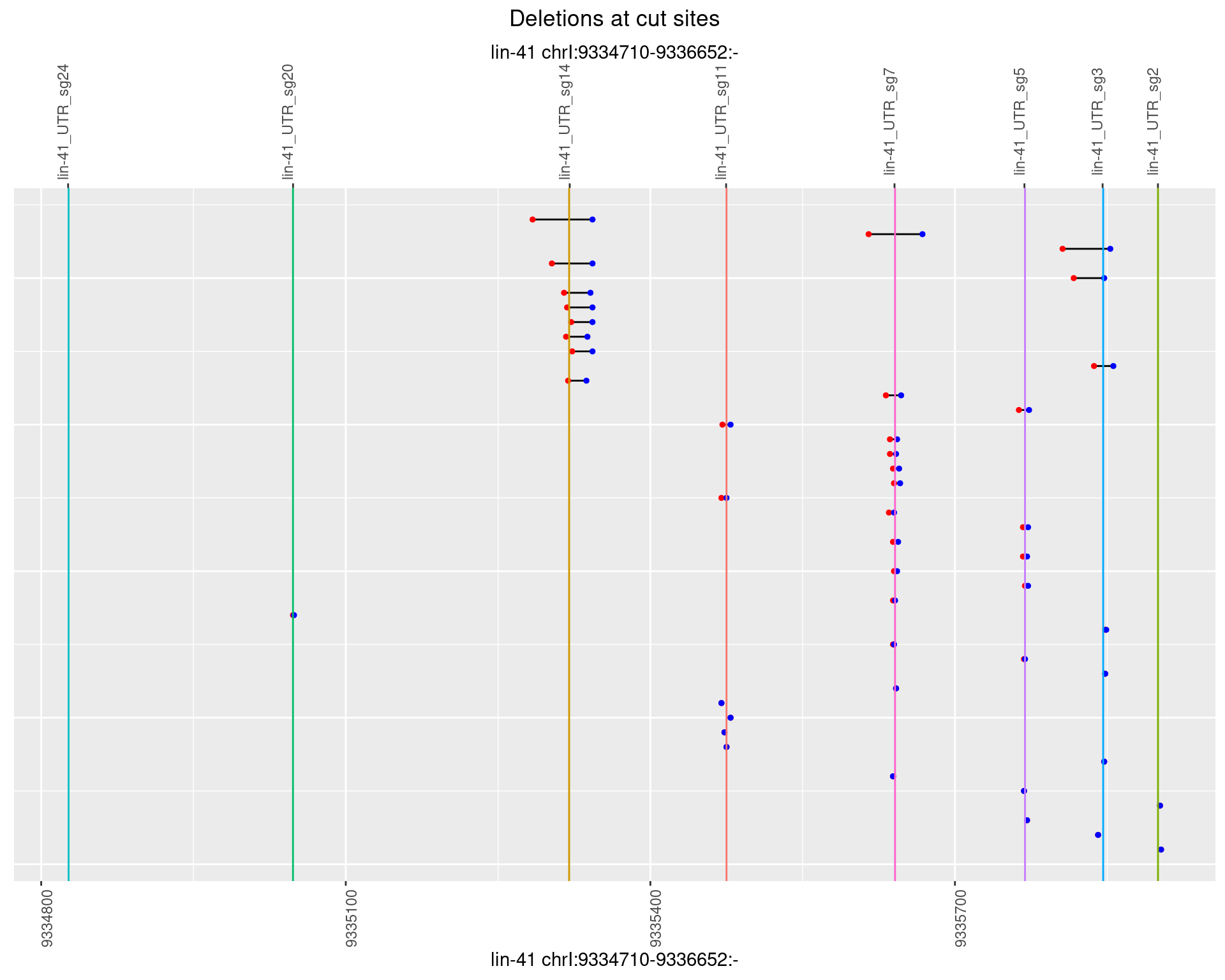
1.96.4 Freq: 0.001
1.96.5 Freq: 0.01
1.96.6 Freq: 0.1
No plot to show
1.97 lin41_RNA_L4_N2_middle_UMI_R2
1.97.1 Freq: 0
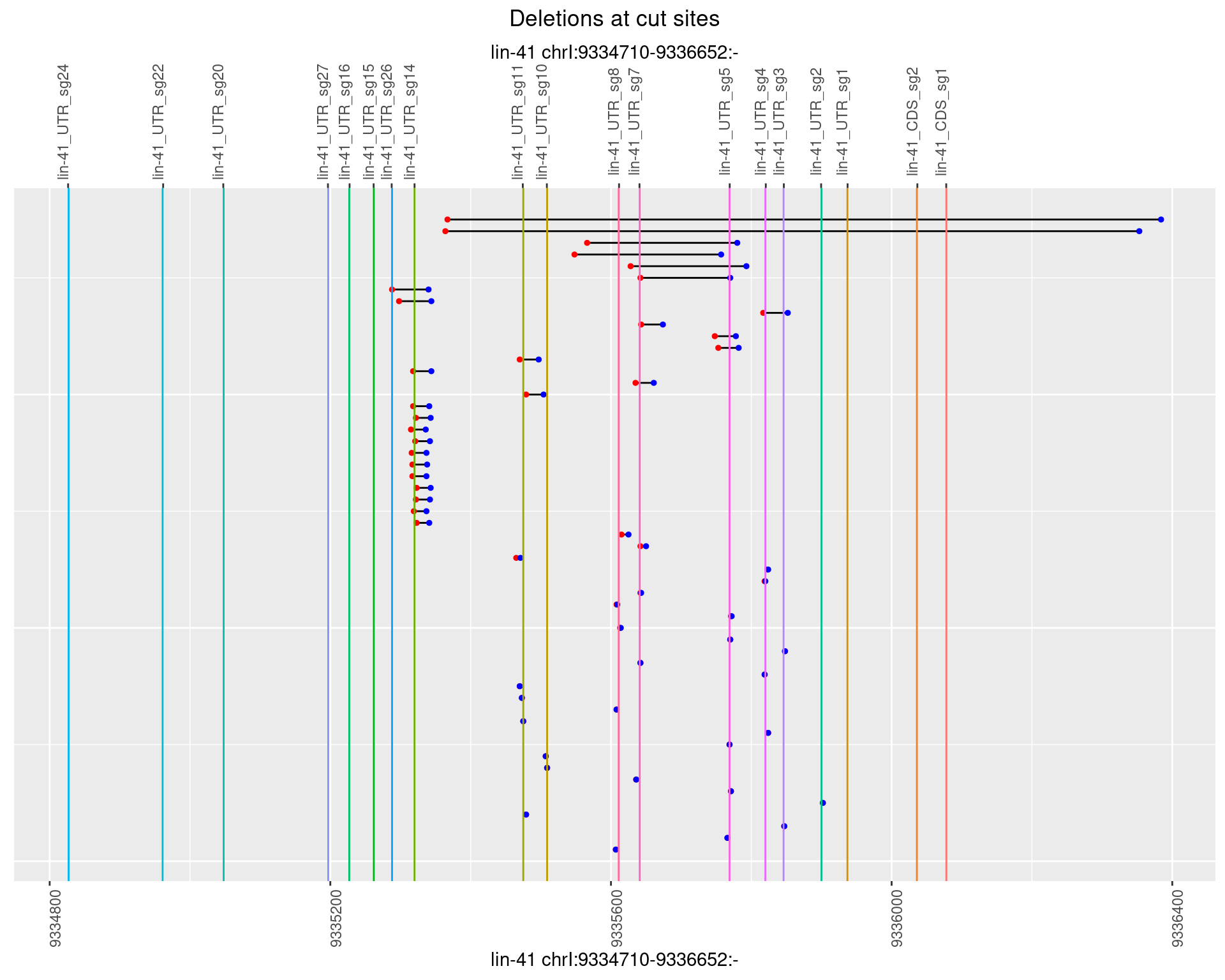
1.97.2 Freq: 1e-05
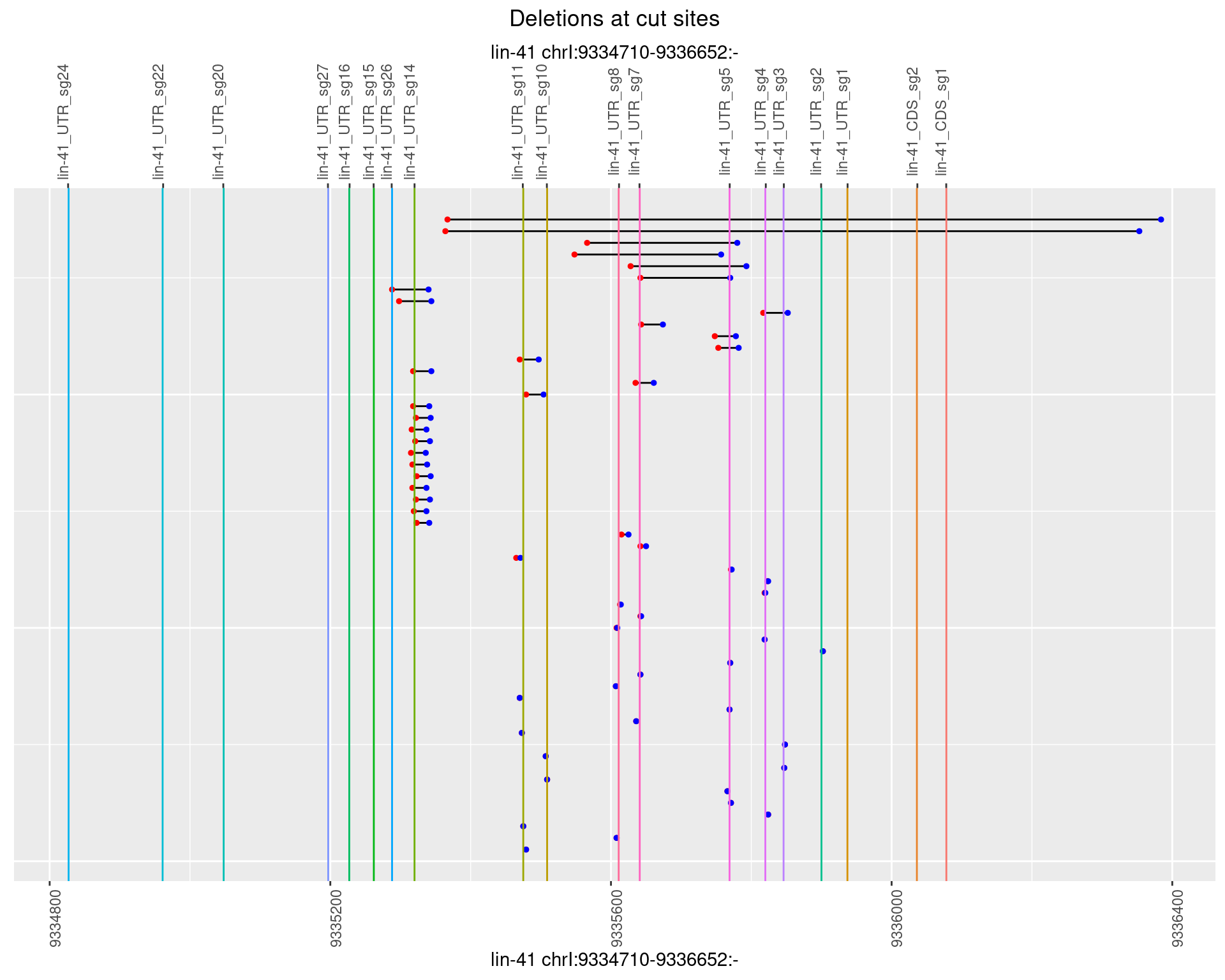
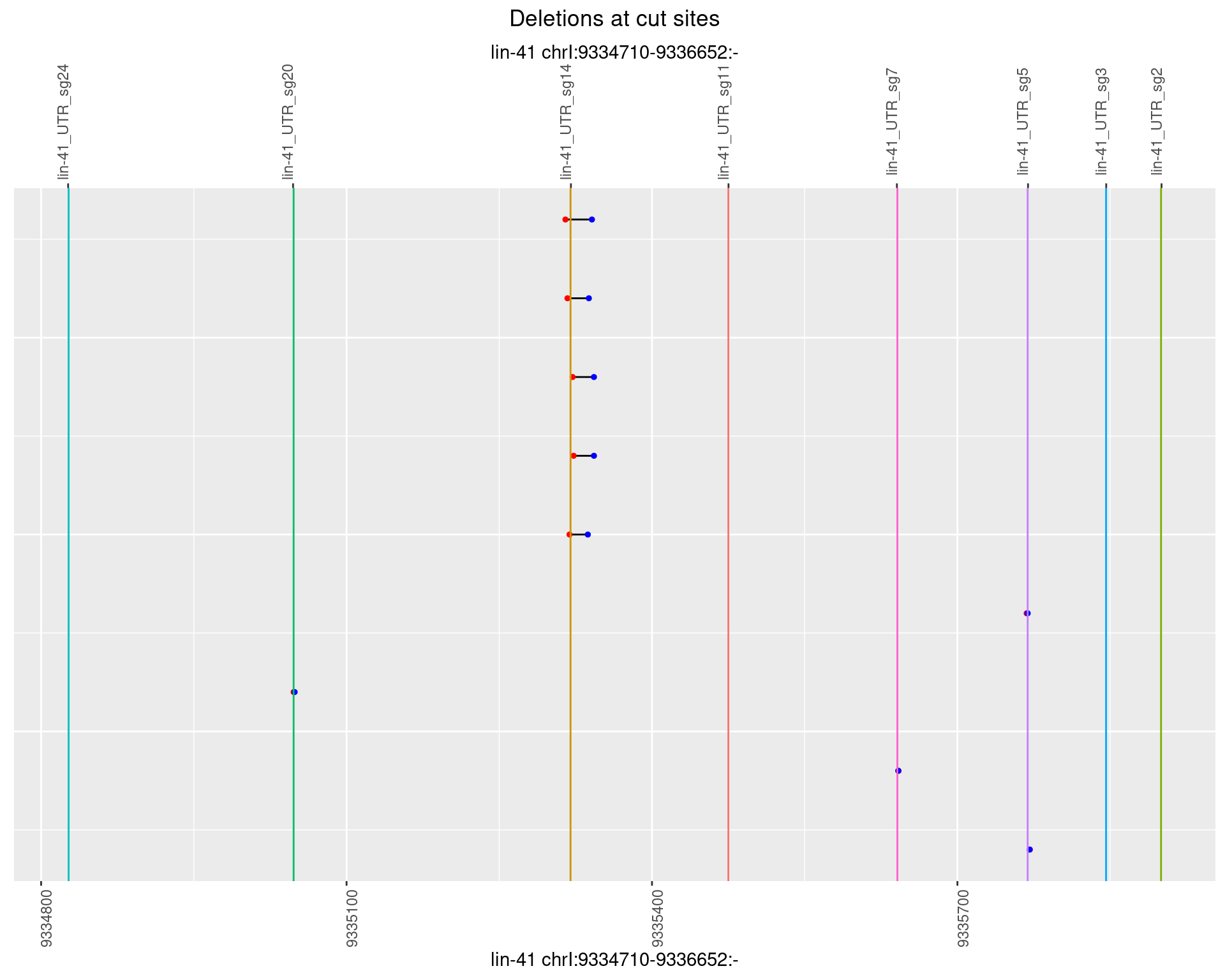
1.97.3 Freq: 1e-04
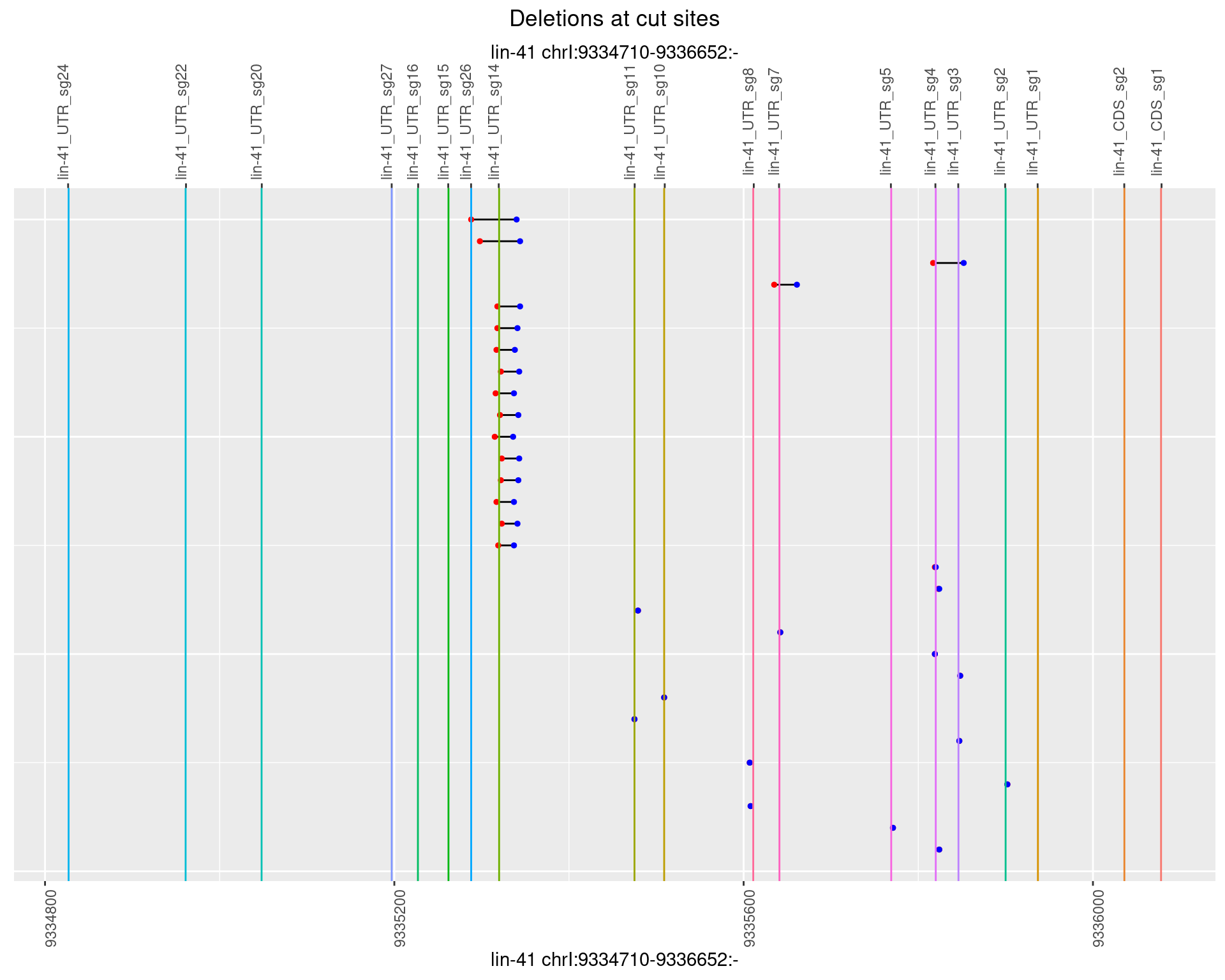
1.97.4 Freq: 0.001
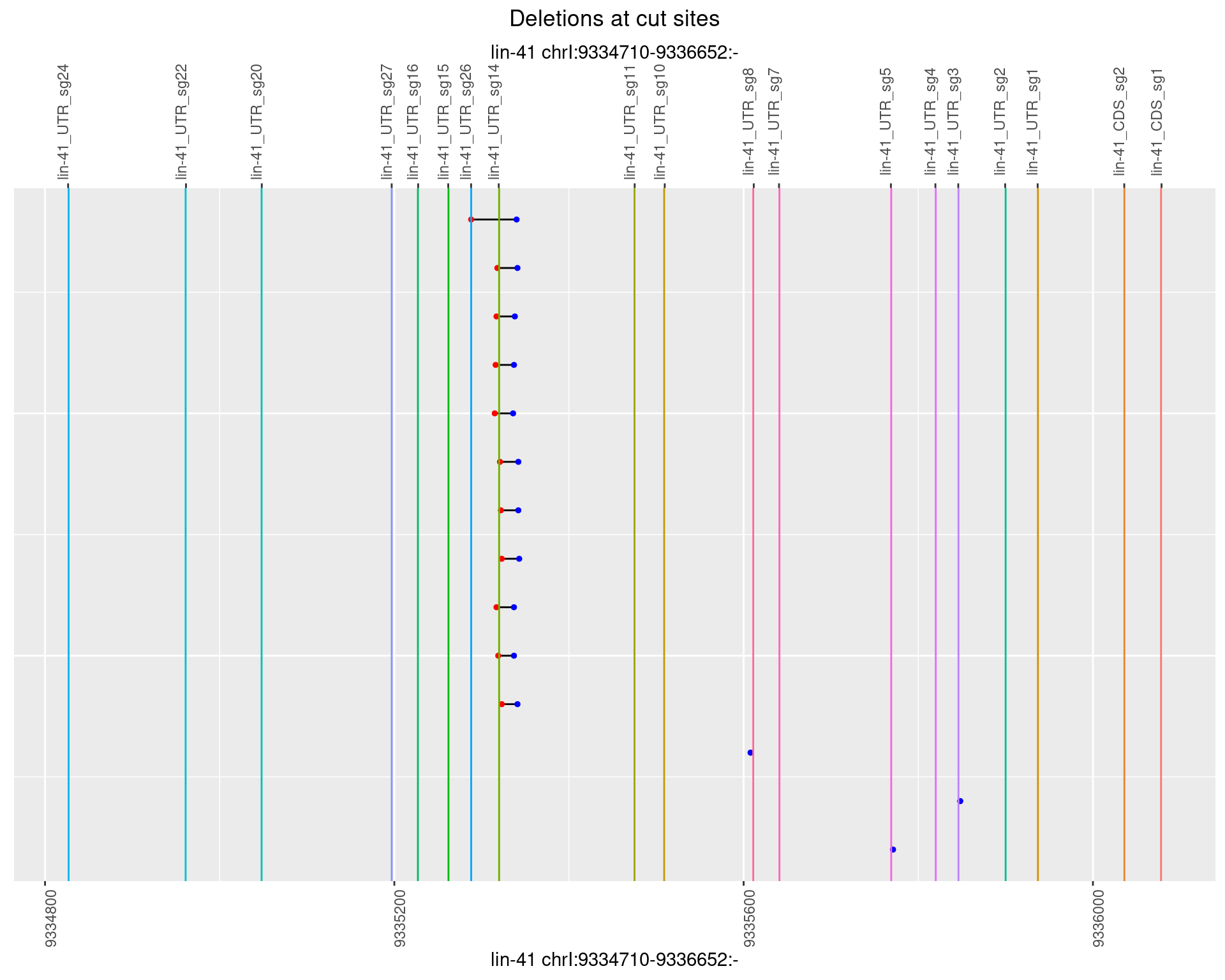
1.97.5 Freq: 0.01
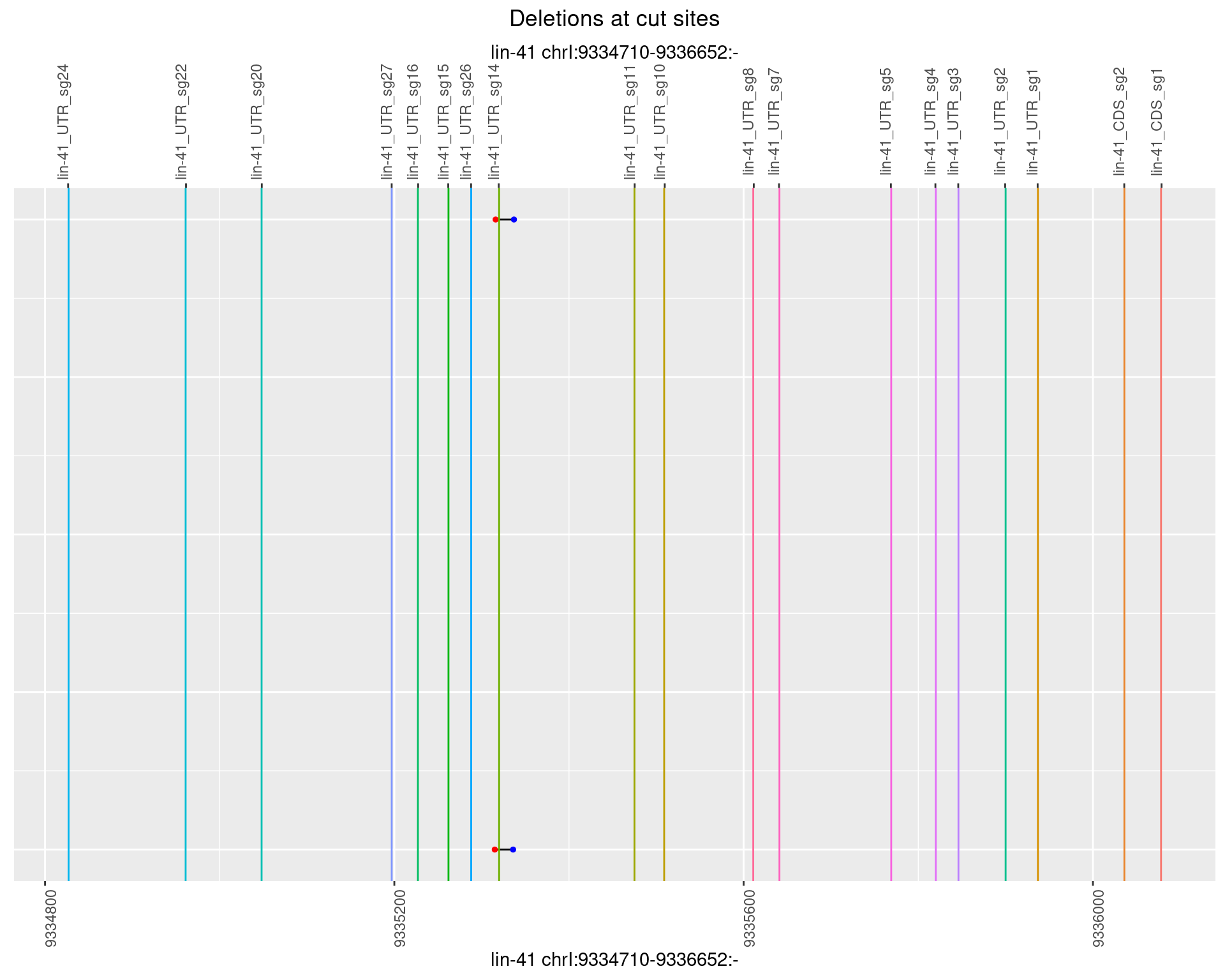
1.97.6 Freq: 0.1
No plot to show
1.98 lin41_RNA_L4_1516_middle_UMI_R2
1.98.1 Freq: 0
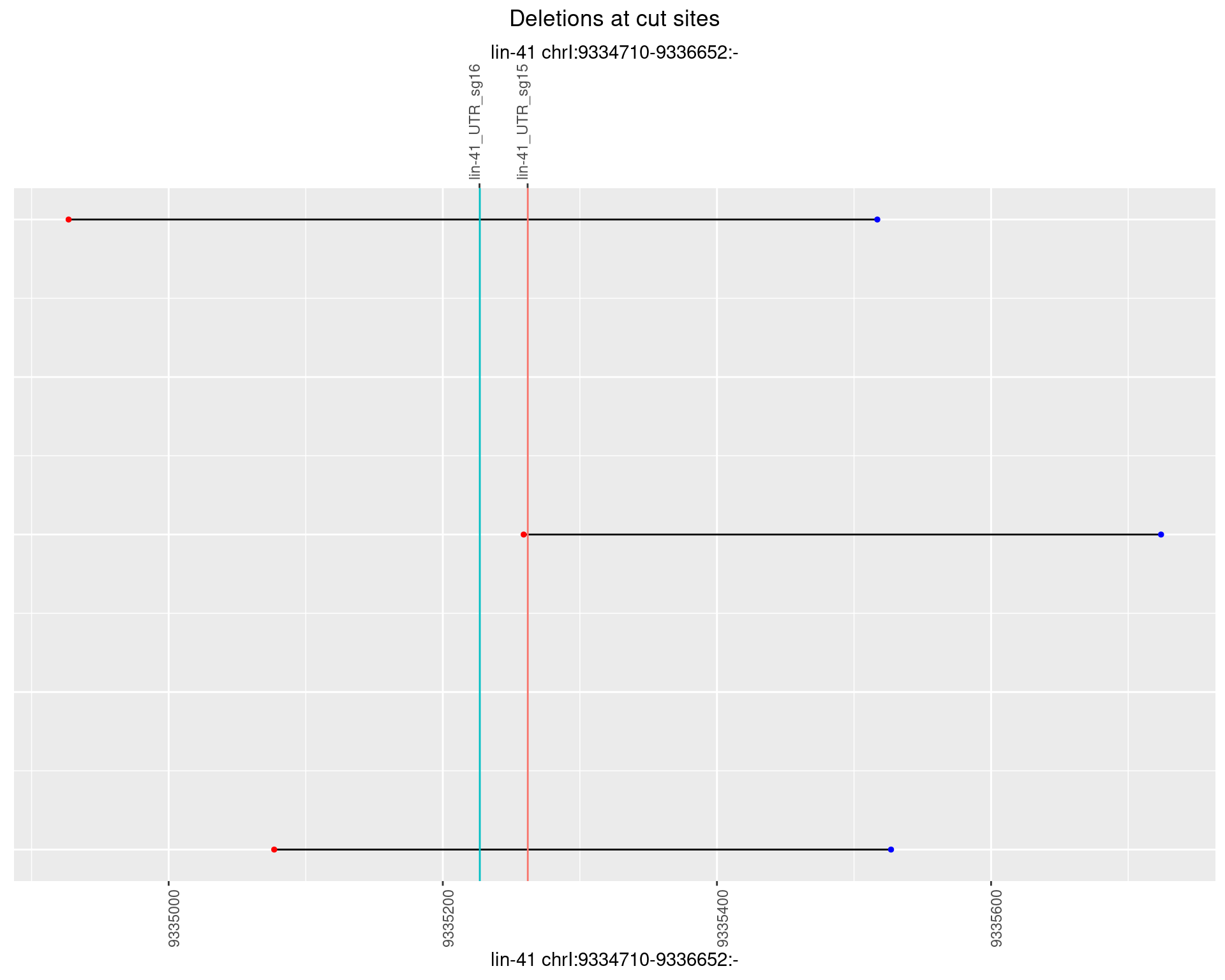
1.98.2 Freq: 1e-05
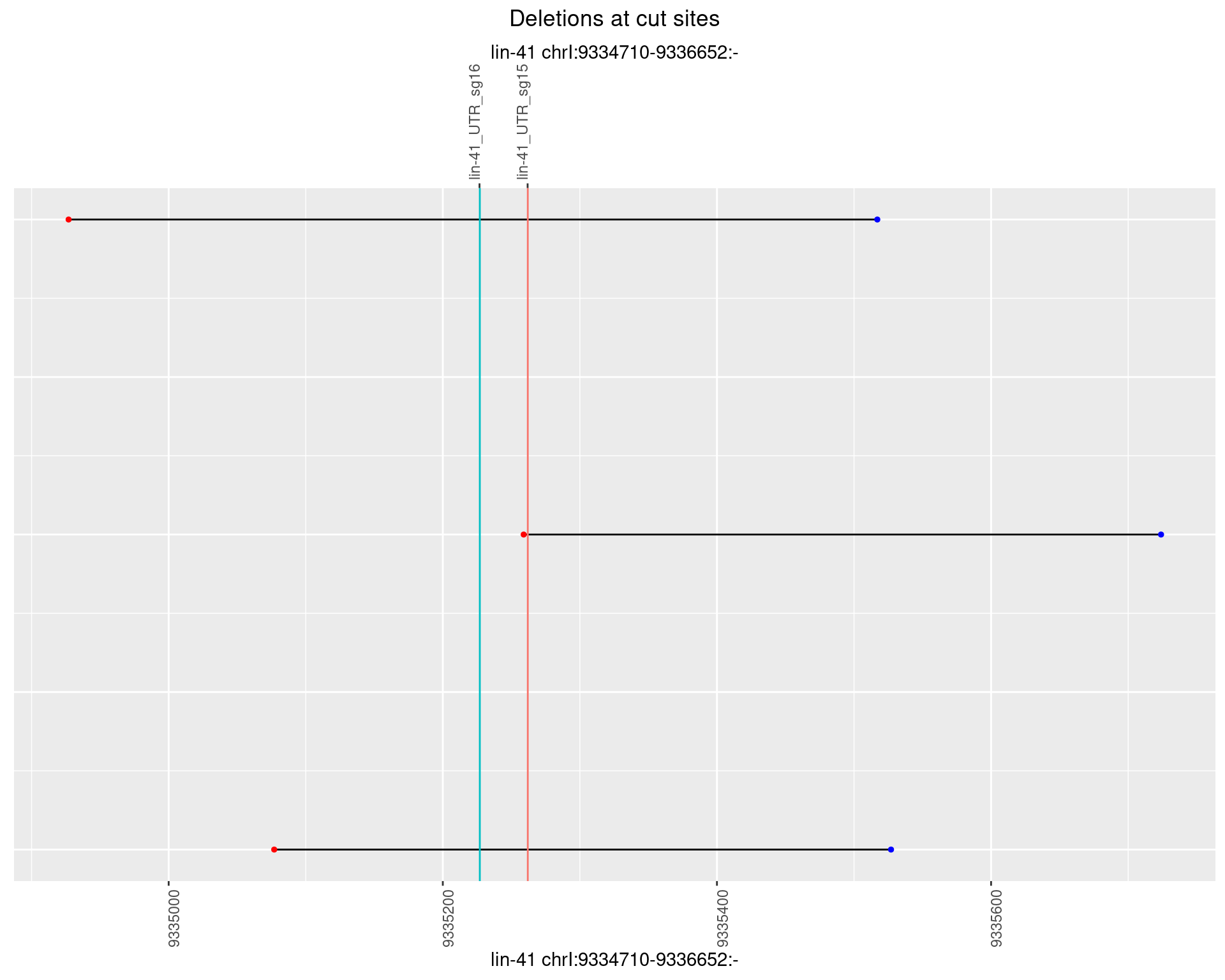
1.98.3 Freq: 1e-04
No plot to show
1.98.4 Freq: 0.001
No plot to show
1.98.5 Freq: 0.01
No plot to show
1.98.6 Freq: 0.1
No plot to show
1.99 lin41_RNA_L4_2627_middle_UMI_R2
1.99.1 Freq: 0
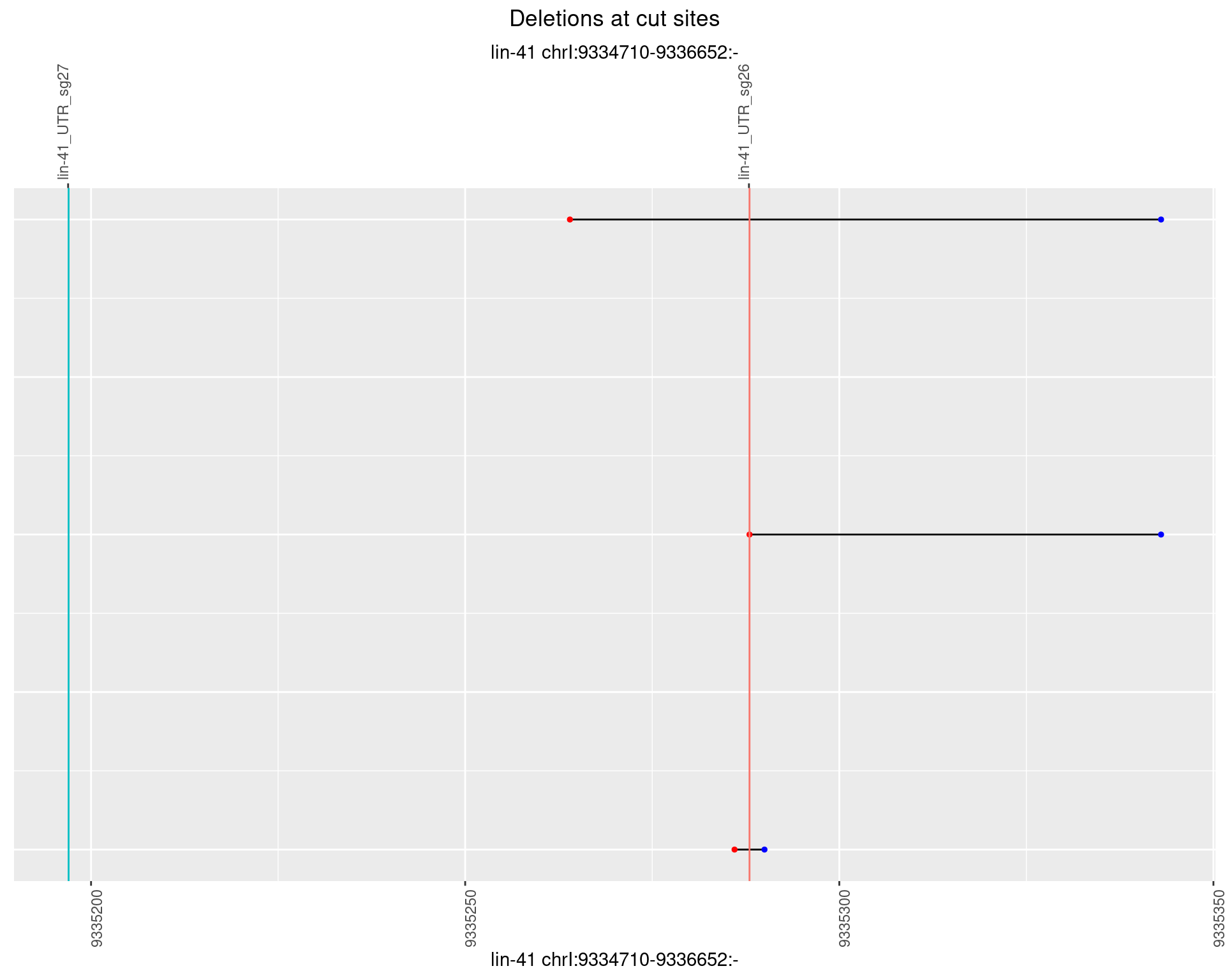
1.99.2 Freq: 1e-05
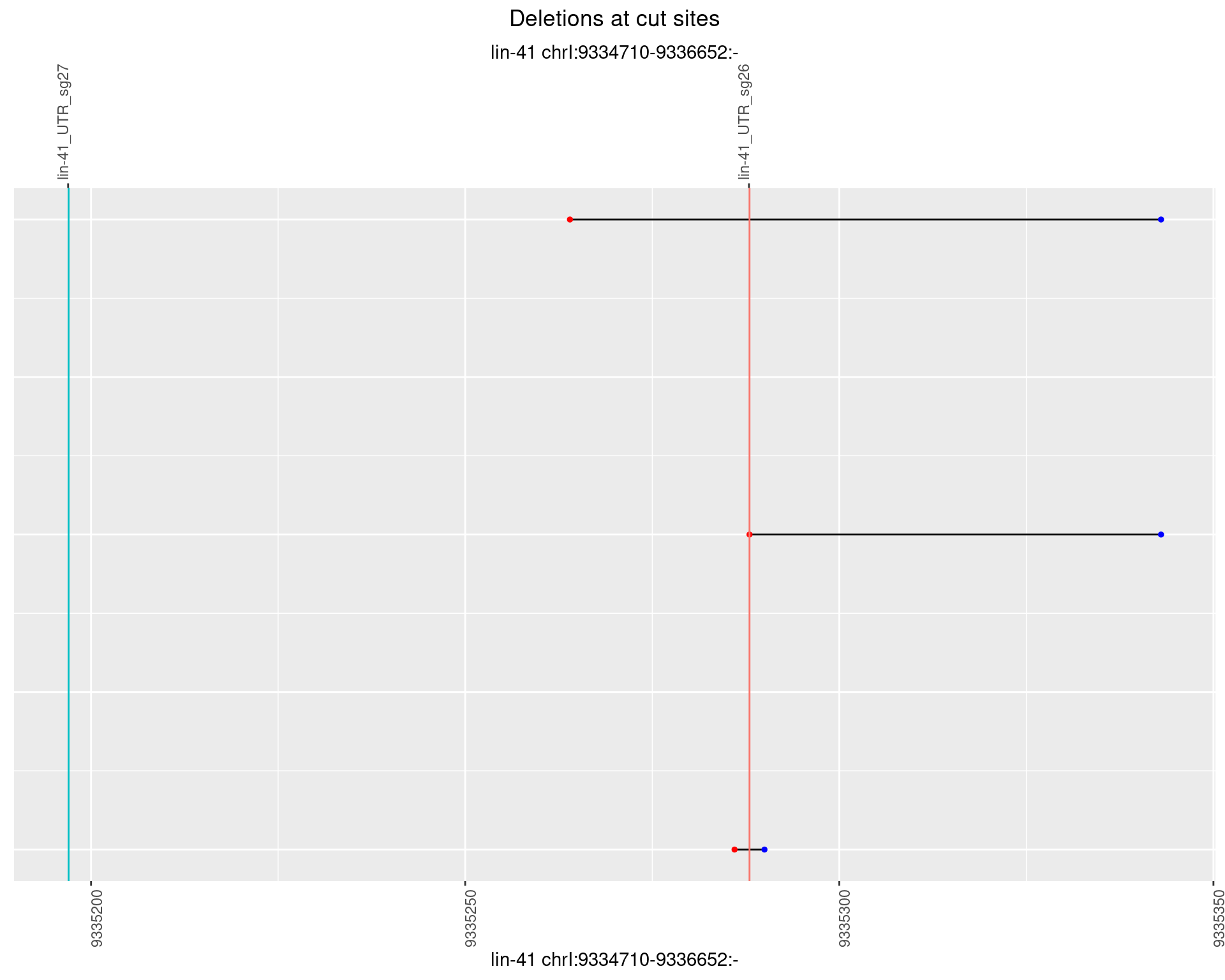
1.99.3 Freq: 1e-04
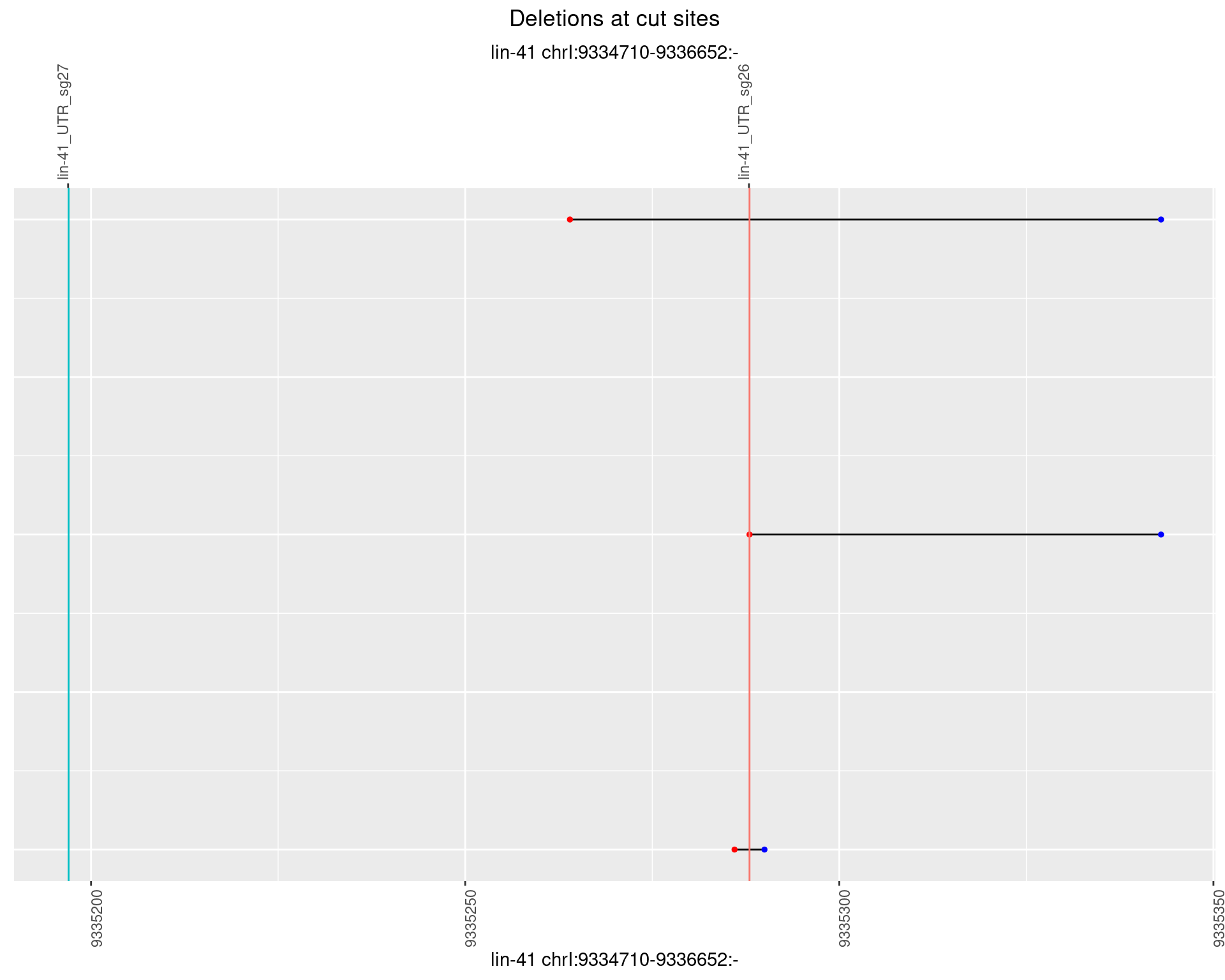
1.99.4 Freq: 0.001
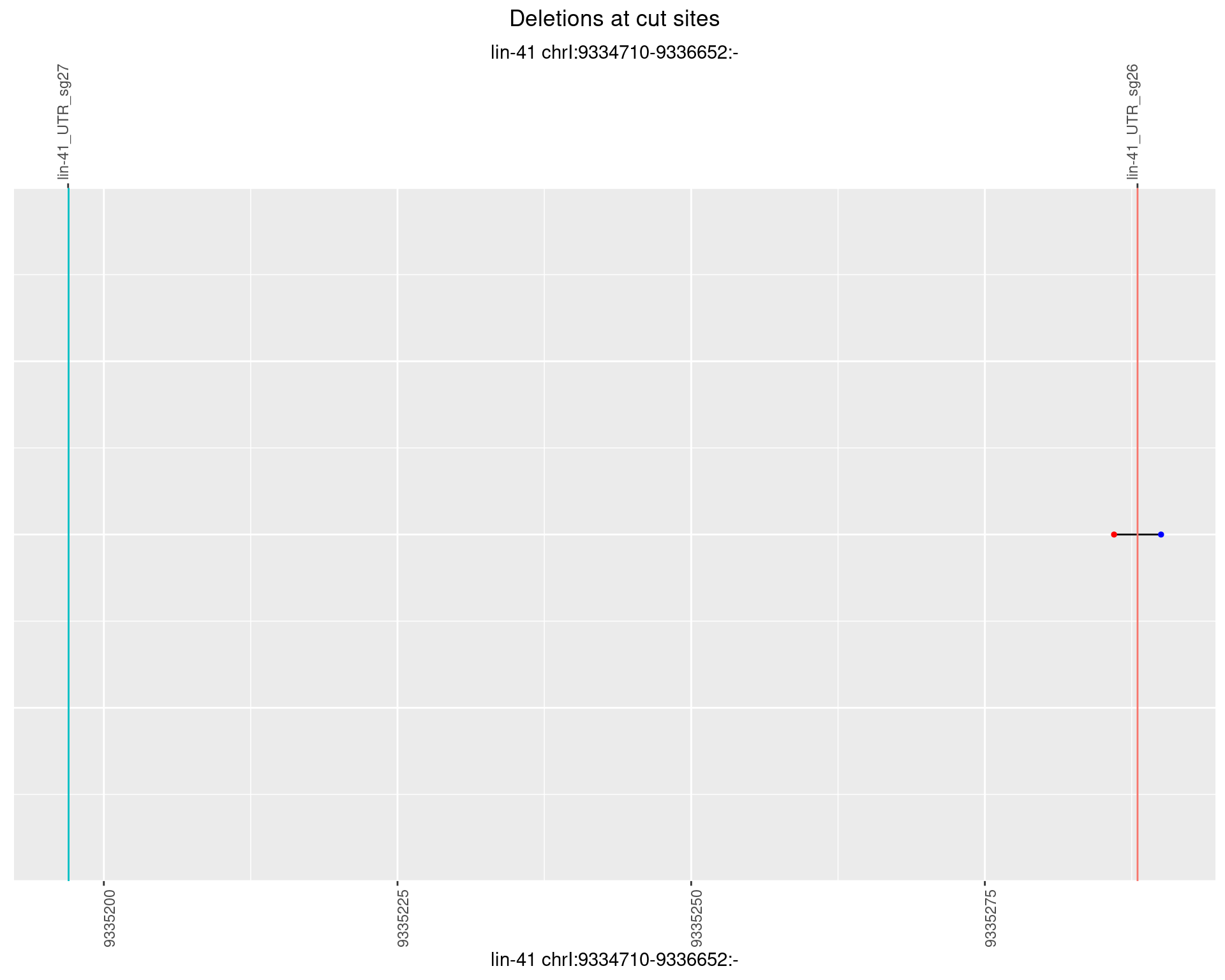
1.99.5 Freq: 0.01
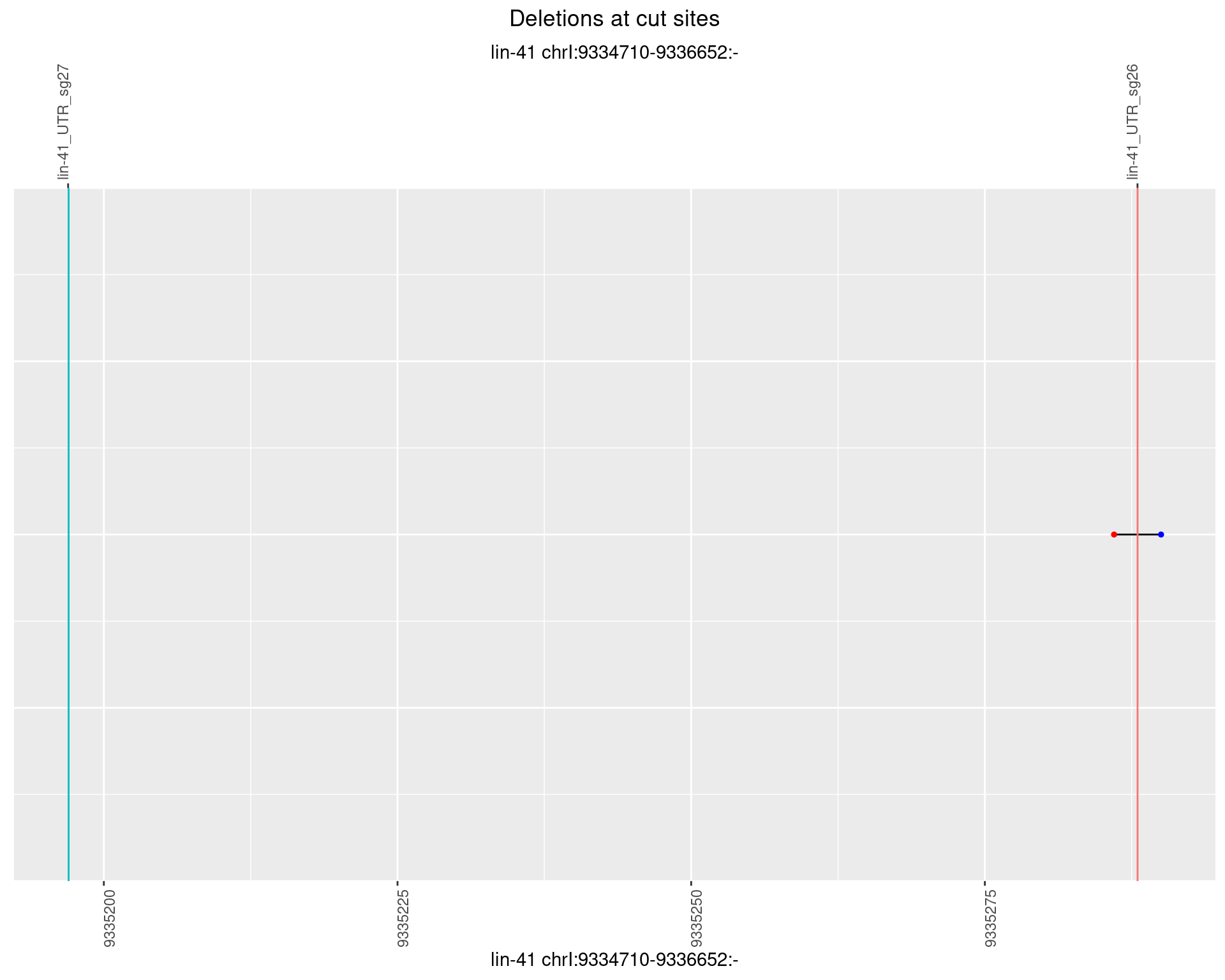
1.99.6 Freq: 0.1
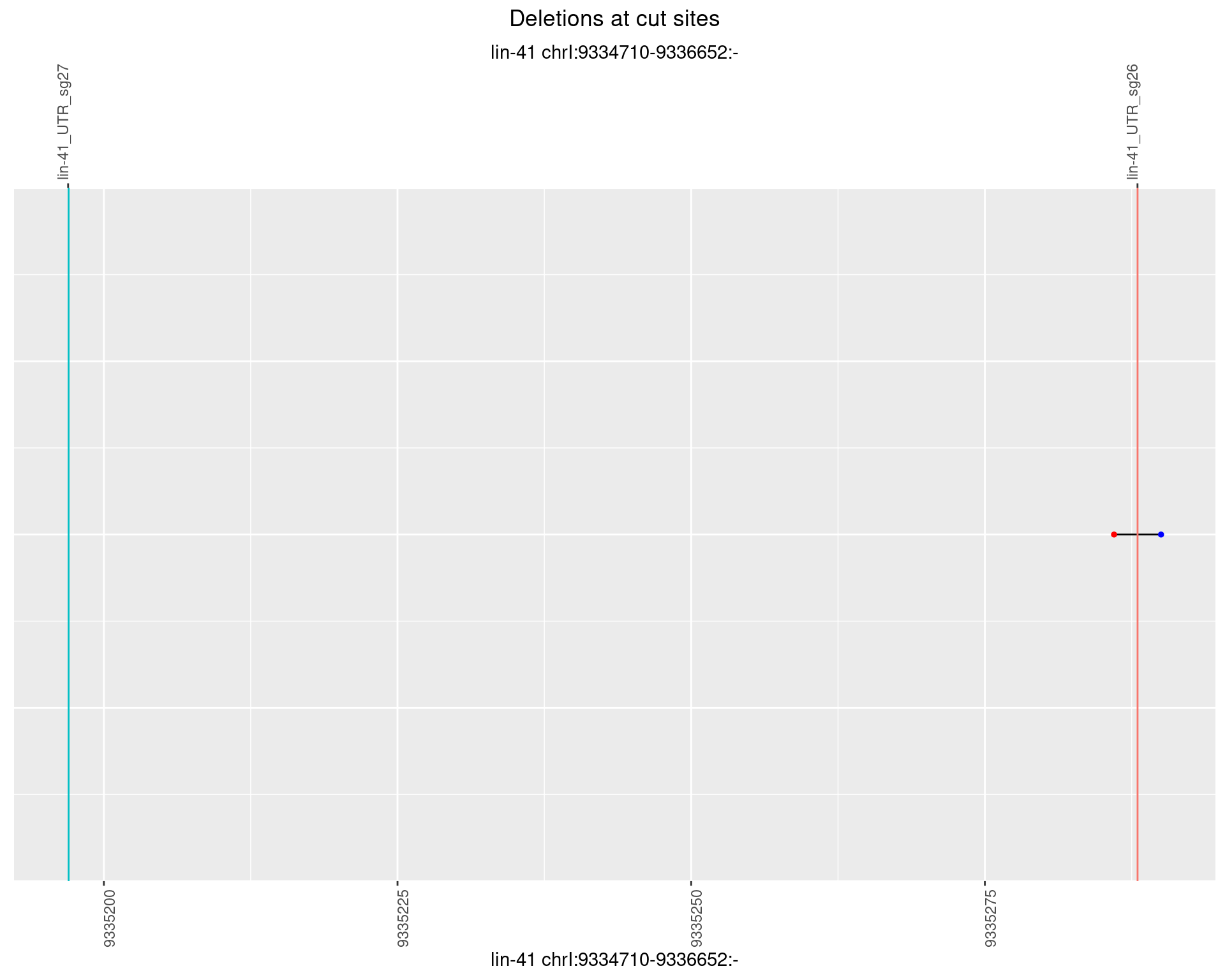
1.100 lin41_RNA_L4_pool2_middle_UMI_R2
1.100.1 Freq: 0
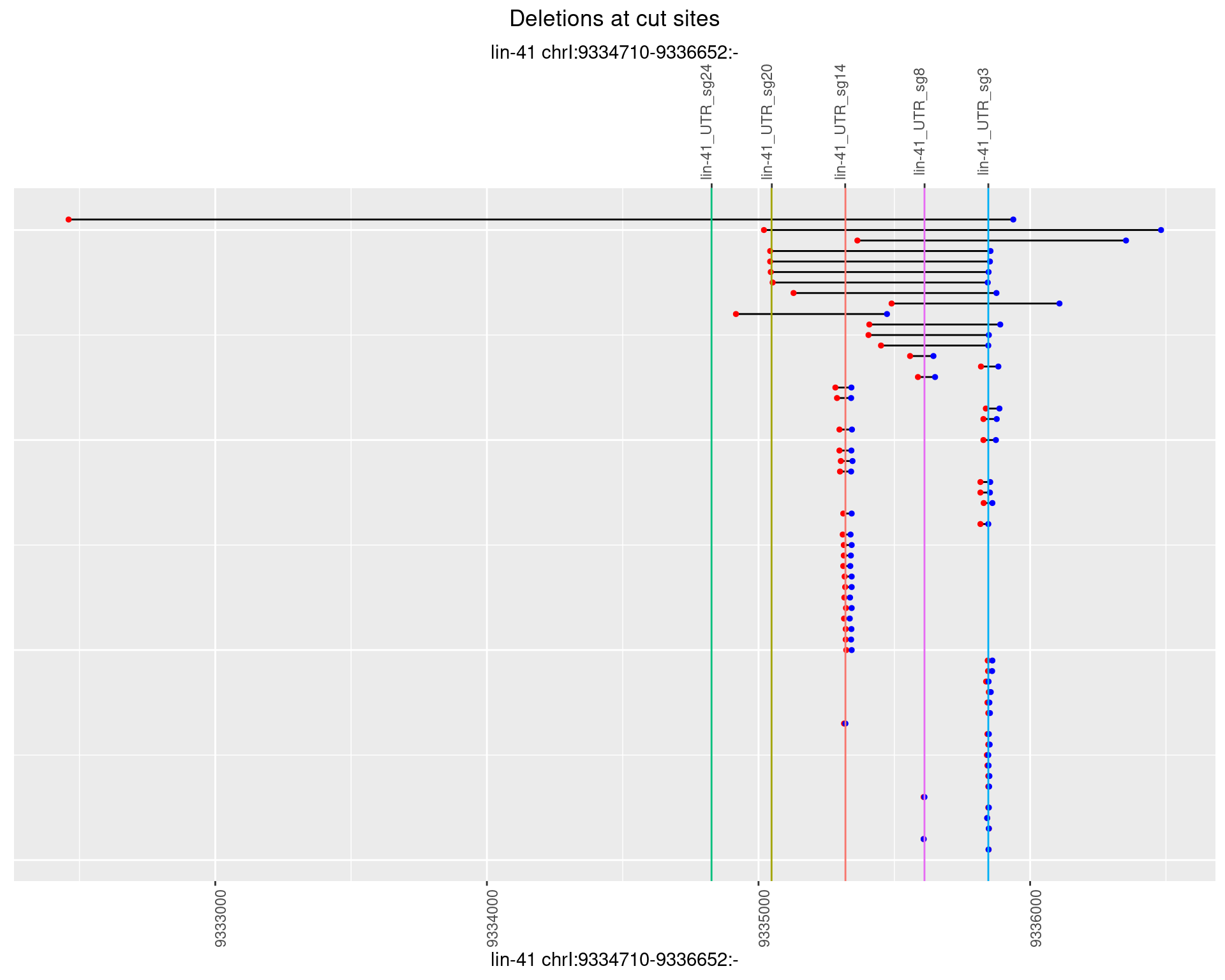
1.100.2 Freq: 1e-05
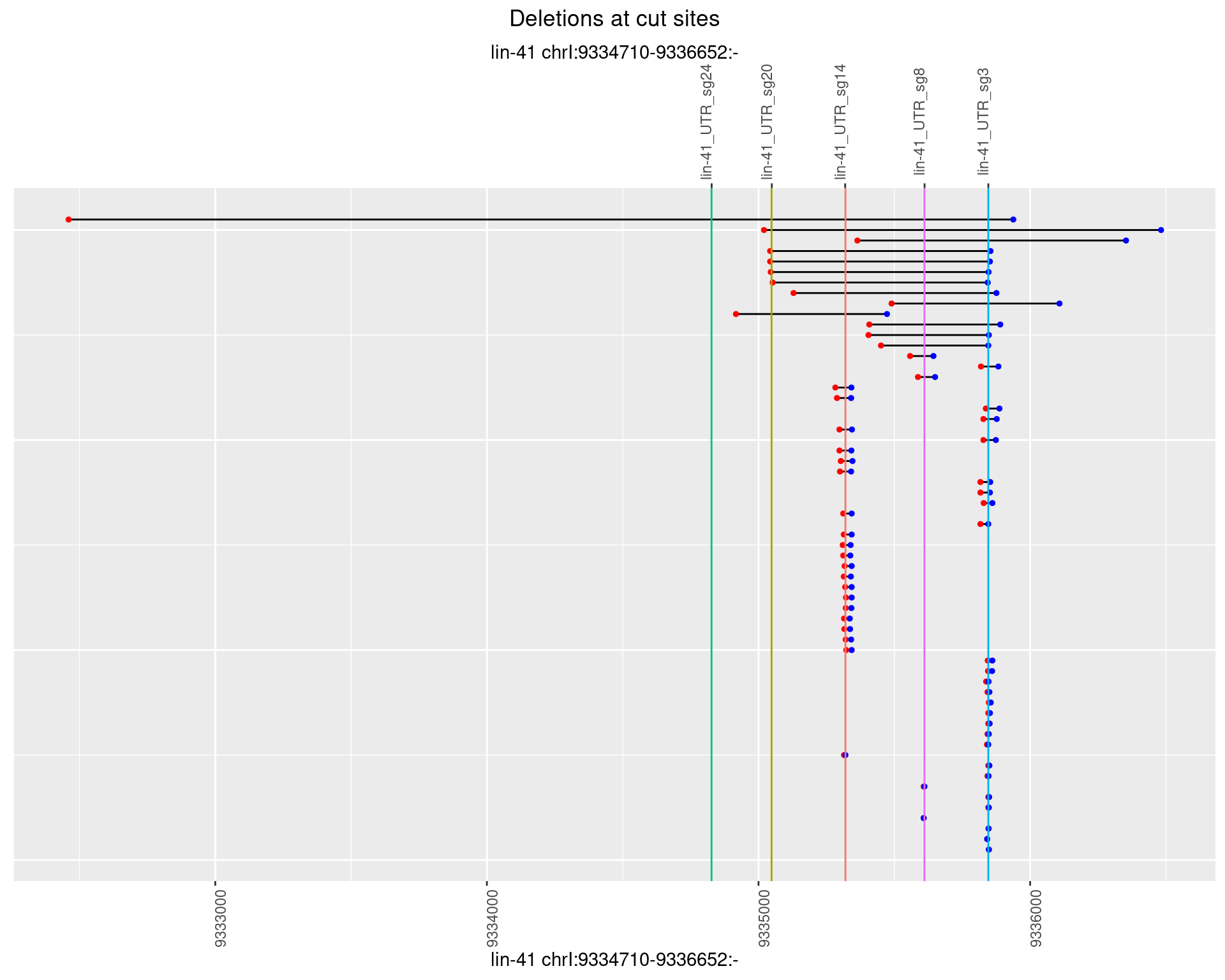
1.100.3 Freq: 1e-04
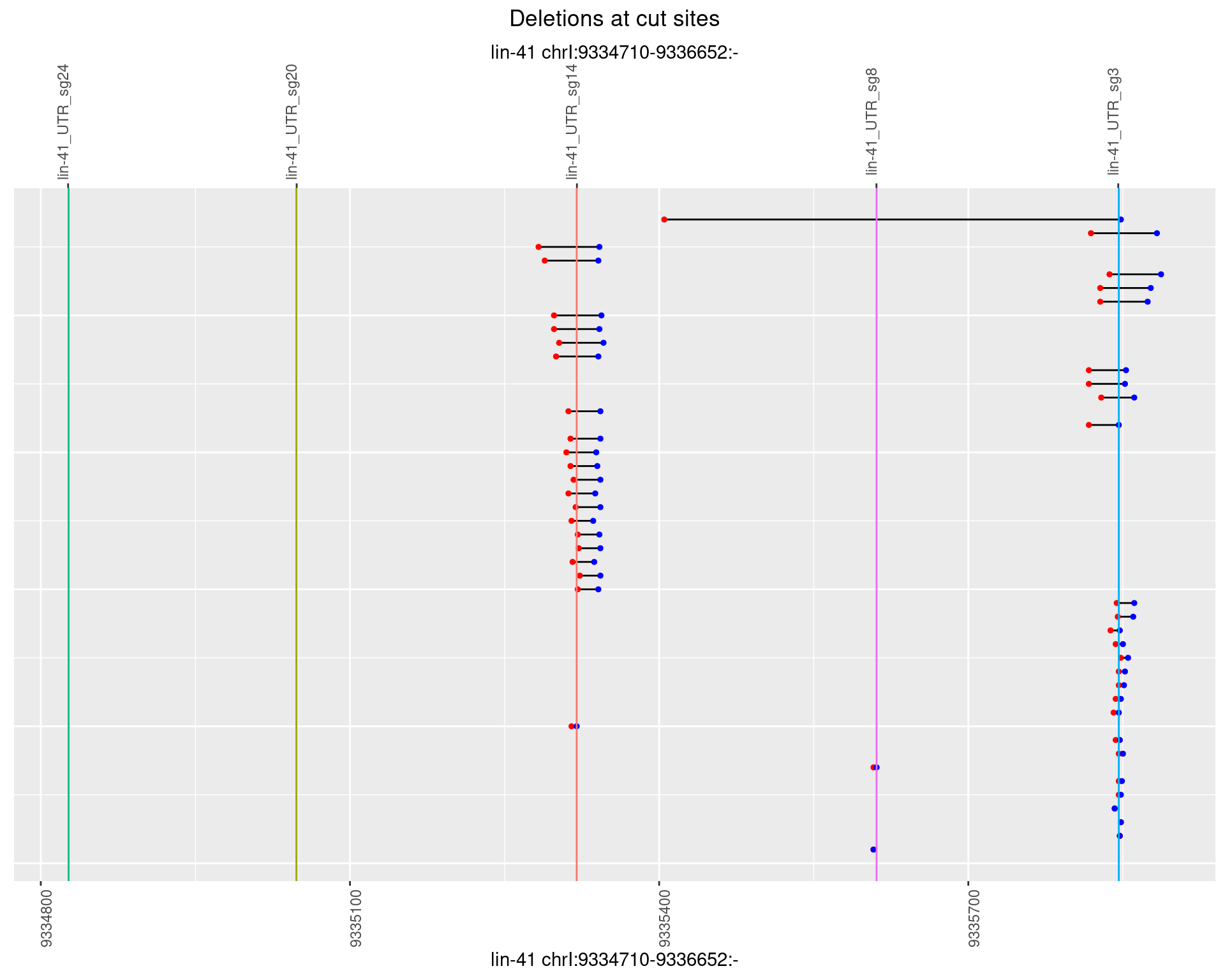
1.100.4 Freq: 0.001
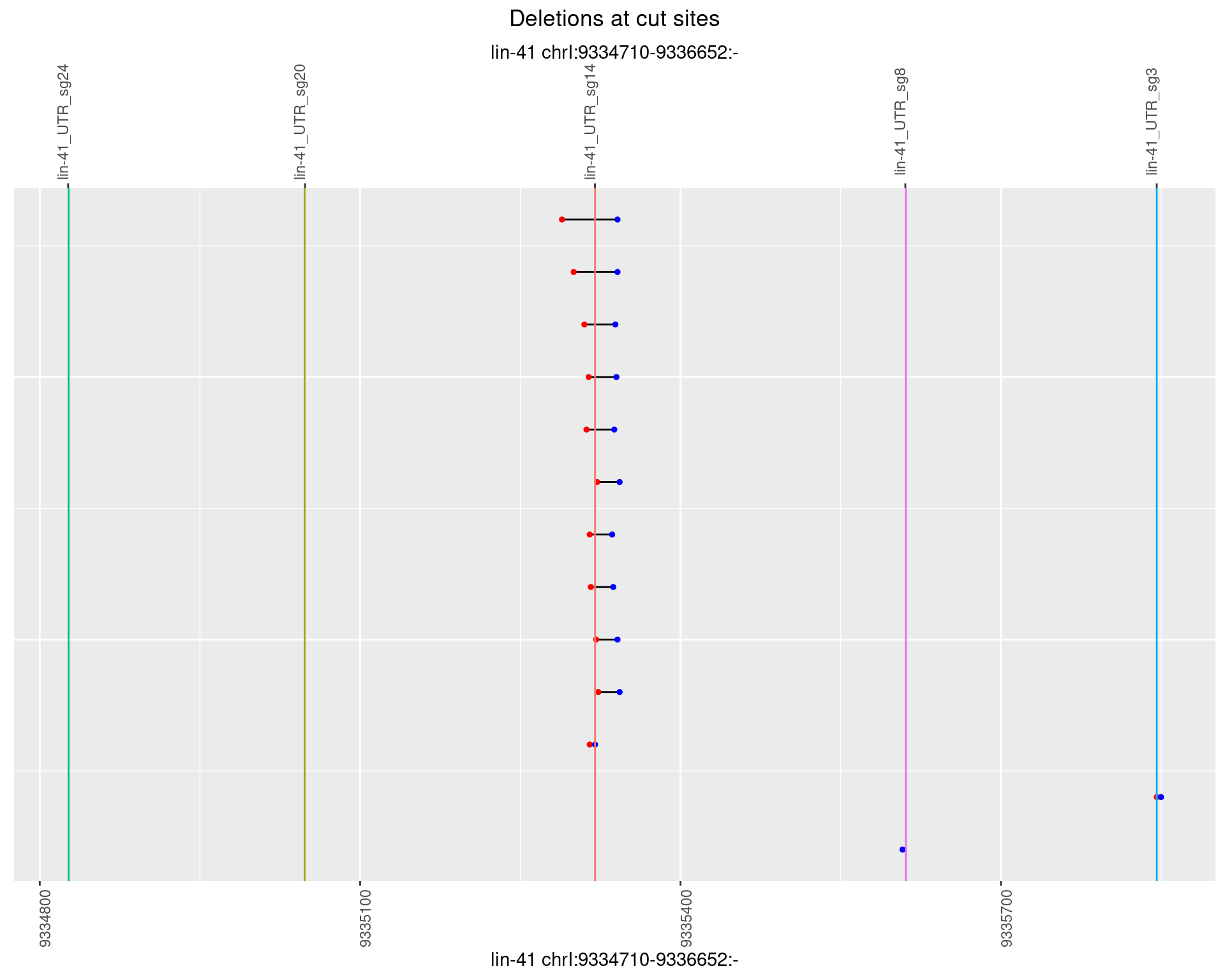
1.100.5 Freq: 0.01
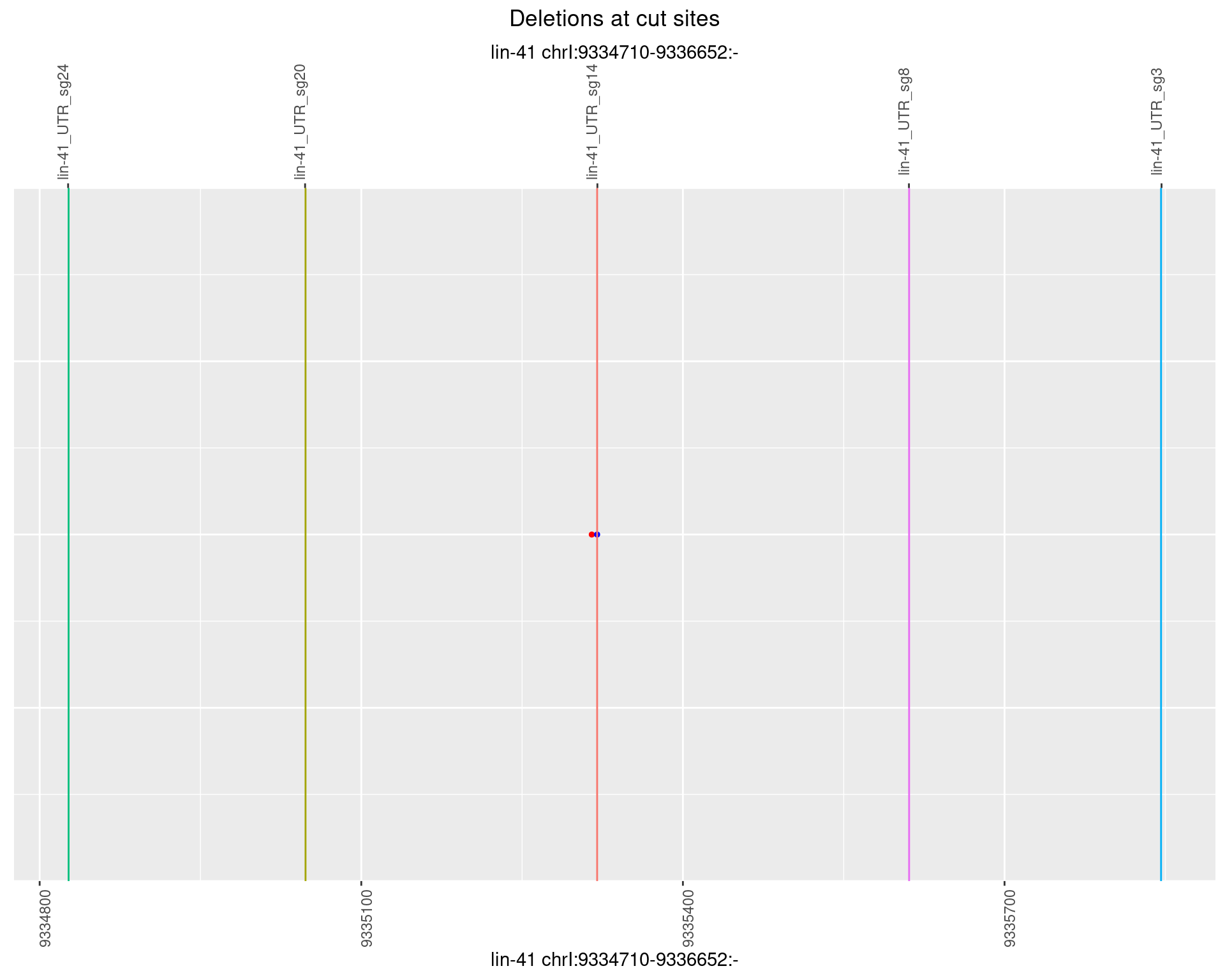
1.100.6 Freq: 0.1
No plot to show
1.101 lin41_RNA_L4_pool3_middle_UMI_R2
1.101.1 Freq: 0
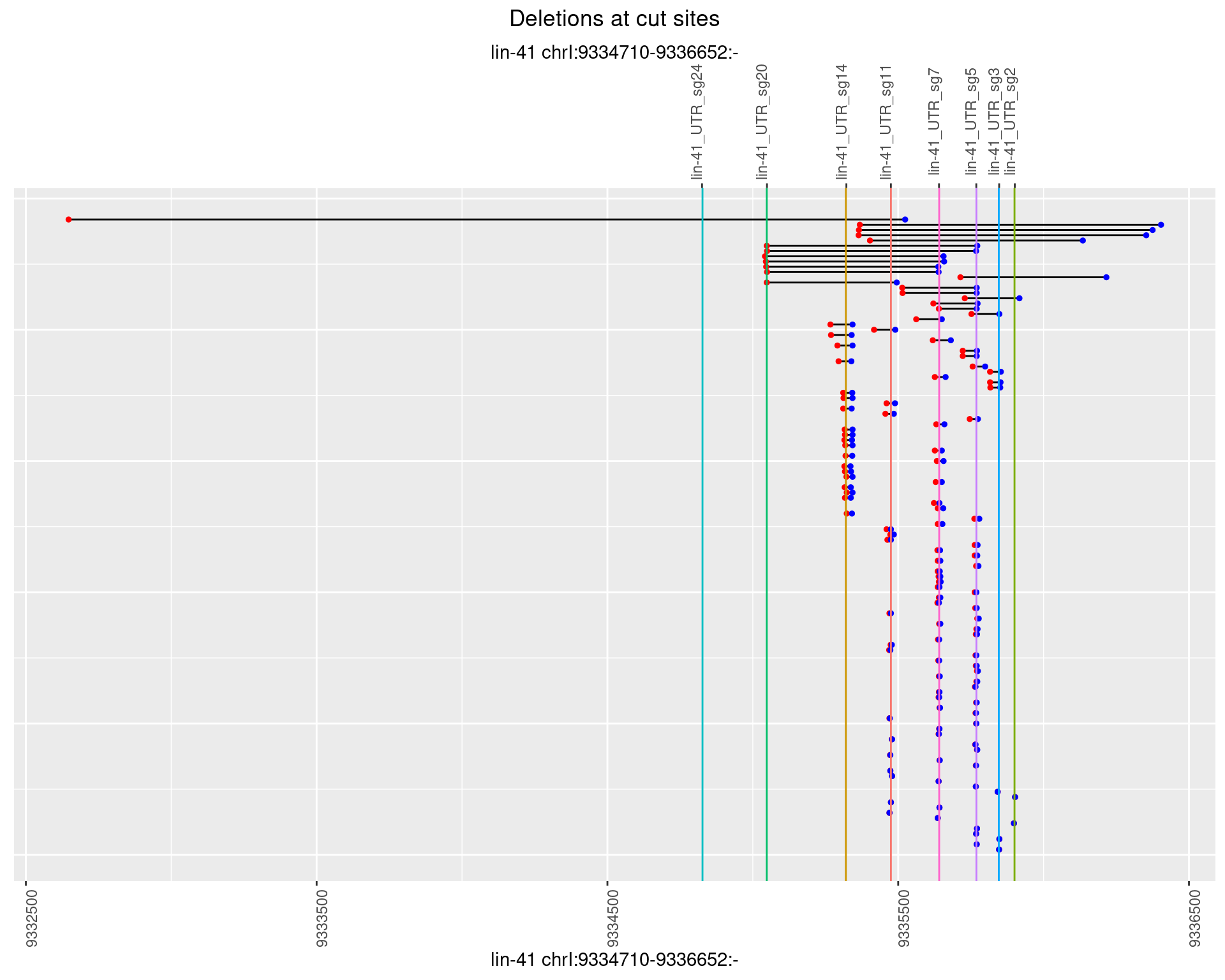
1.101.2 Freq: 1e-05
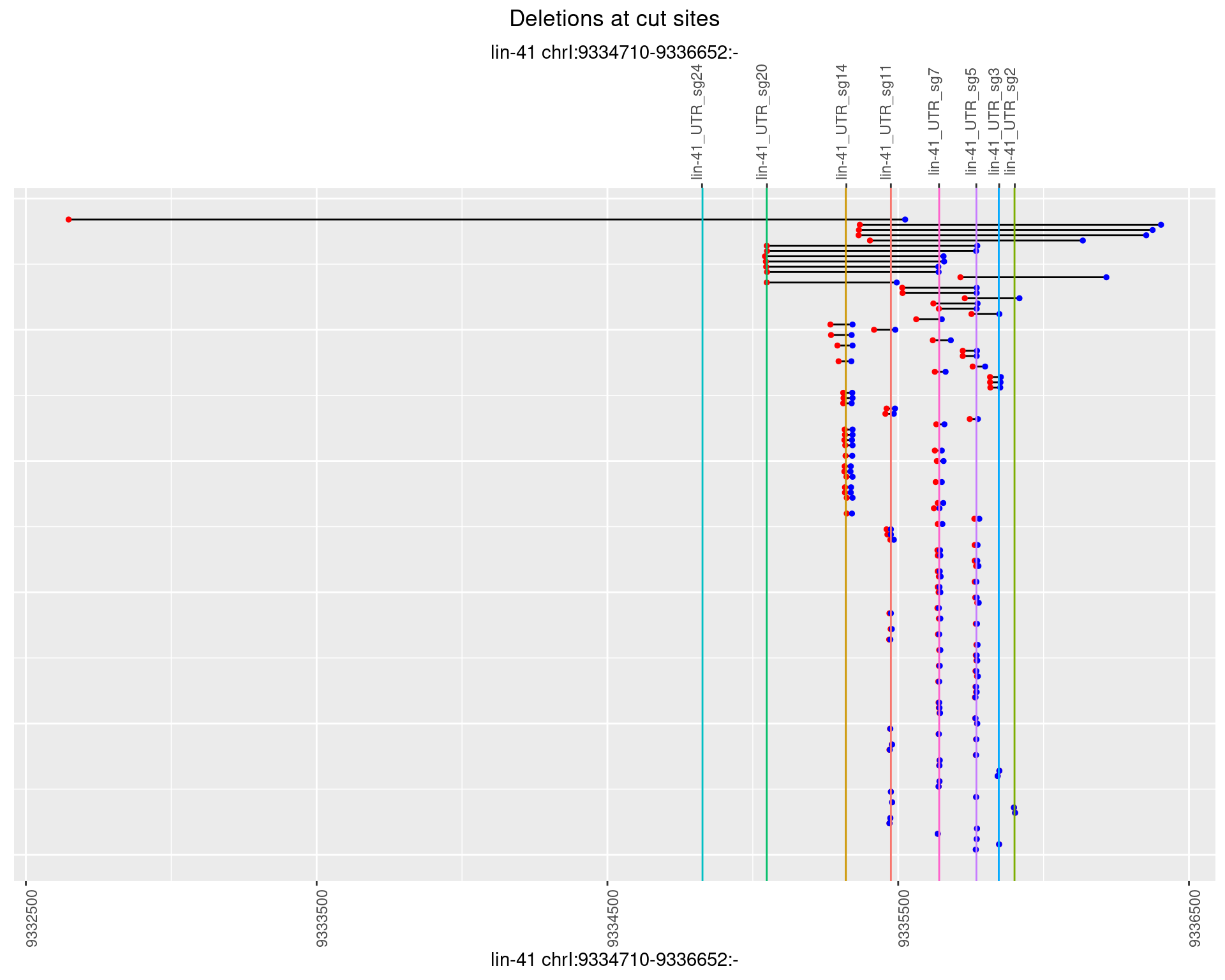
1.101.3 Freq: 1e-04
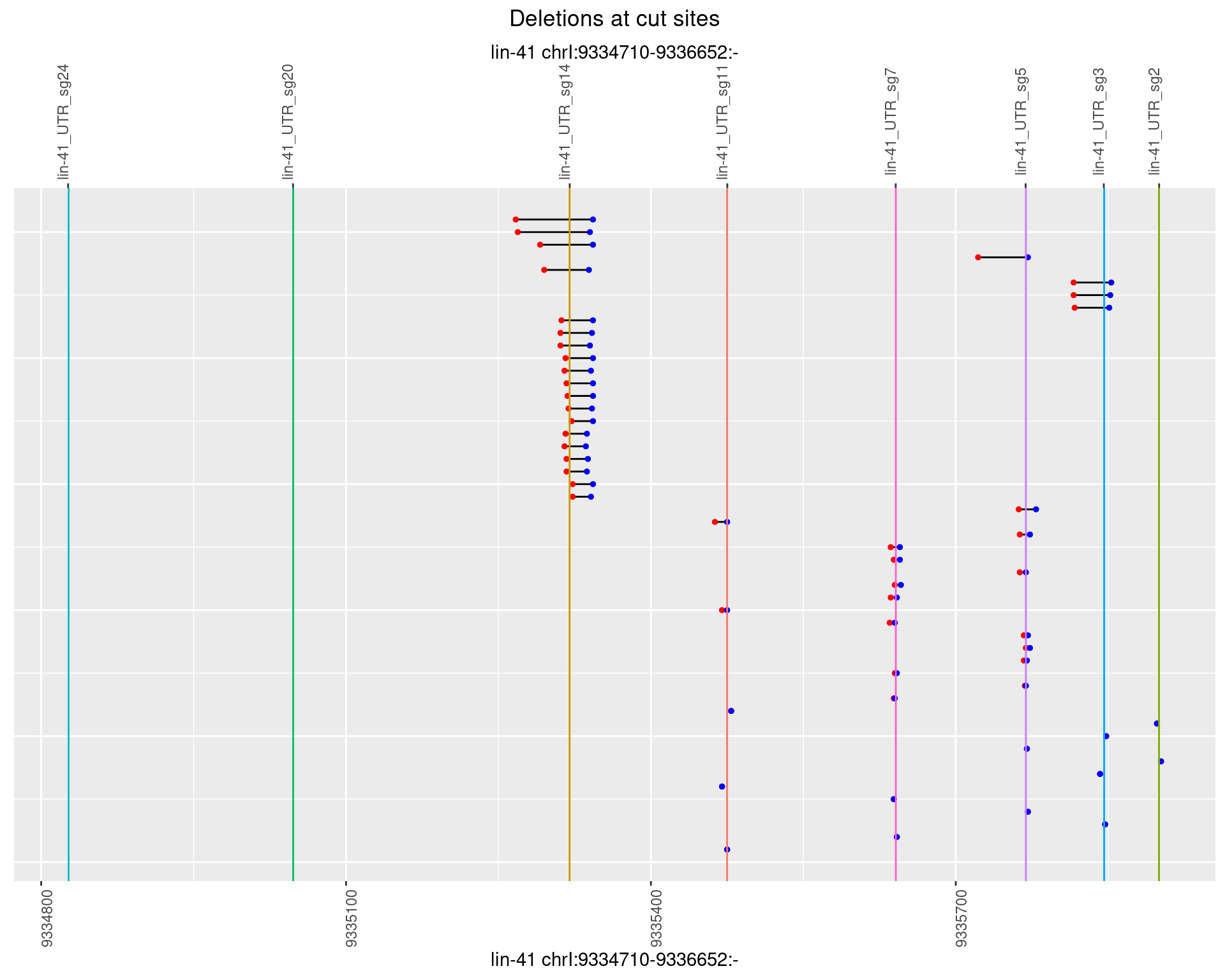
1.101.4 Freq: 0.001
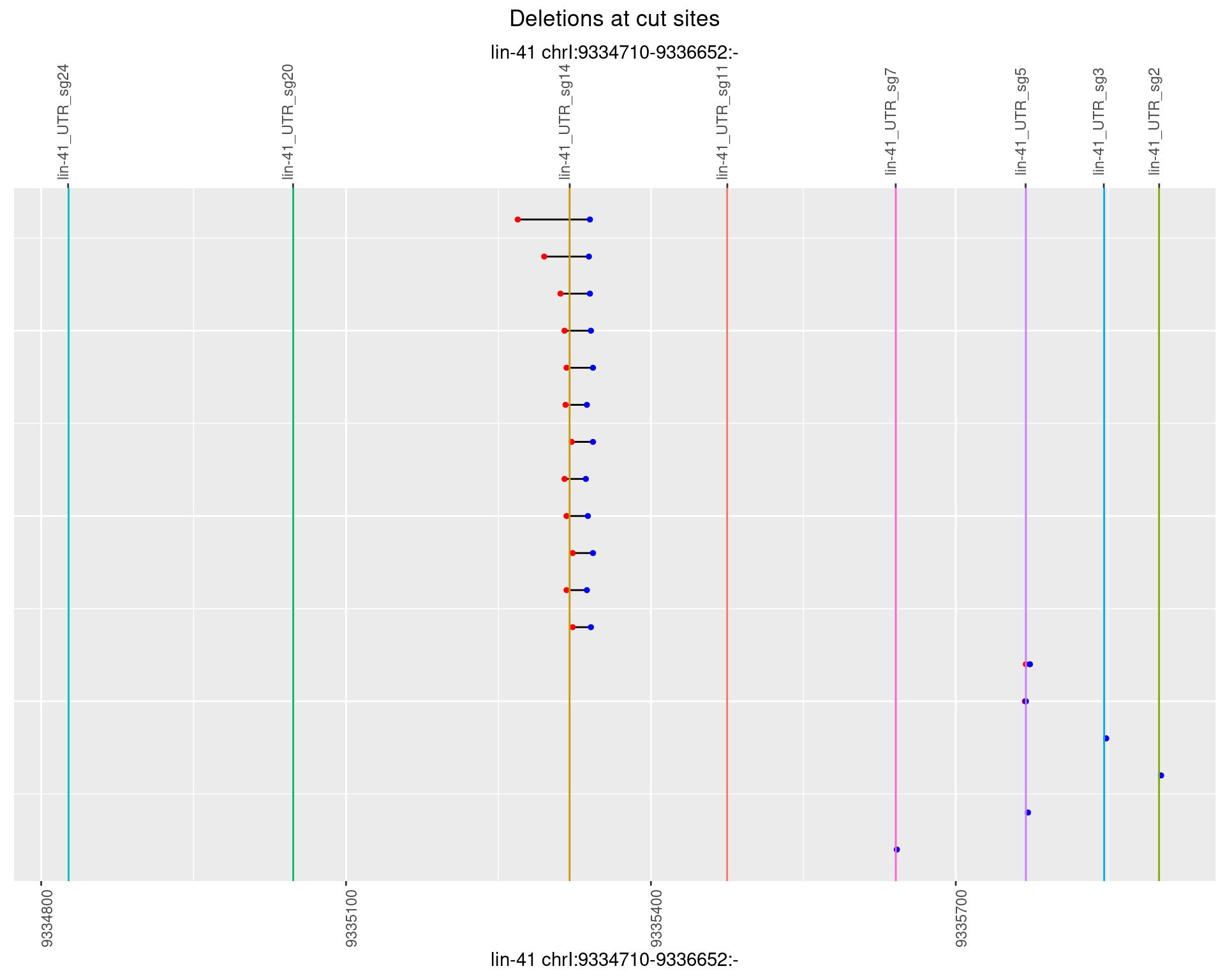
1.101.5 Freq: 0.01
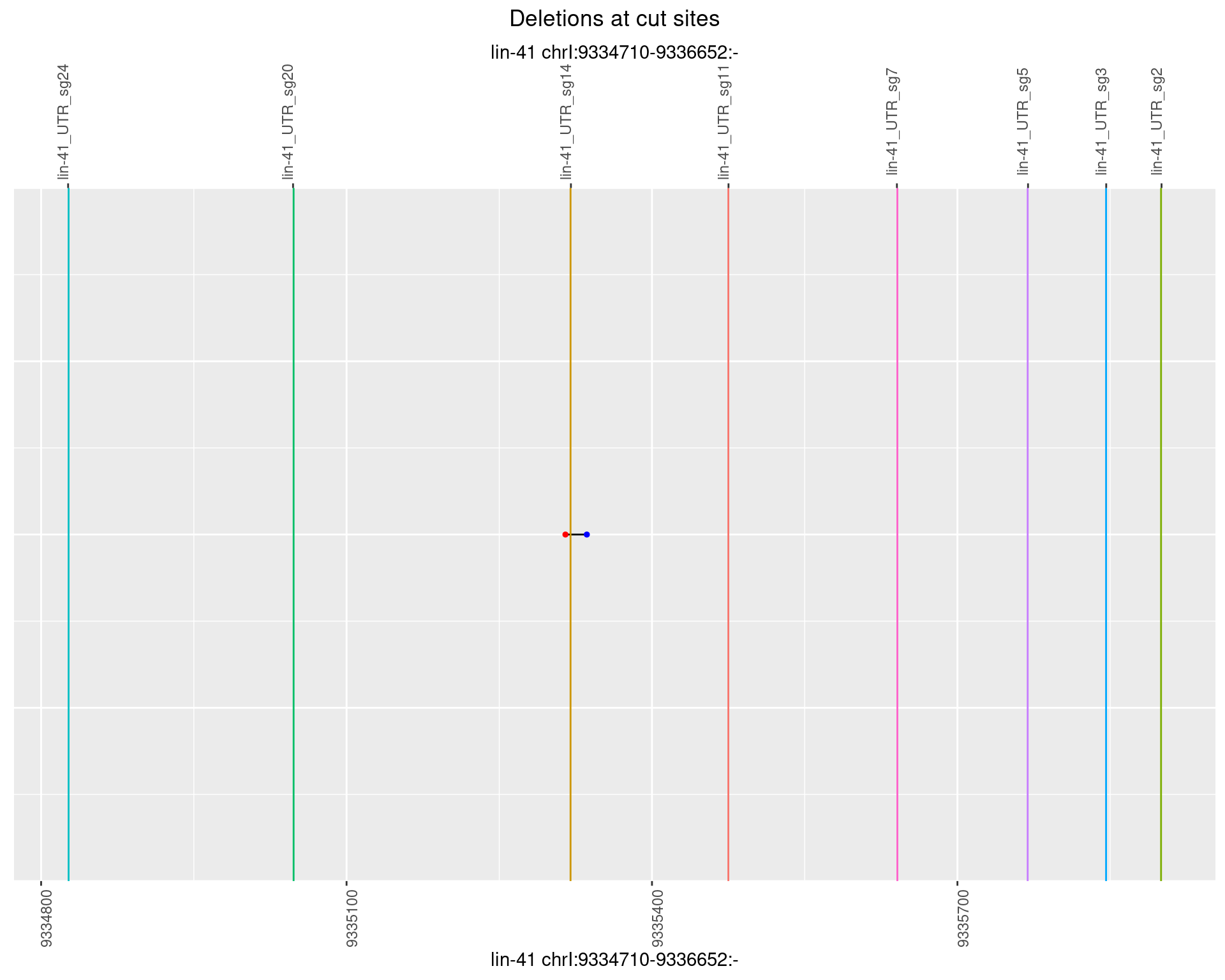
1.101.6 Freq: 0.1
No plot to show
1.102 lin41_RNApacbio_L1_1516
1.102.1 Freq: 0
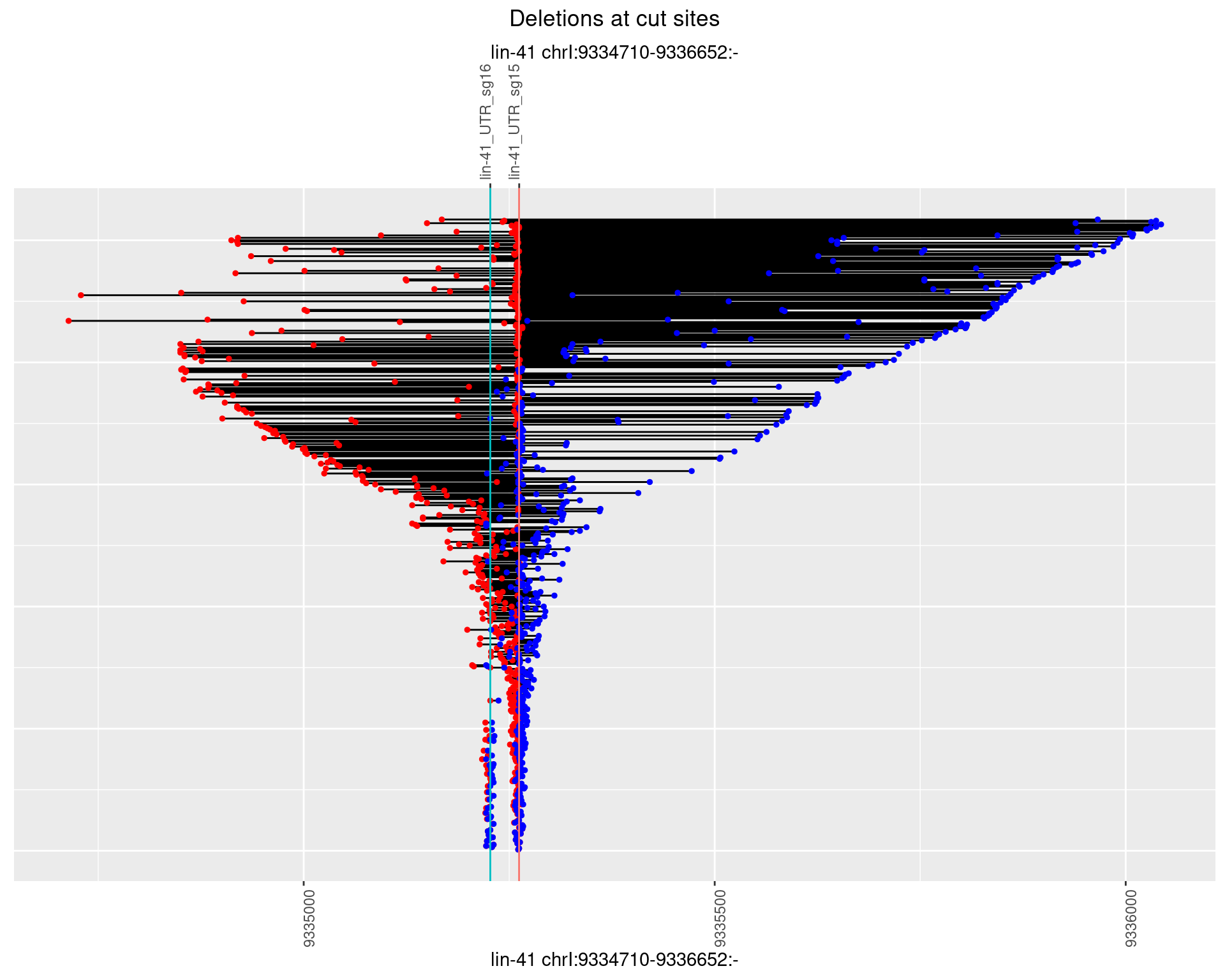
1.102.2 Freq: 1e-05
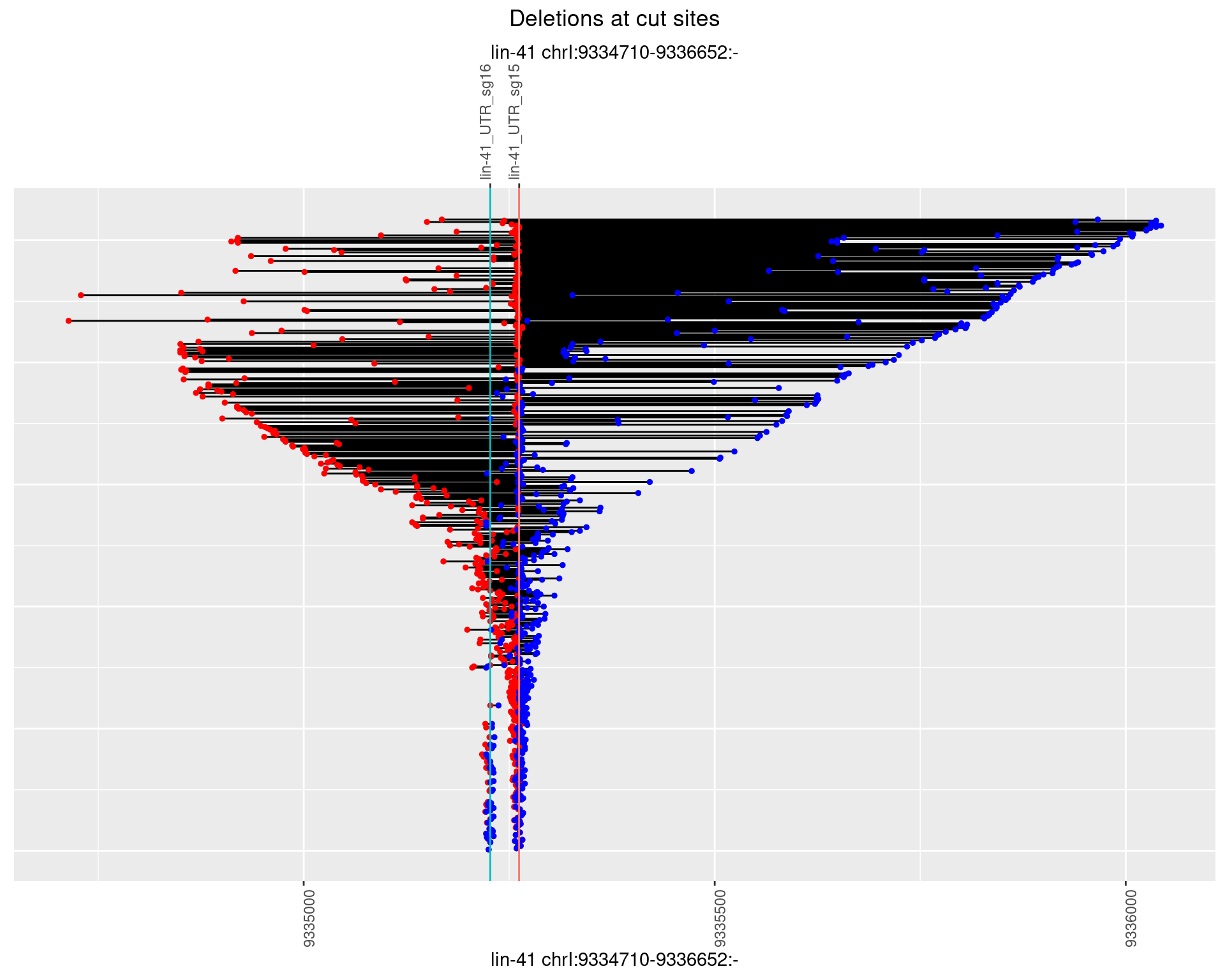
1.102.3 Freq: 1e-04
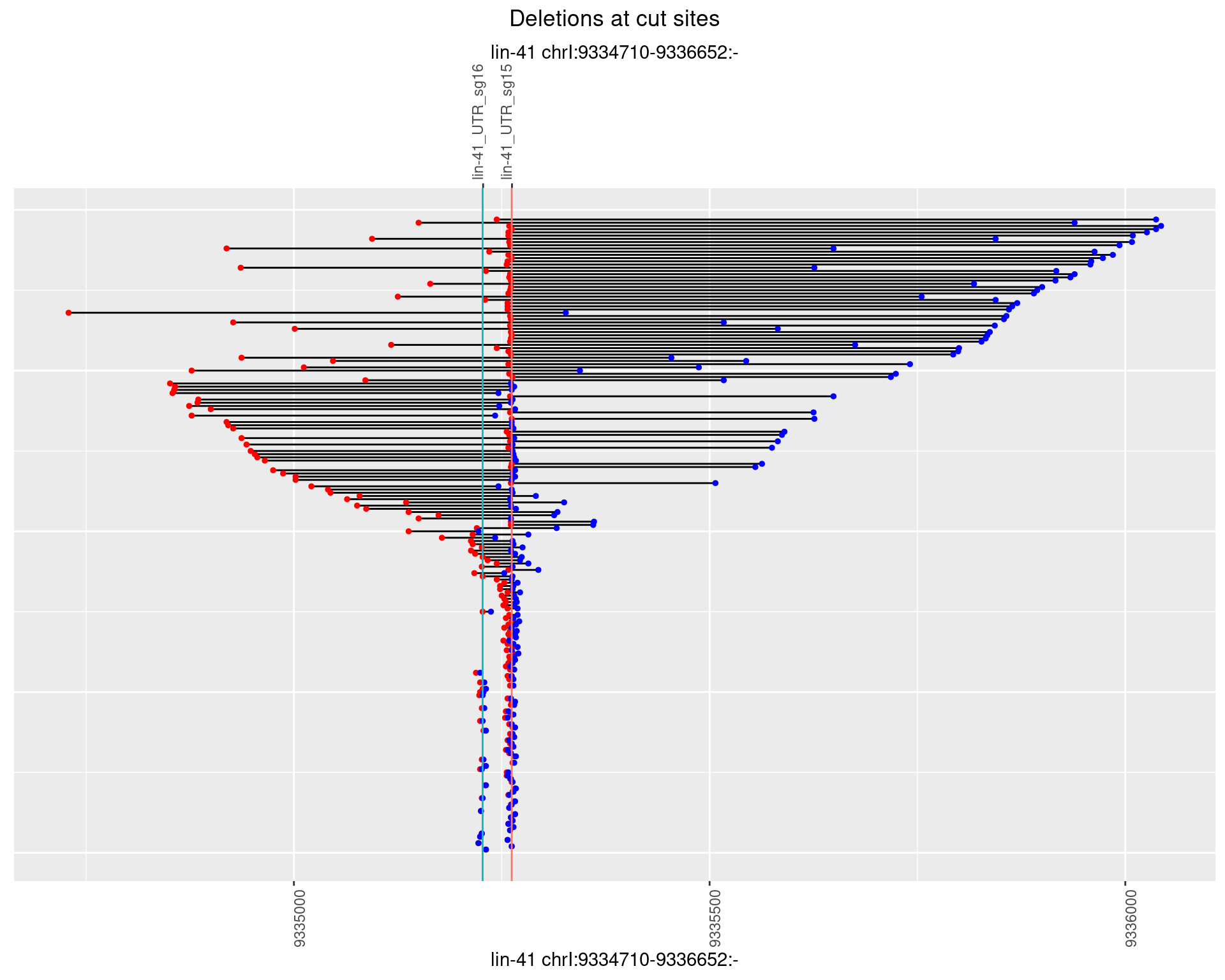
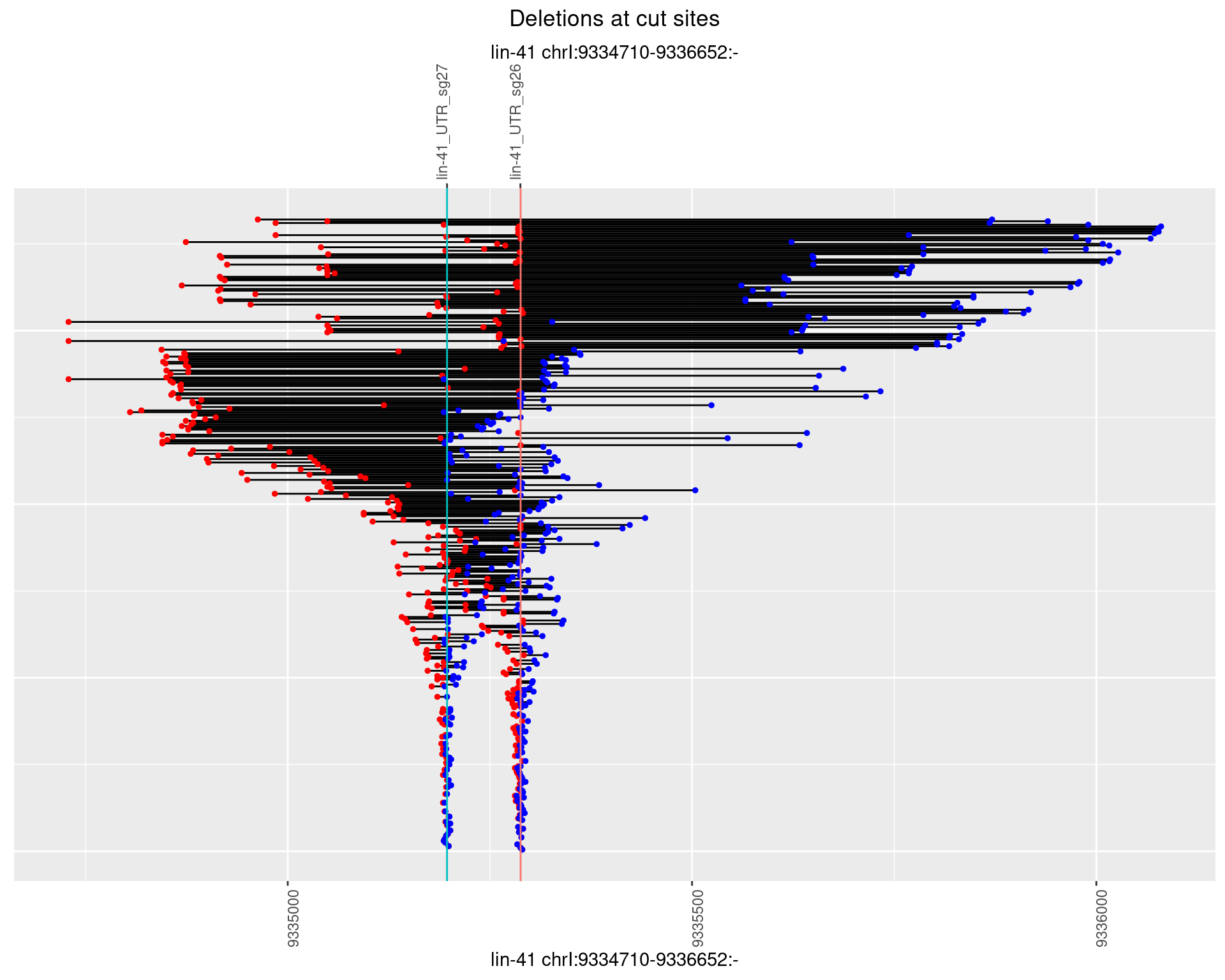
1.102.4 Freq: 0.001
1.102.5 Freq: 0.01
1.102.6 Freq: 0.1
No plot to show
1.103 lin41_RNApacbio_L1_2627
1.103.1 Freq: 0
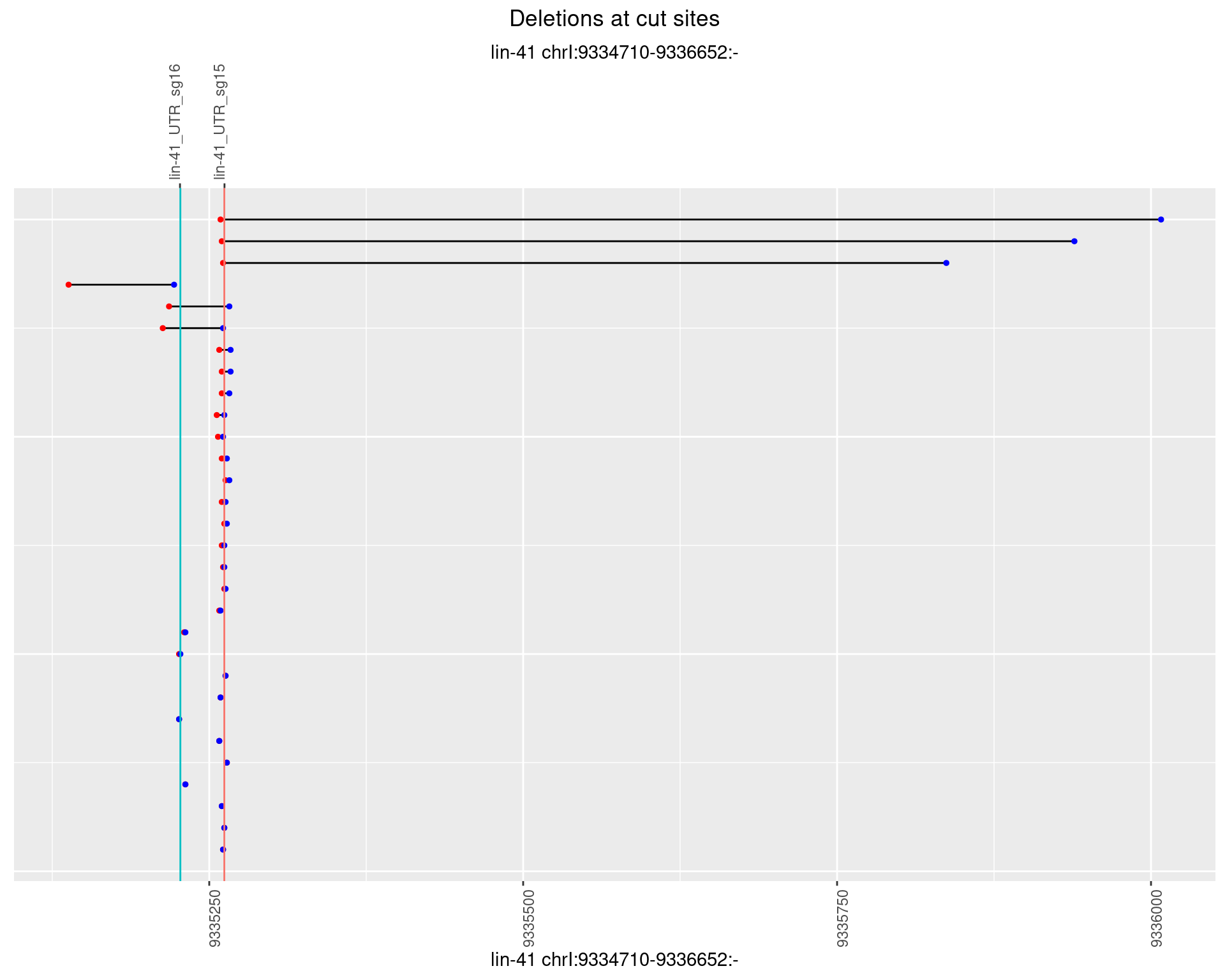
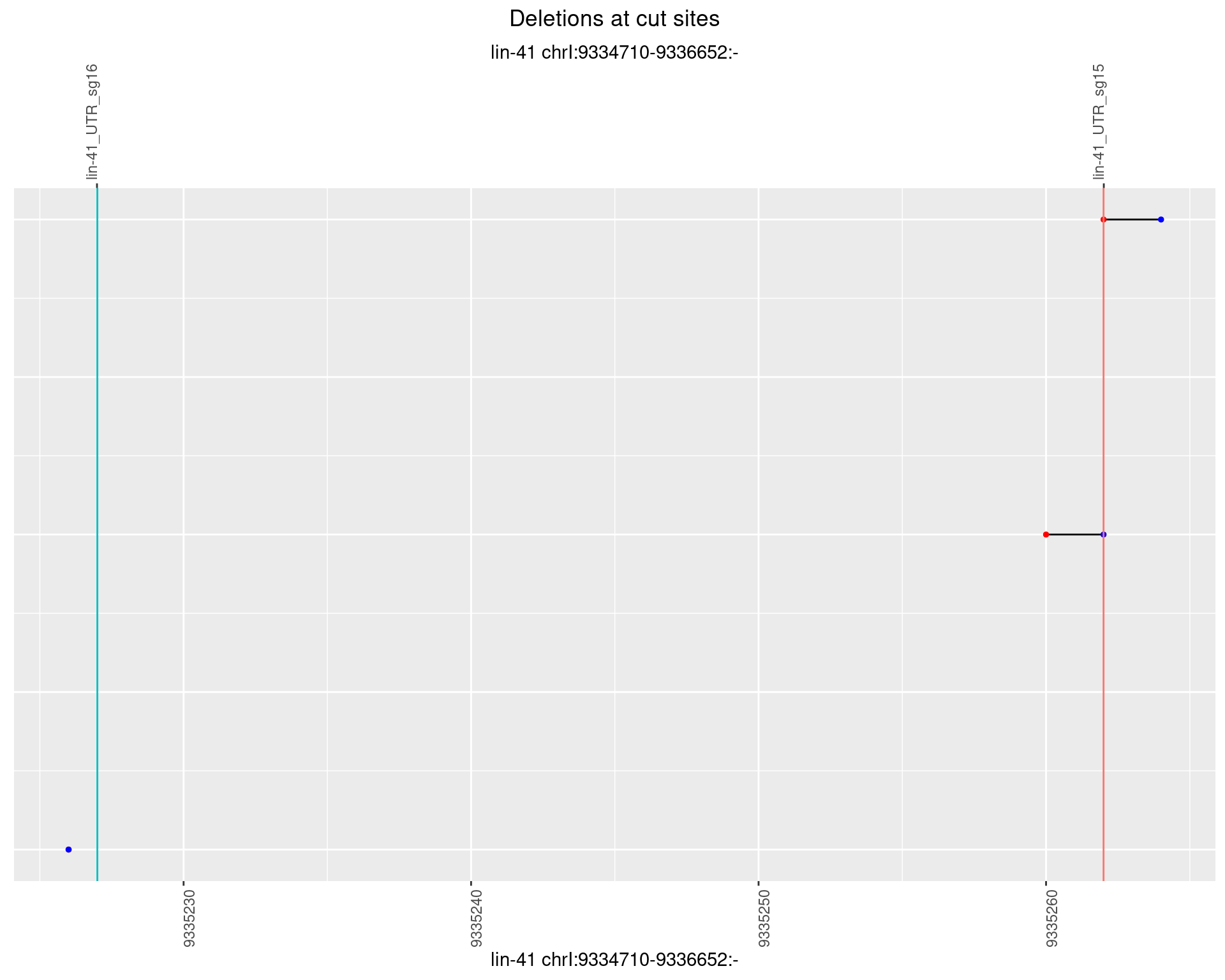
1.103.2 Freq: 1e-05
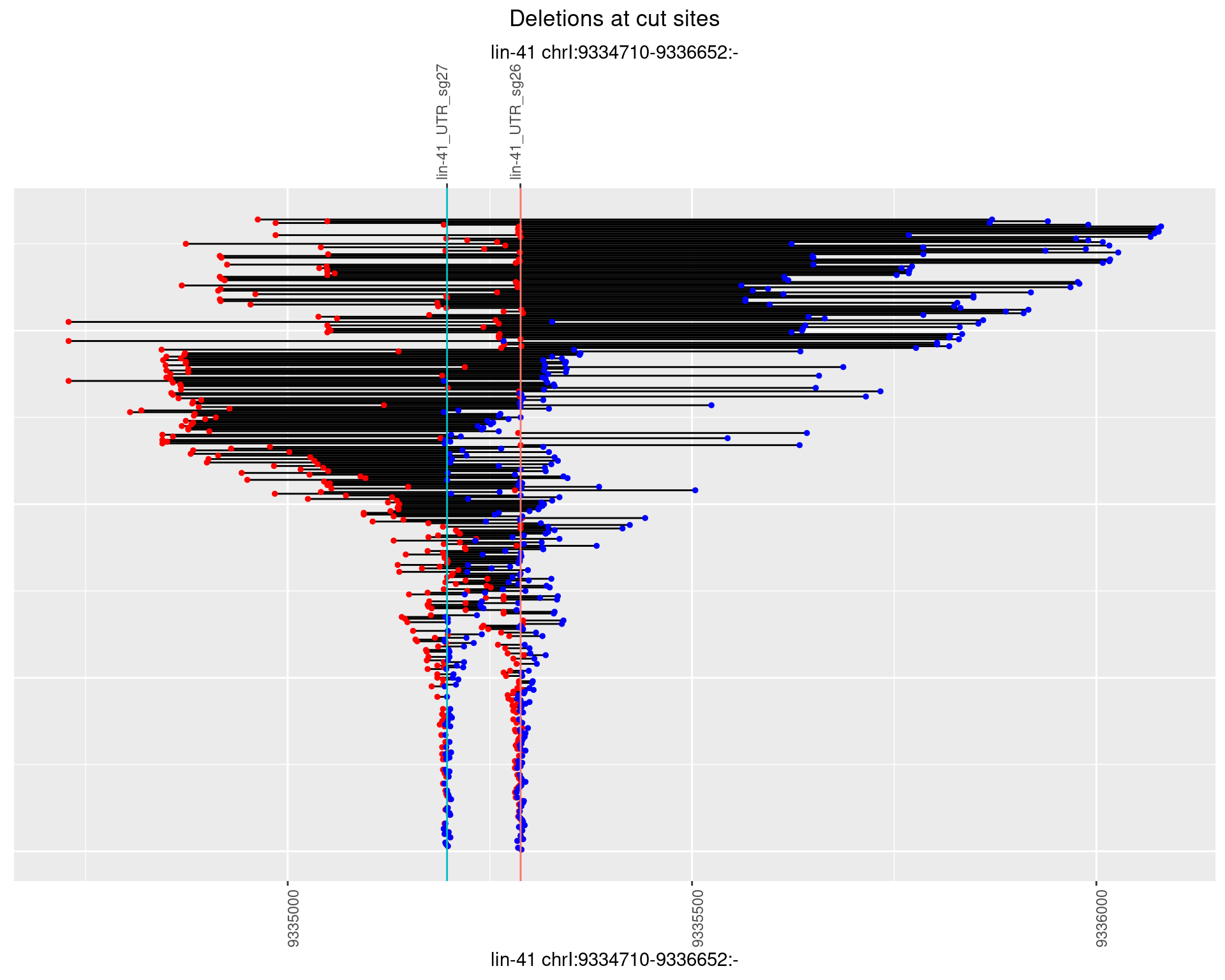
1.103.3 Freq: 1e-04
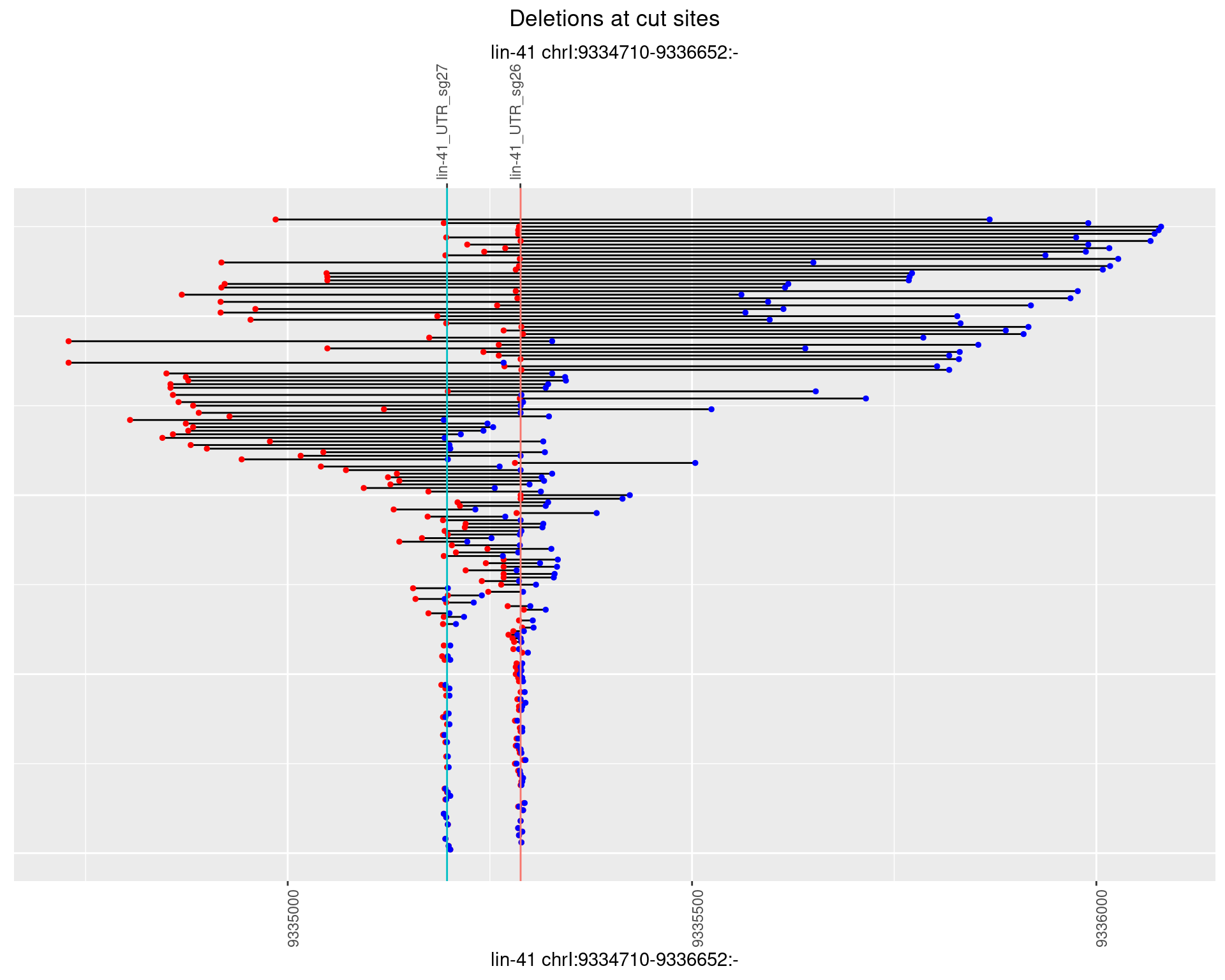
1.103.4 Freq: 0.001
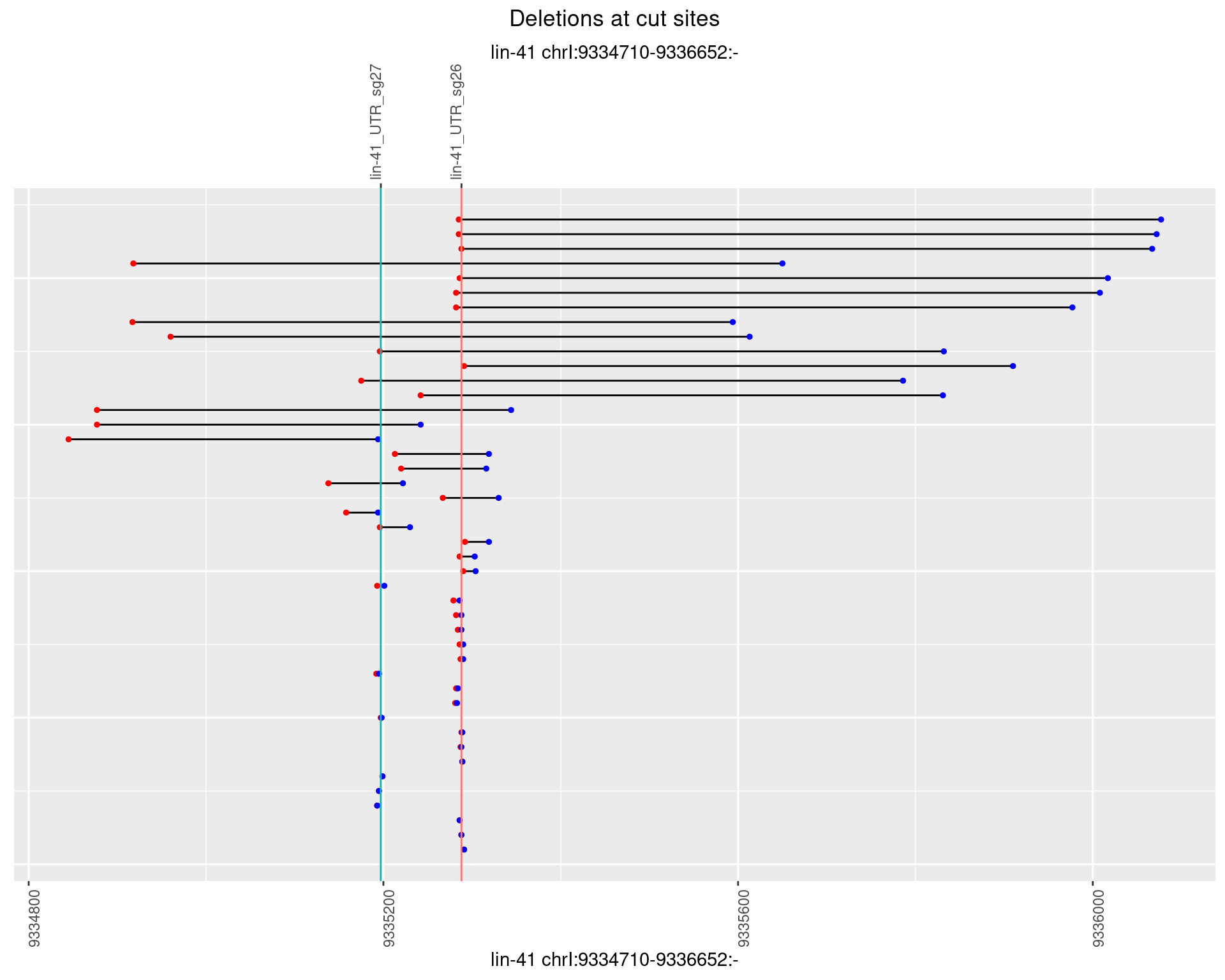
1.103.5 Freq: 0.01
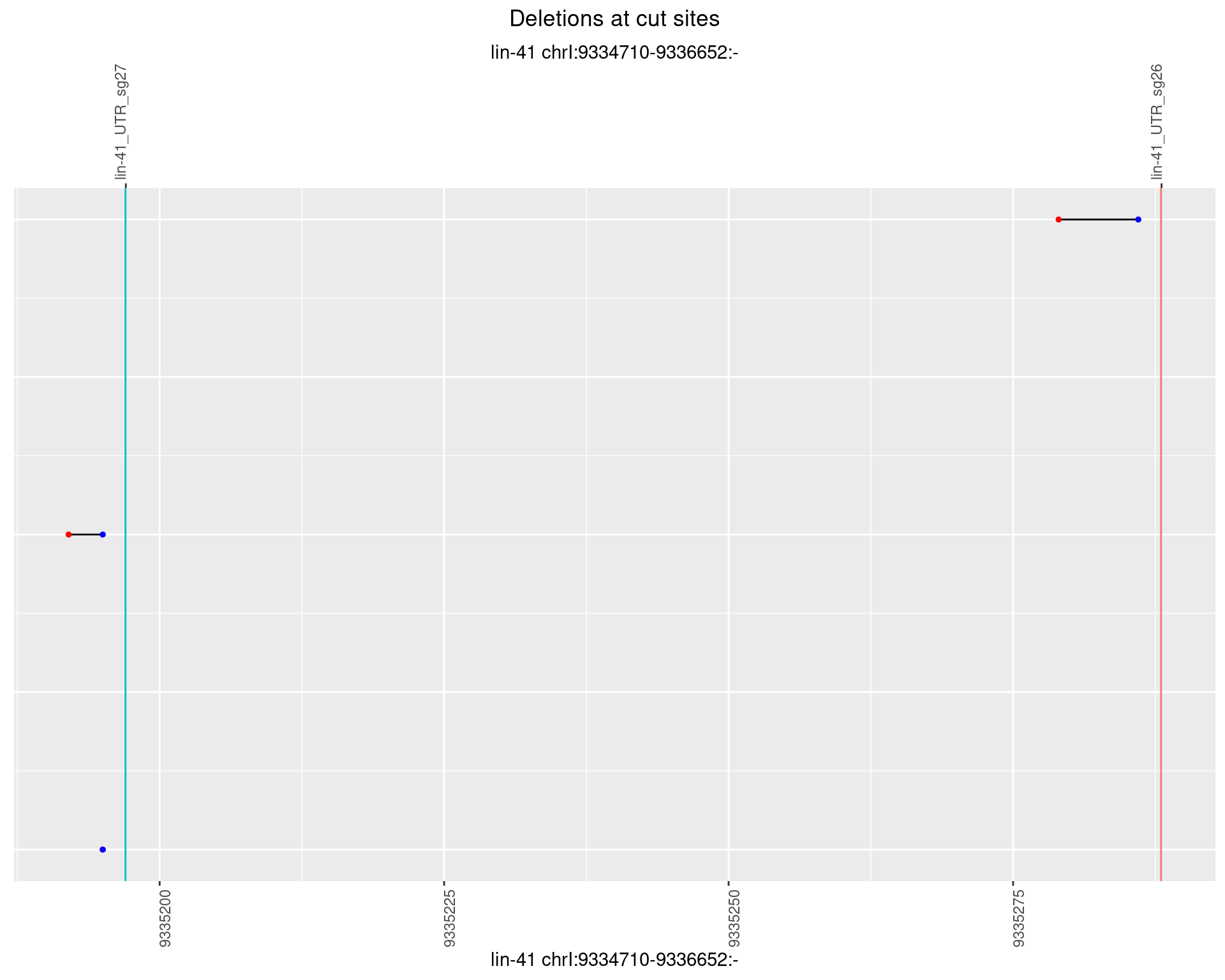
1.103.6 Freq: 0.1
No plot to show
1.104 lin41_RNApacbio_L1_pool3
1.104.1 Freq: 0
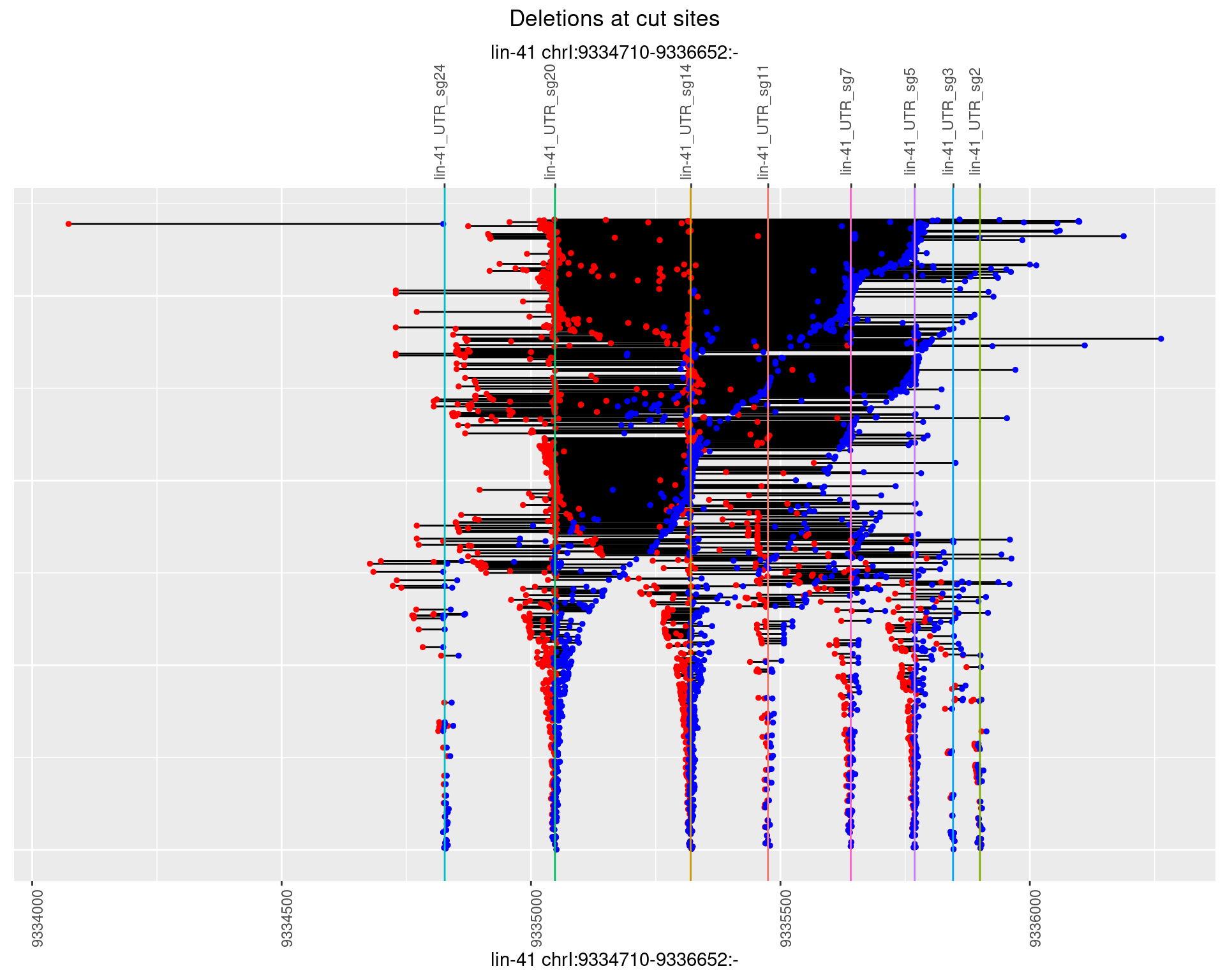
1.104.2 Freq: 1e-05
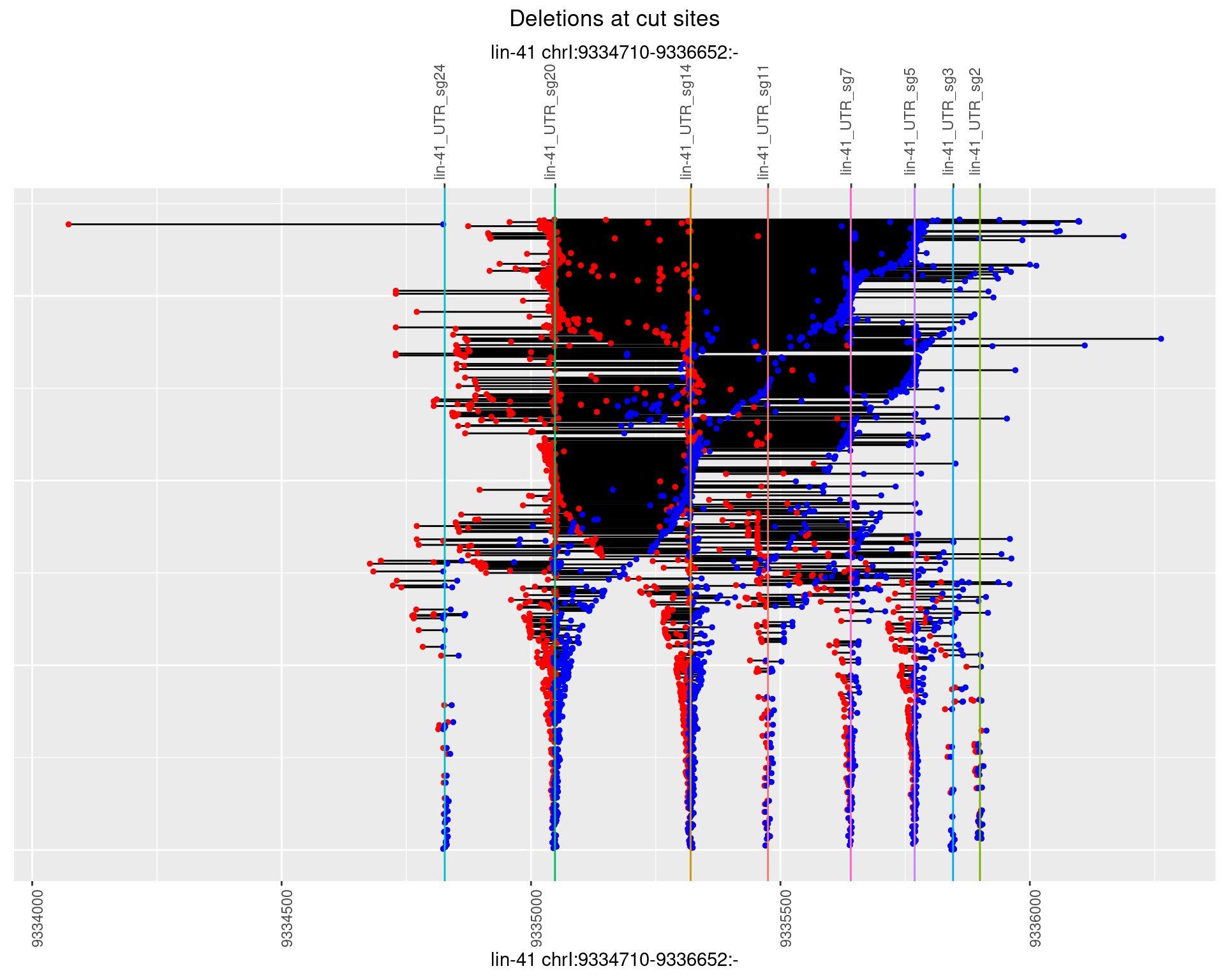
1.104.3 Freq: 1e-04
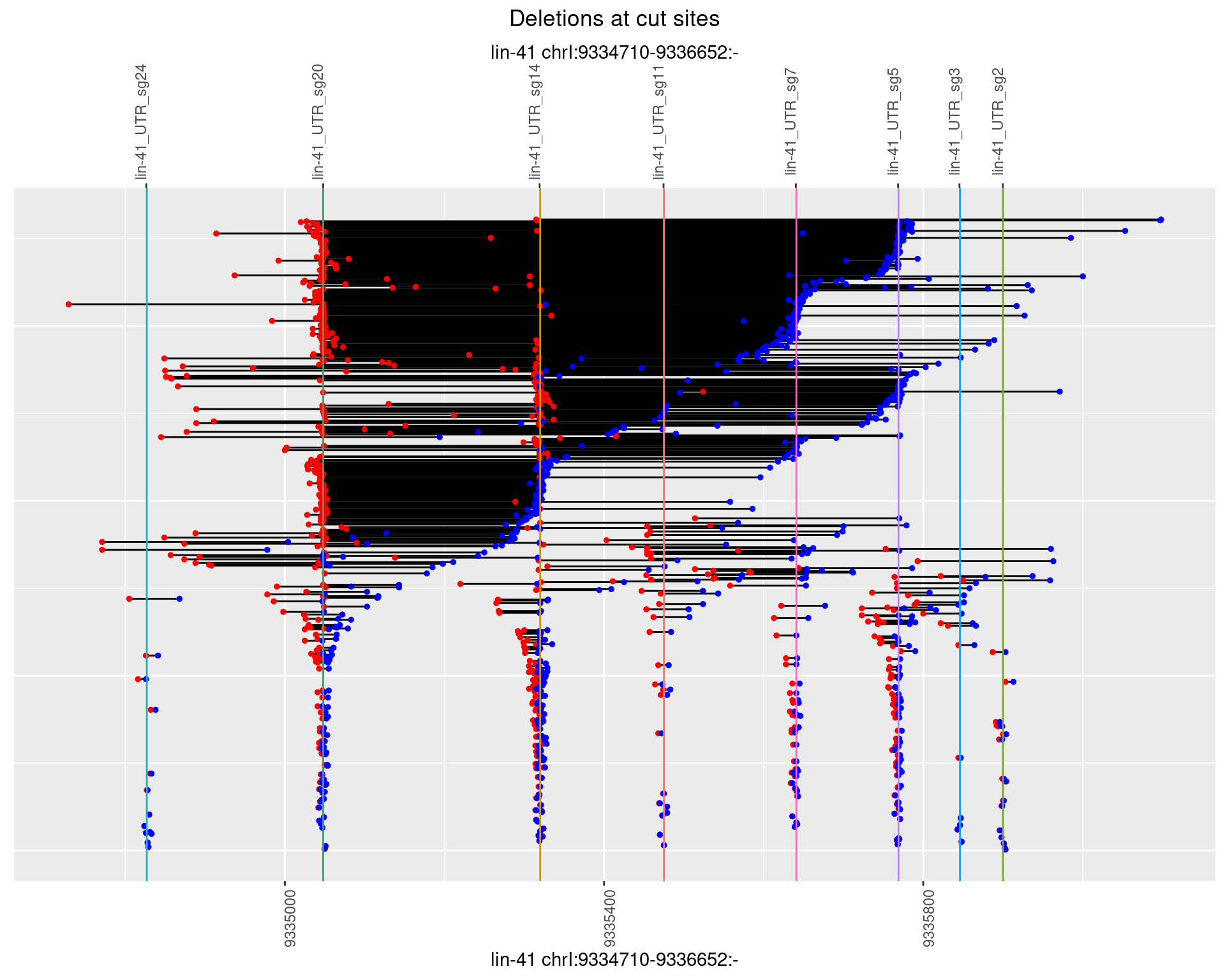
1.104.4 Freq: 0.001
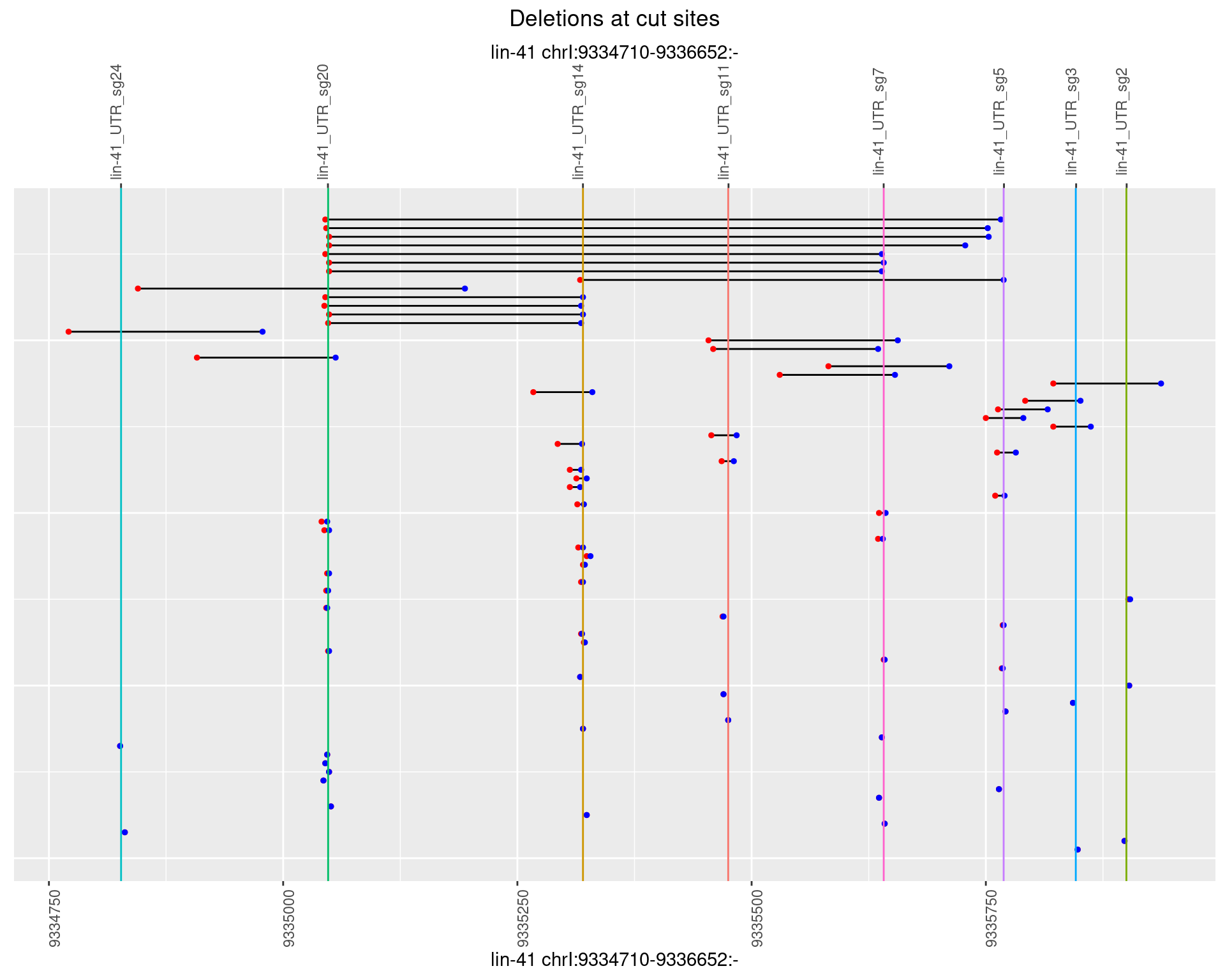
1.104.5 Freq: 0.01
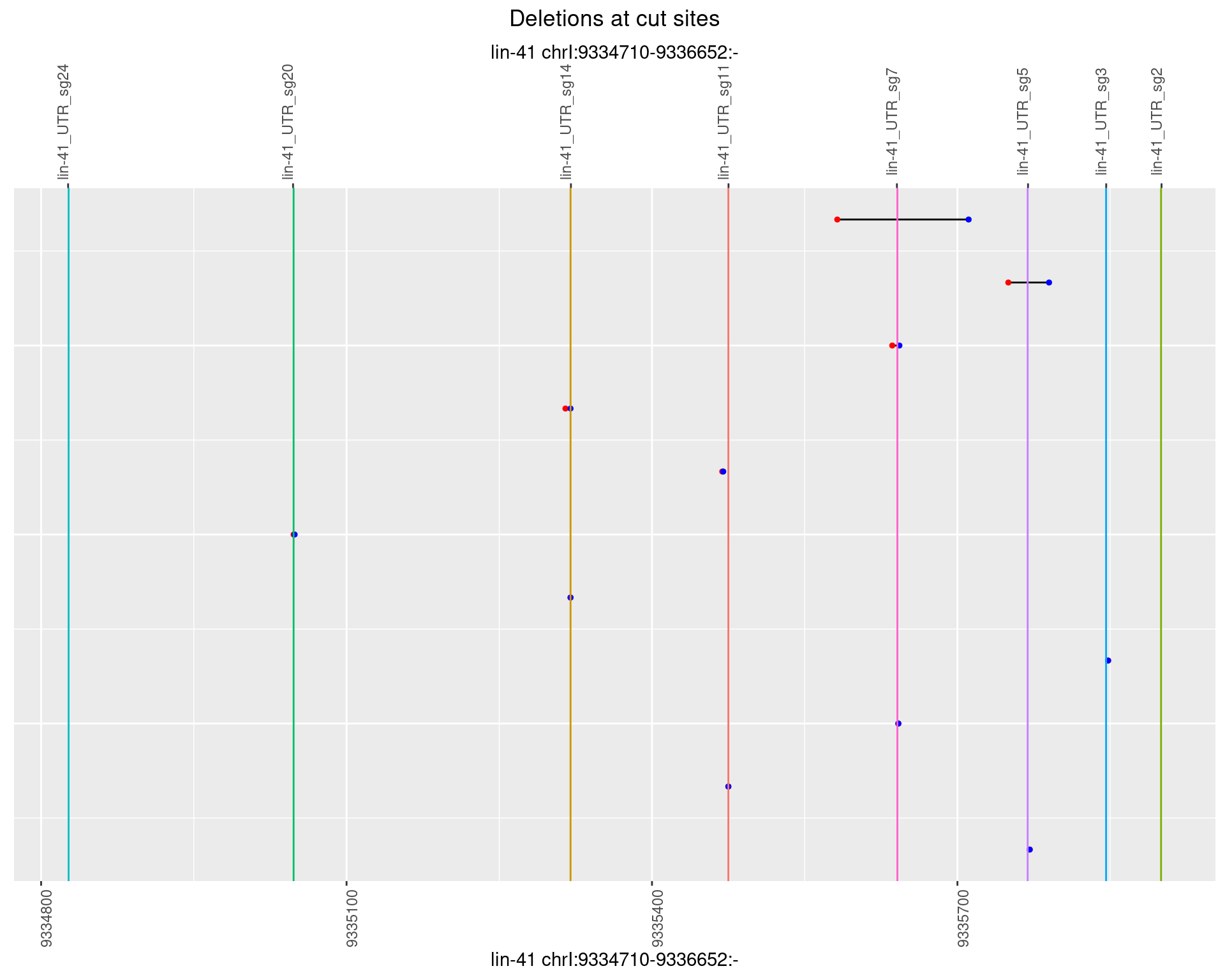
1.104.6 Freq: 0.1
No plot to show
1.105 lin41_RNApacbio_L1_N2
1.105.1 Freq: 0
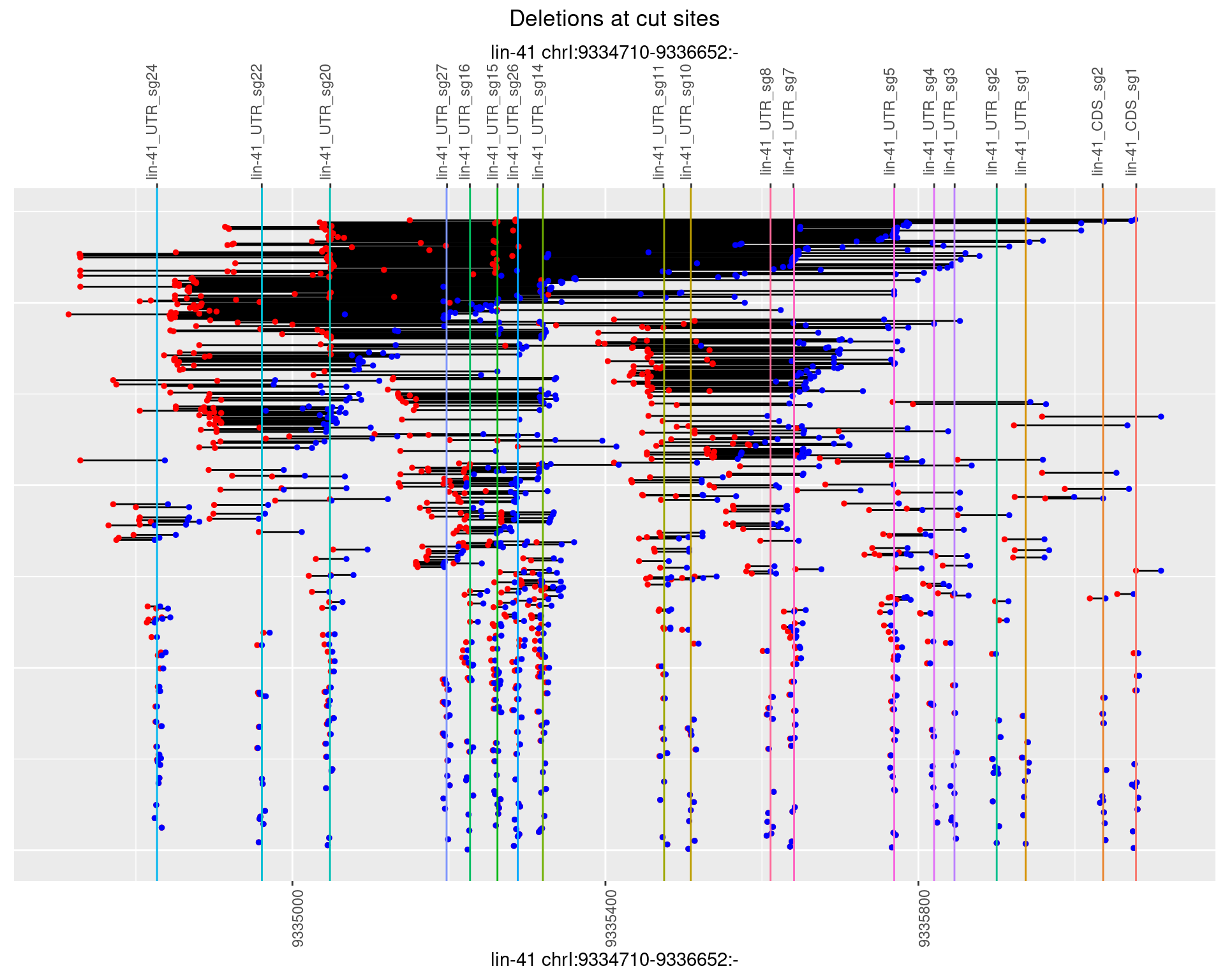
1.105.2 Freq: 1e-05
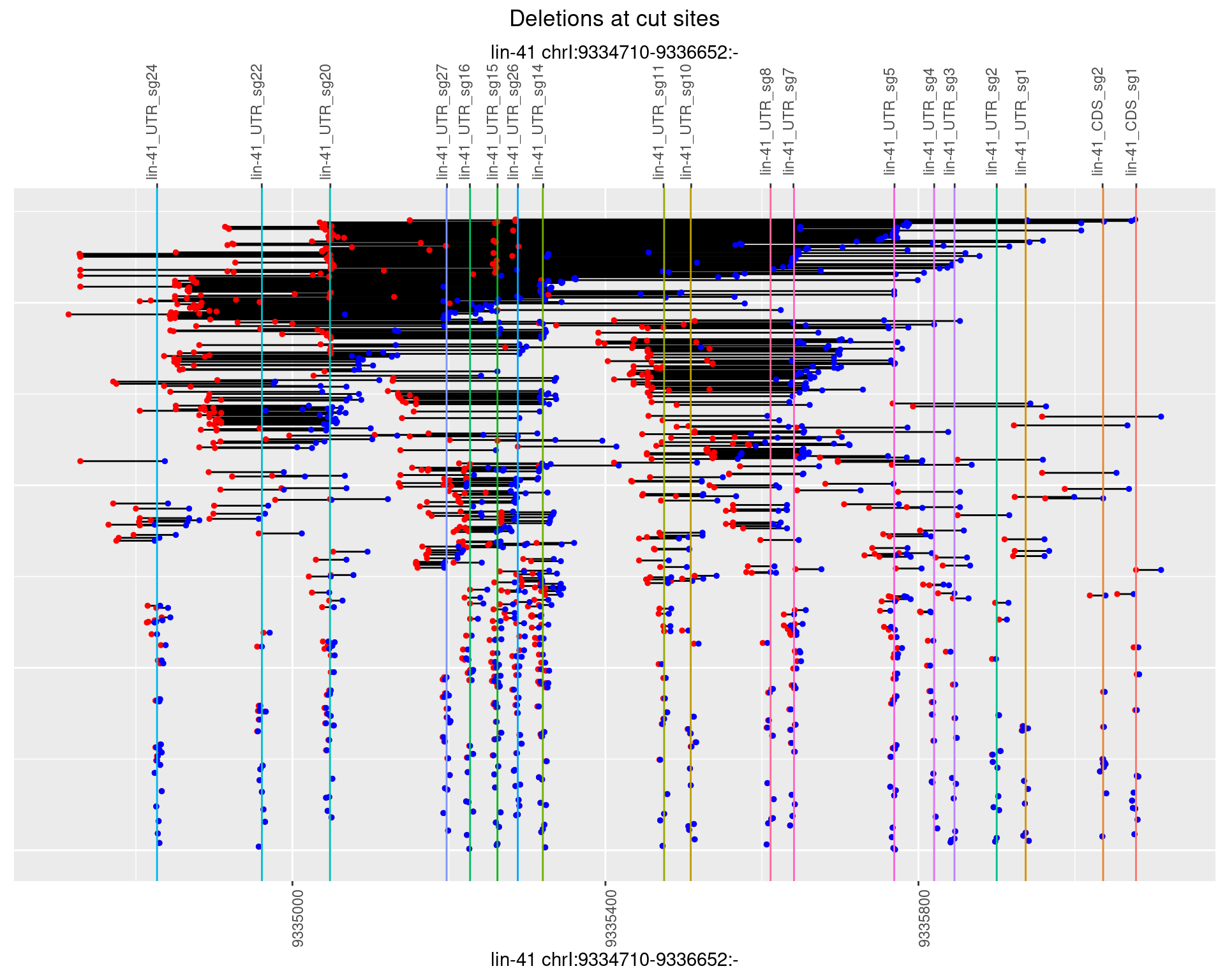
1.105.3 Freq: 1e-04
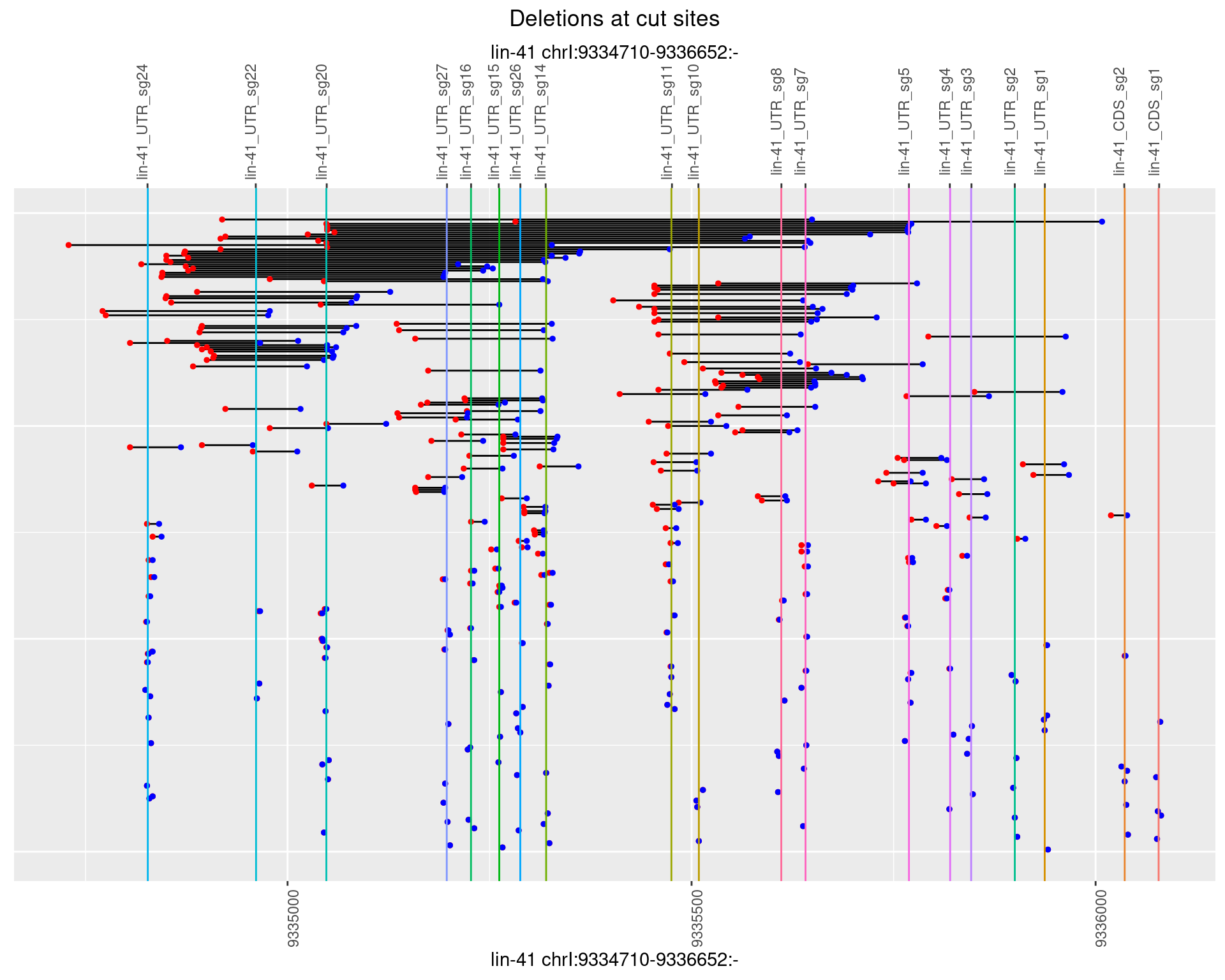
1.105.4 Freq: 0.001
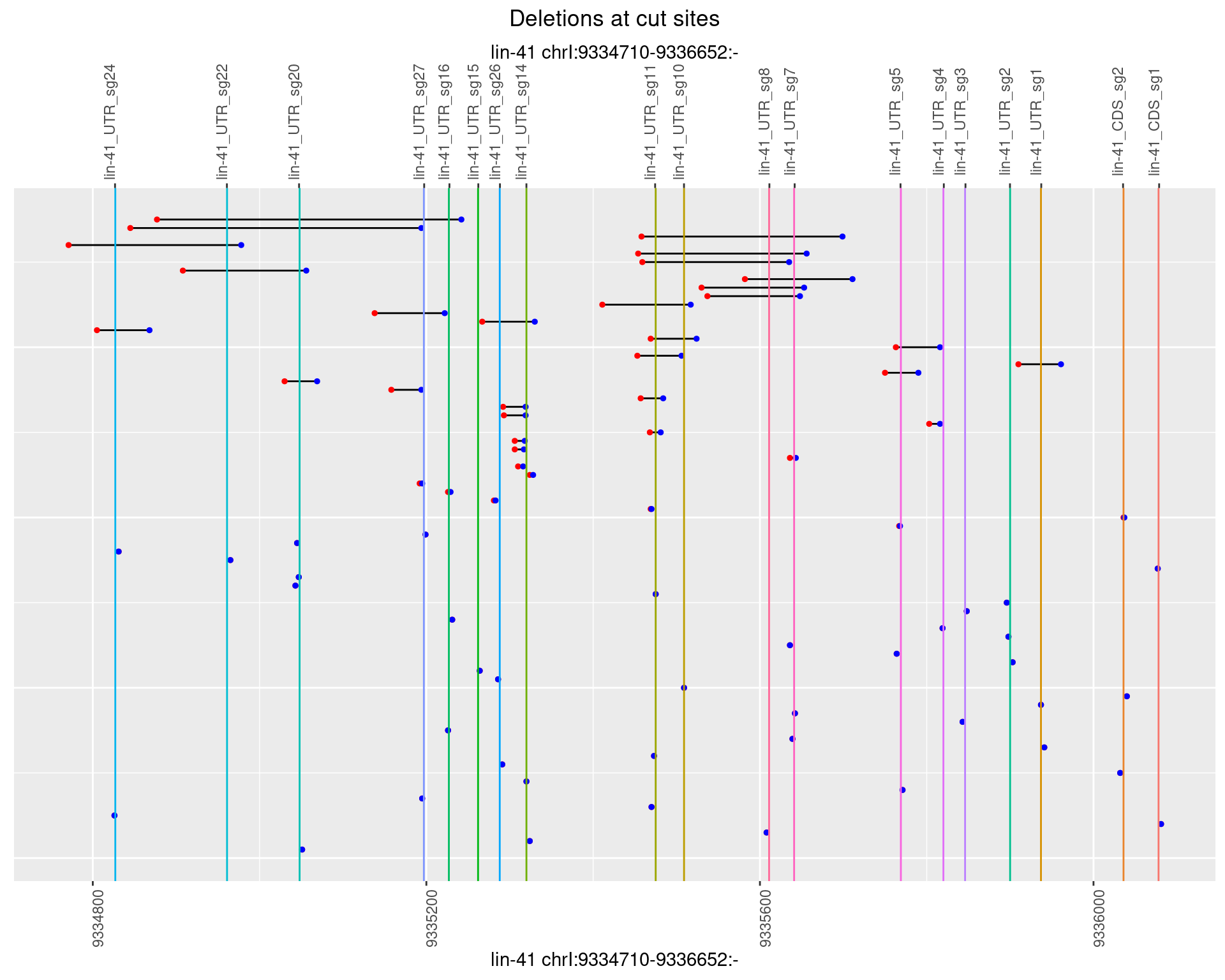
1.105.5 Freq: 0.01
1.105.6 Freq: 0.1
1.106 lin41_RNApacbio_L4_1516
1.106.1 Freq: 0
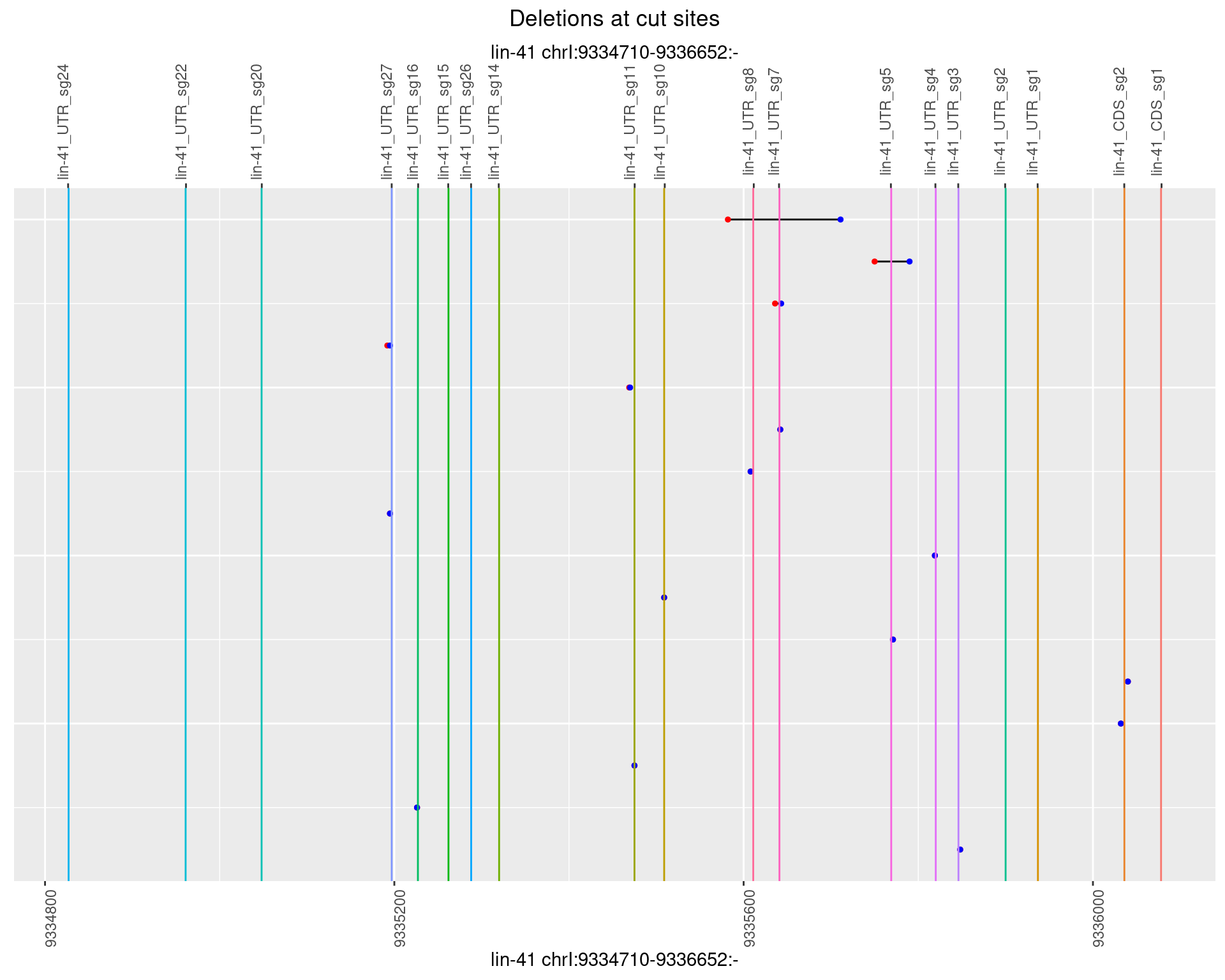
1.106.2 Freq: 1e-05
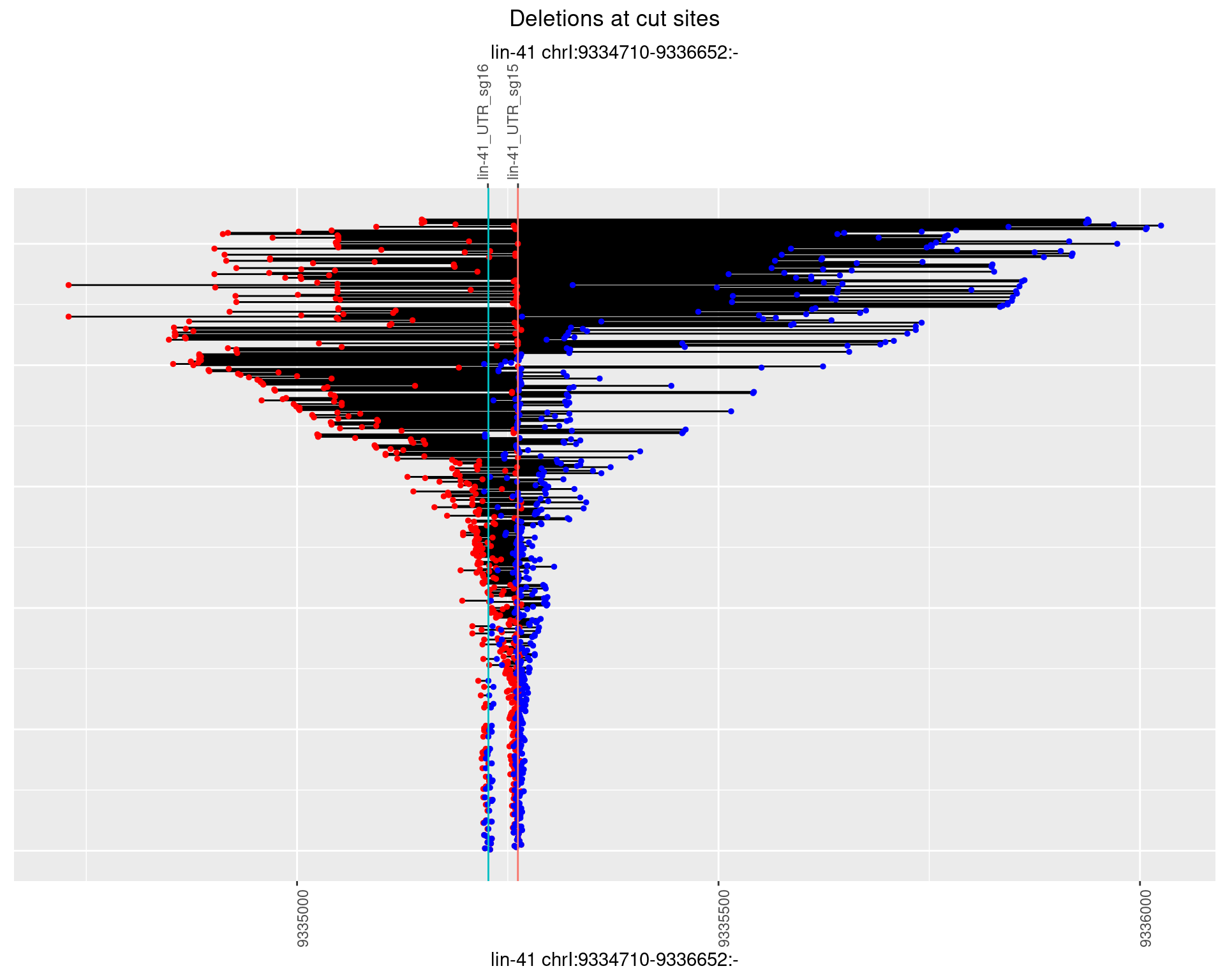
1.106.3 Freq: 1e-04
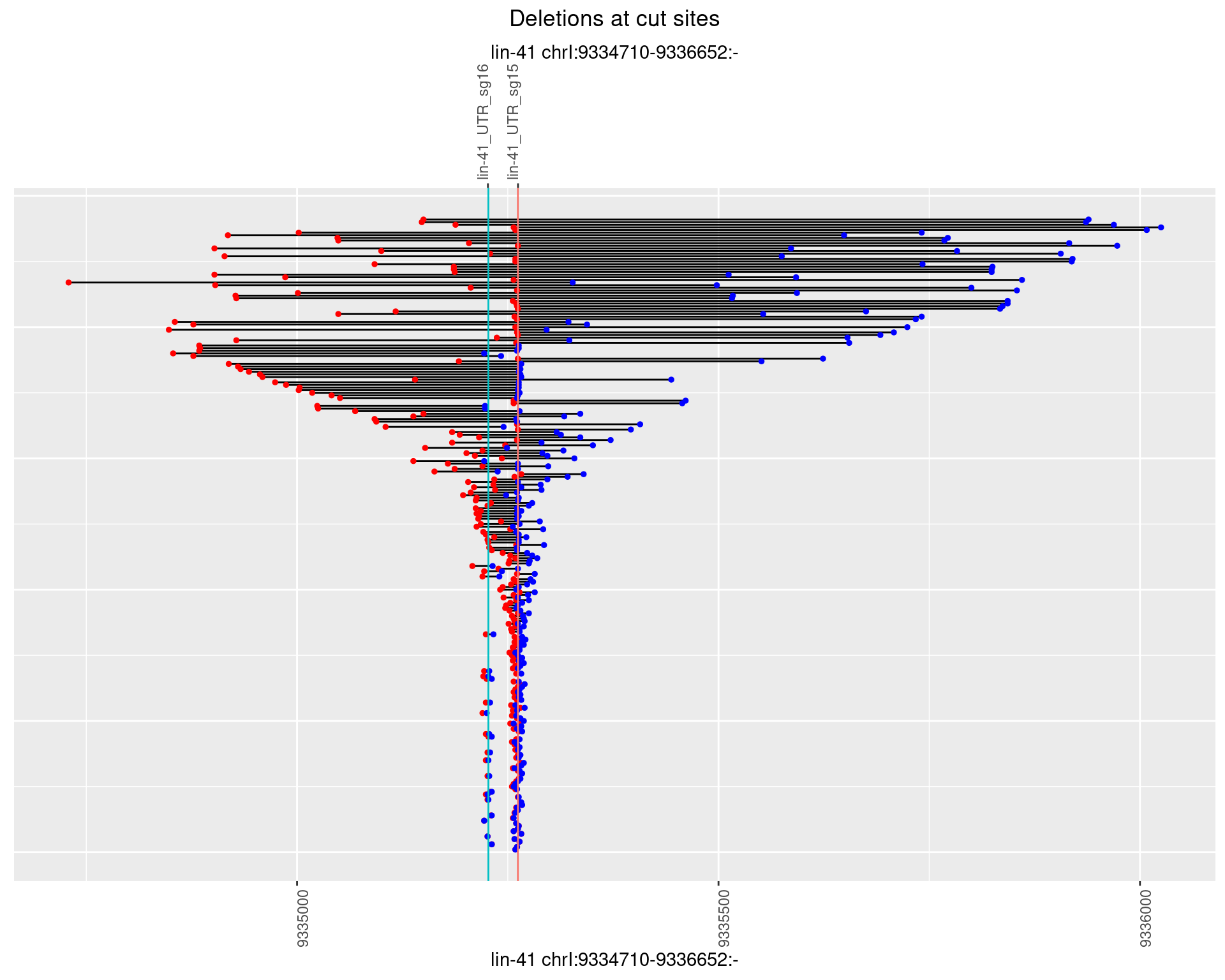
1.106.4 Freq: 0.001
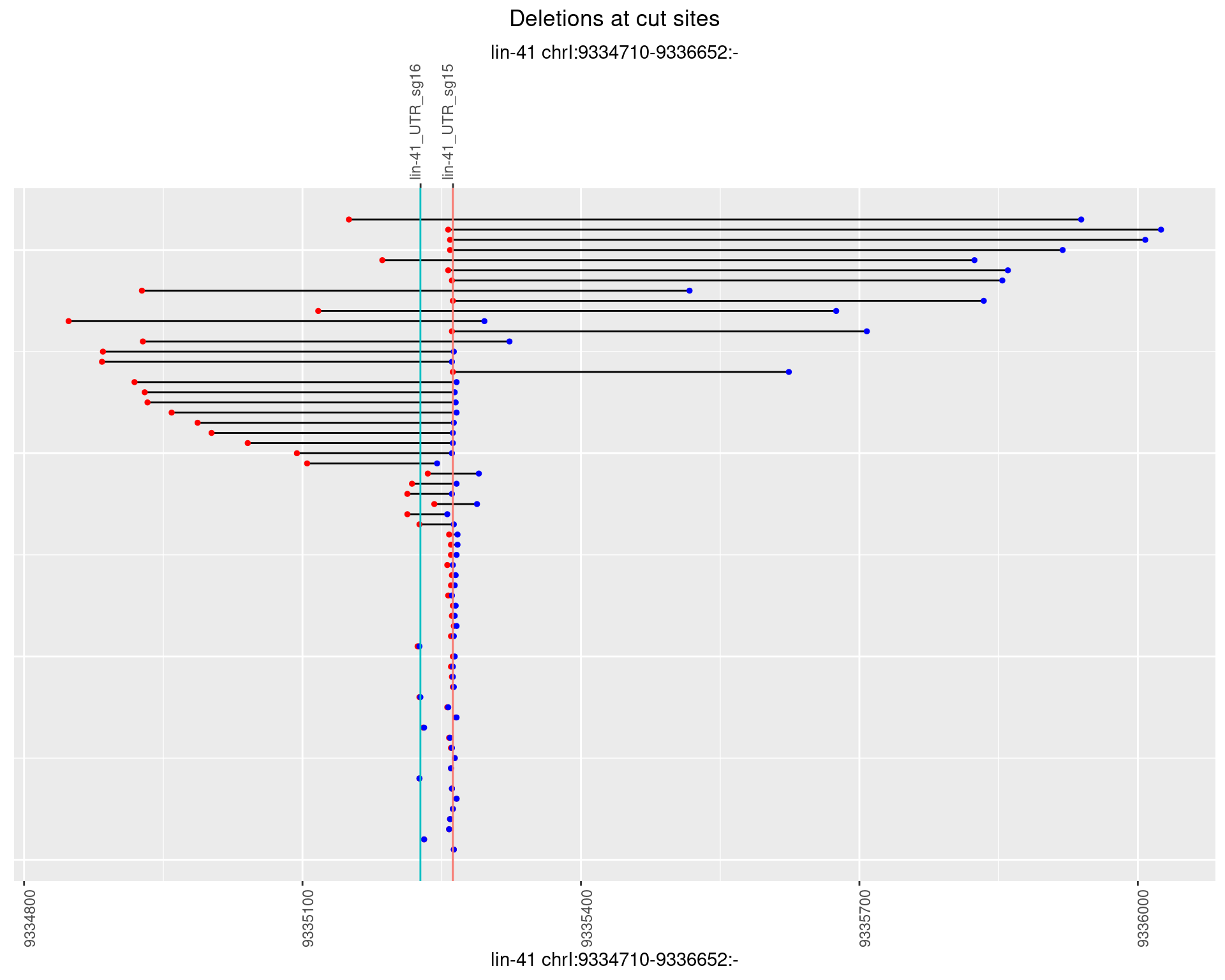
1.106.5 Freq: 0.01
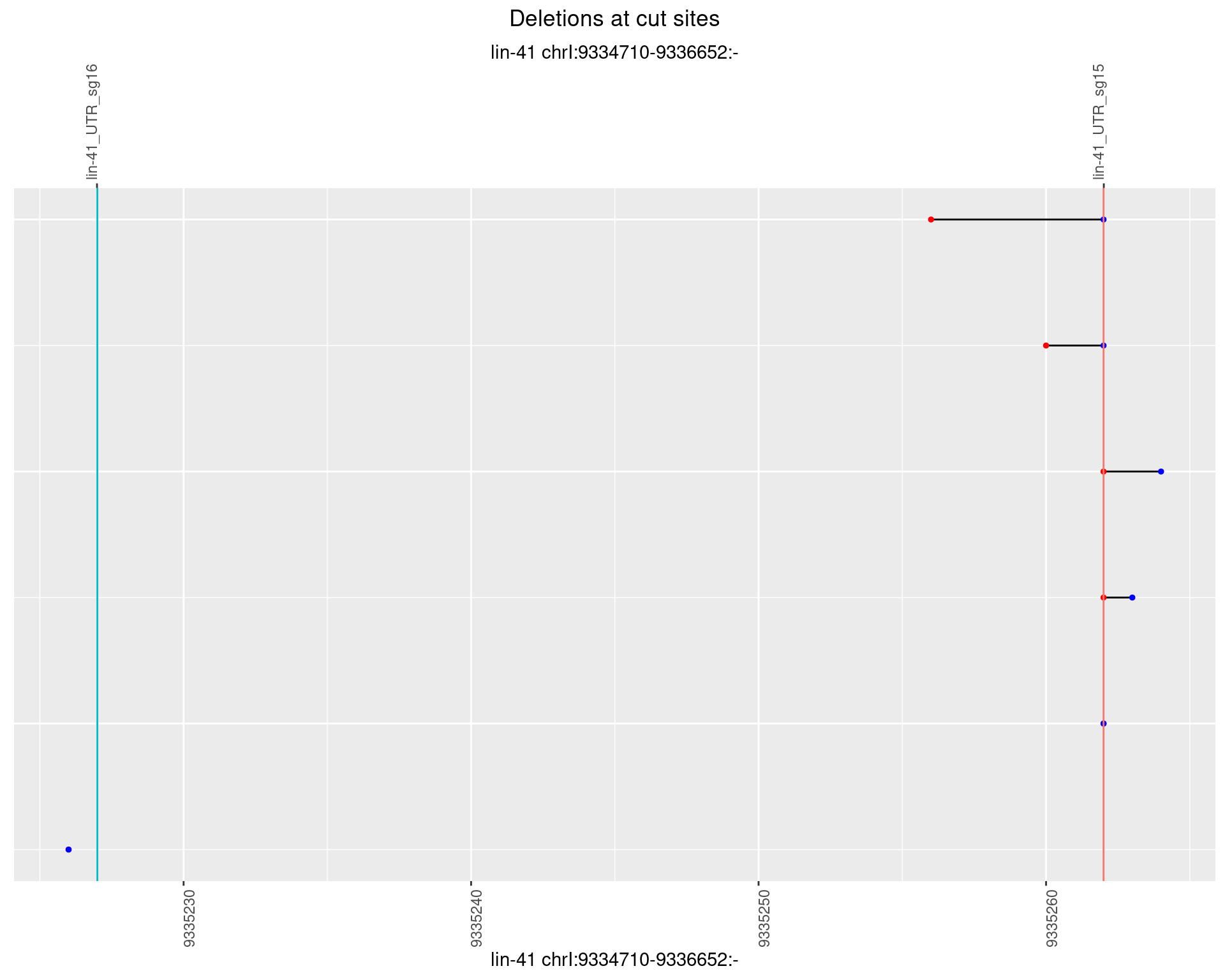
1.106.6 Freq: 0.1
No plot to show
1.107 lin41_RNApacbio_L4_2627
1.107.1 Freq: 0
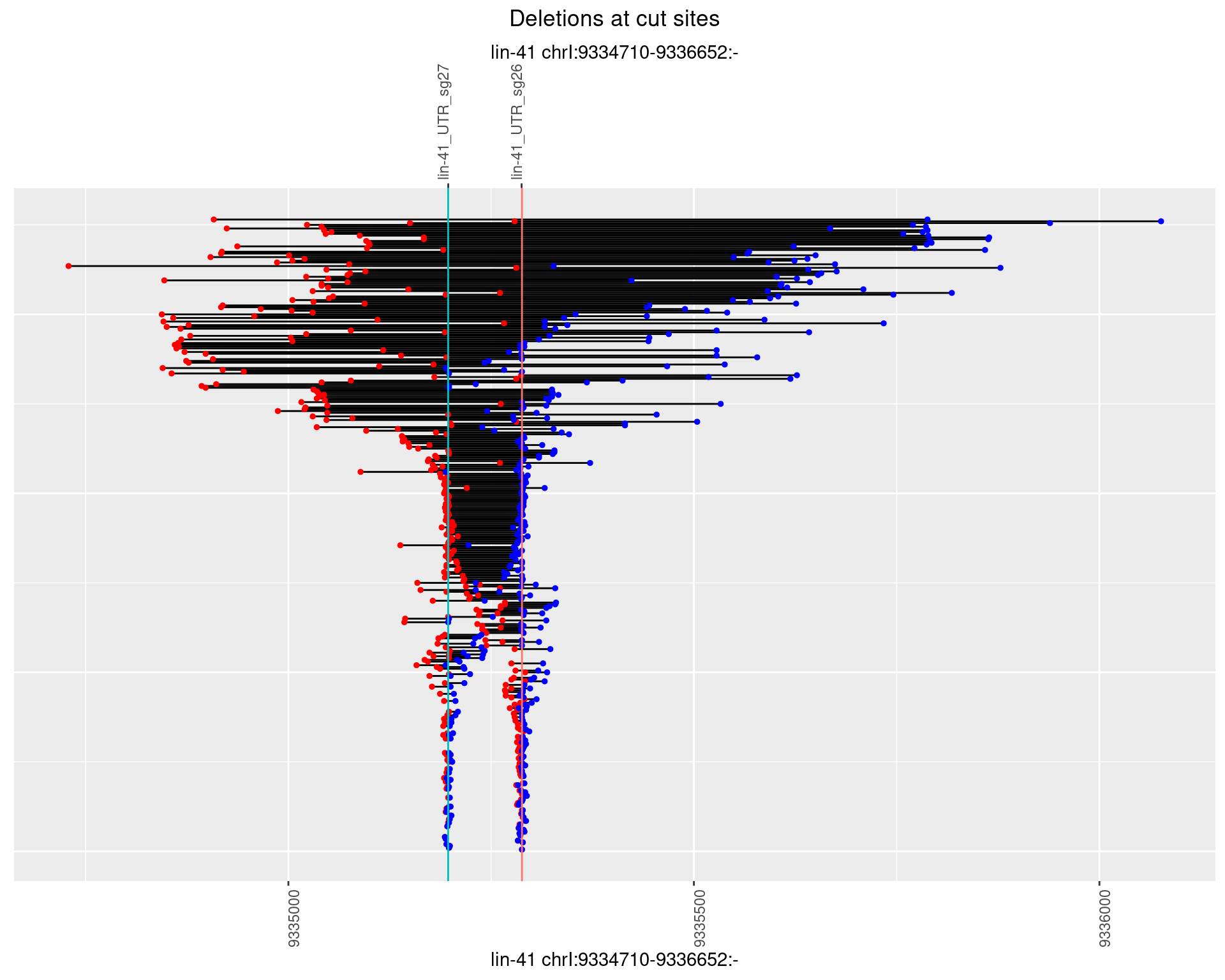
1.107.2 Freq: 1e-05
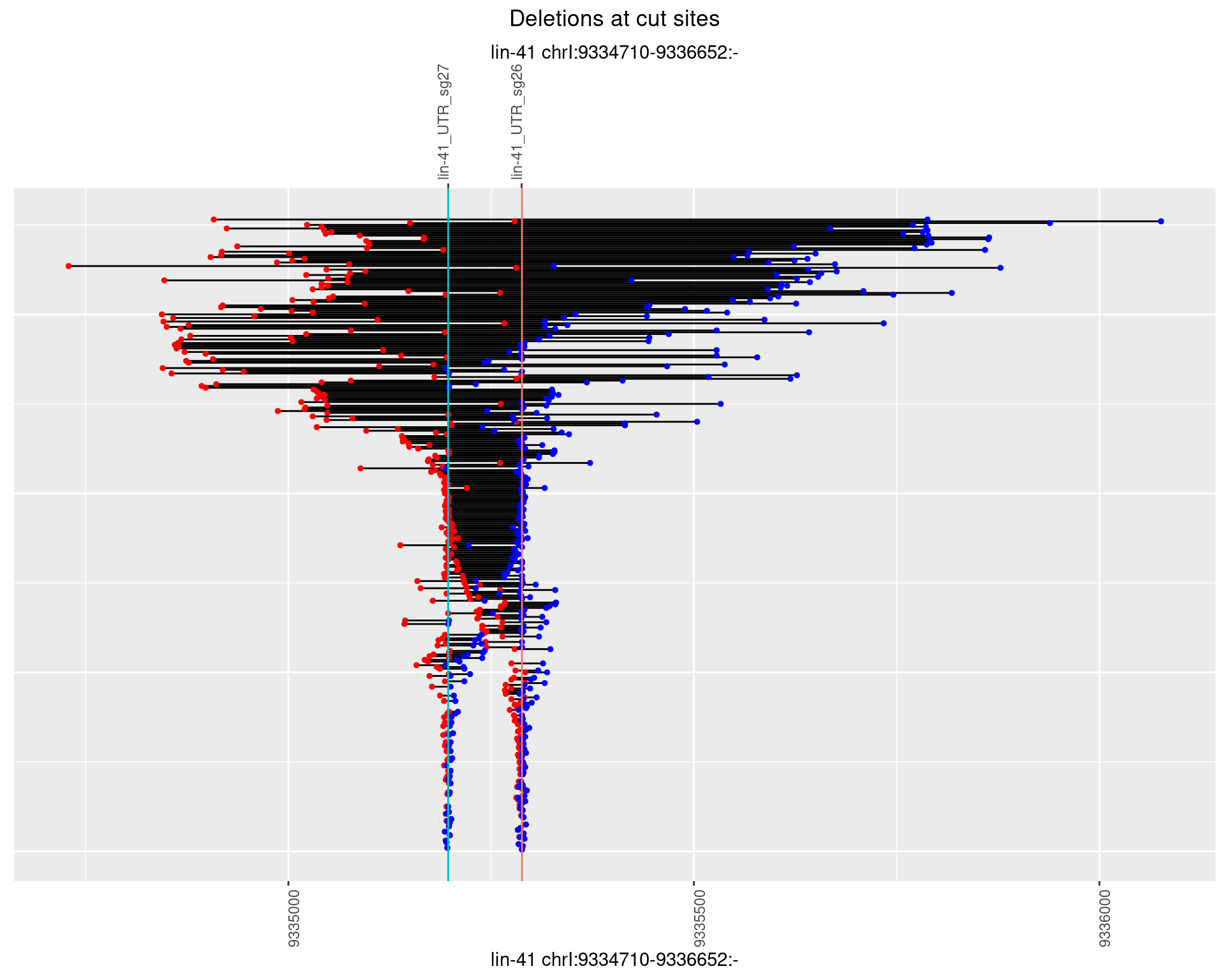
1.107.3 Freq: 1e-04
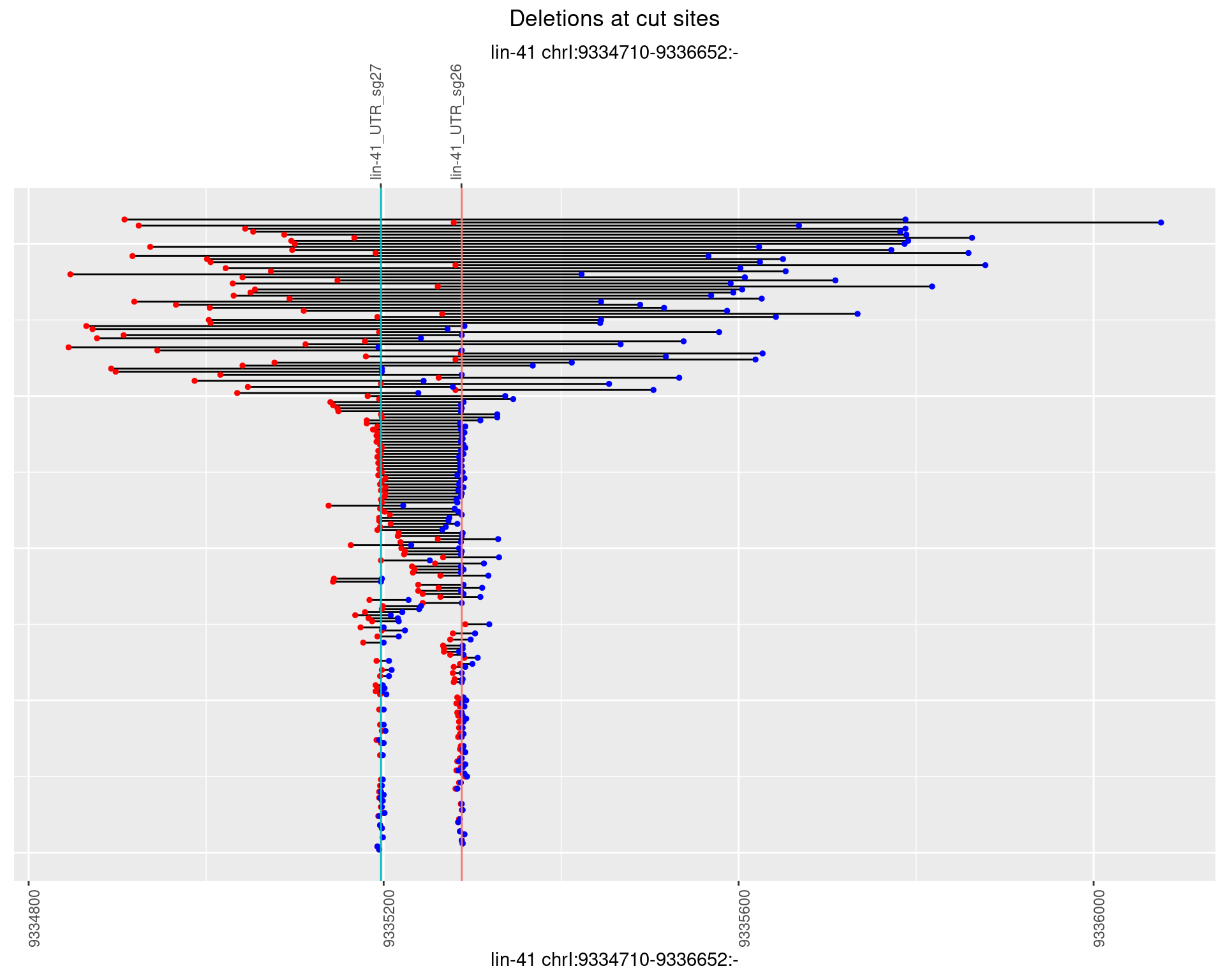
1.107.4 Freq: 0.001
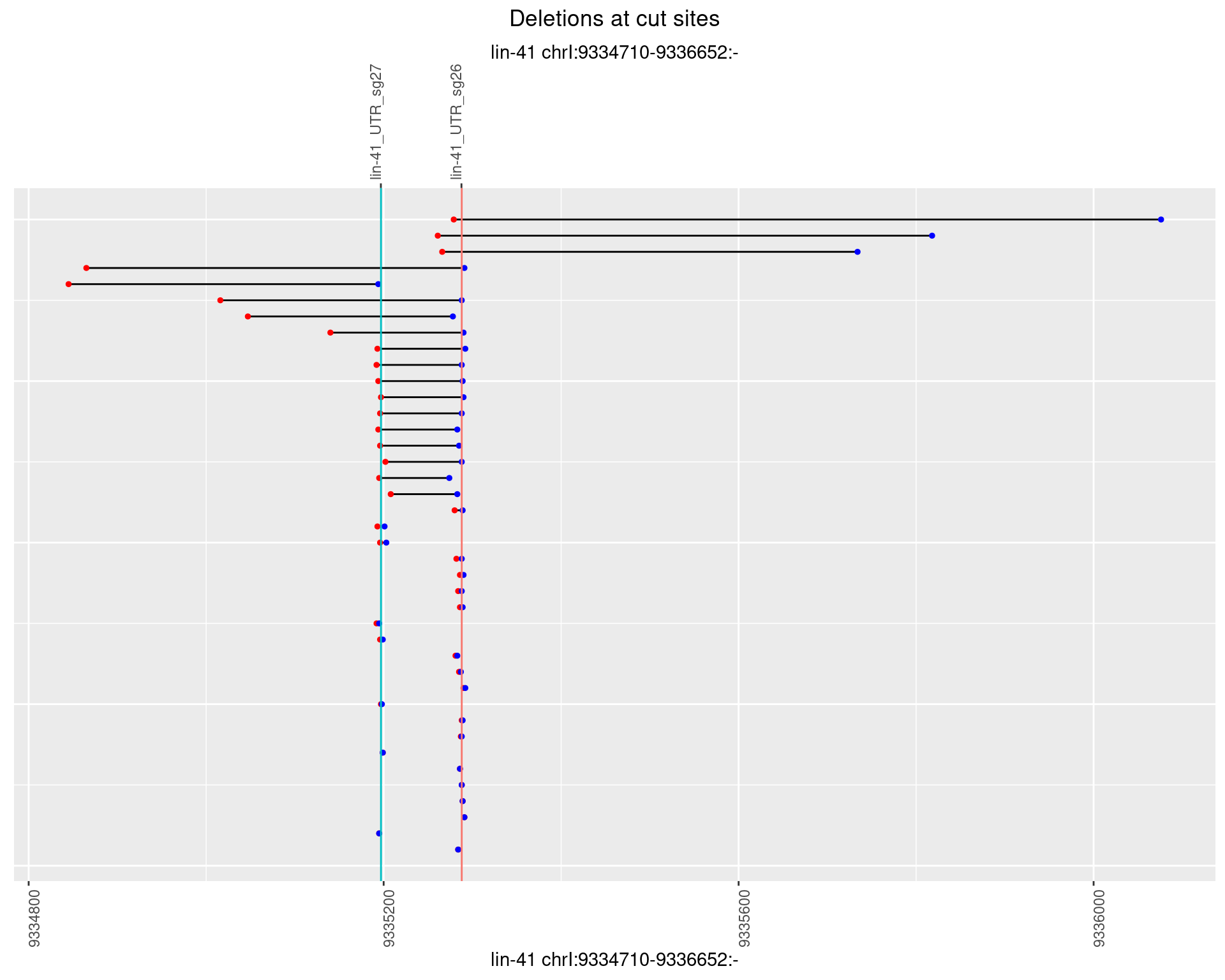
1.107.5 Freq: 0.01
1.107.6 Freq: 0.1
No plot to show
1.108 lin41_RNApacbio_L4_pool3
1.108.1 Freq: 0
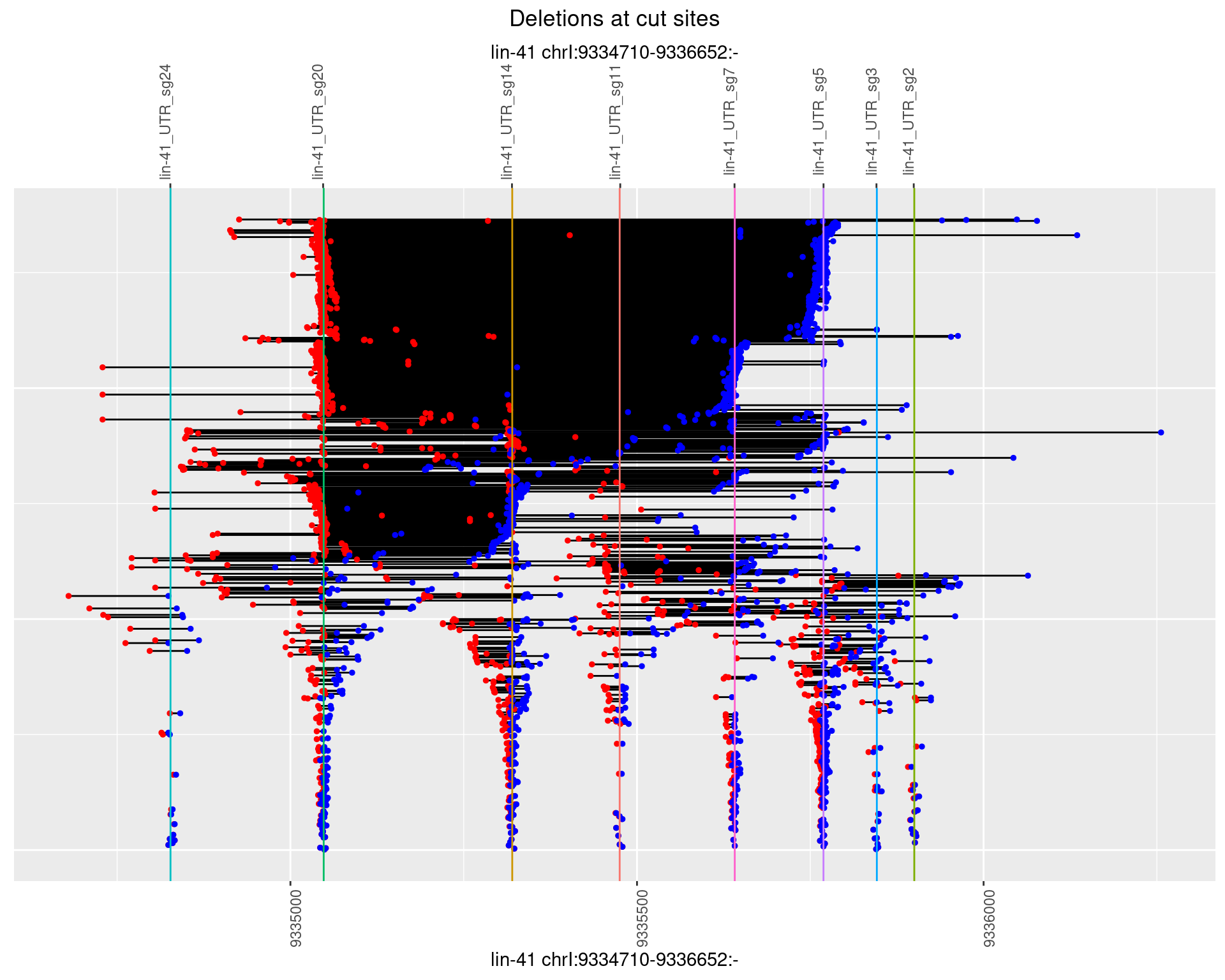
1.108.2 Freq: 1e-05
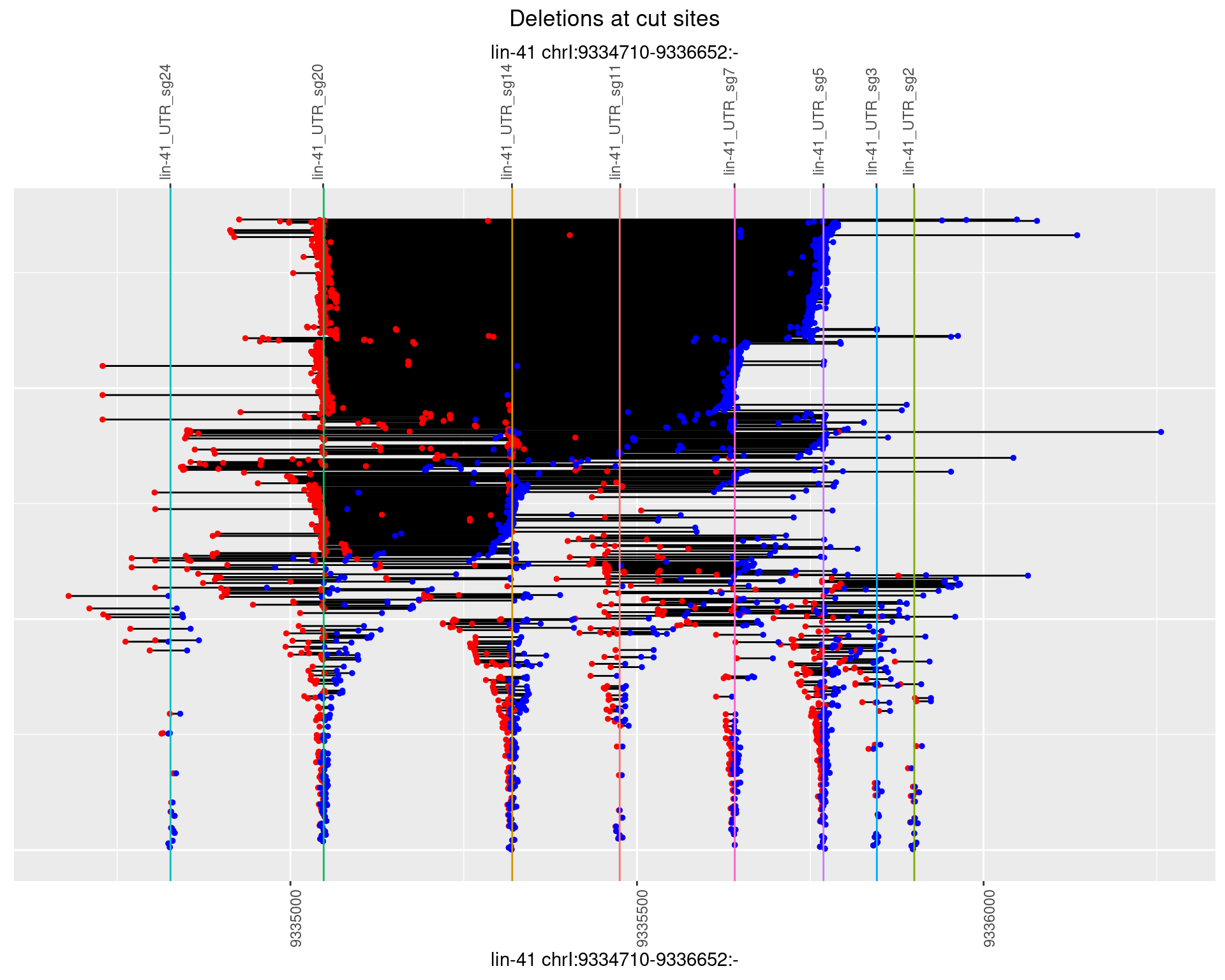
1.108.3 Freq: 1e-04
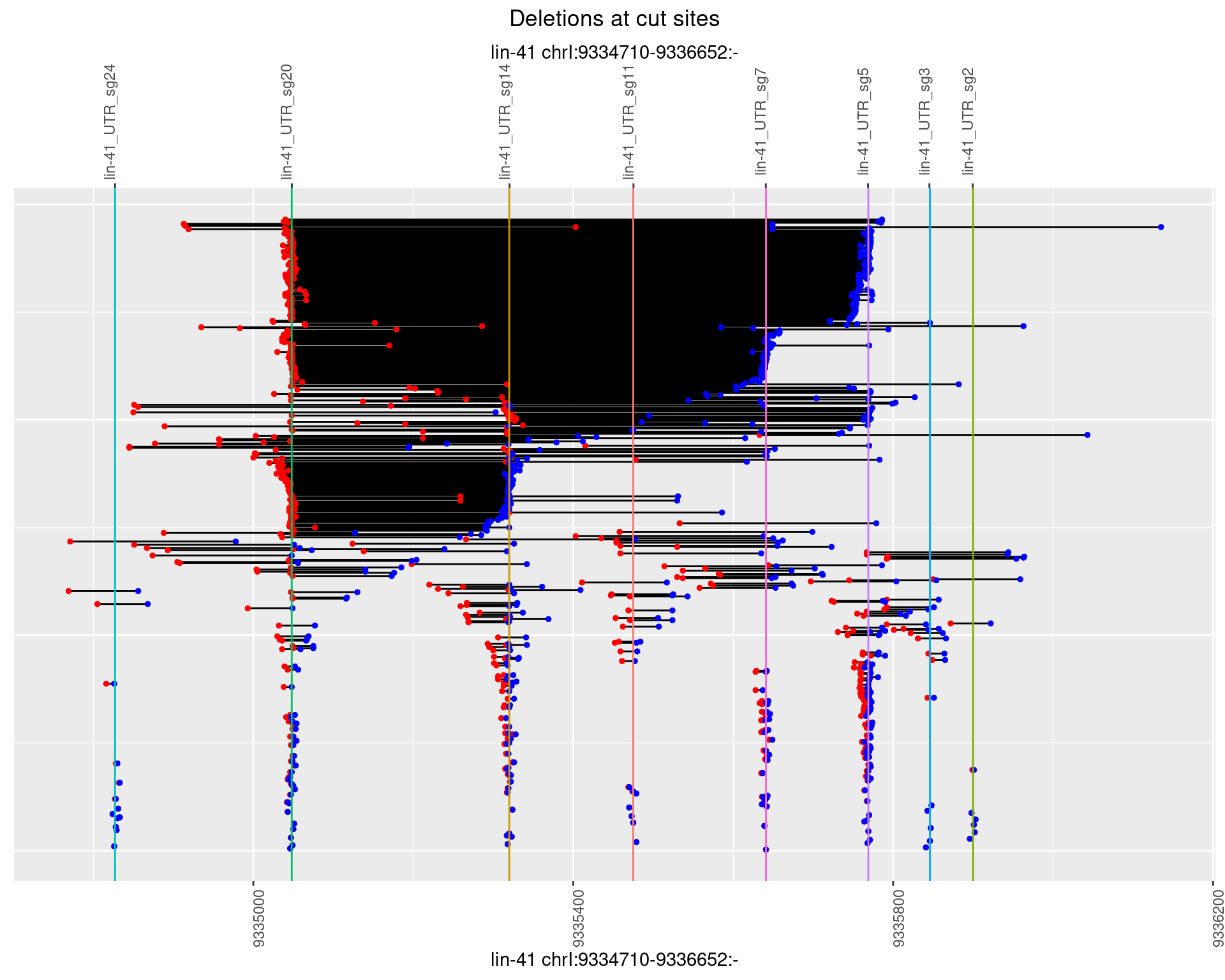
1.108.4 Freq: 0.001
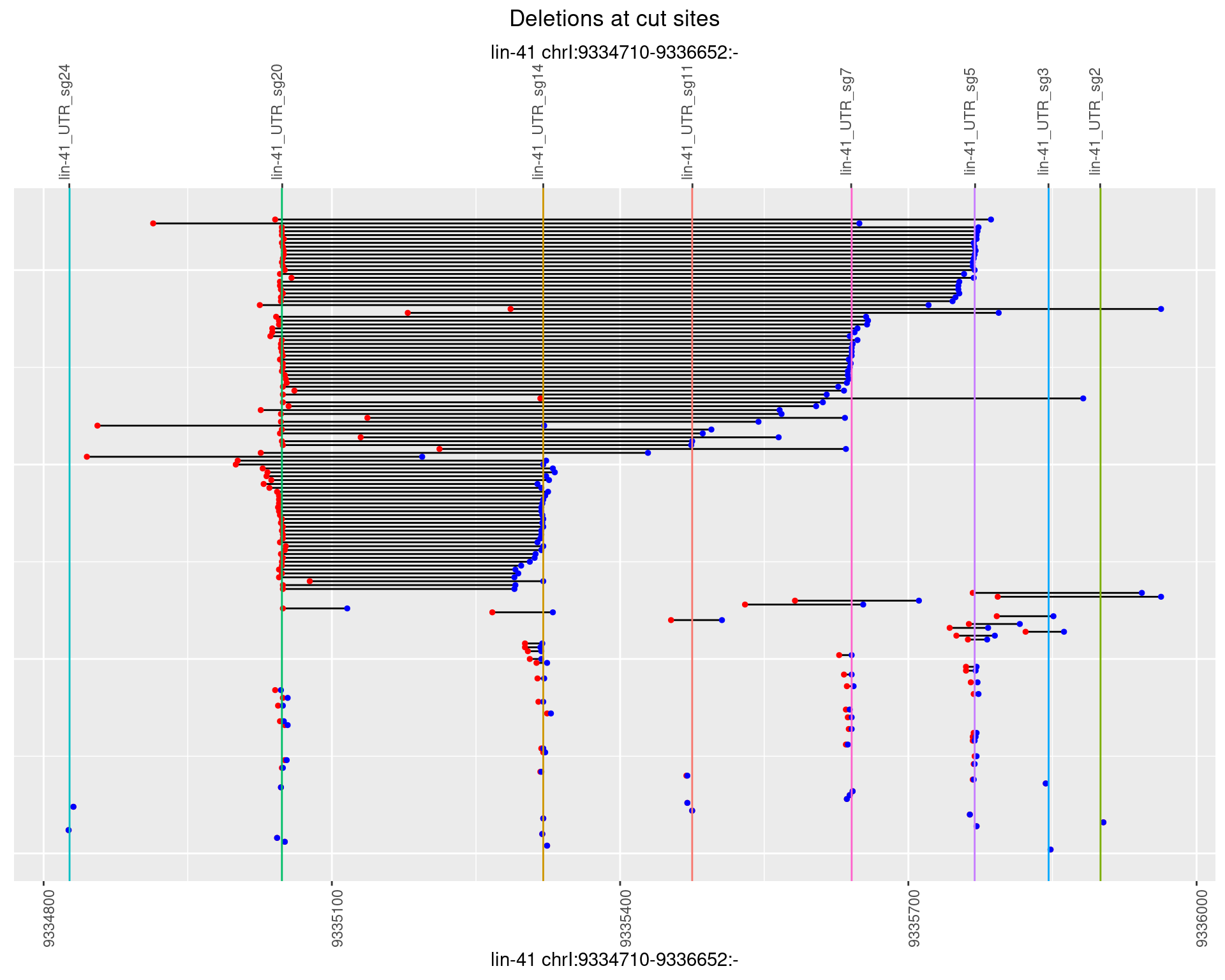
1.108.5 Freq: 0.01
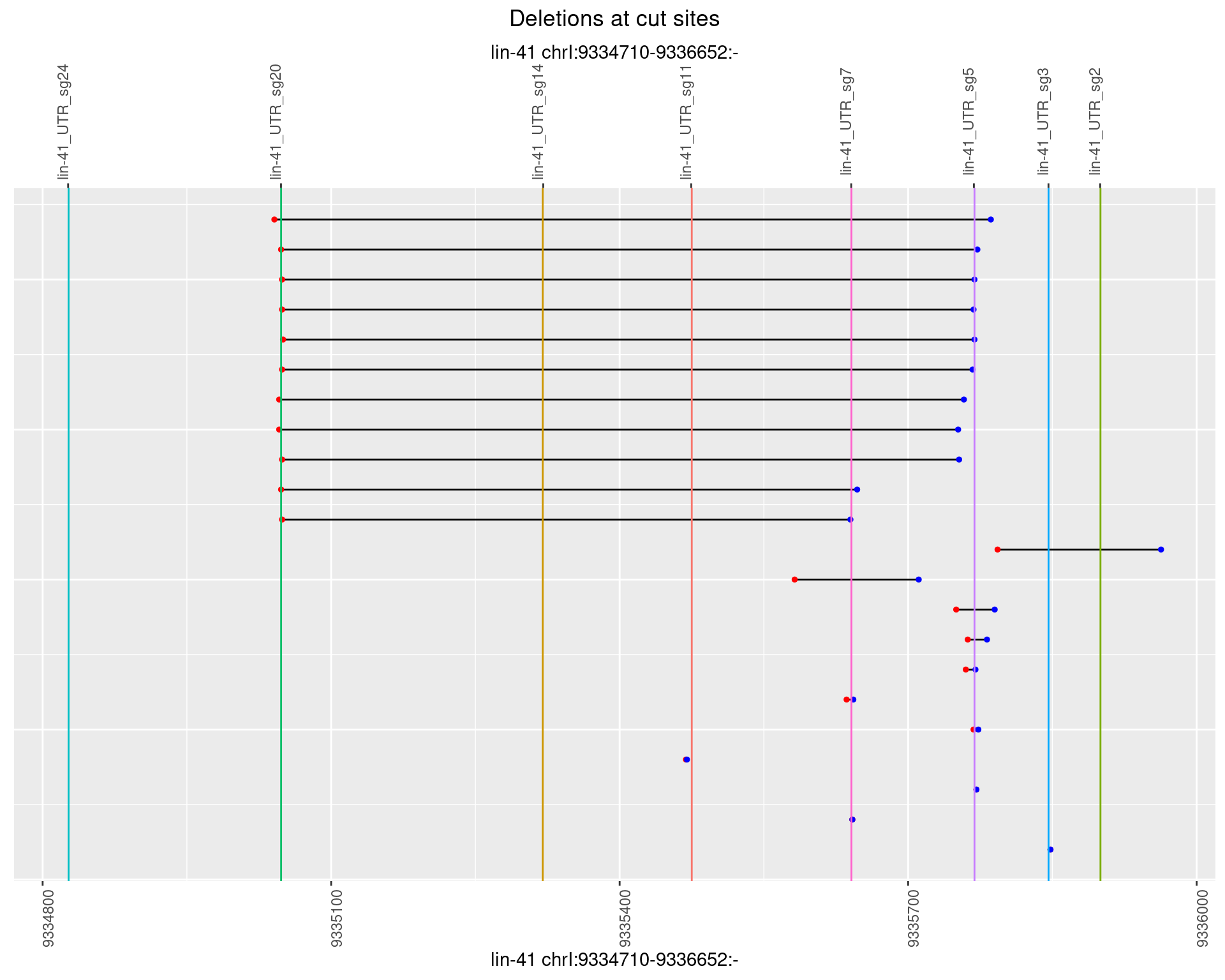
1.108.6 Freq: 0.1
No plot to show
1.109 lin41_RNApacbio_L4_N2
1.109.1 Freq: 0
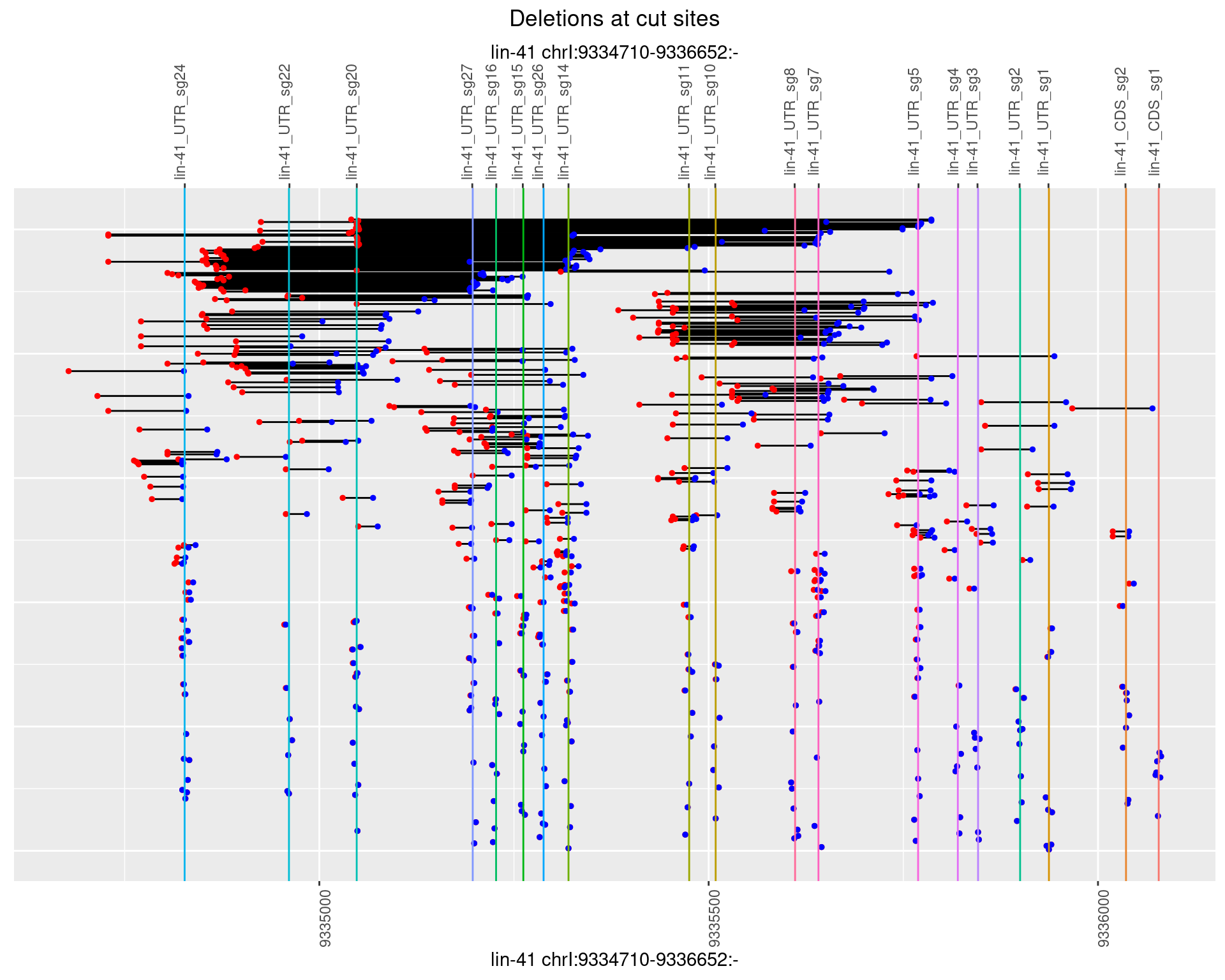
1.109.2 Freq: 1e-05
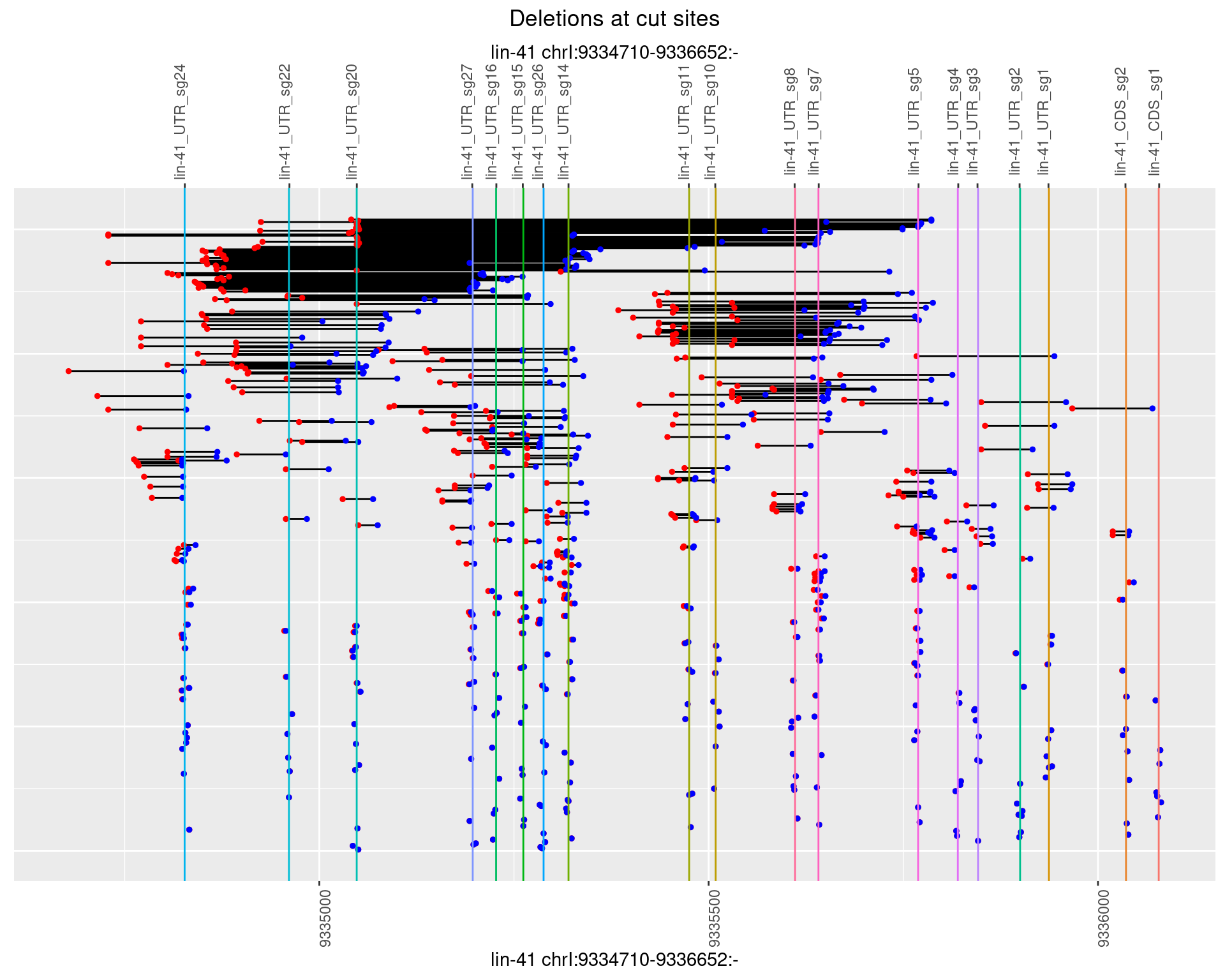
1.109.3 Freq: 1e-04
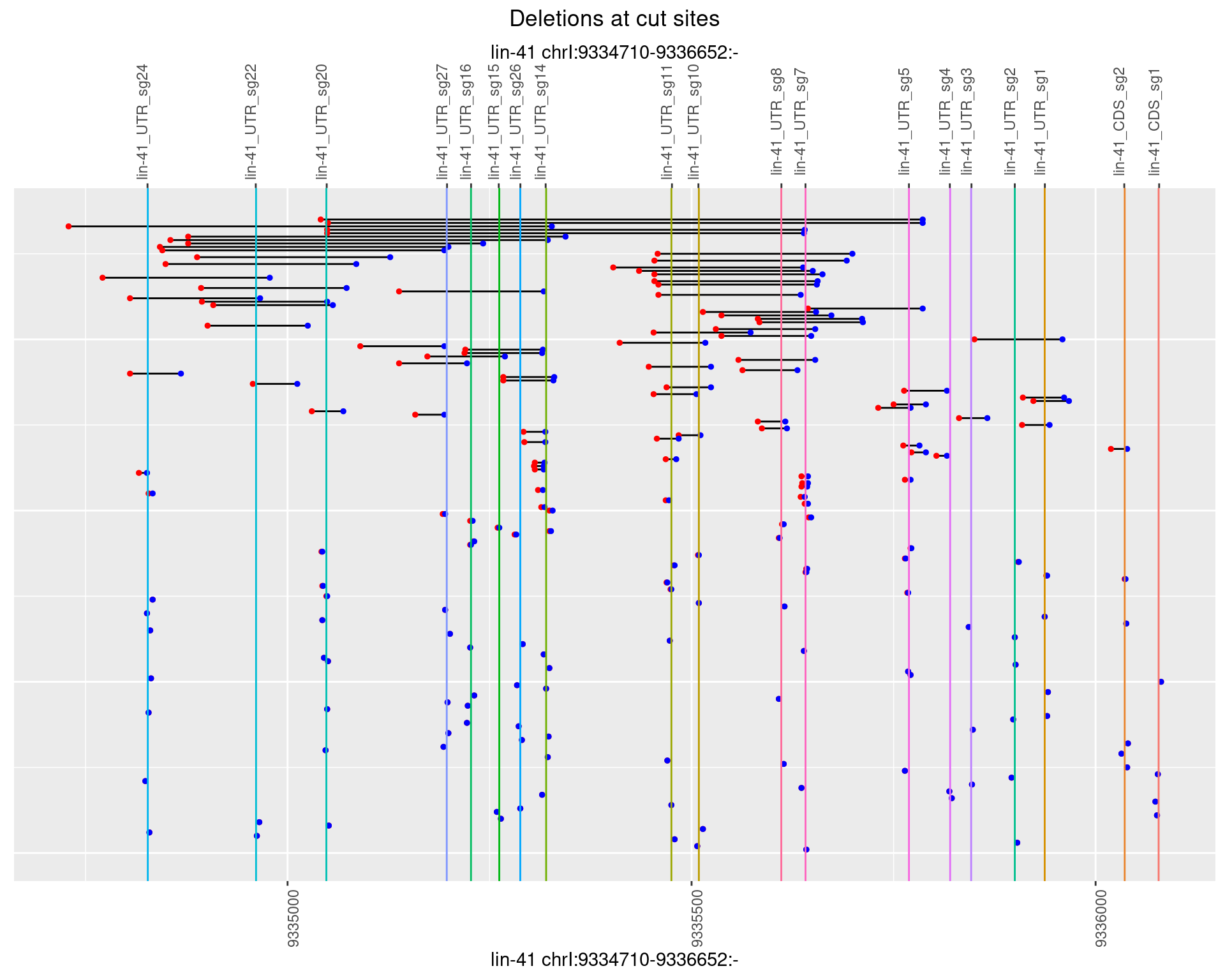
1.109.4 Freq: 0.001
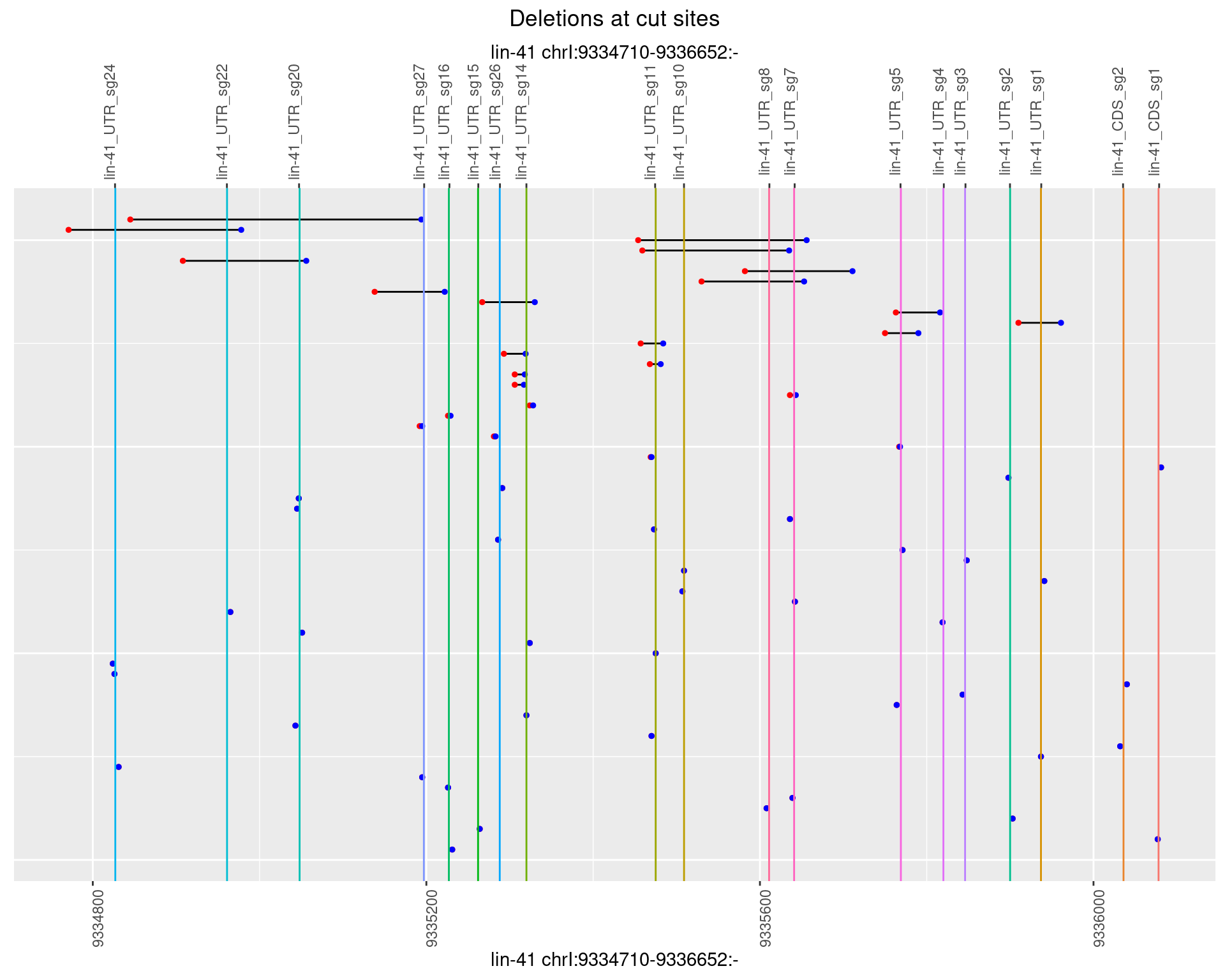
1.109.5 Freq: 0.01
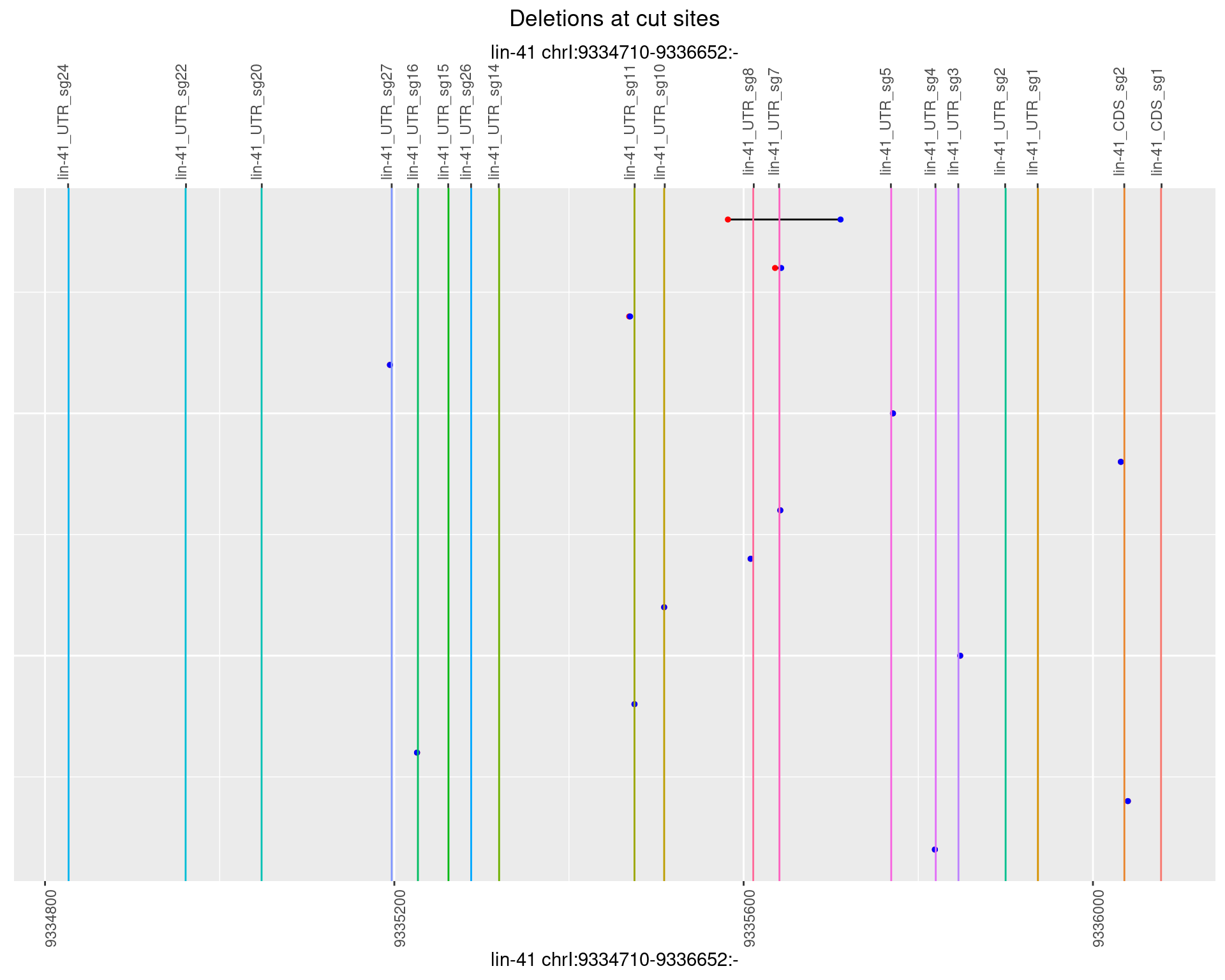
1.109.6 Freq: 0.1
1.110 lin41_RNApacbio_L1_all
1.110.1 Freq: 0
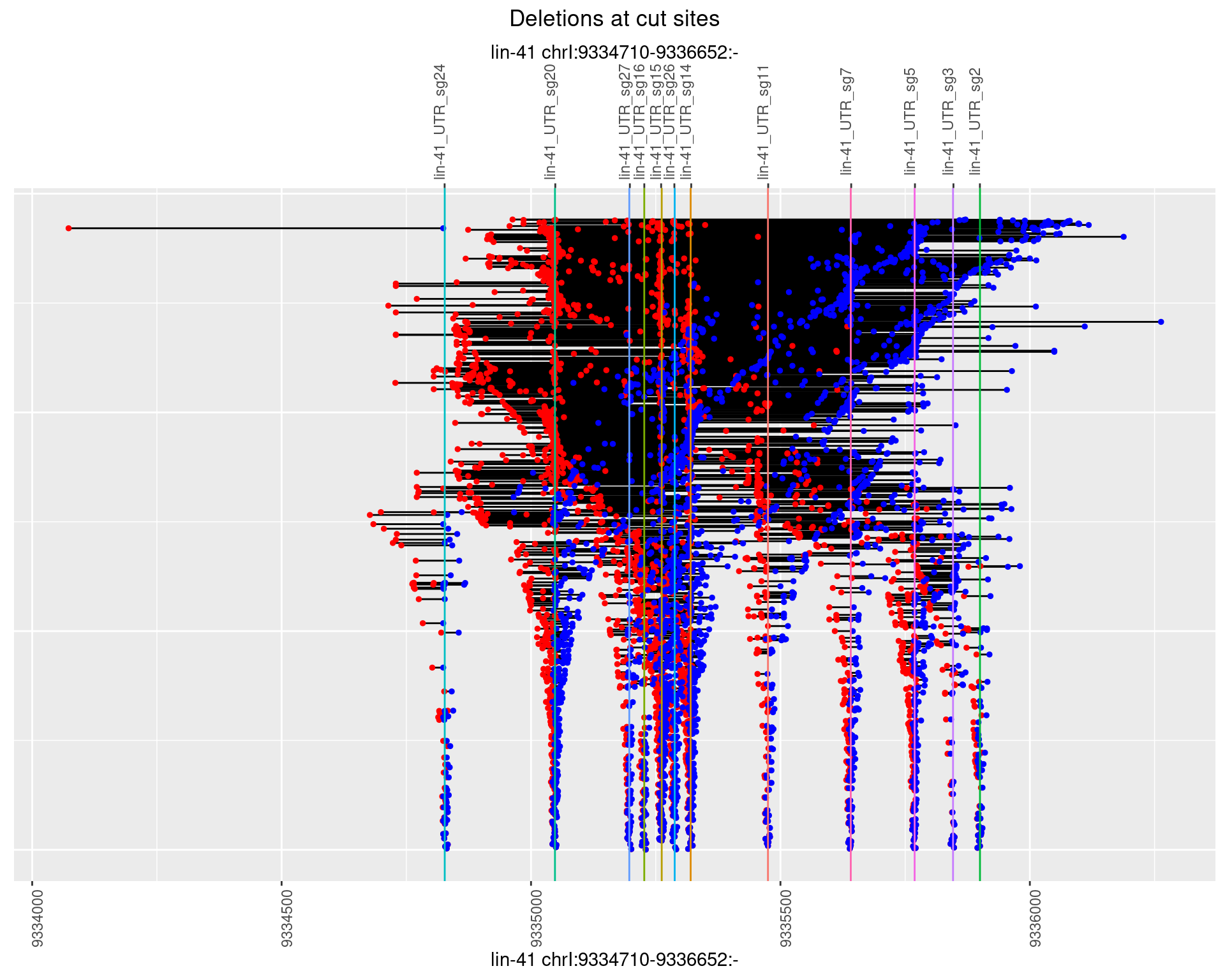
1.110.2 Freq: 1e-05
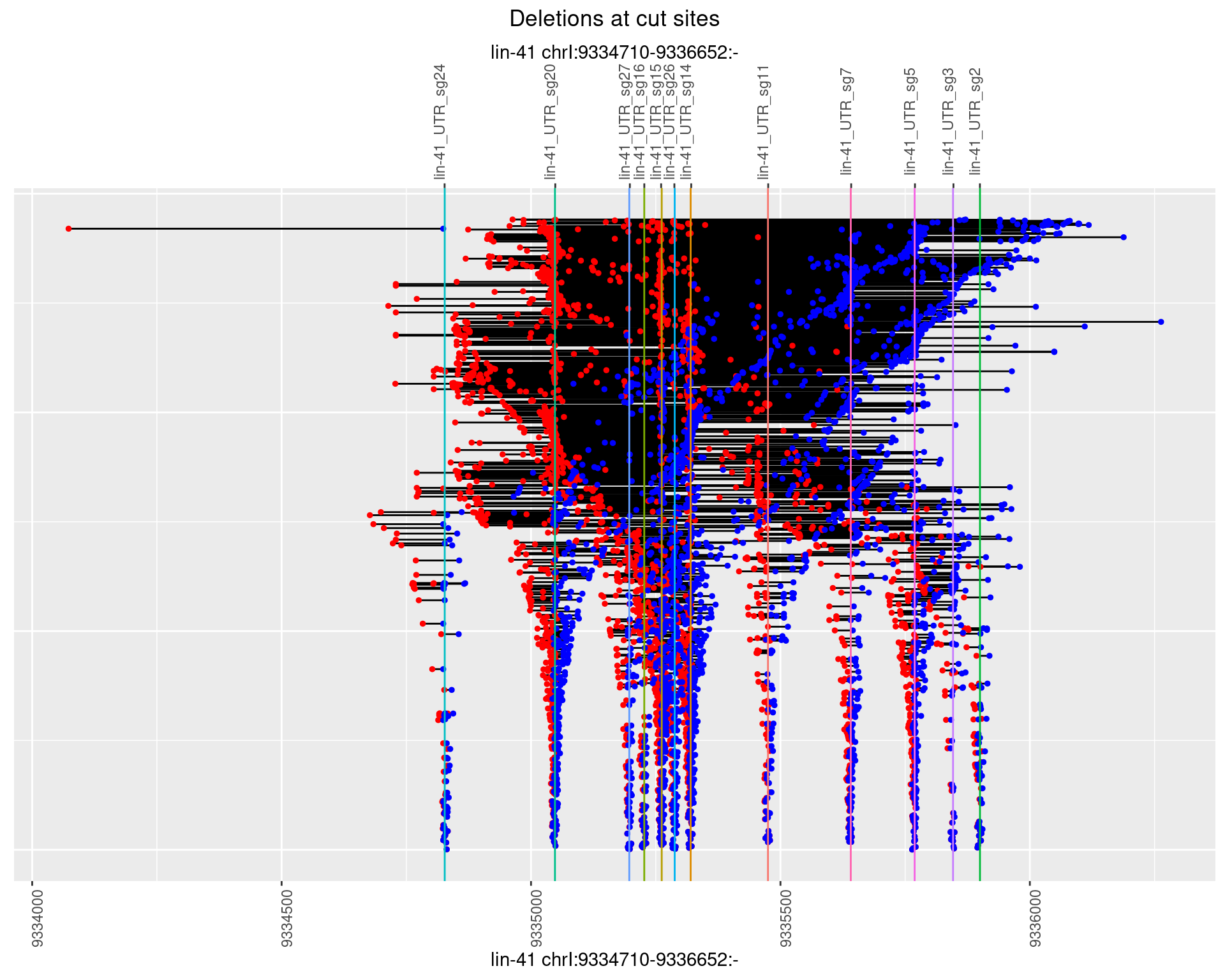
1.110.3 Freq: 1e-04
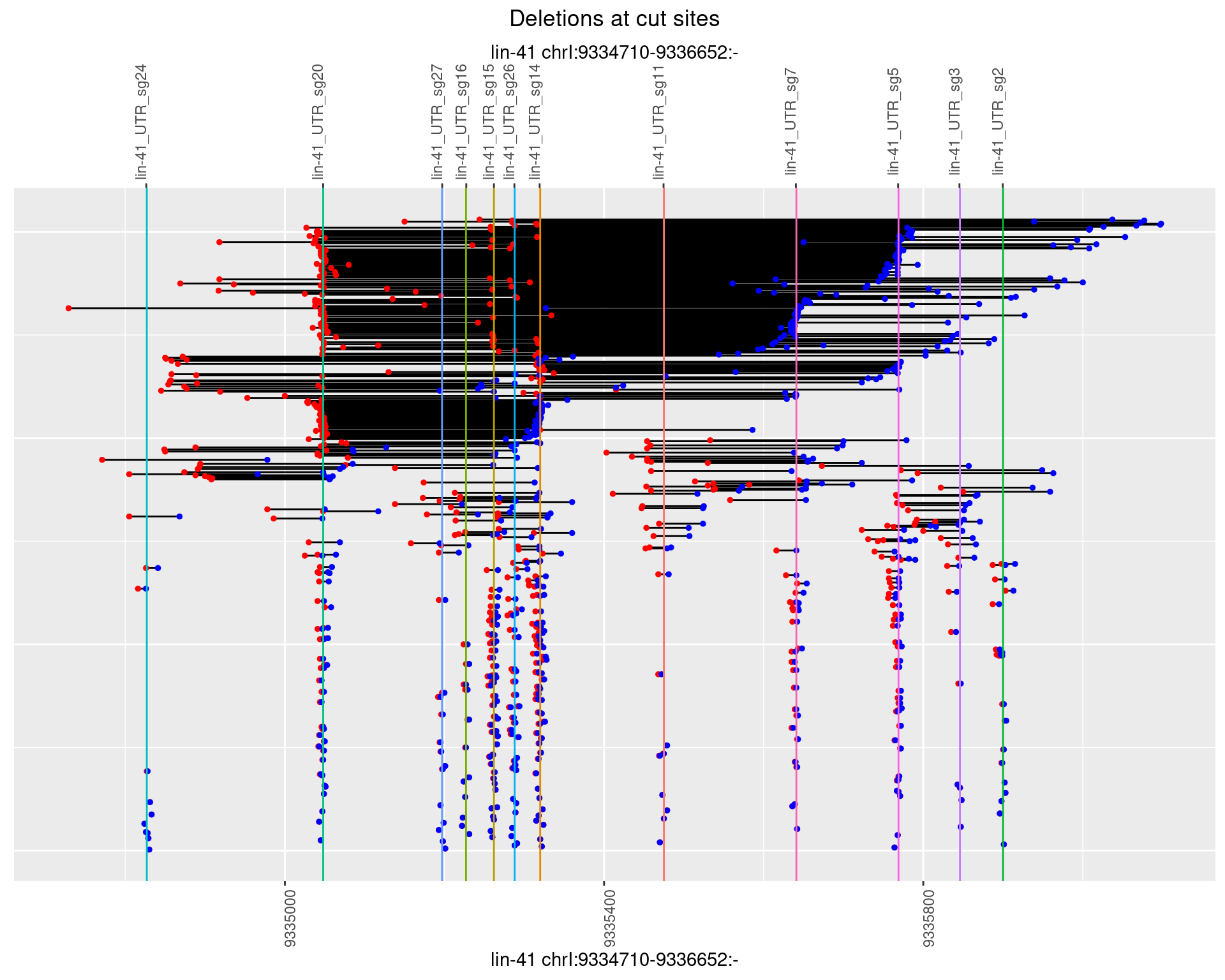
1.110.4 Freq: 0.001
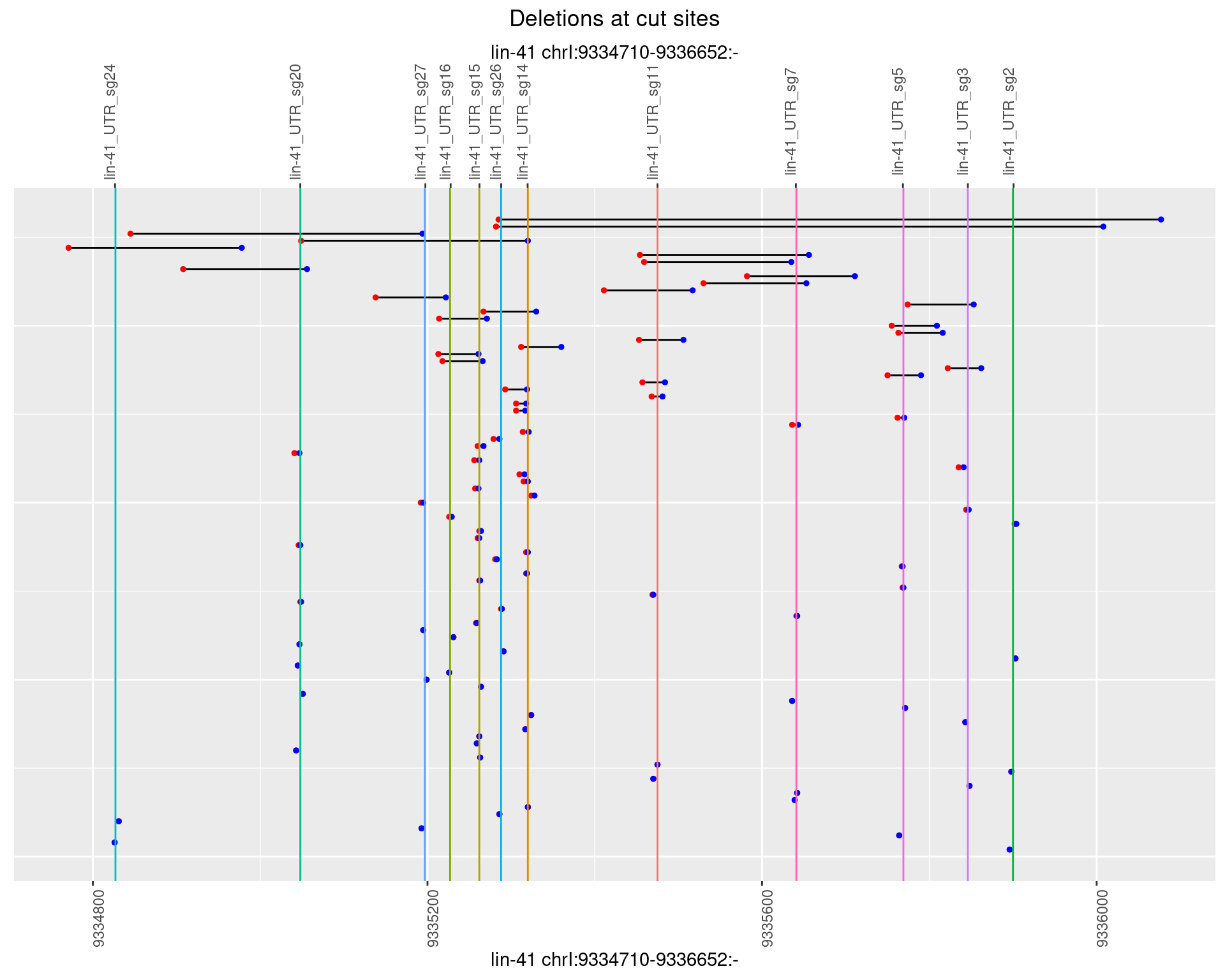
1.110.5 Freq: 0.01
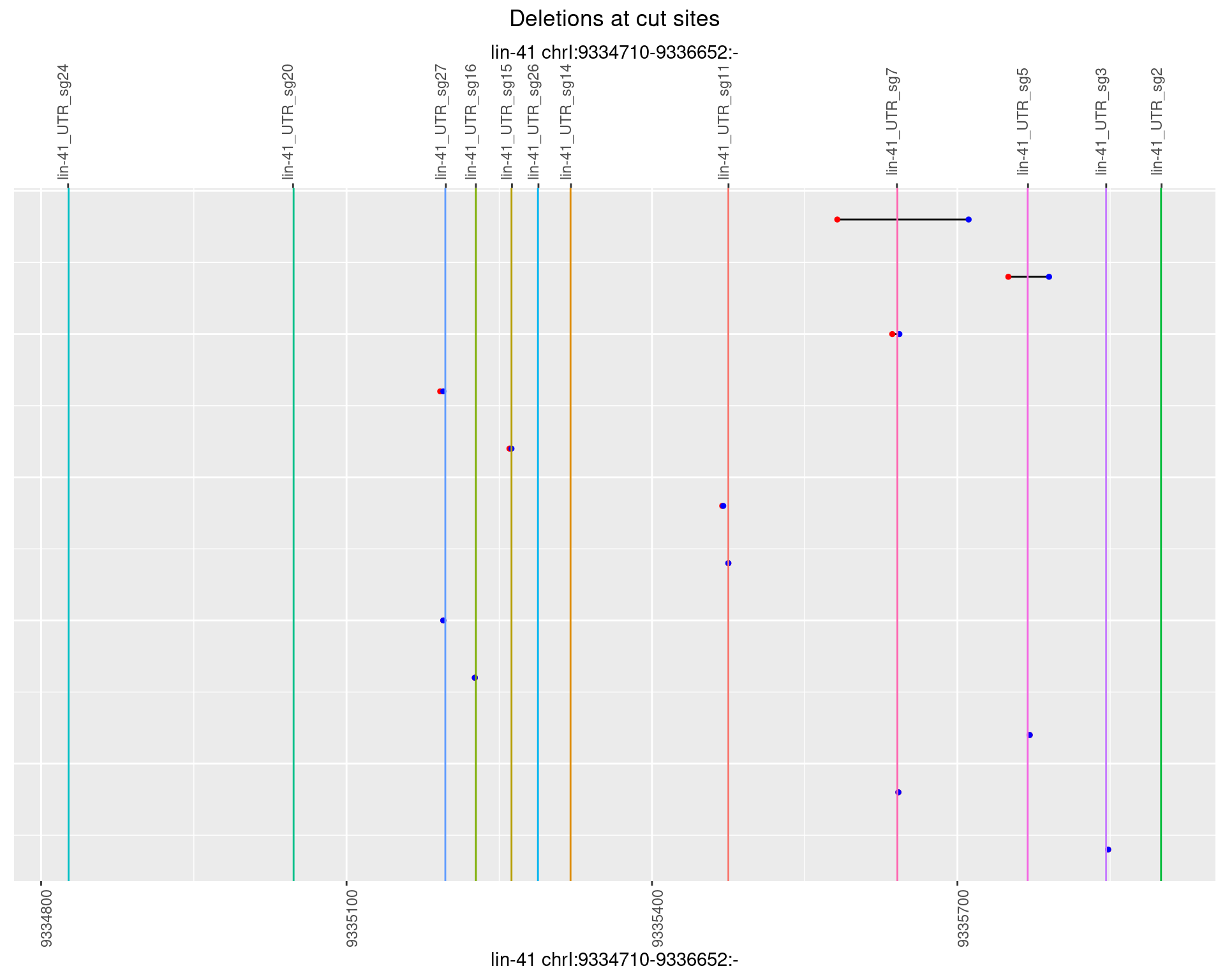
1.110.6 Freq: 0.1
No plot to show
1.111 lin41_RNApacbio_L4_all
1.111.1 Freq: 0
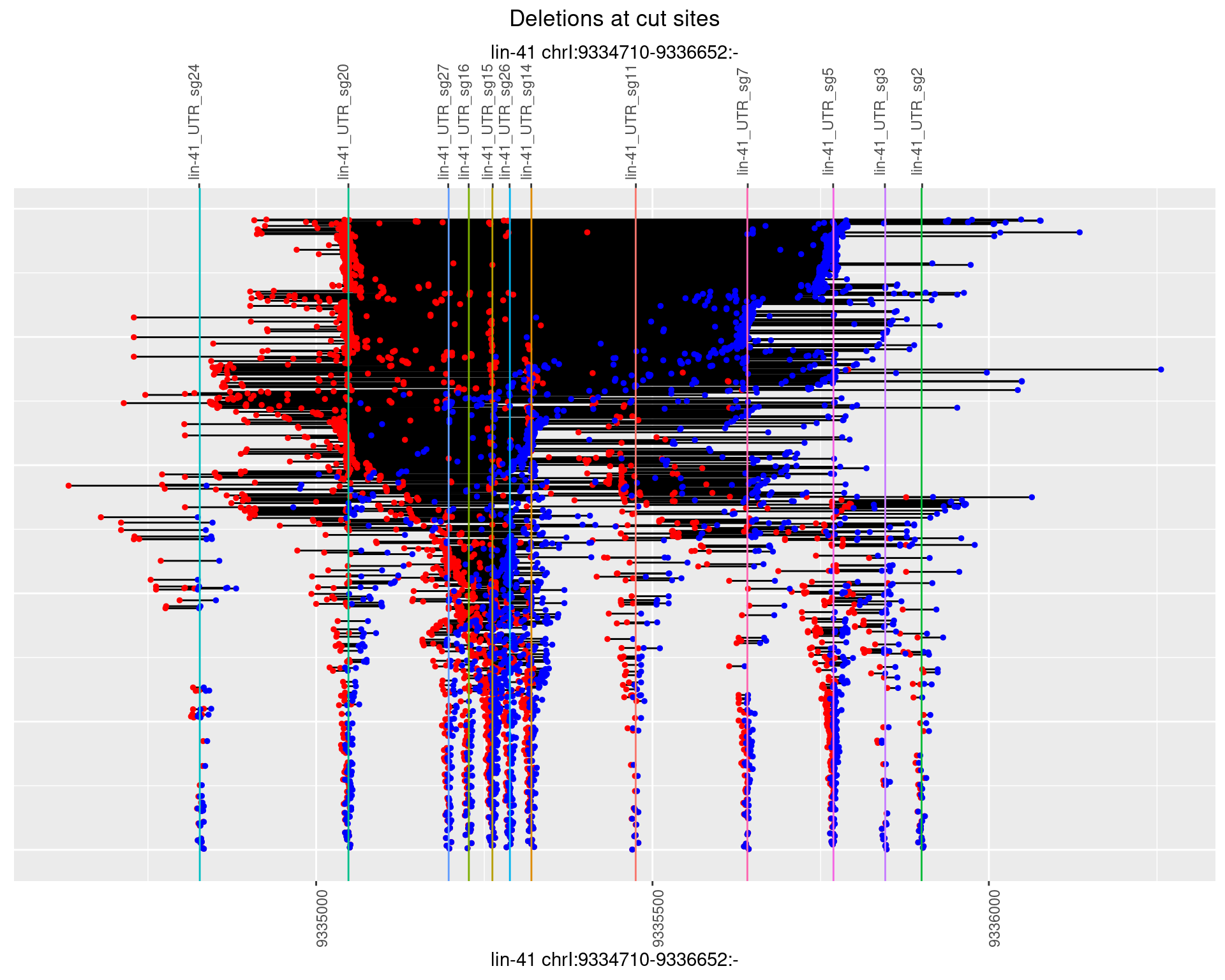
1.111.2 Freq: 1e-05
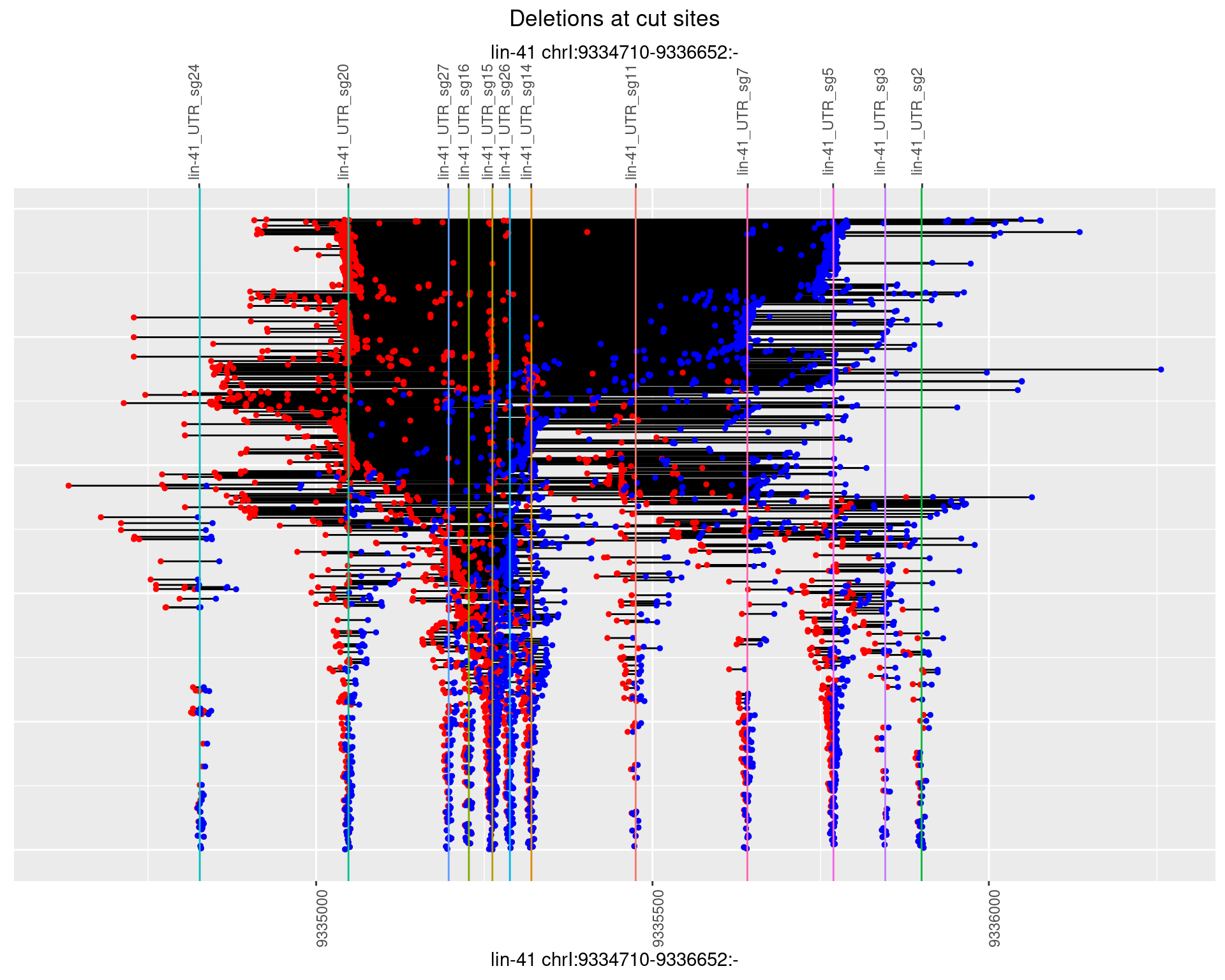
1.111.3 Freq: 1e-04
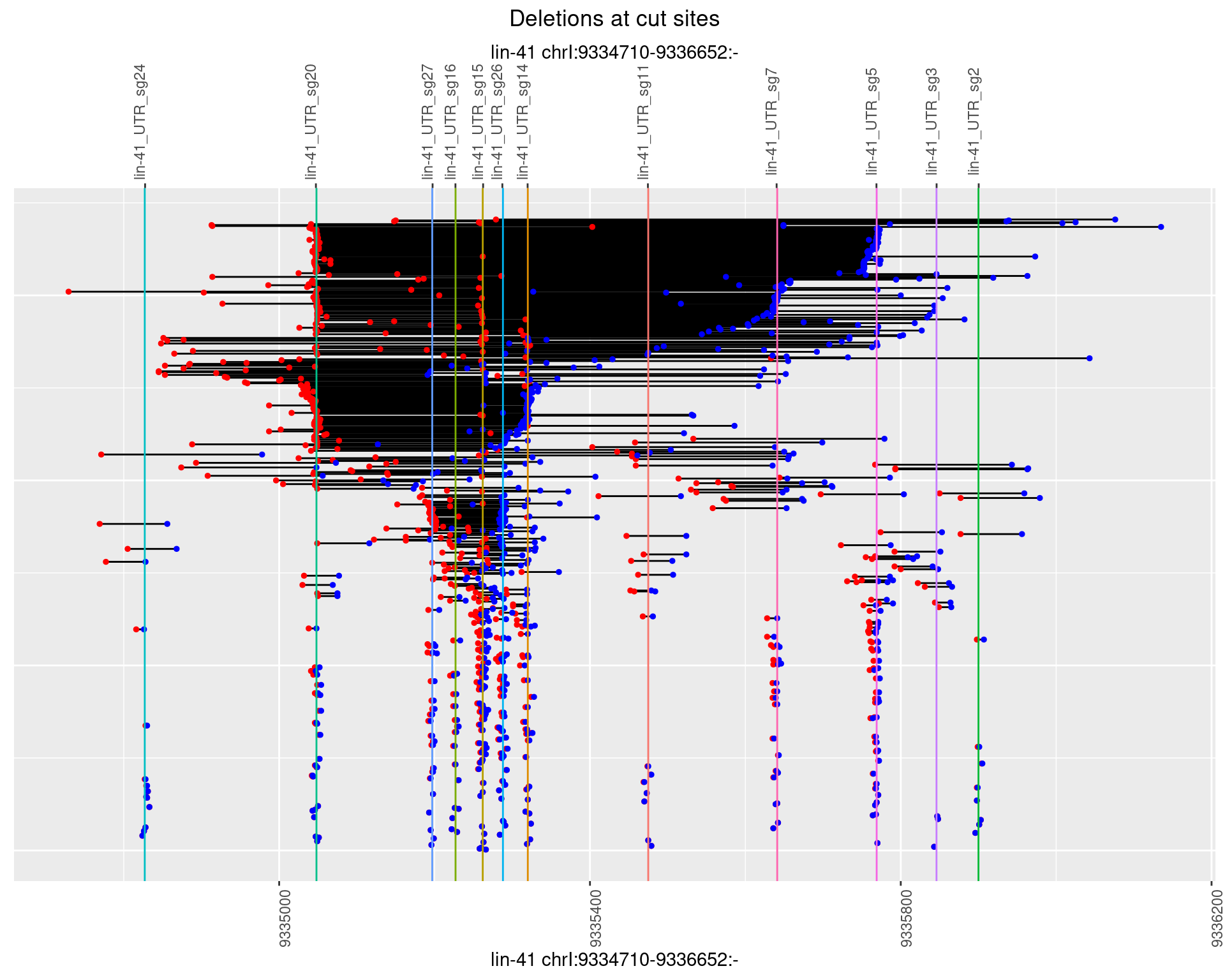
1.111.4 Freq: 0.001
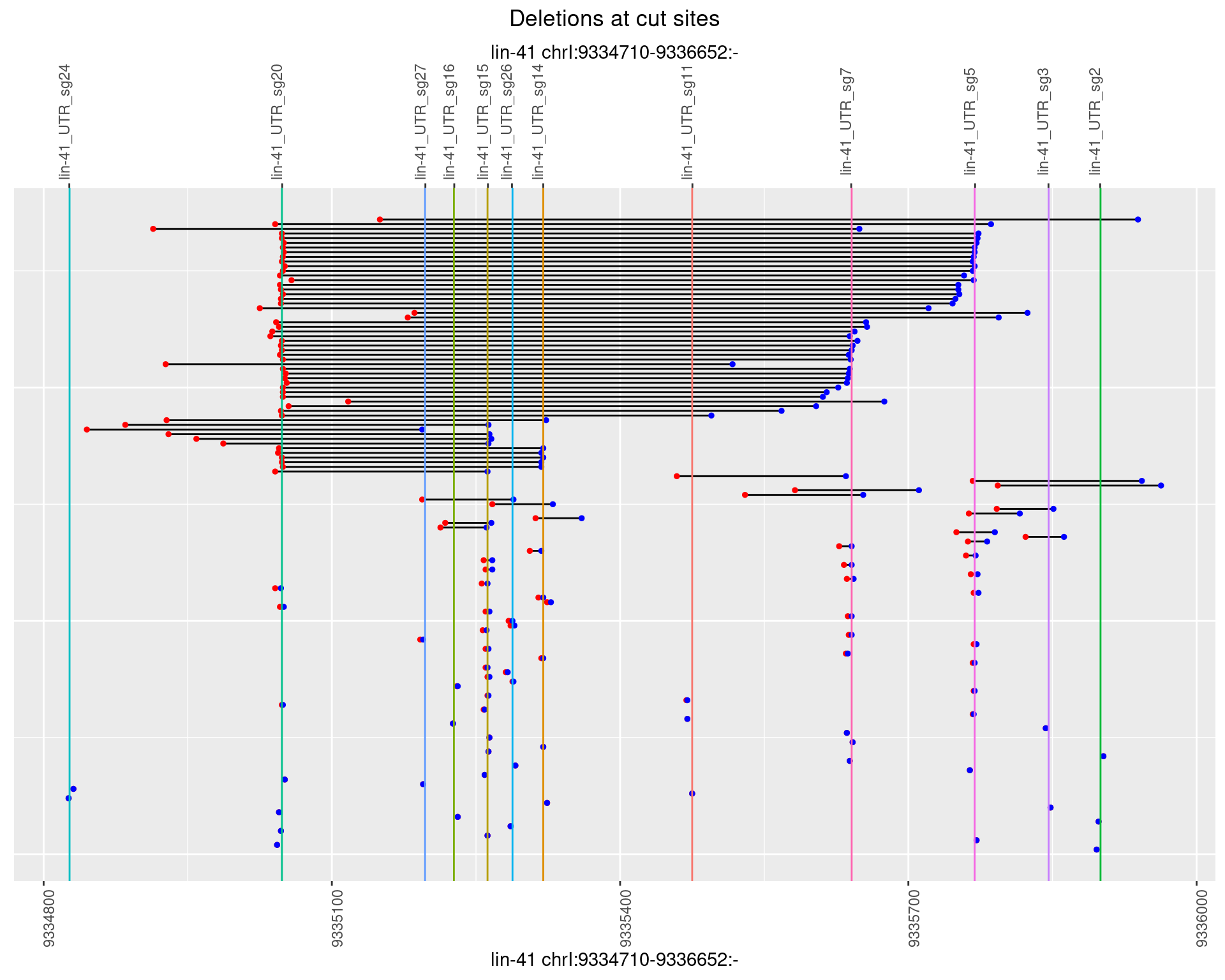
1.111.5 Freq: 0.01
1.111.6 Freq: 0.1
No plot to show
1.112 lin41_DNA_sg1516_2627_pool3
1.112.1 Freq: 0
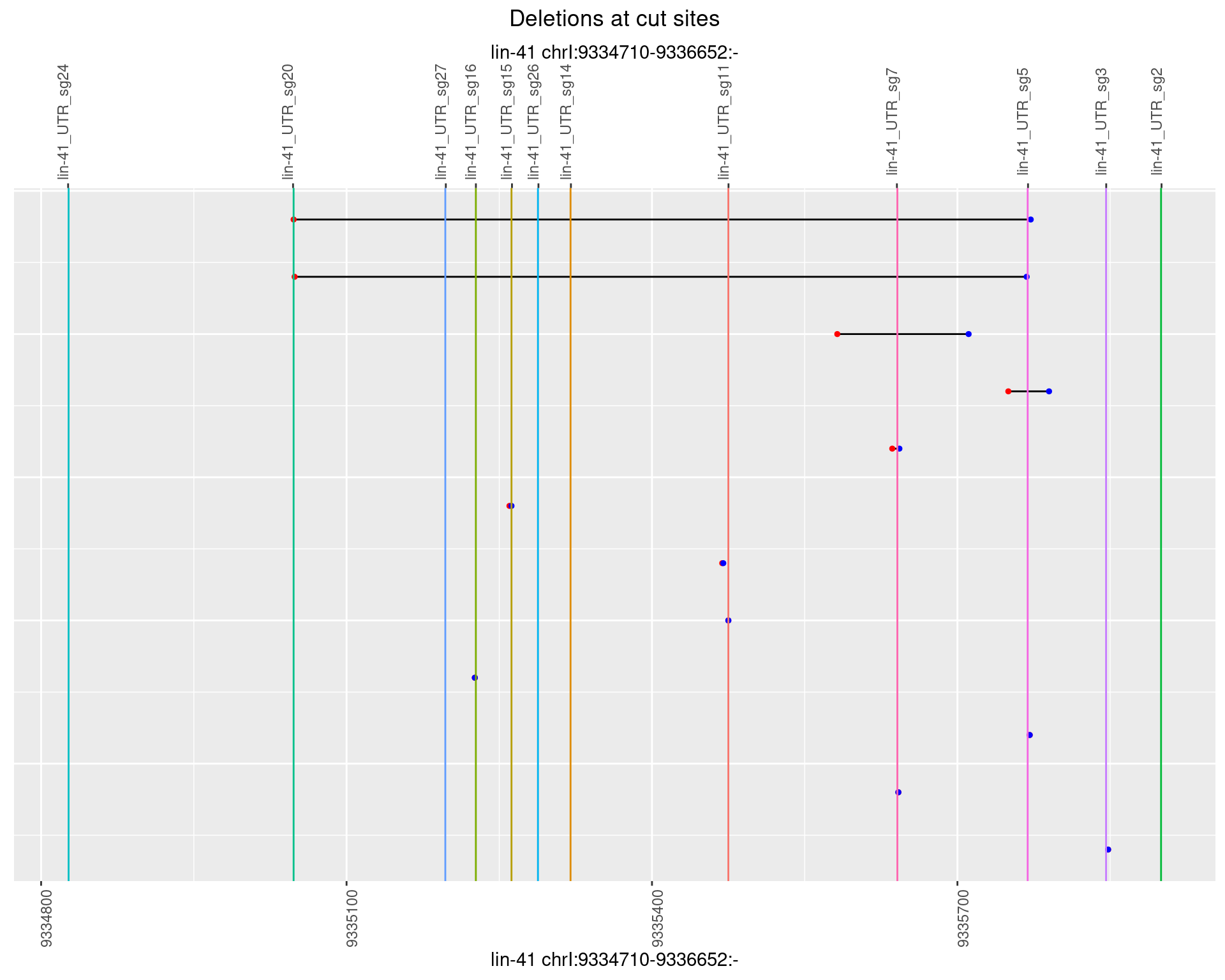
1.112.2 Freq: 1e-05
1.112.3 Freq: 1e-04
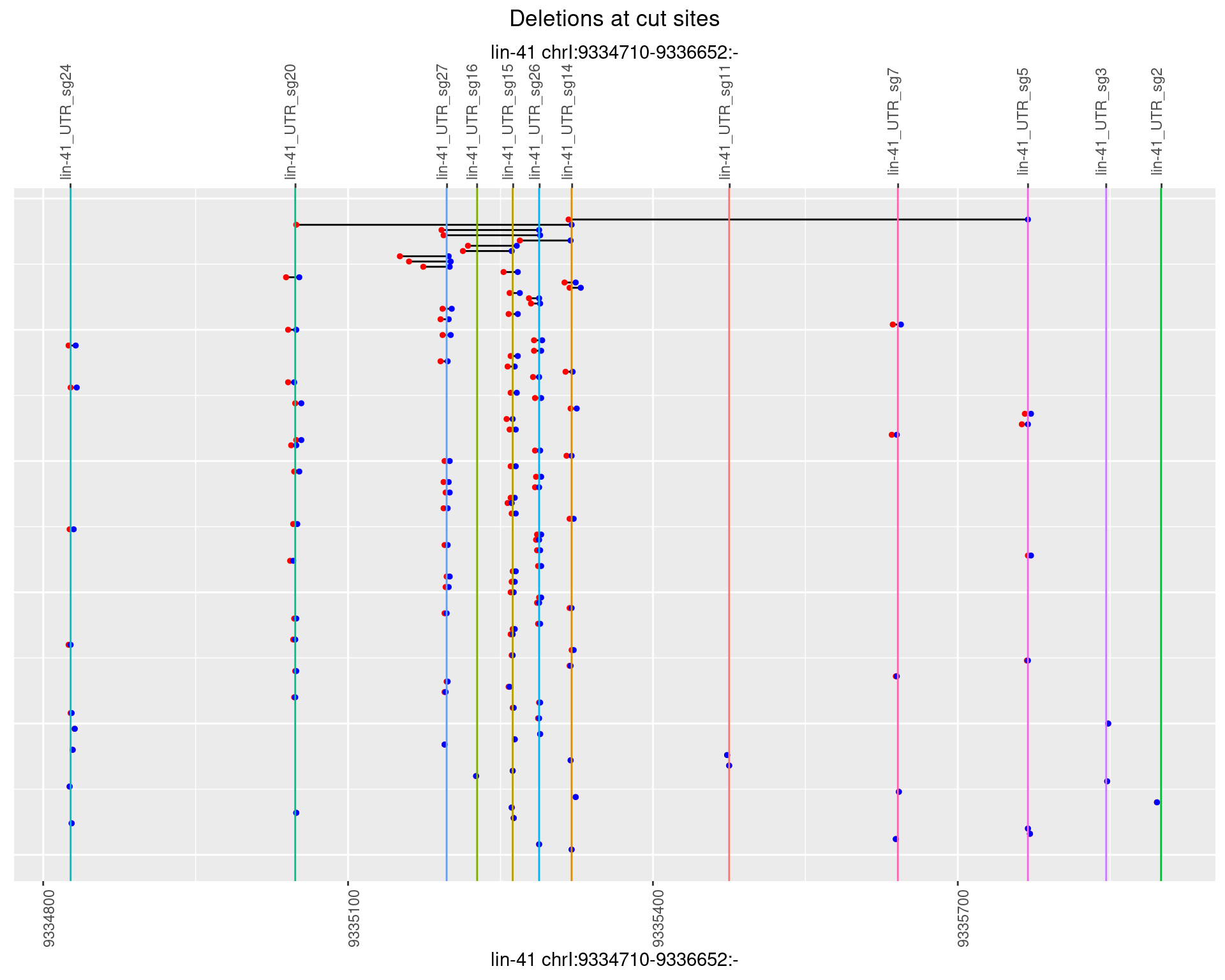
1.112.4 Freq: 0.001
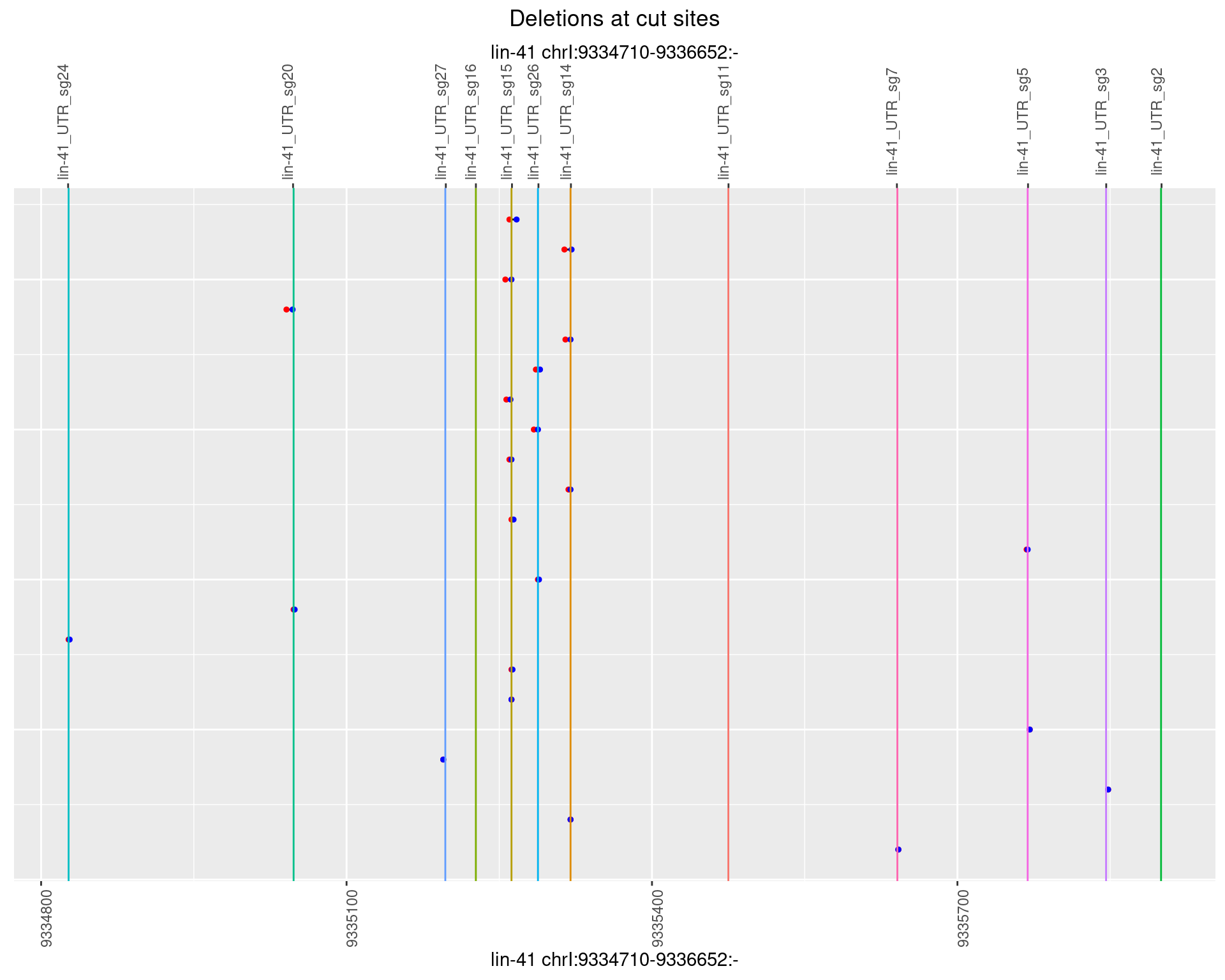
1.112.5 Freq: 0.01
No plot to show
1.112.6 Freq: 0.1
No plot to show
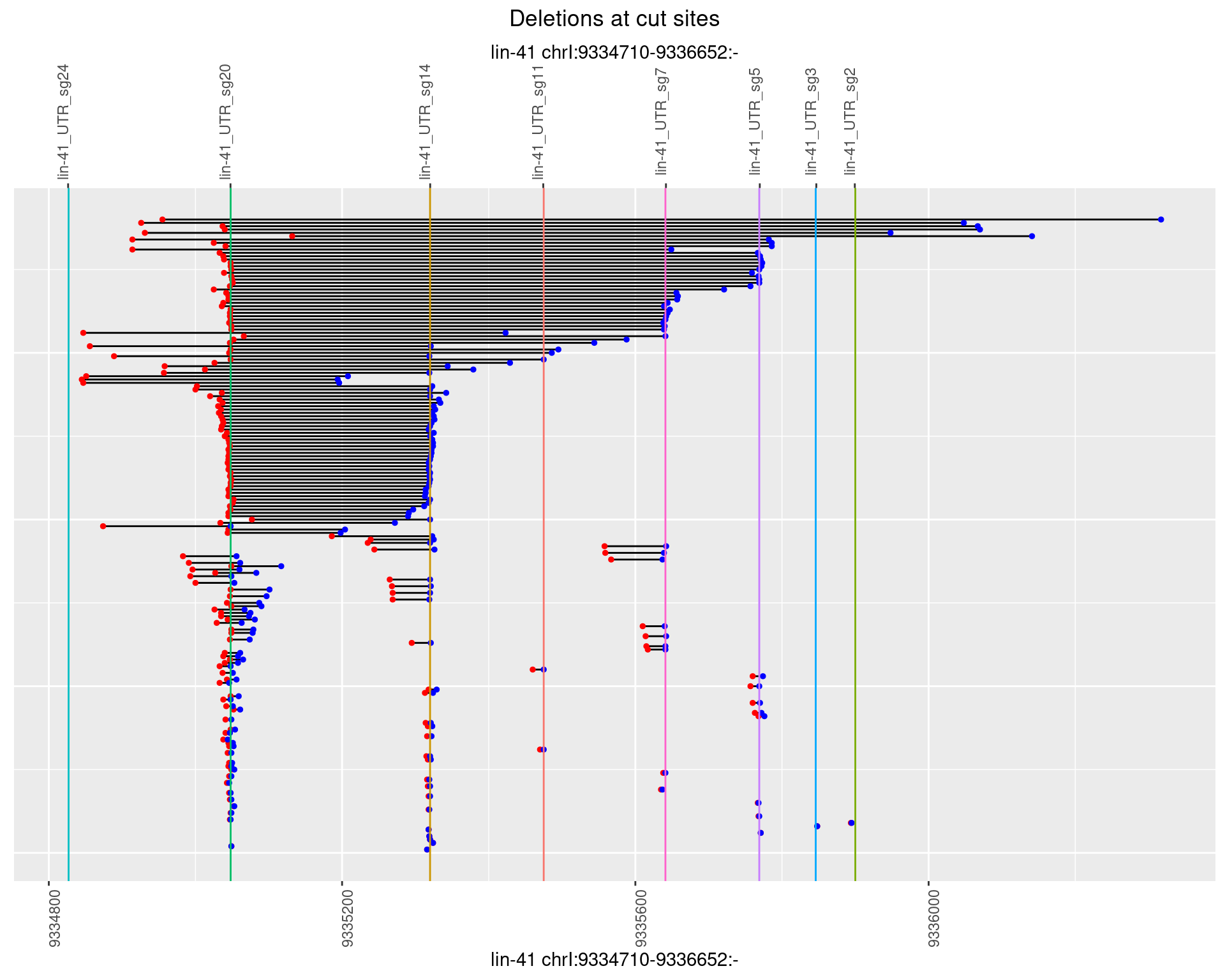
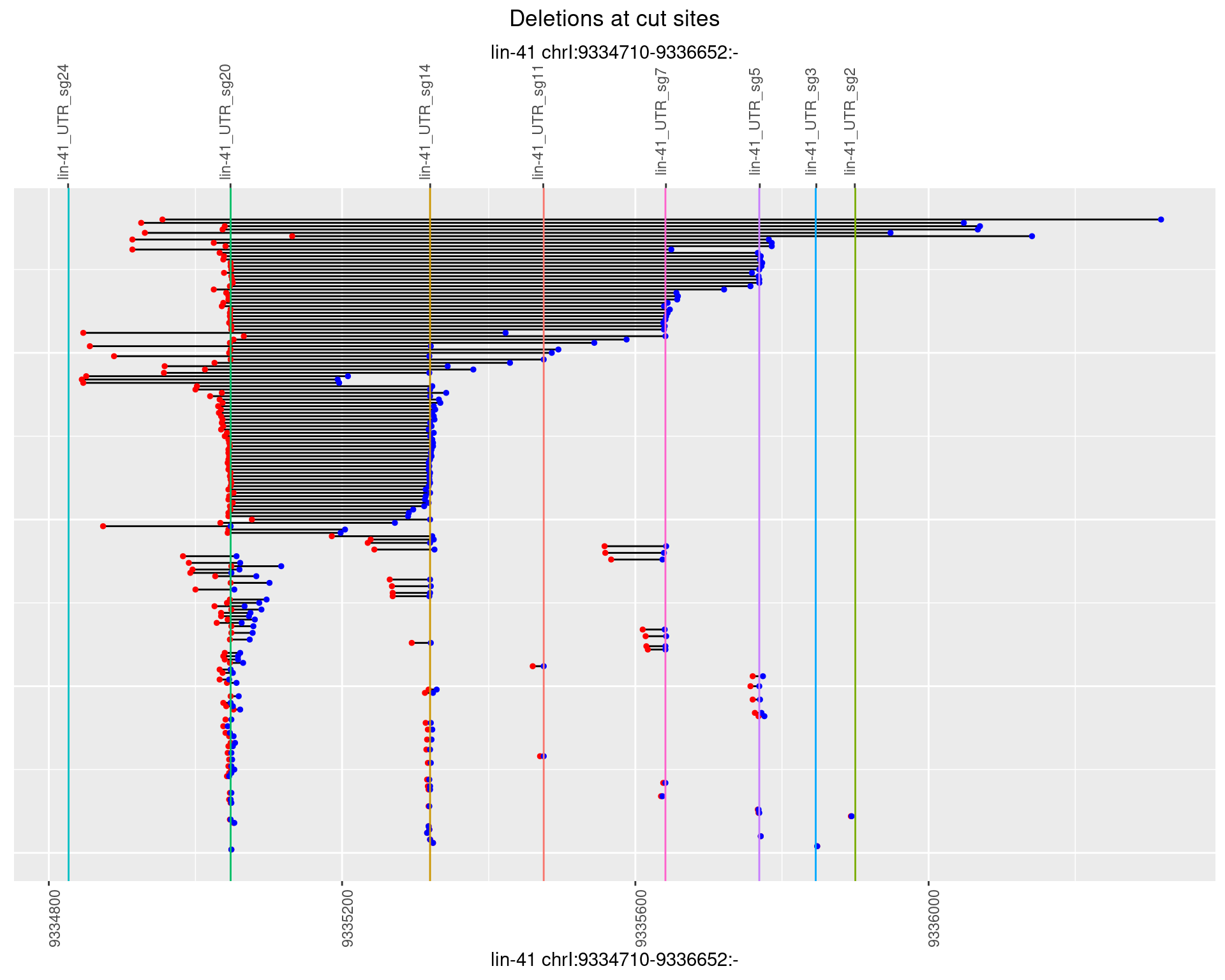
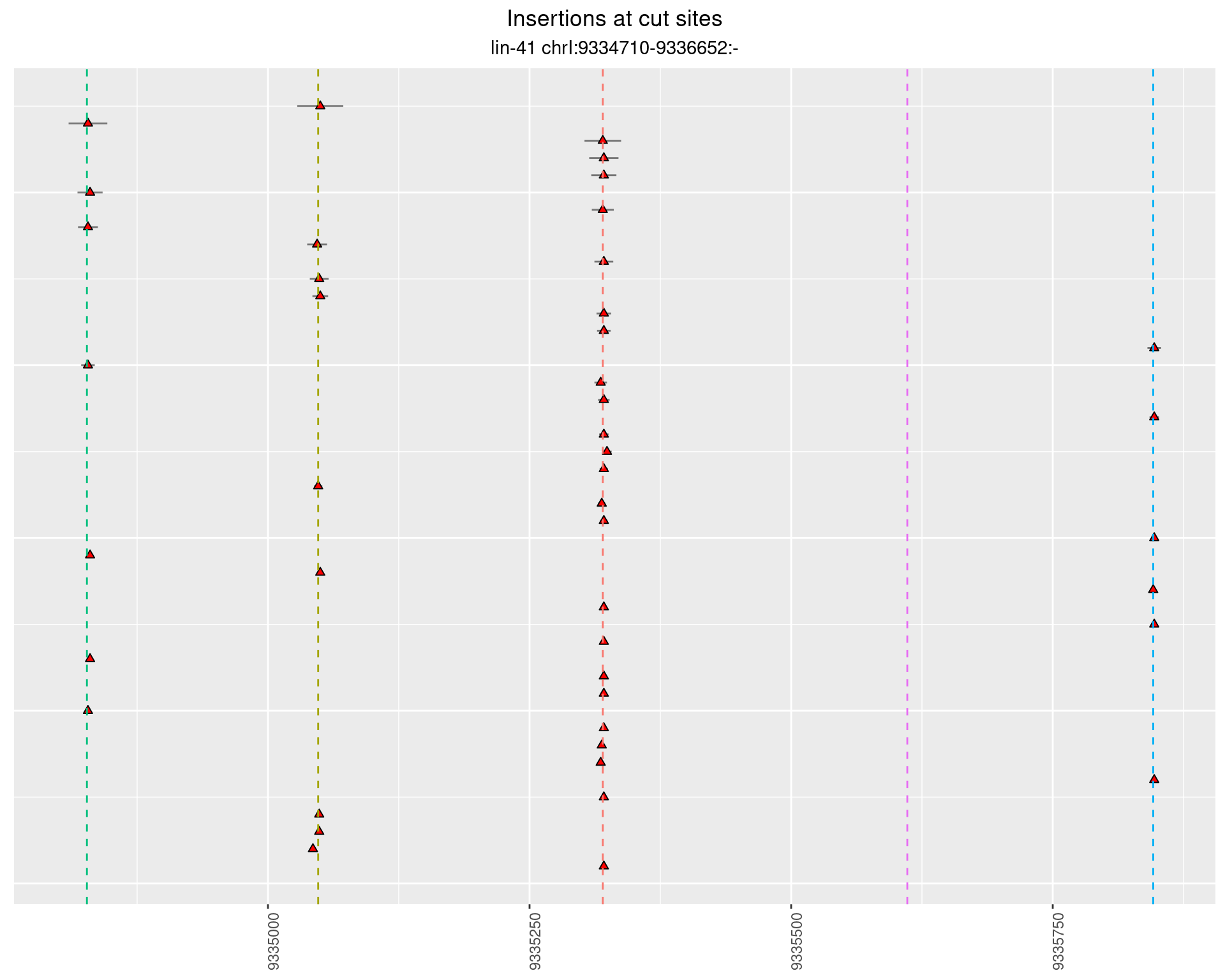
2 Insertion diversity at cut sites
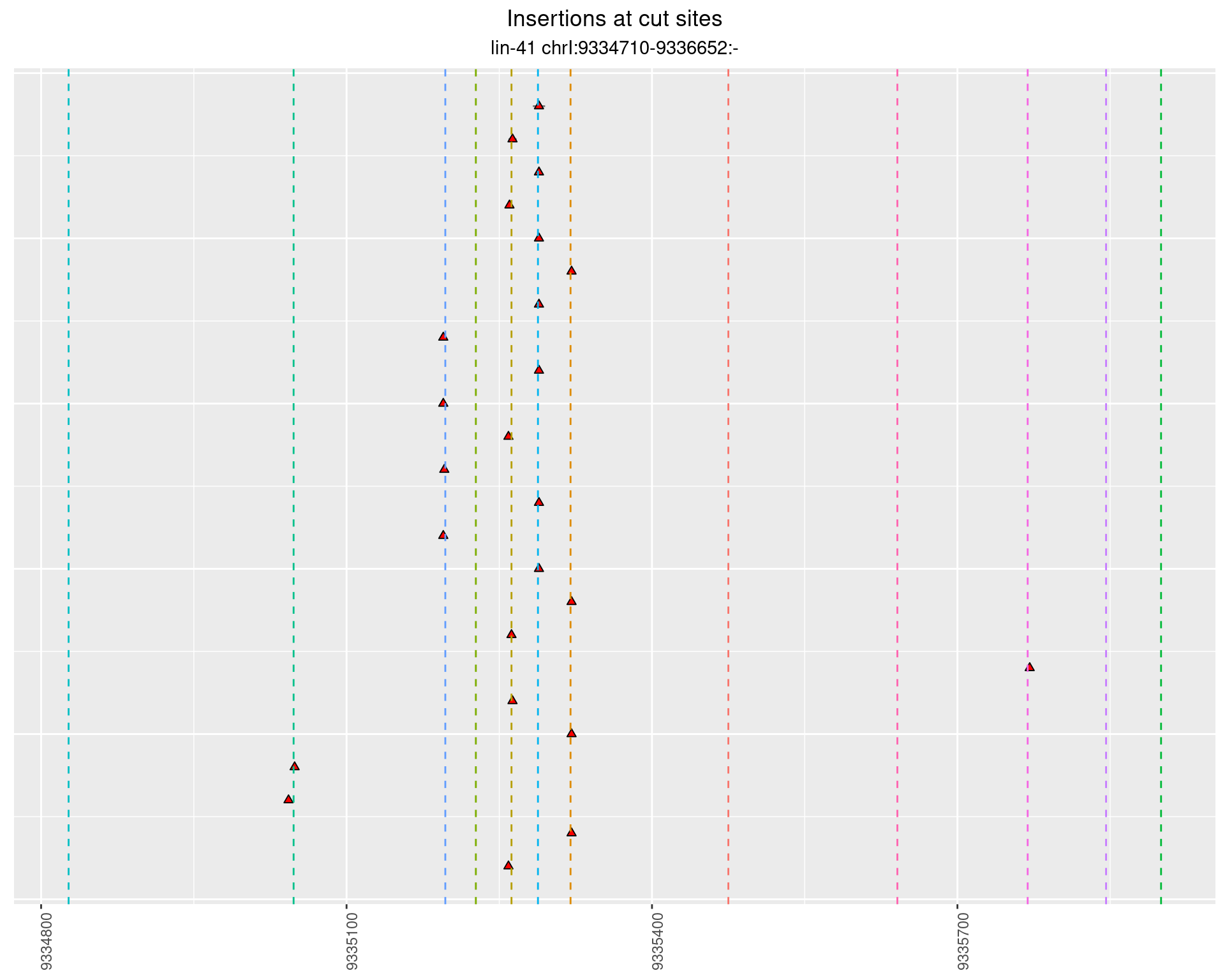
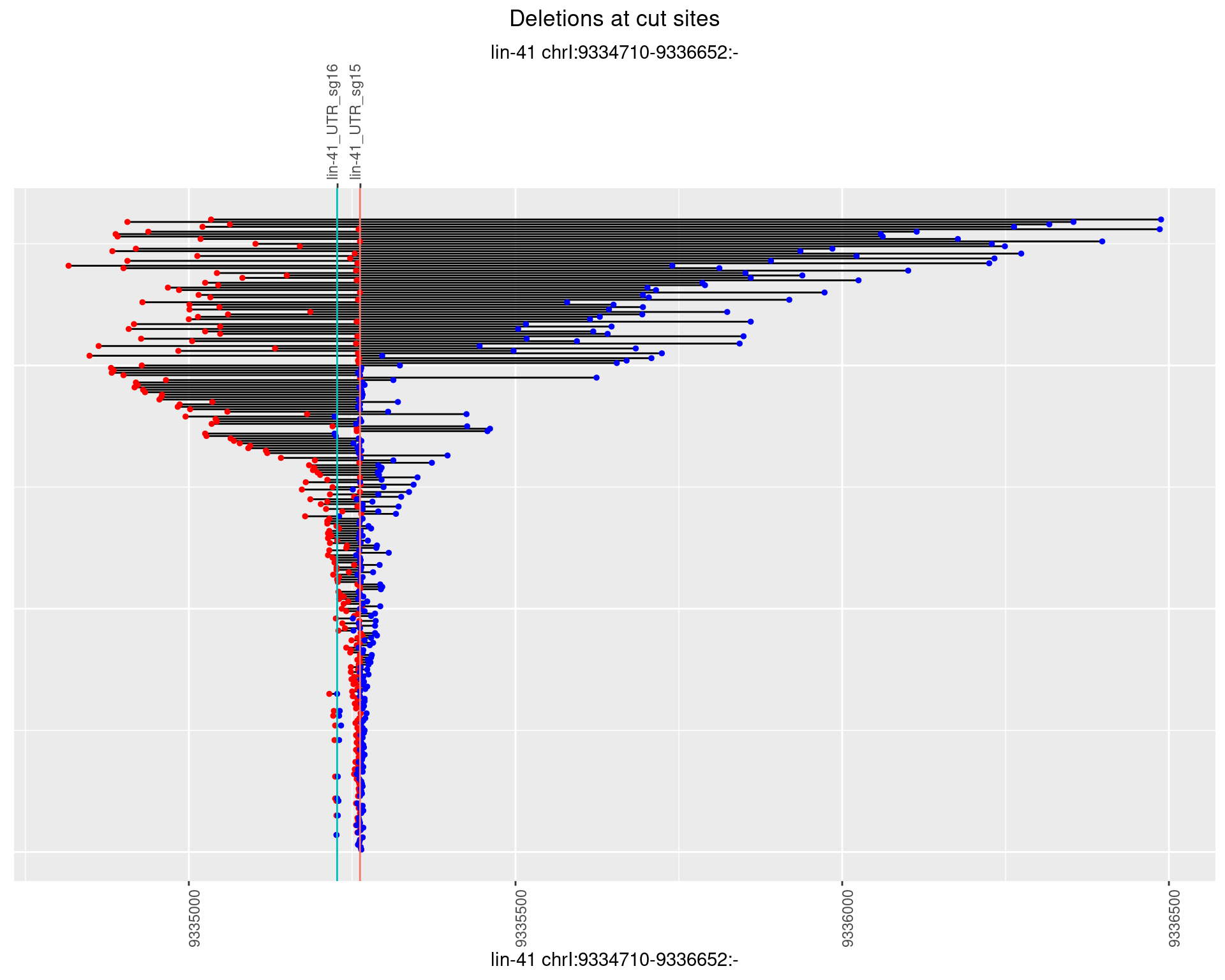
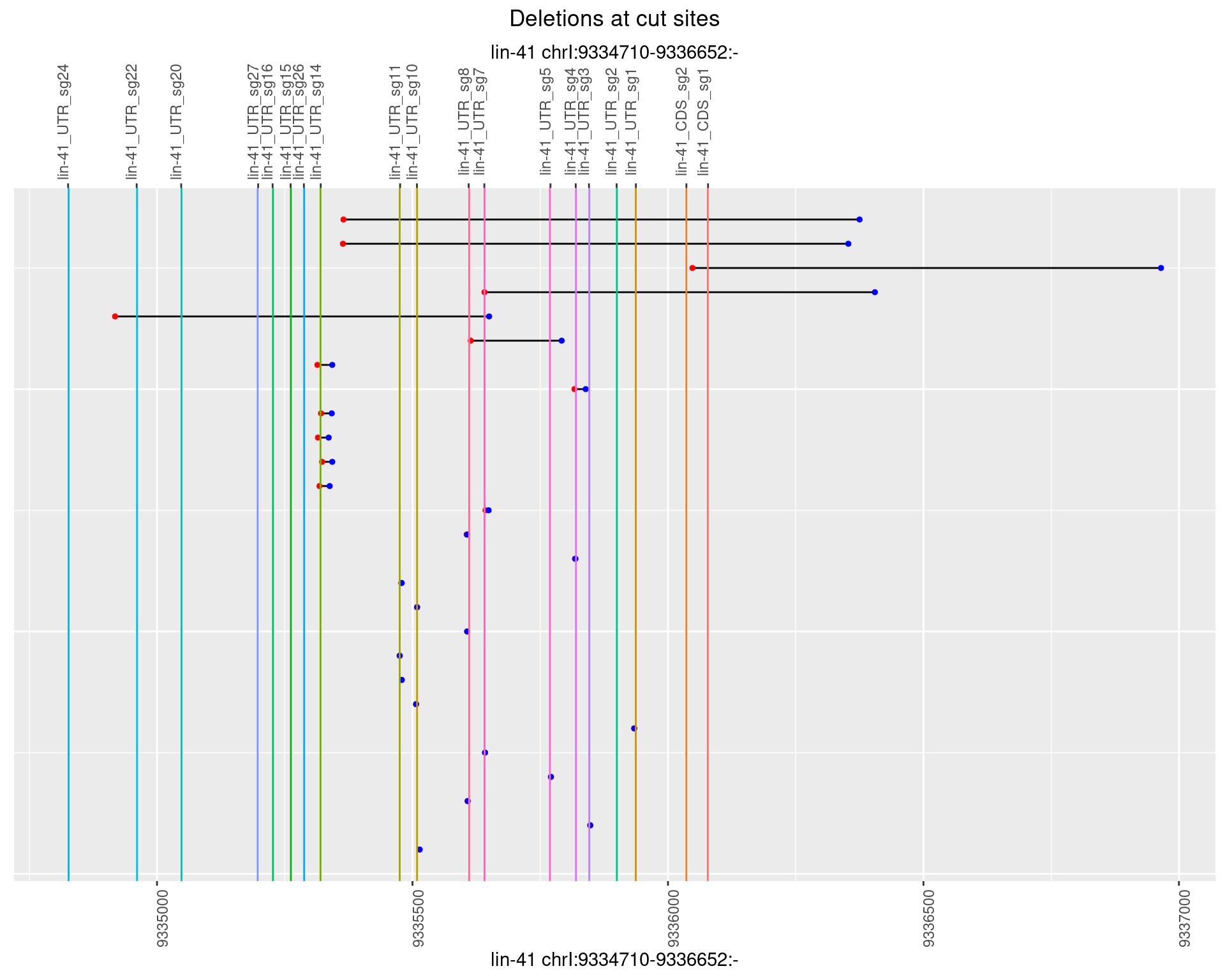
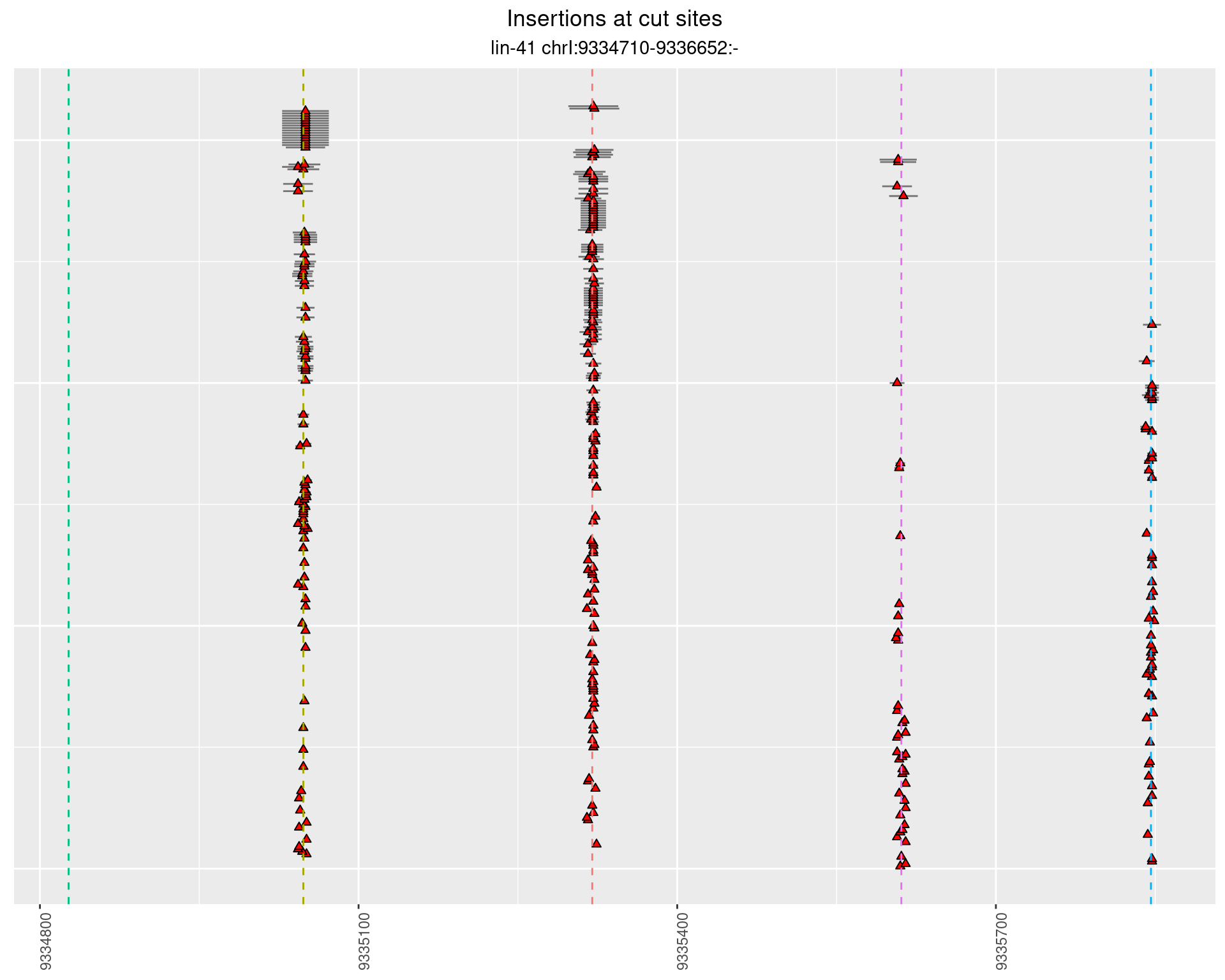
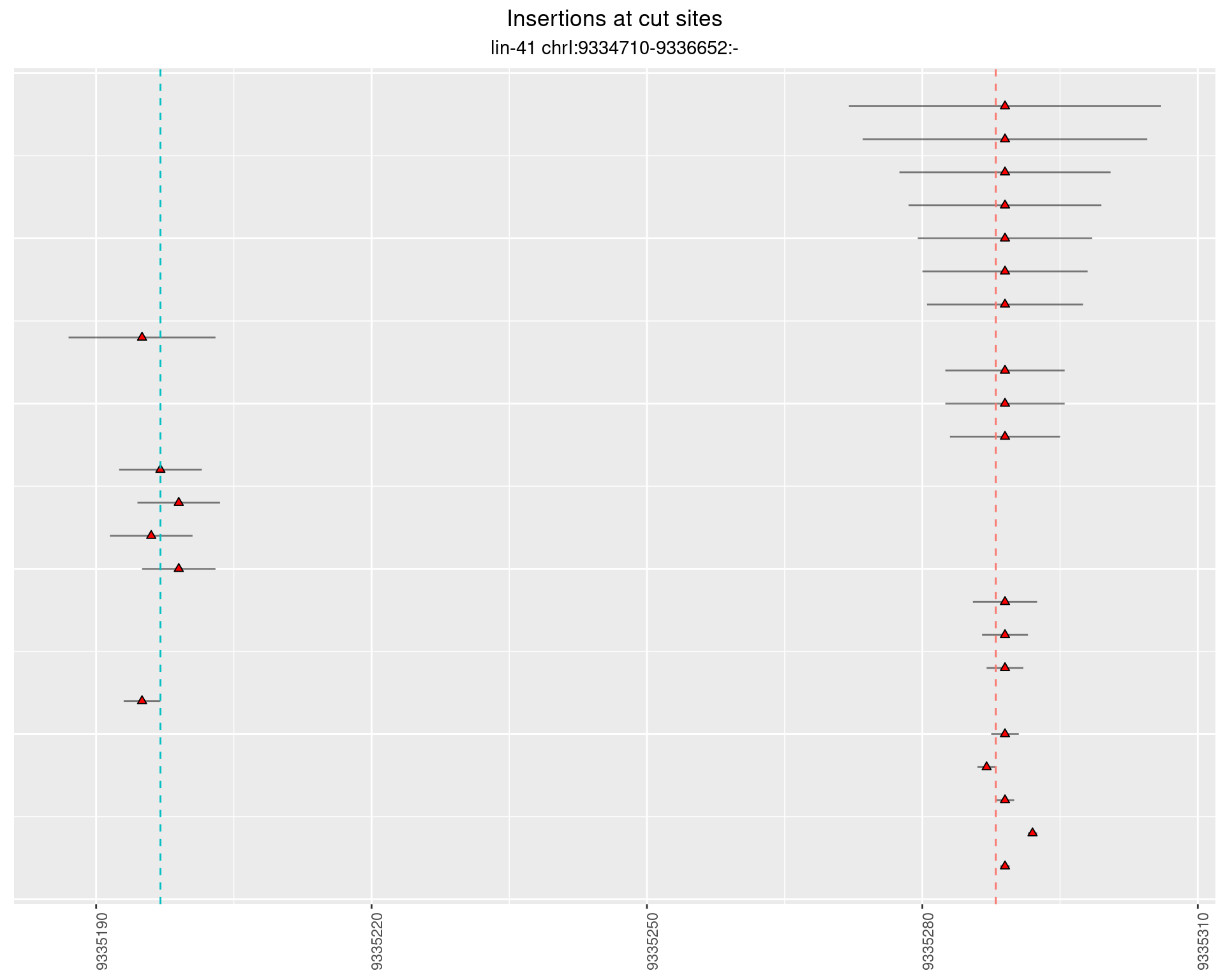
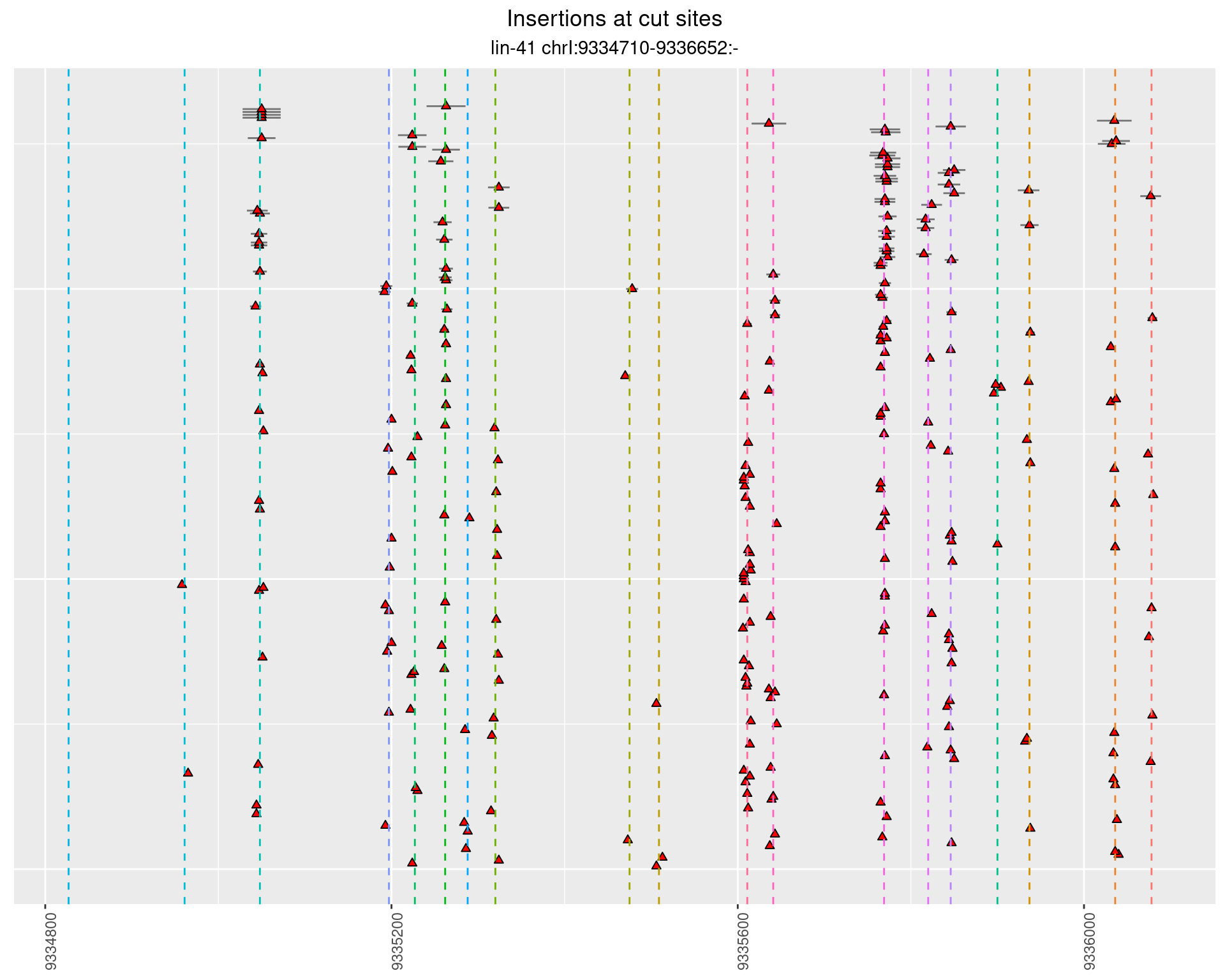
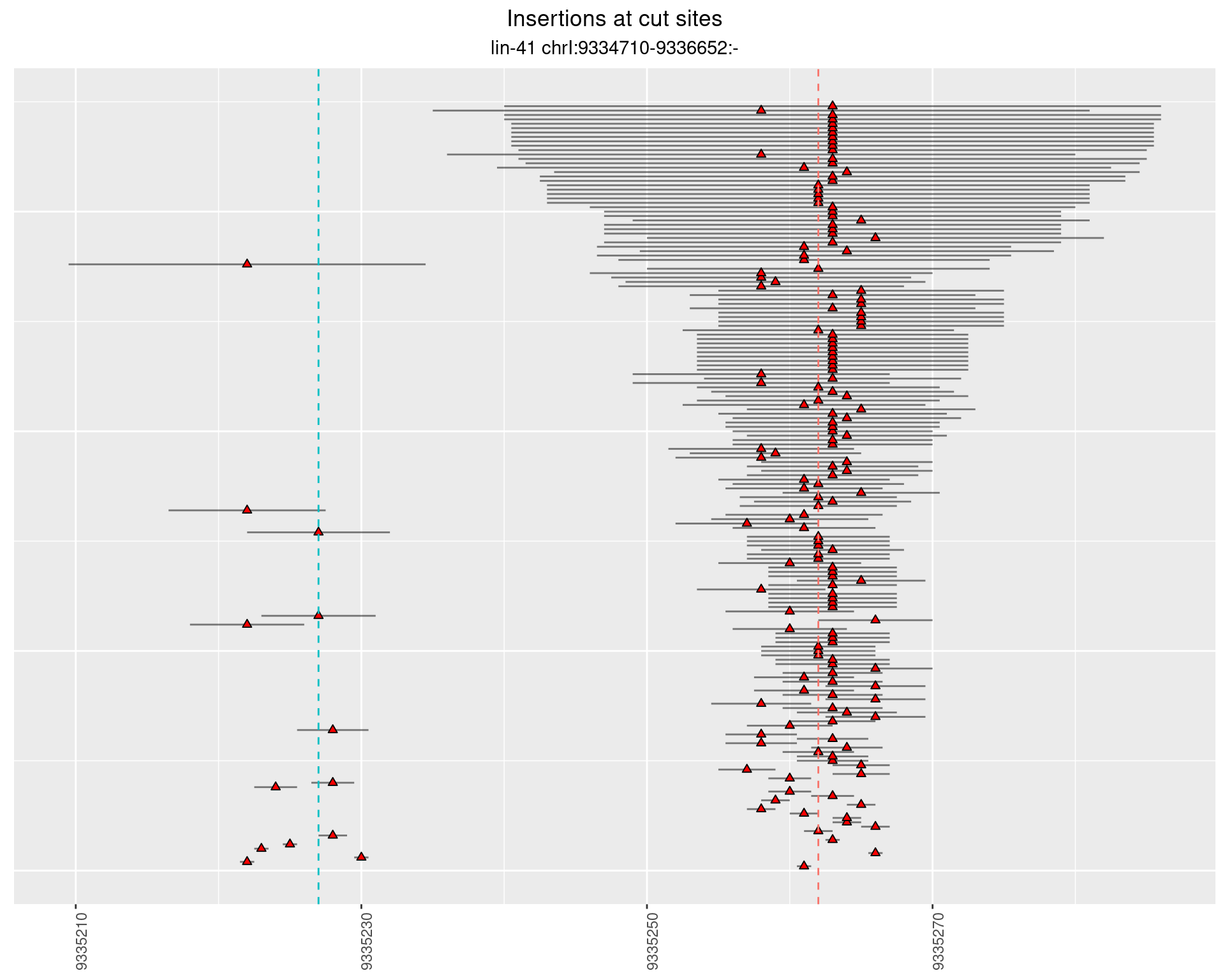
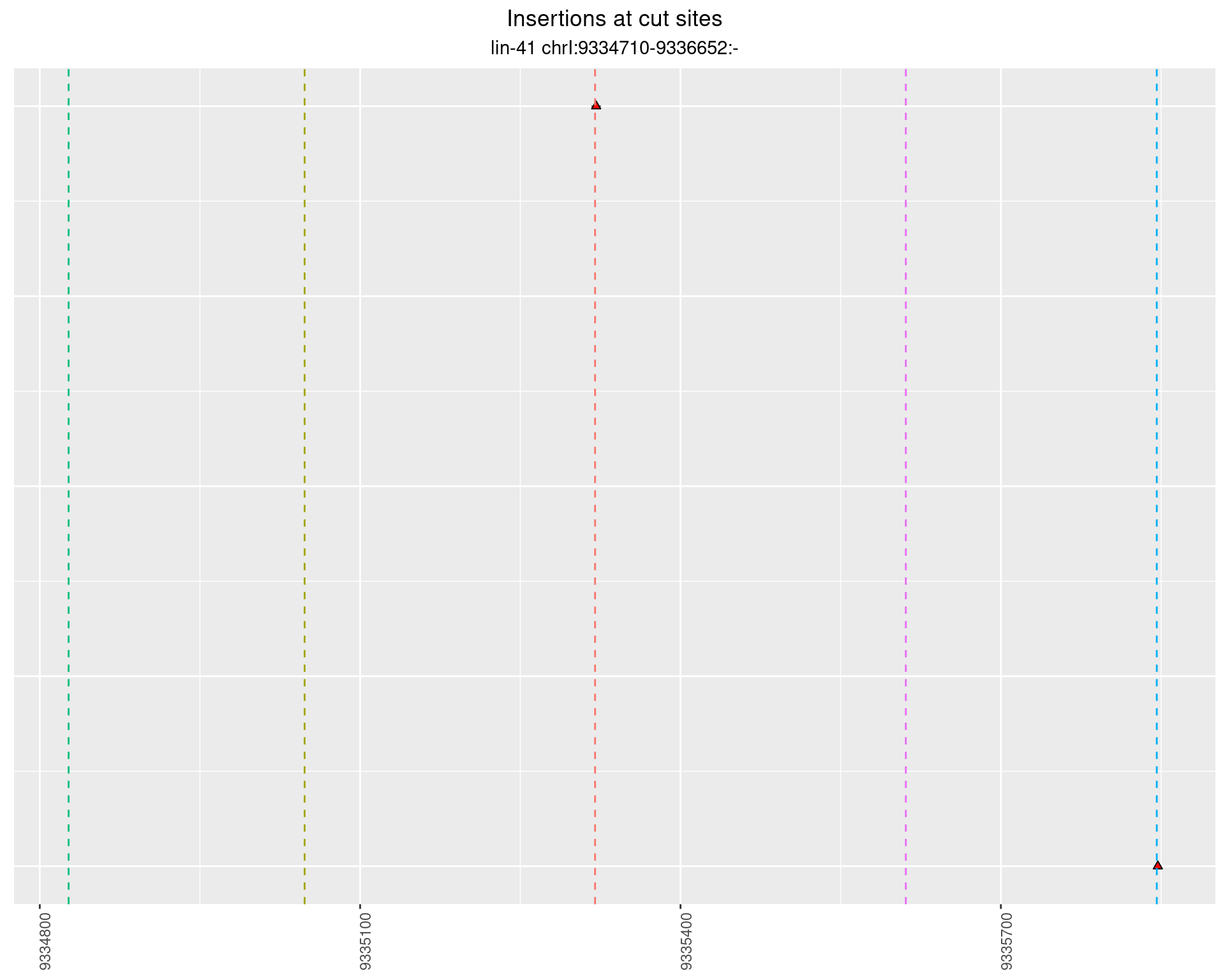
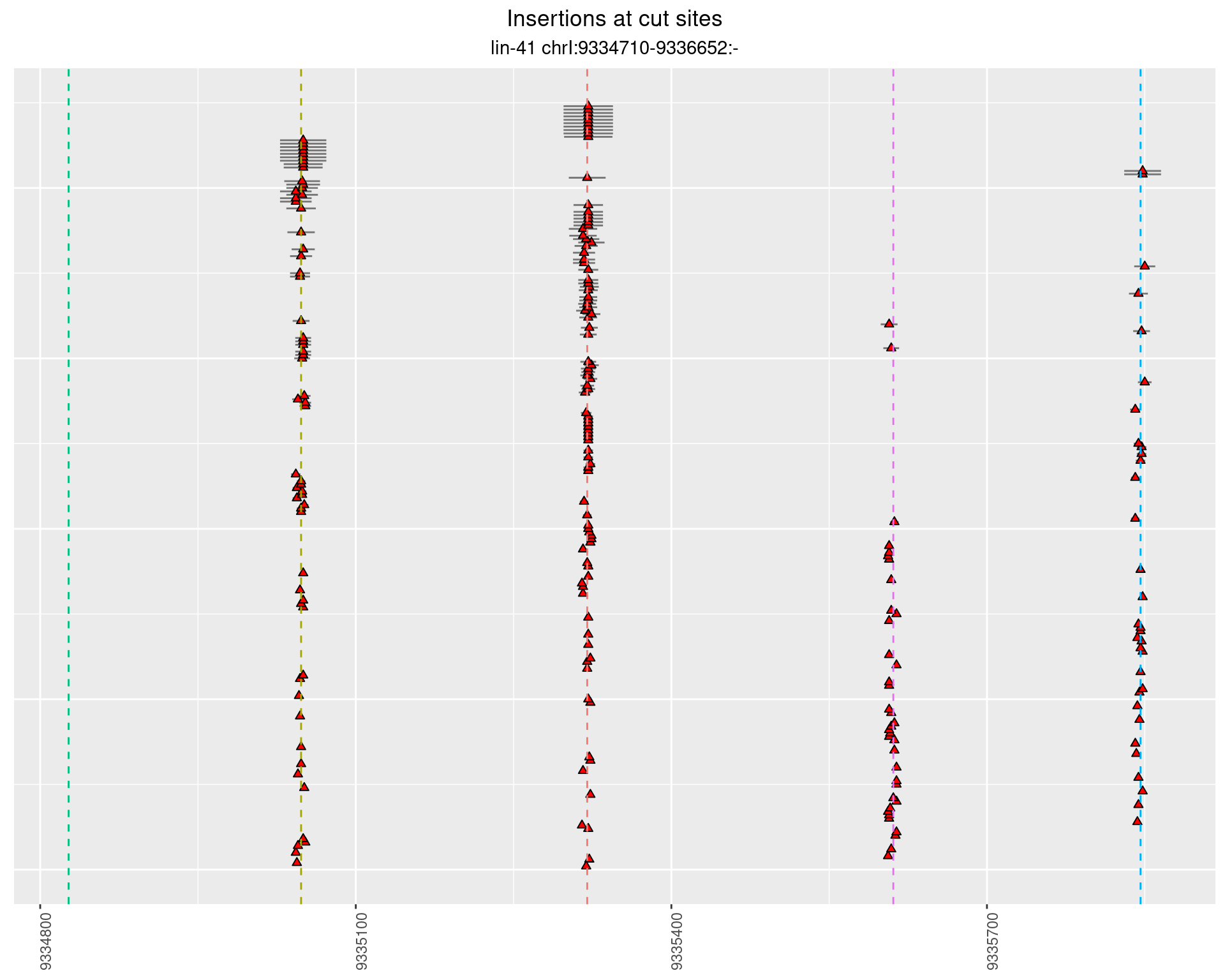
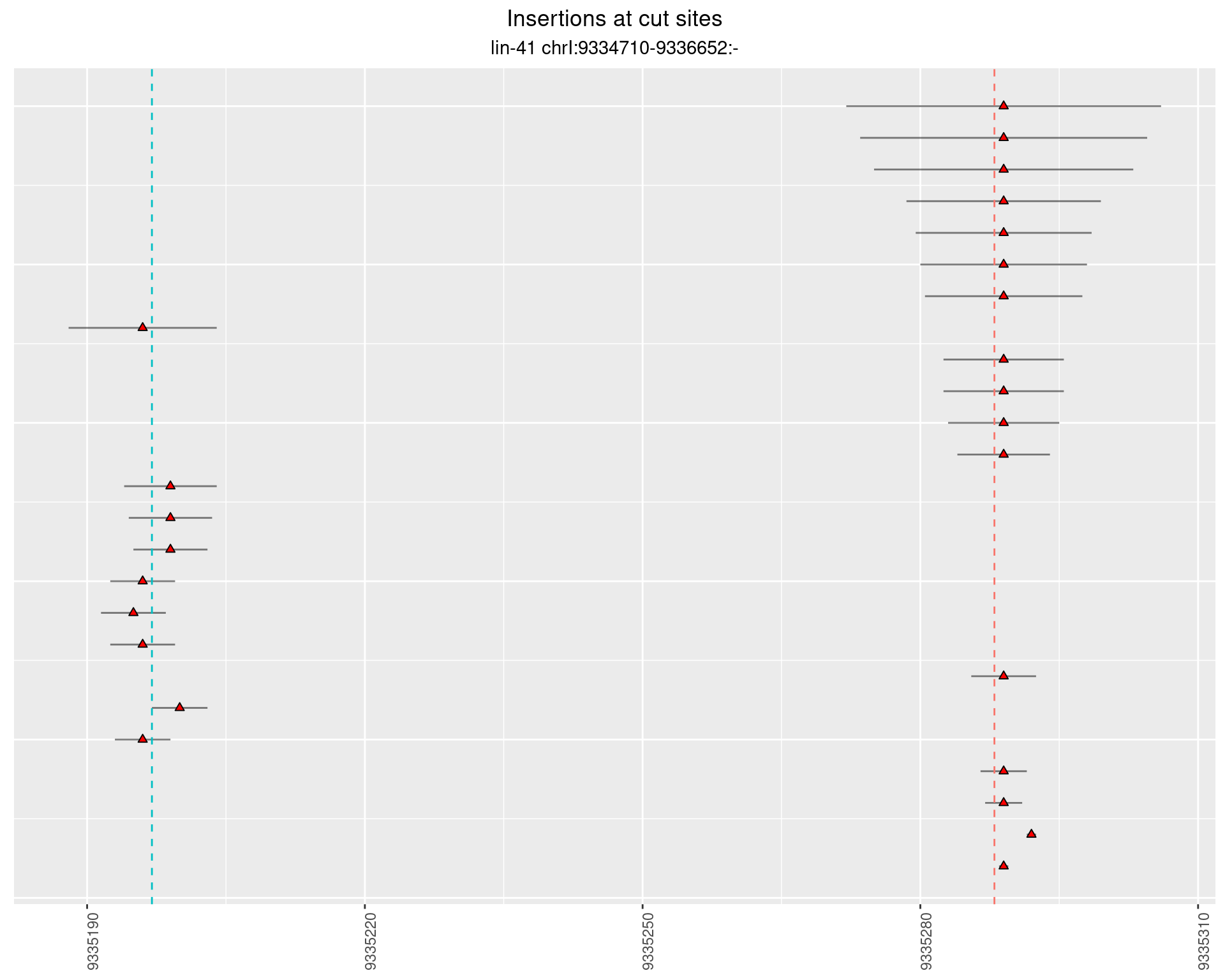
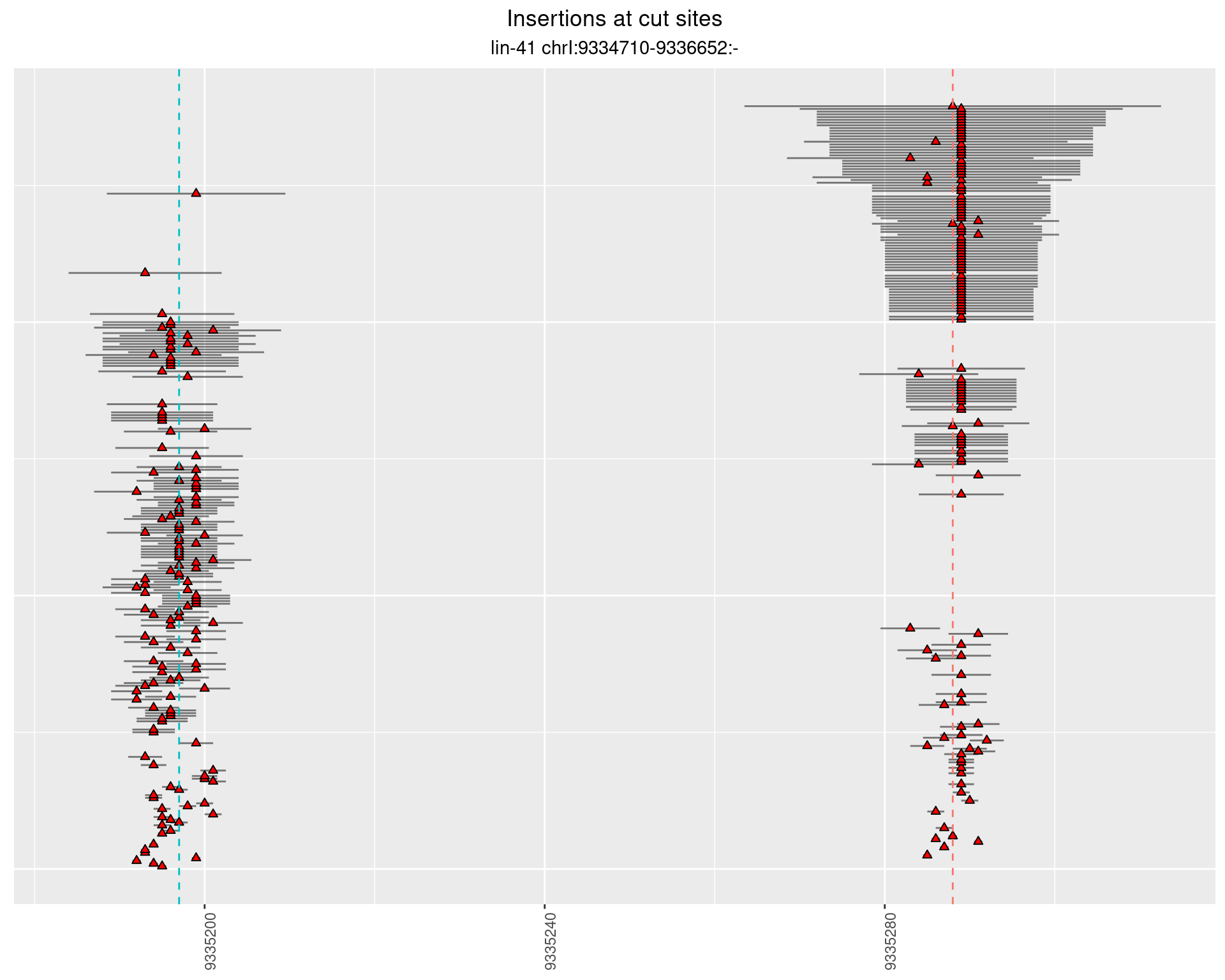
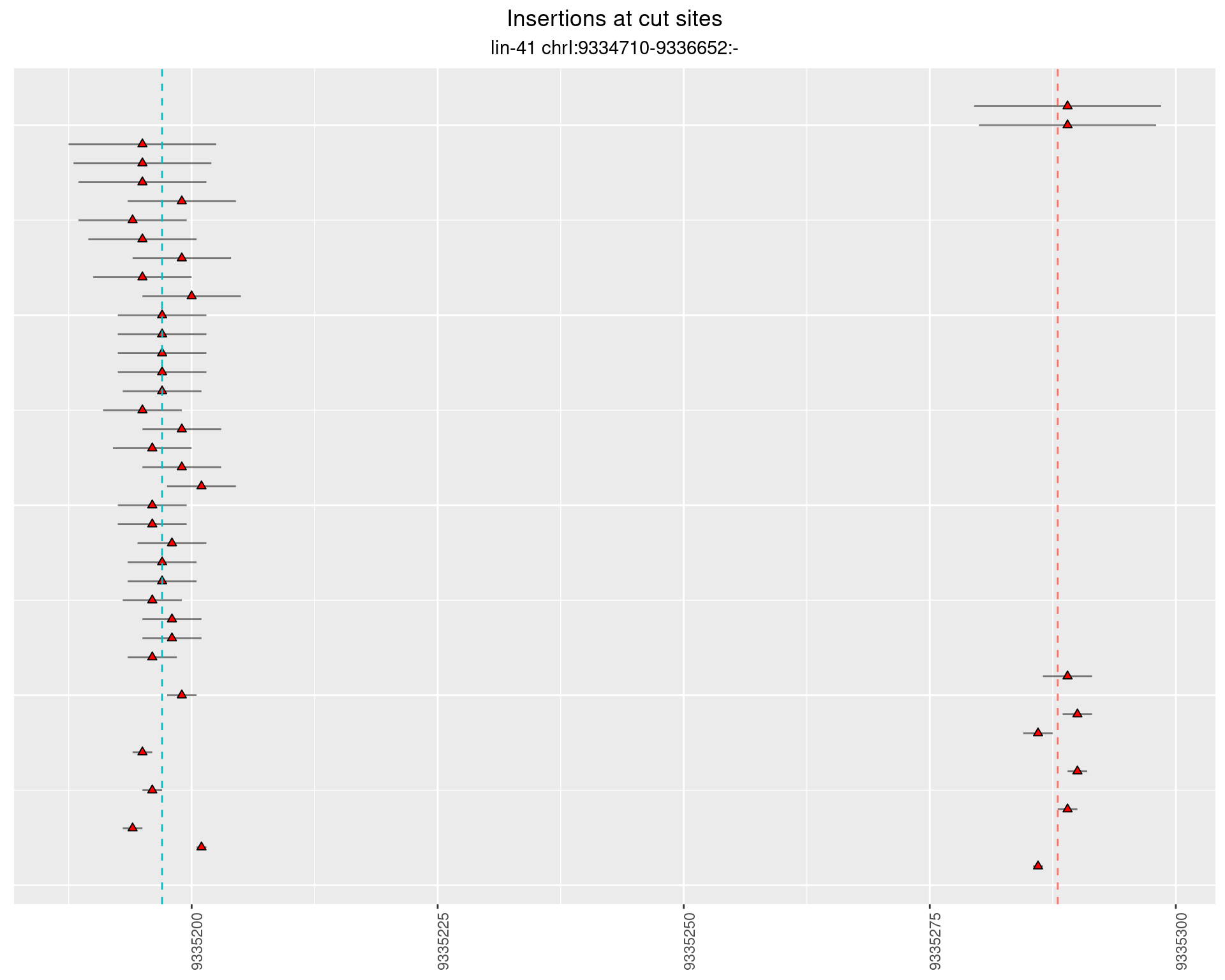
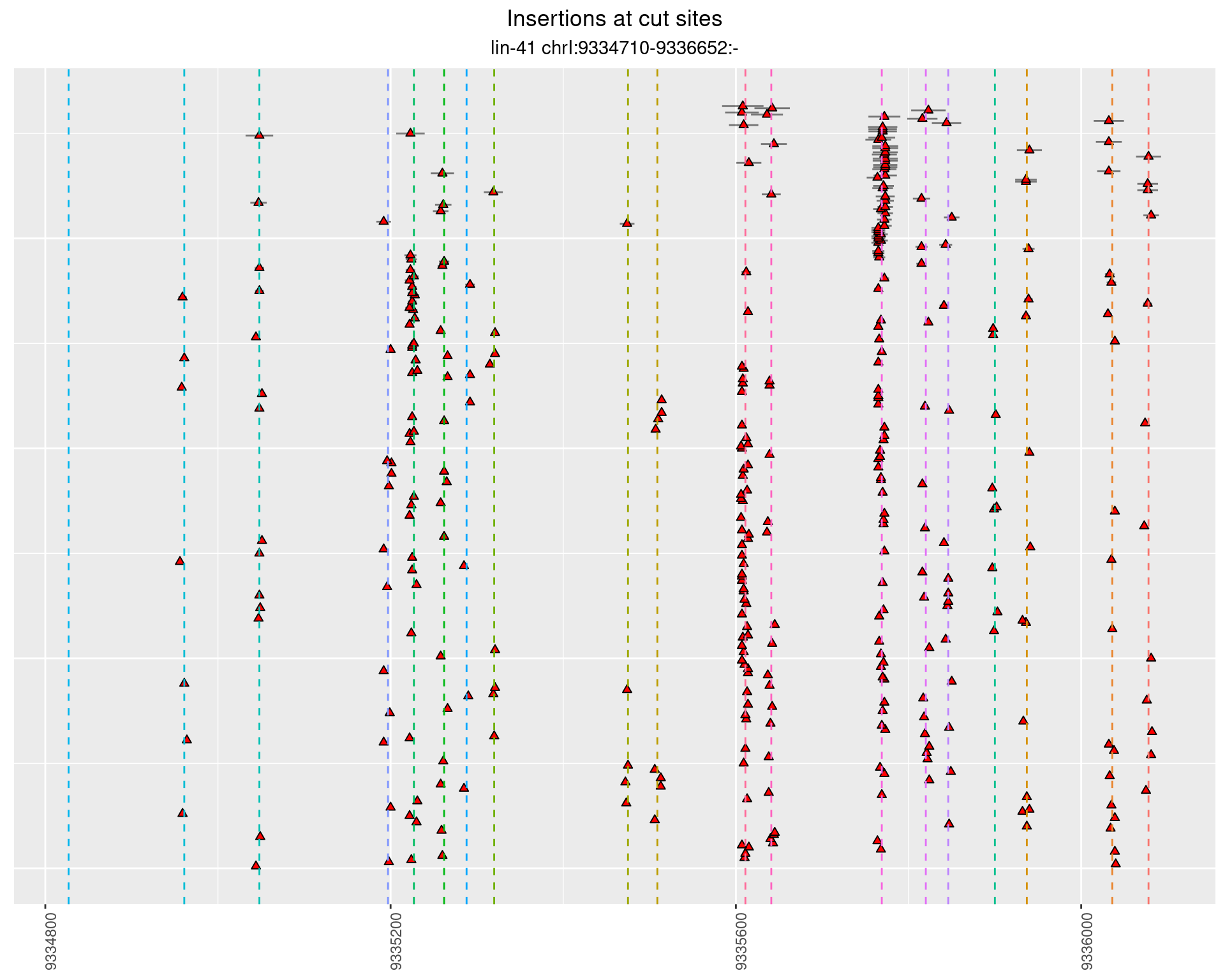
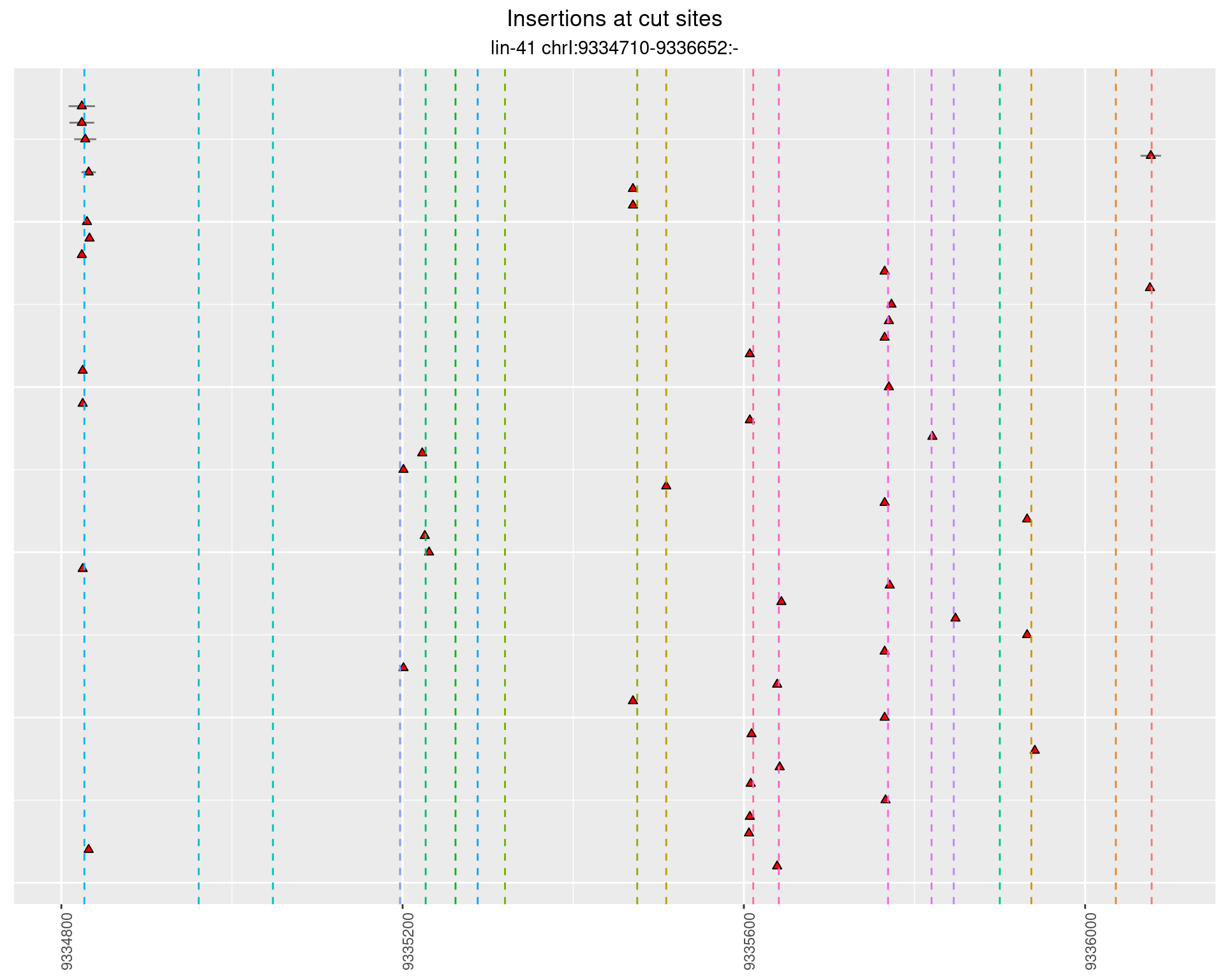
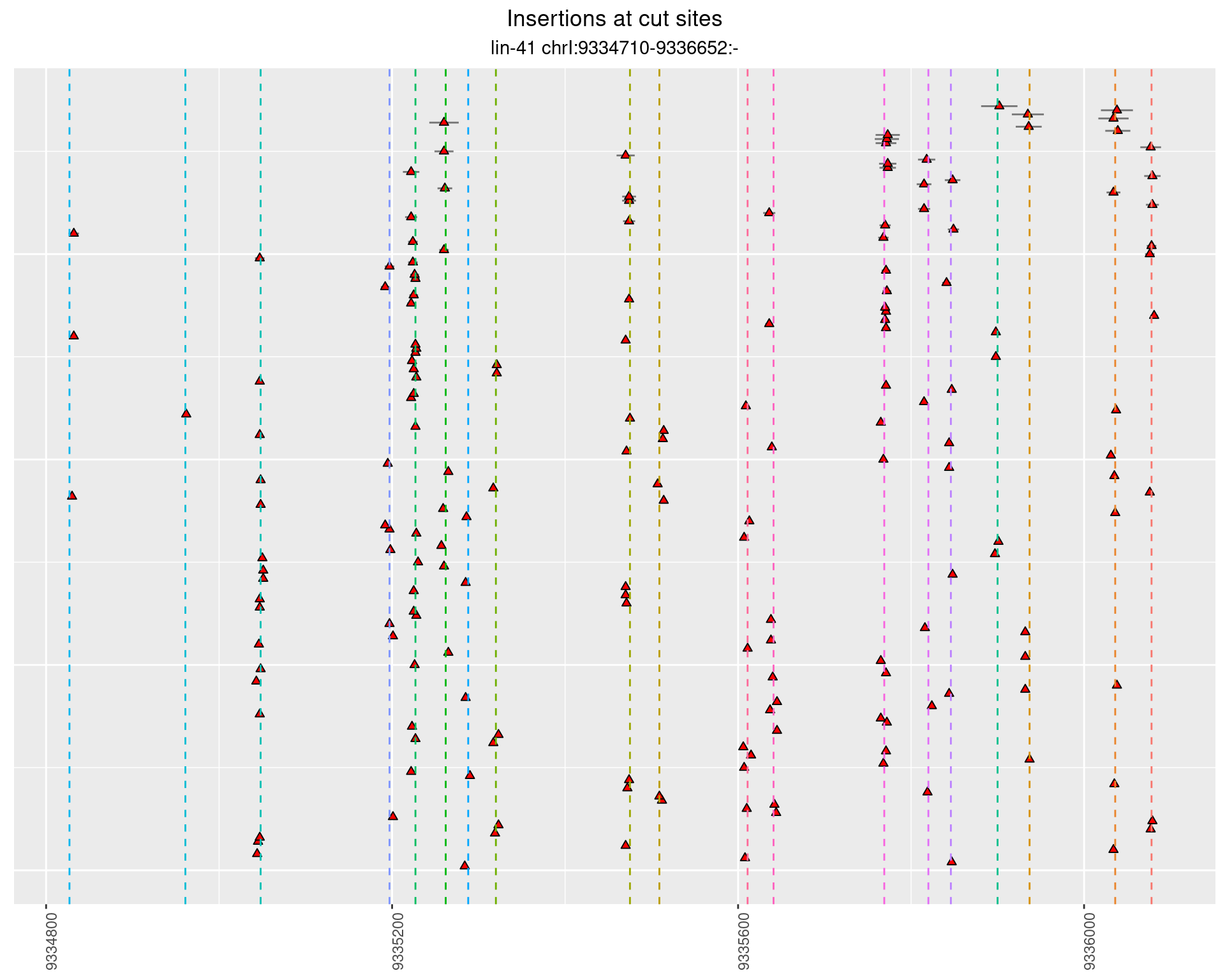
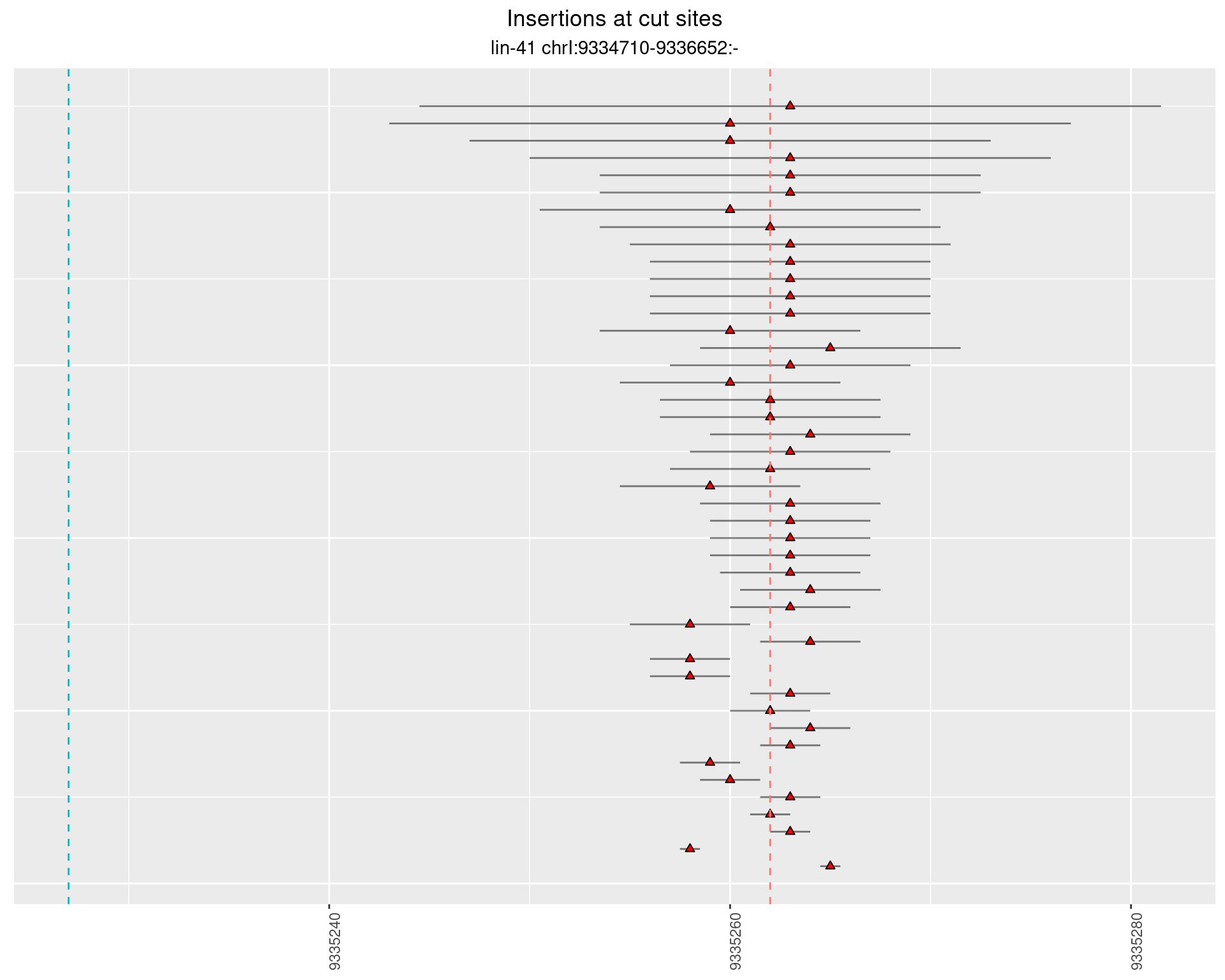
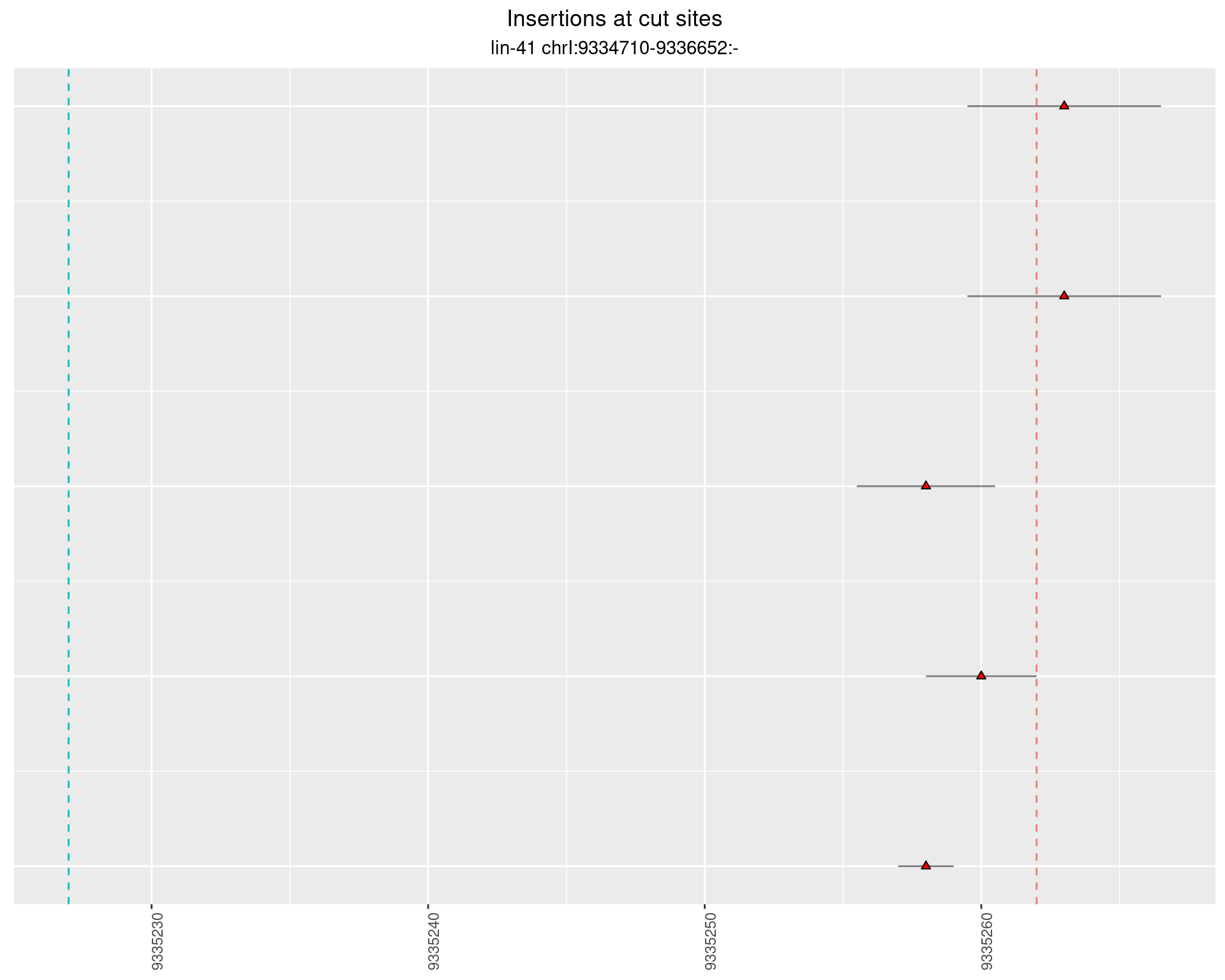
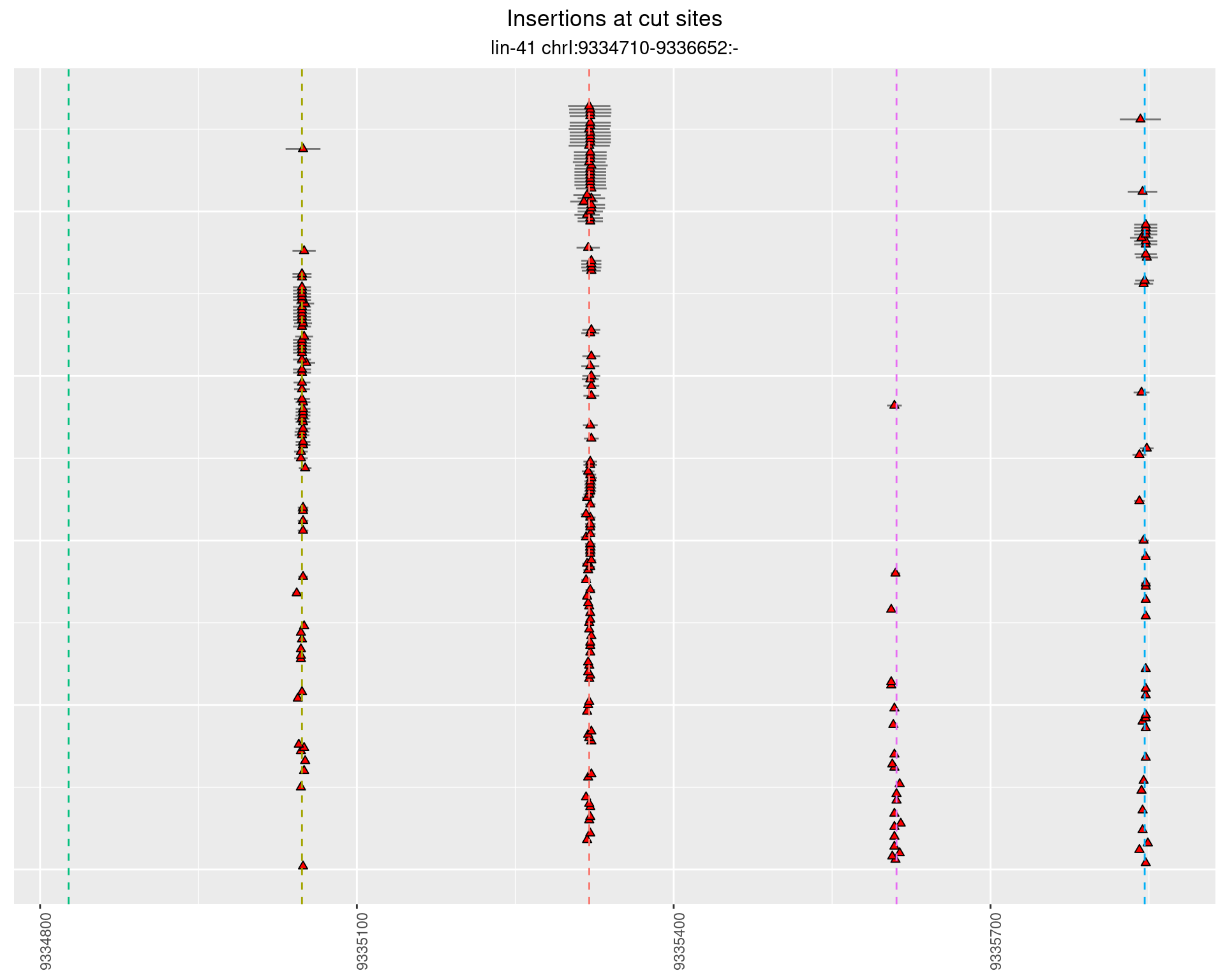
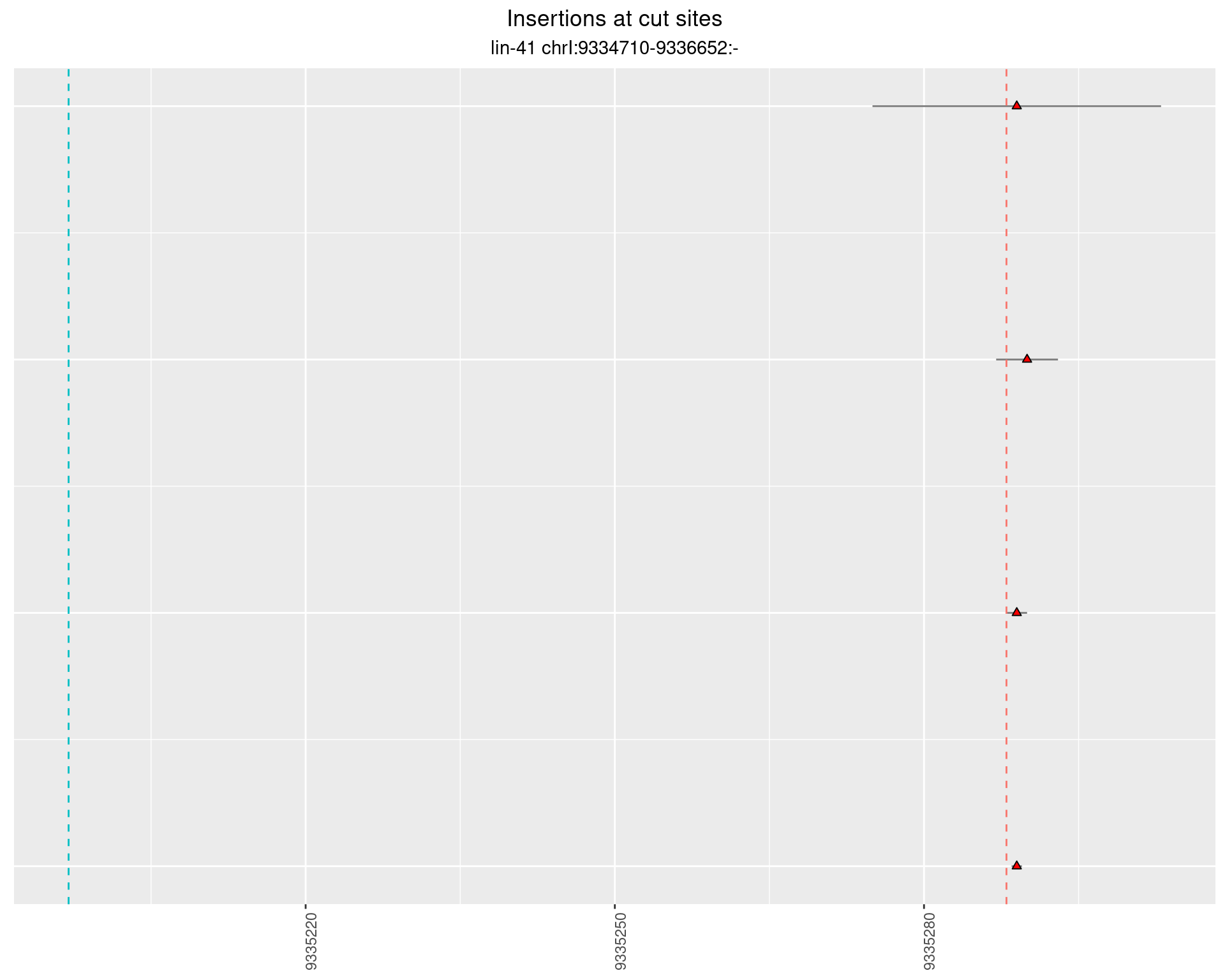
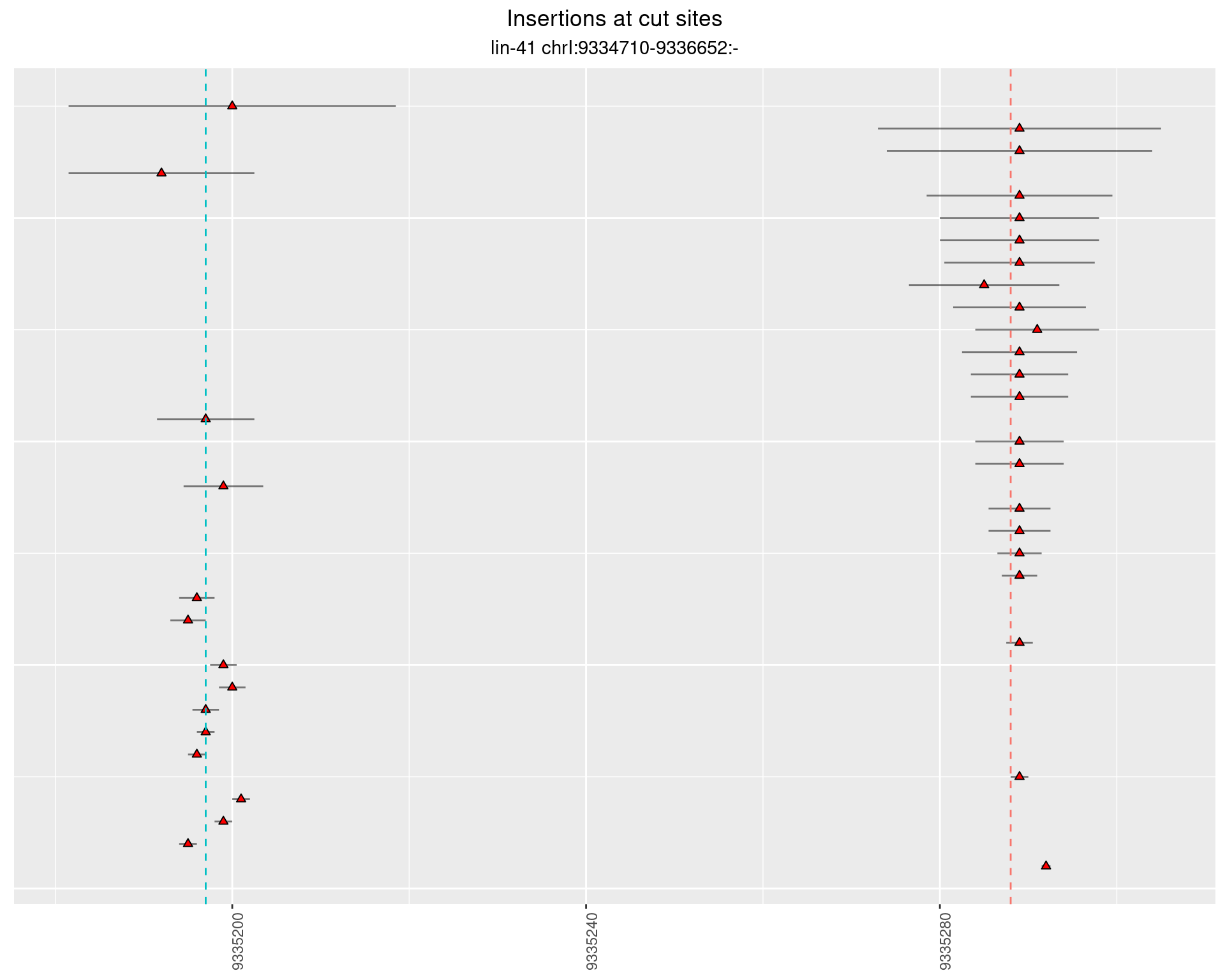
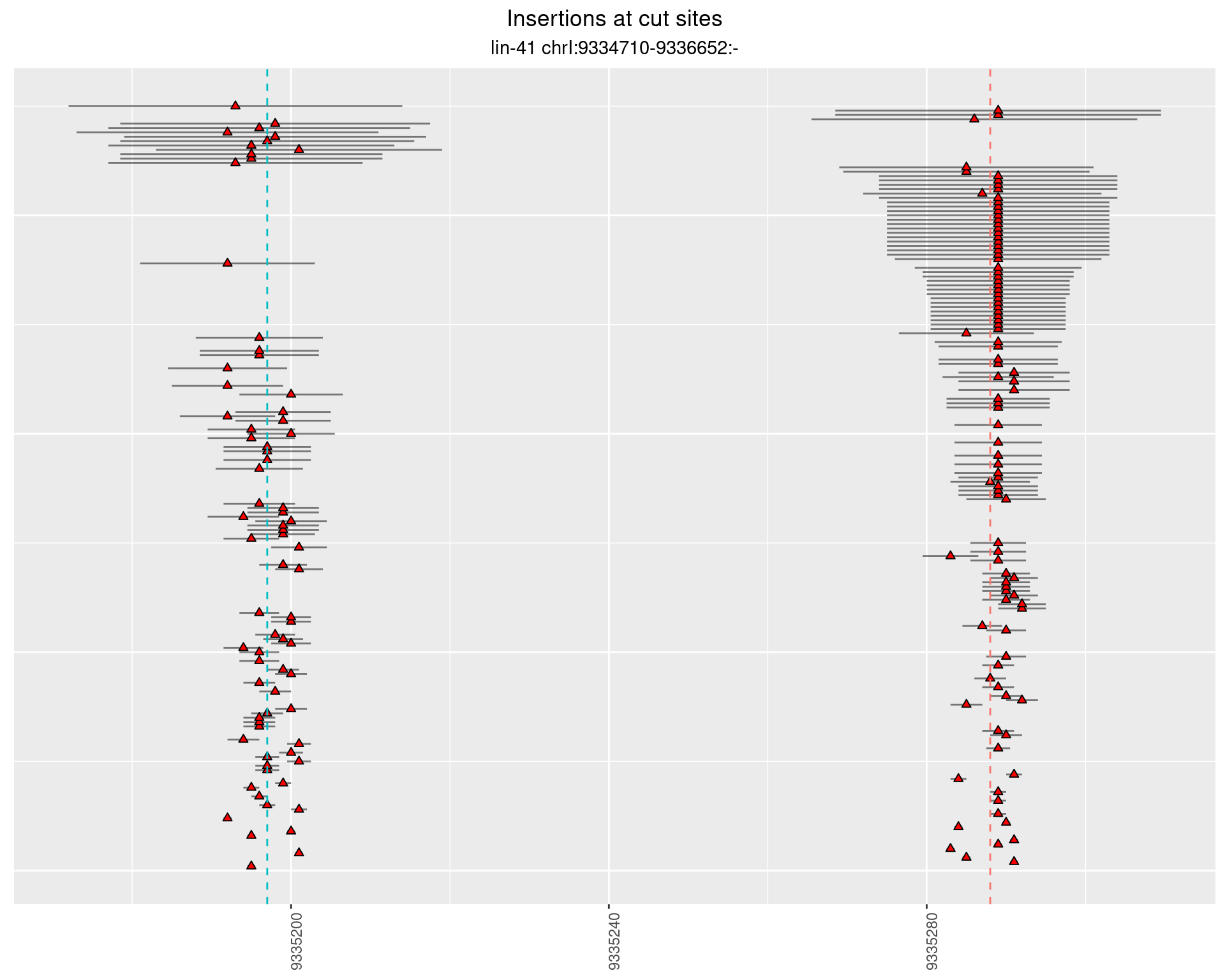
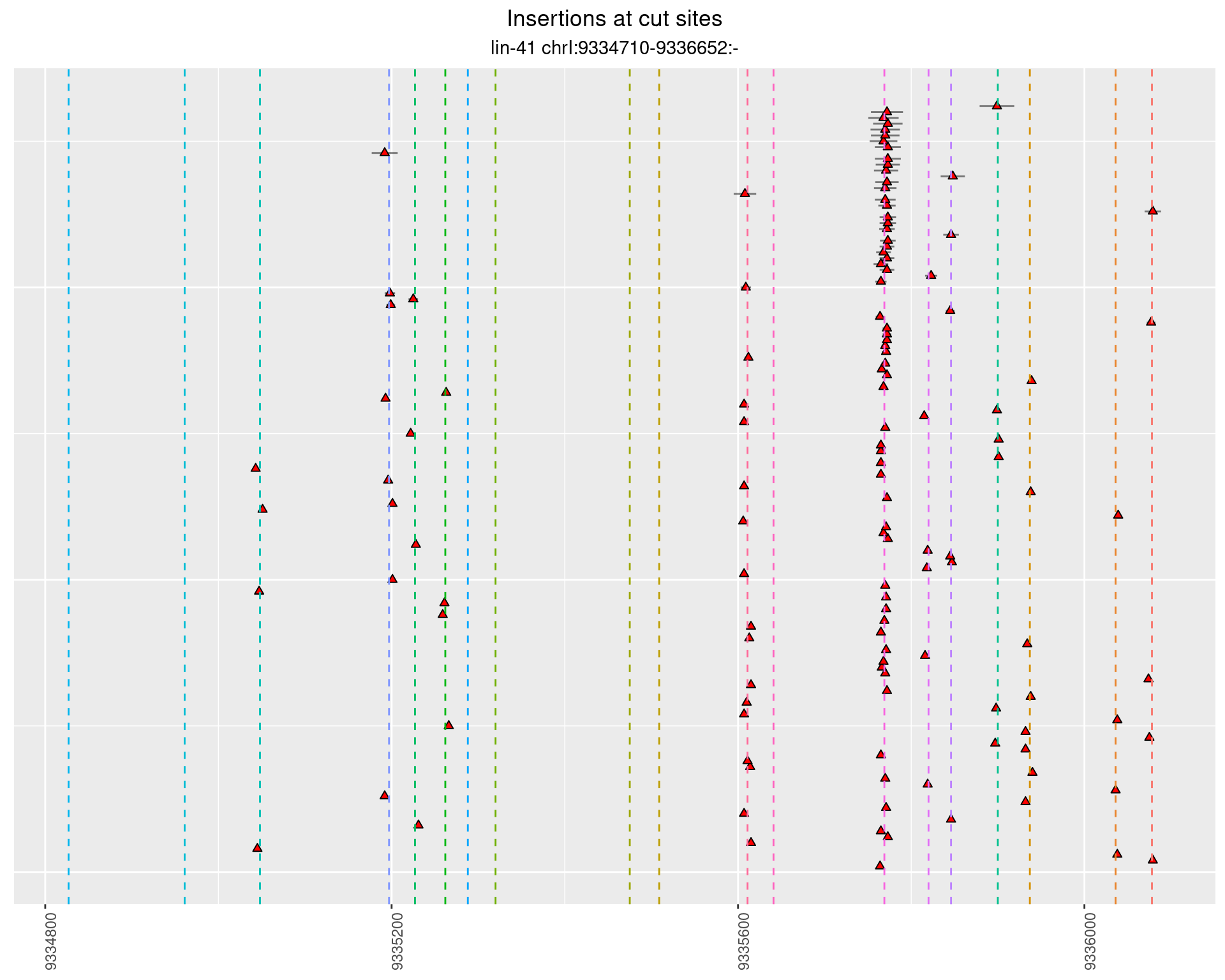
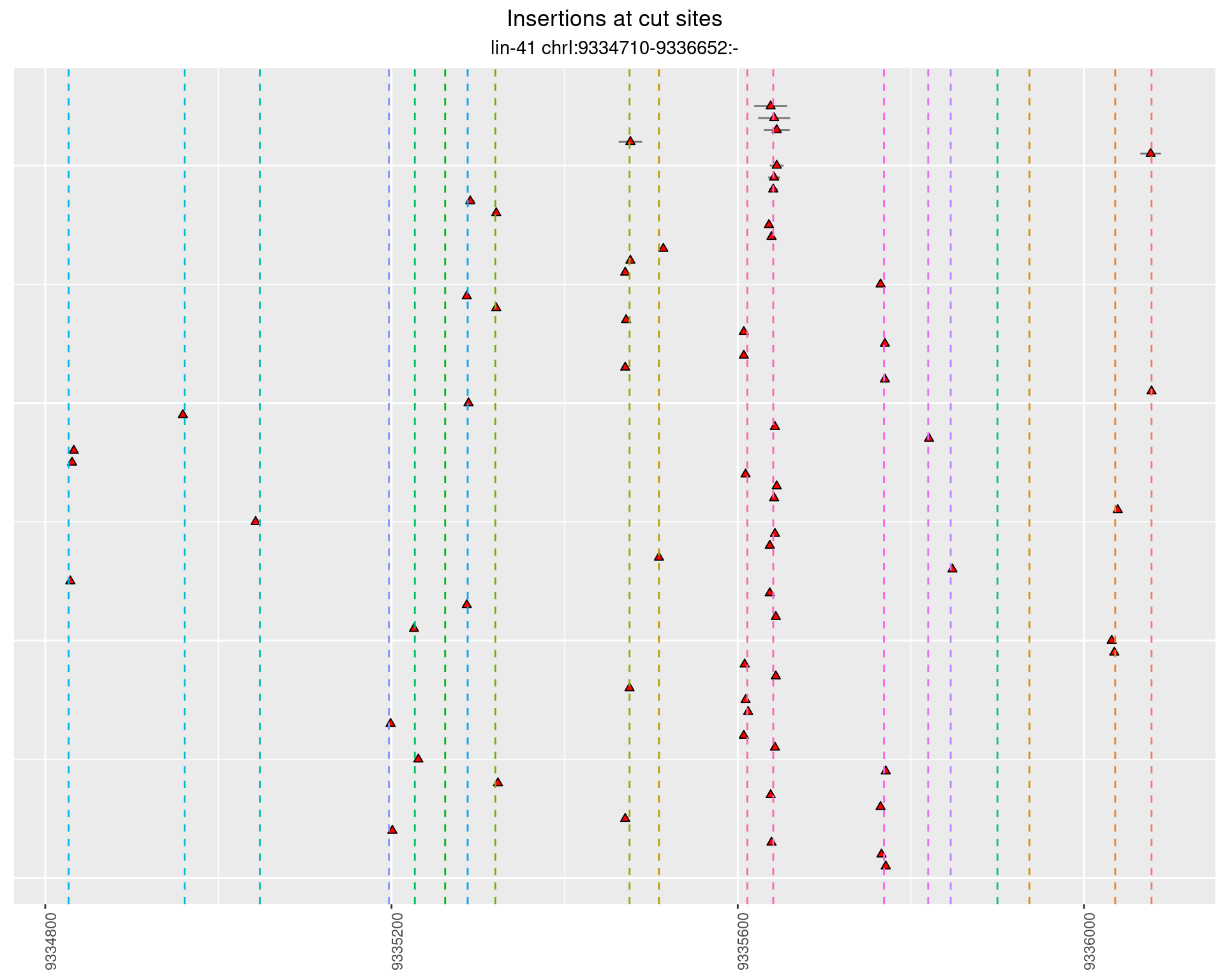
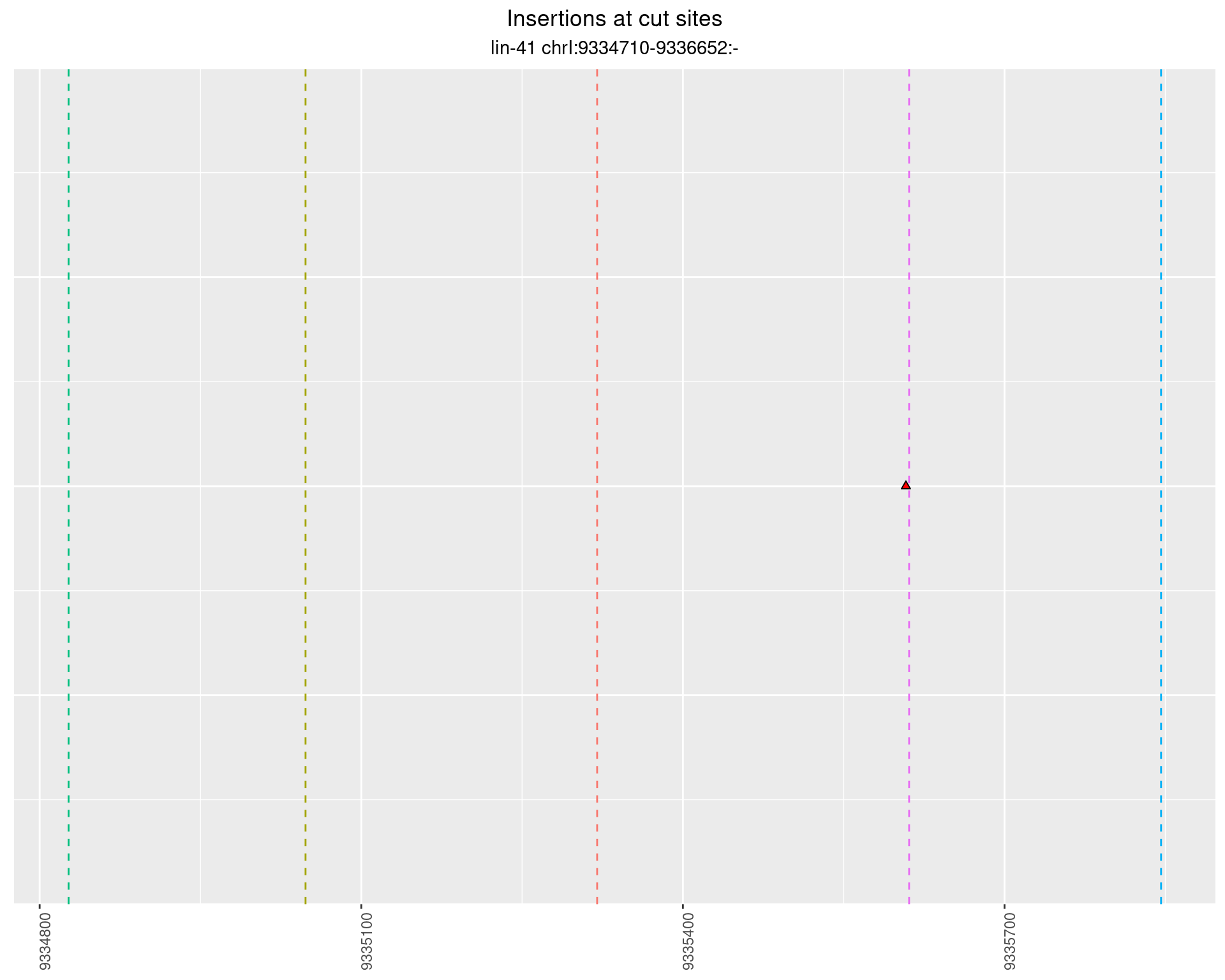
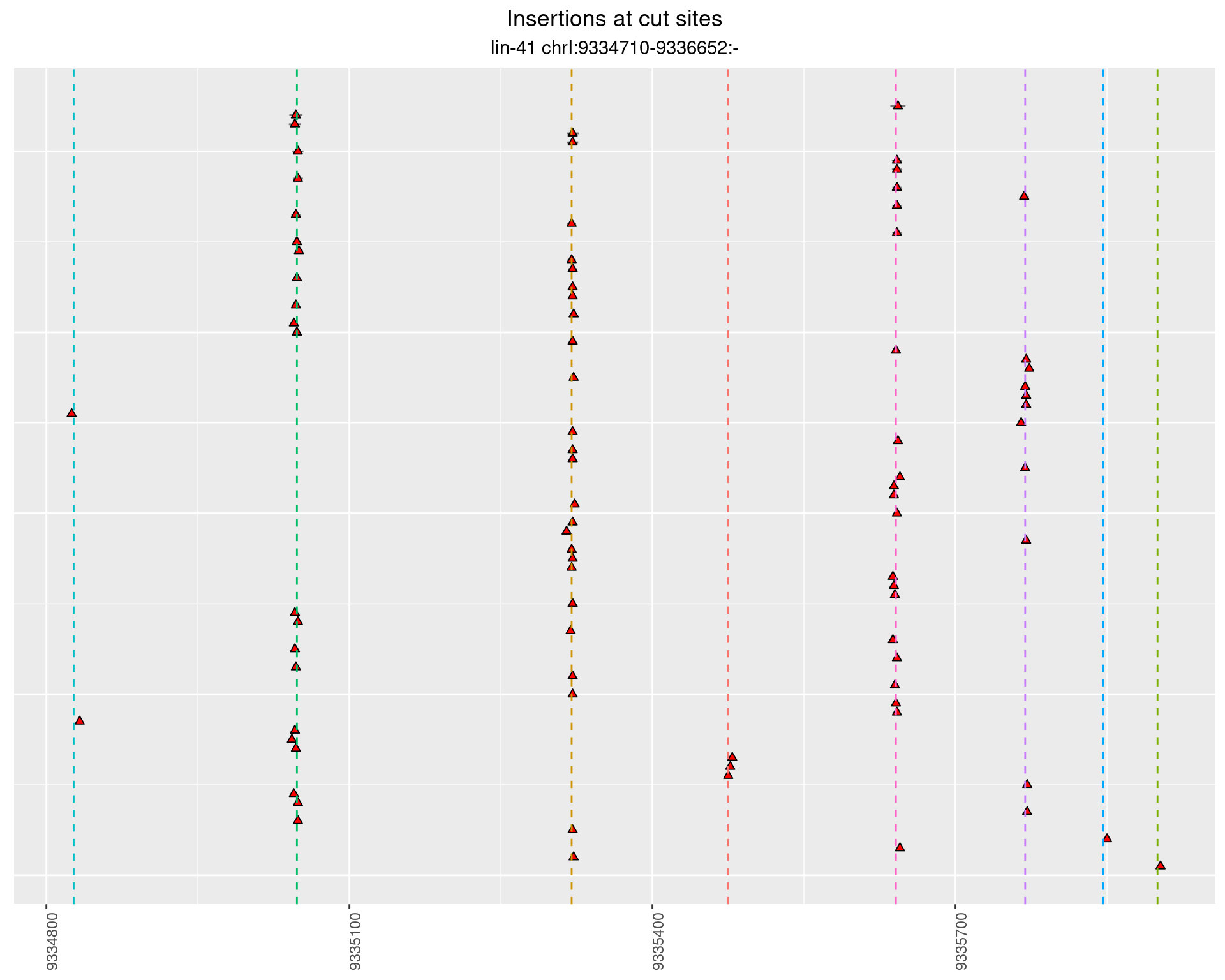
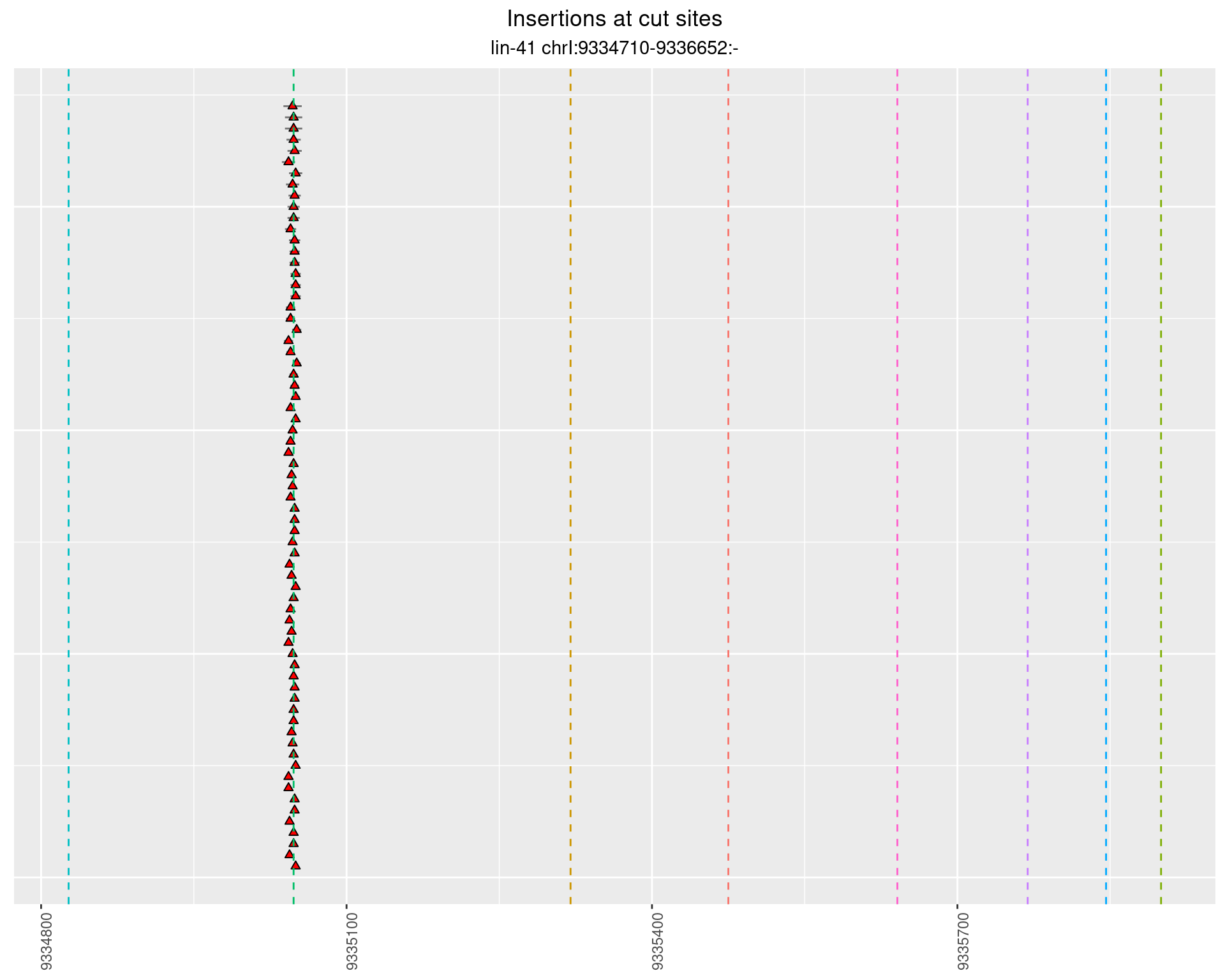
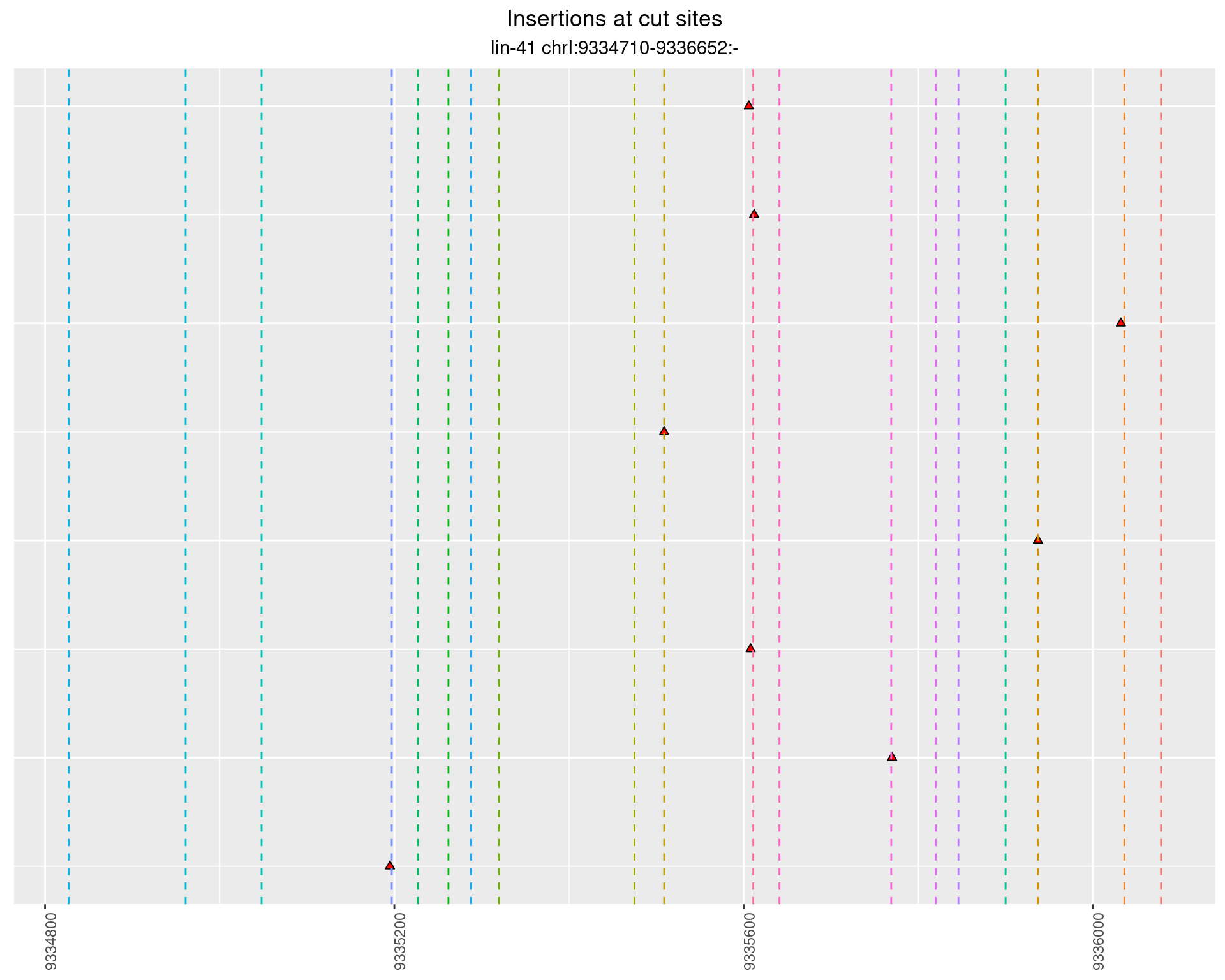
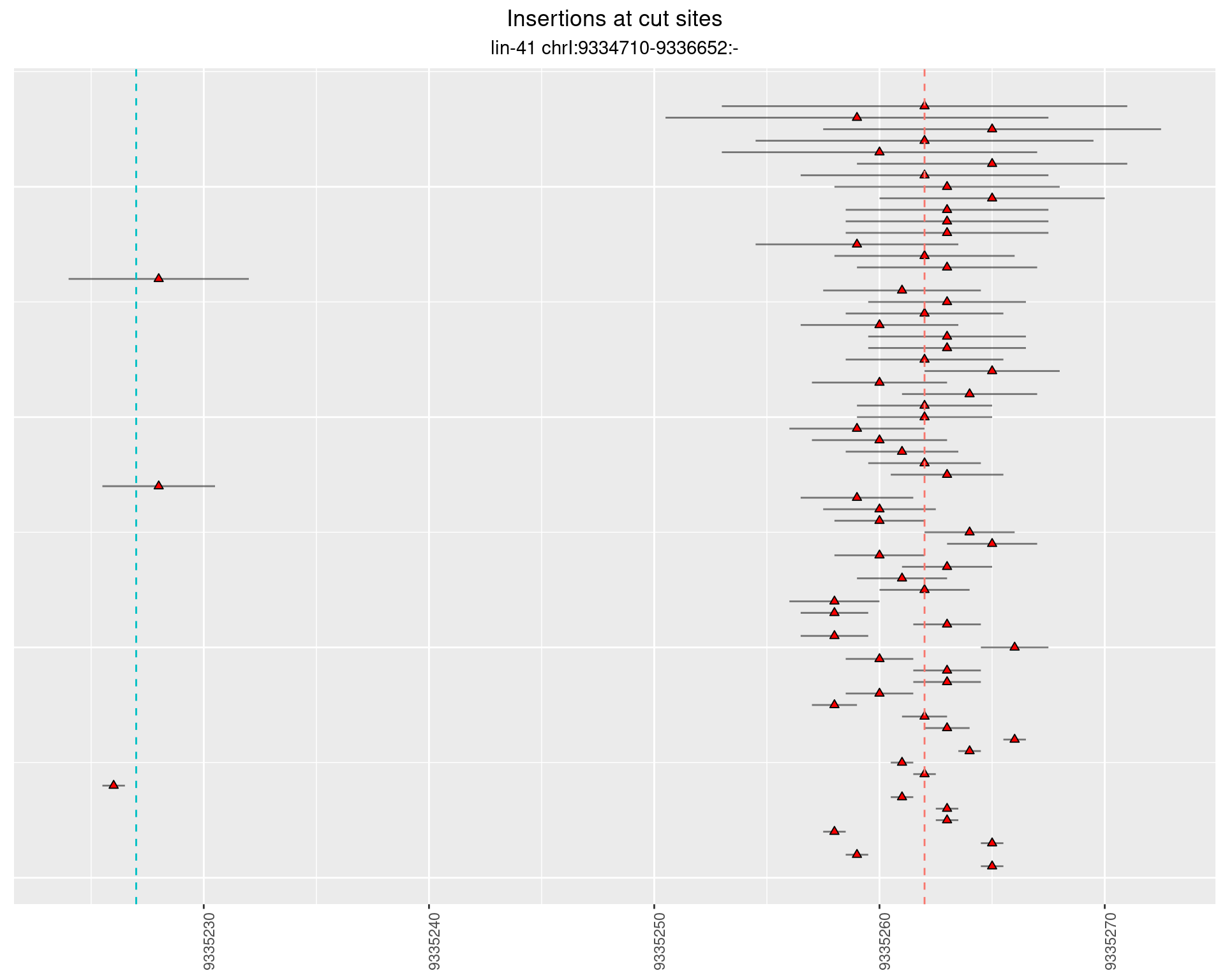
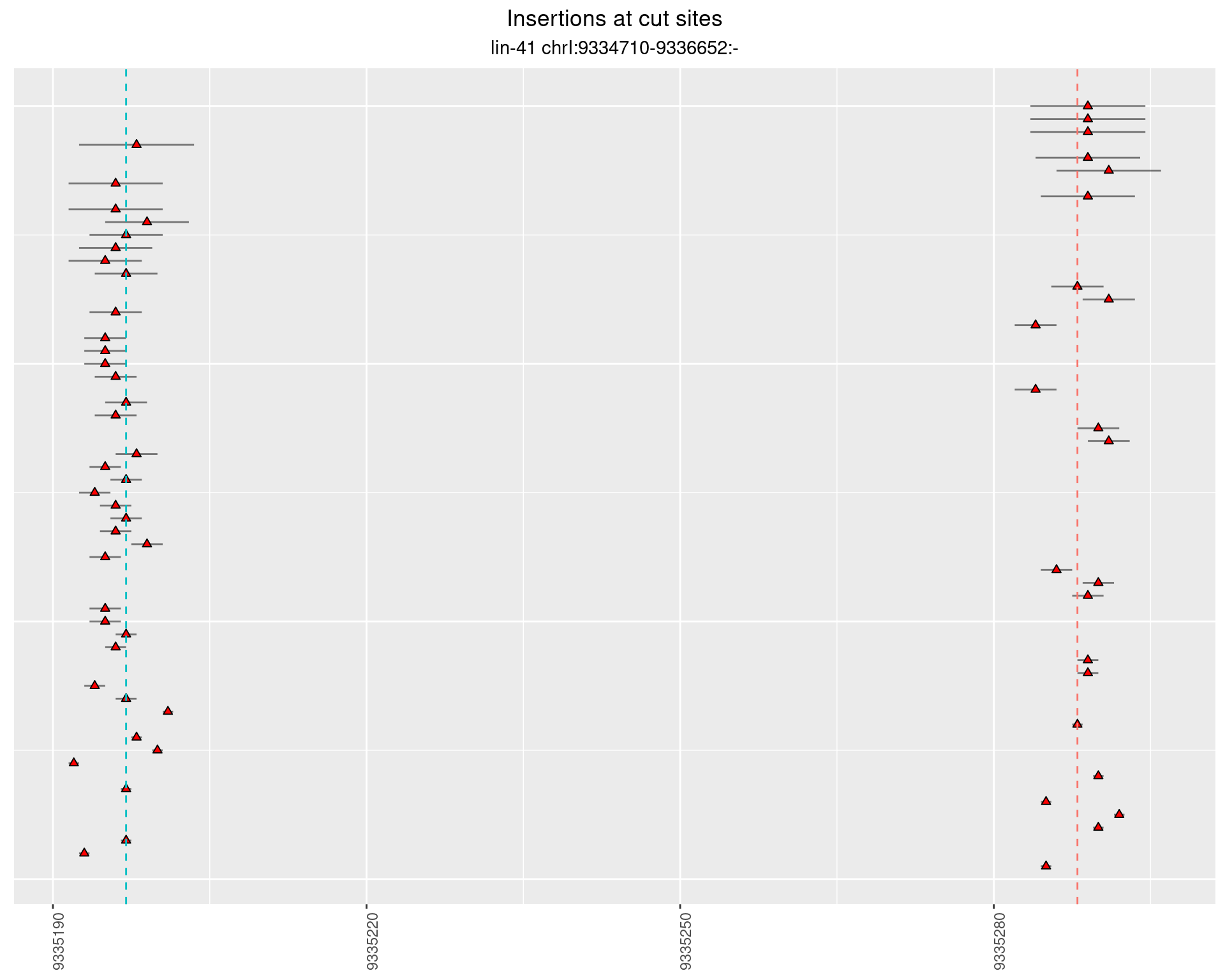
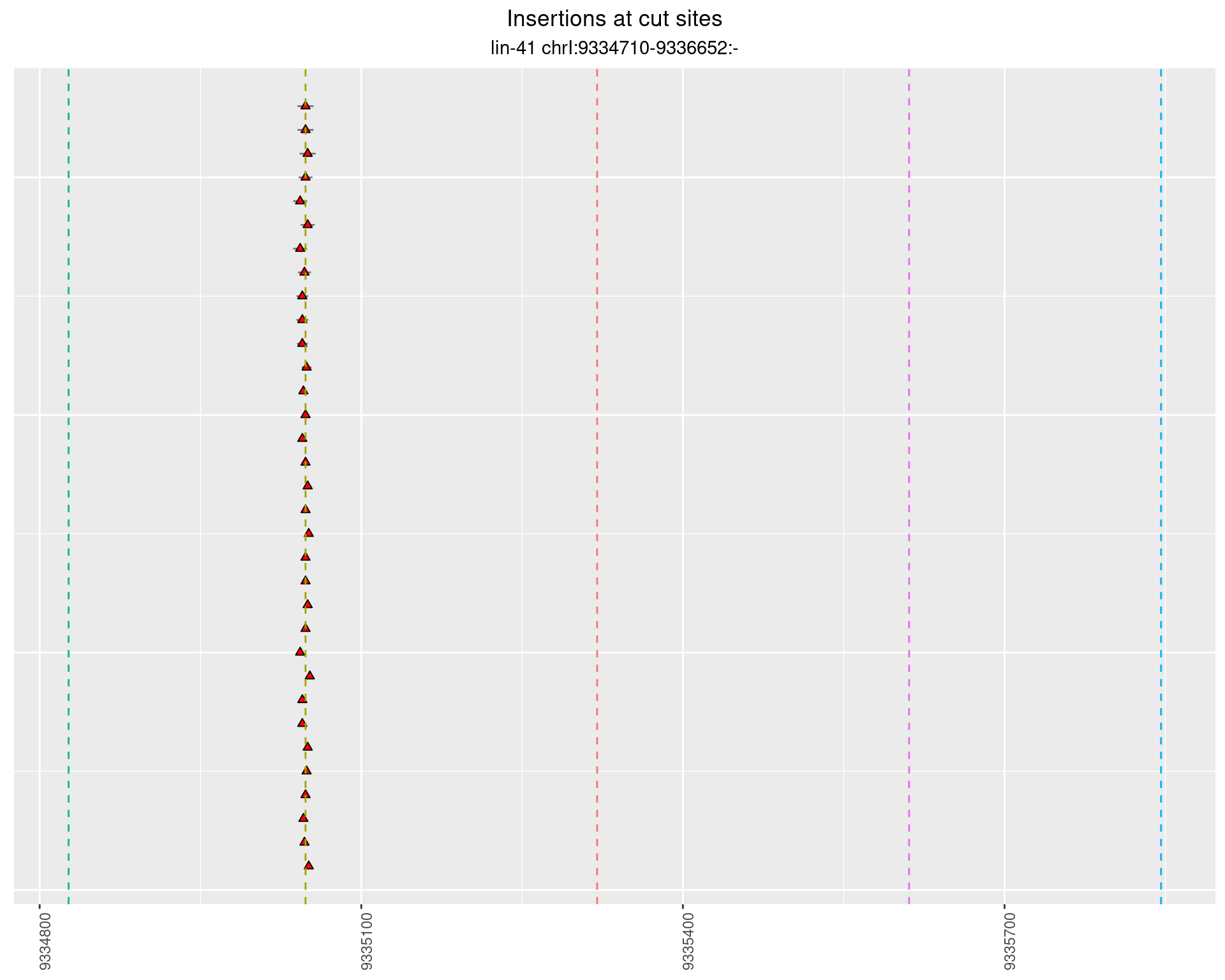
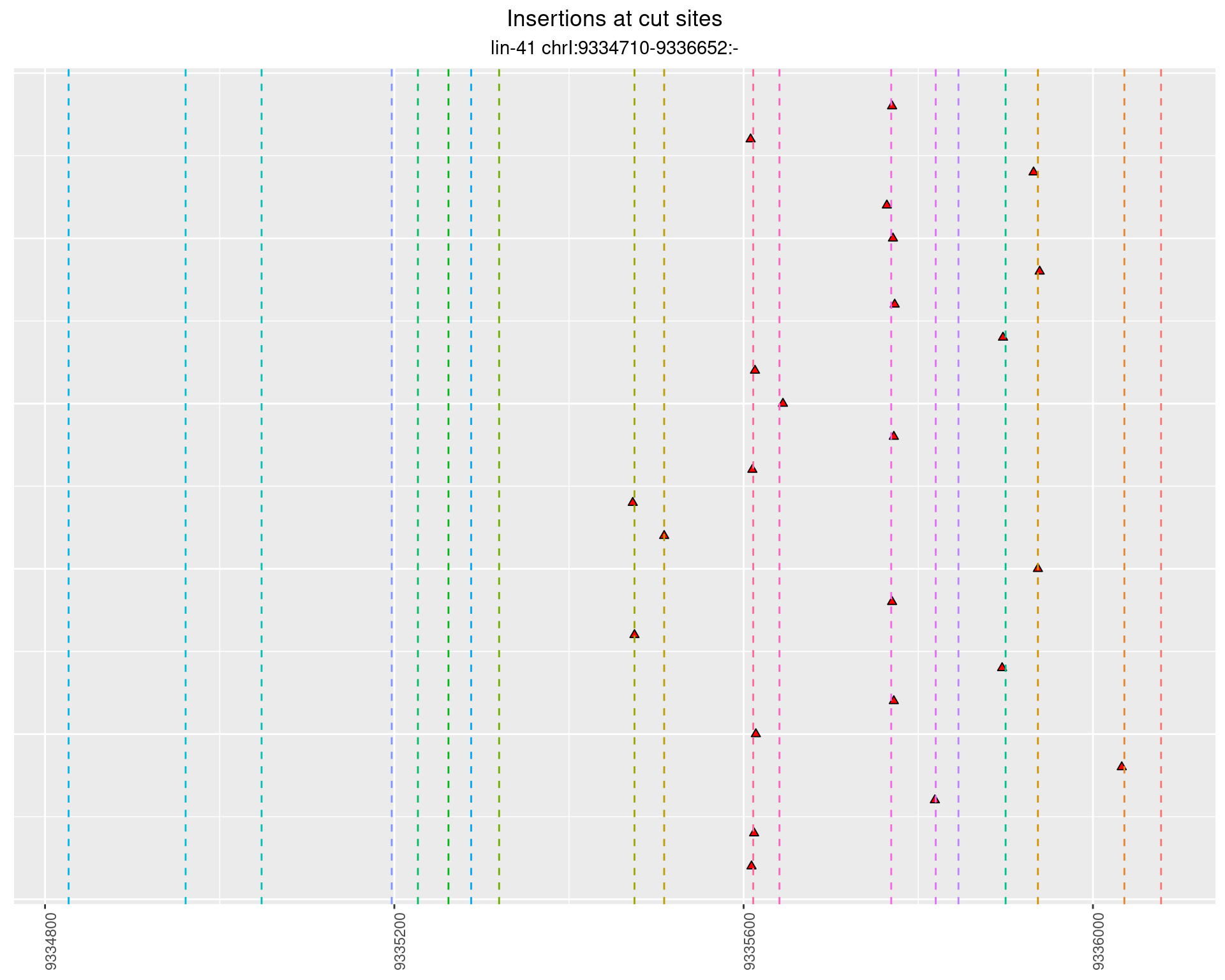
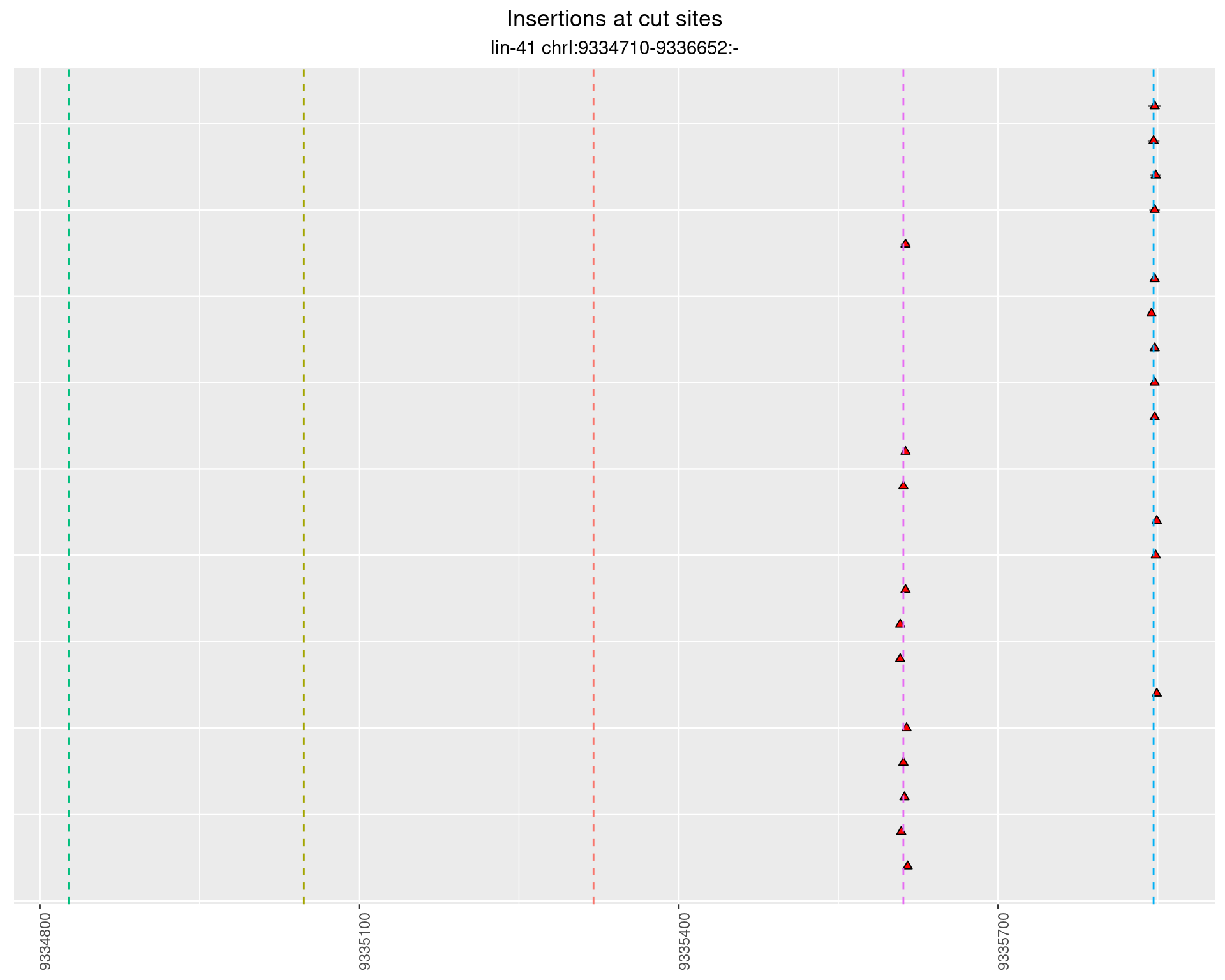
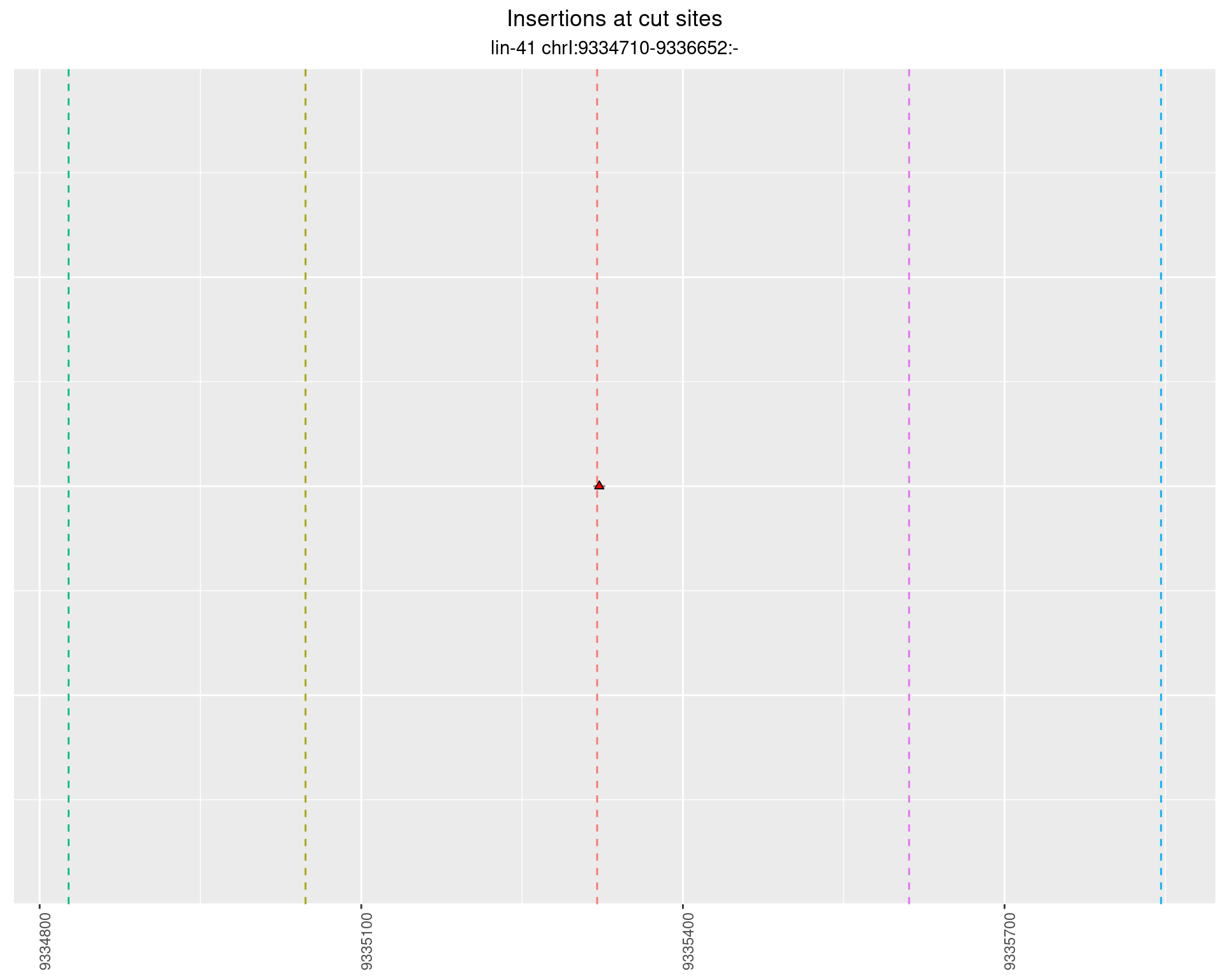
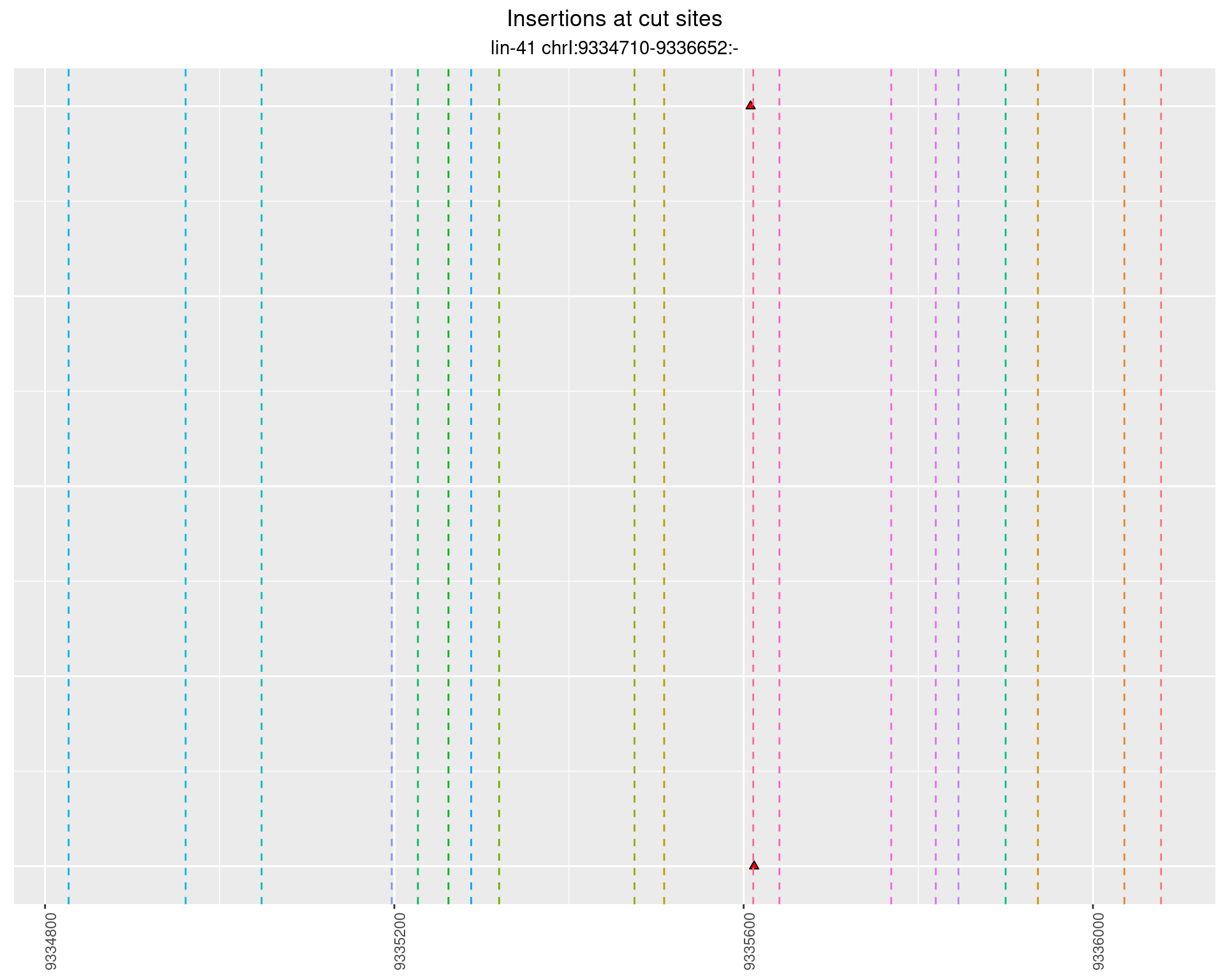
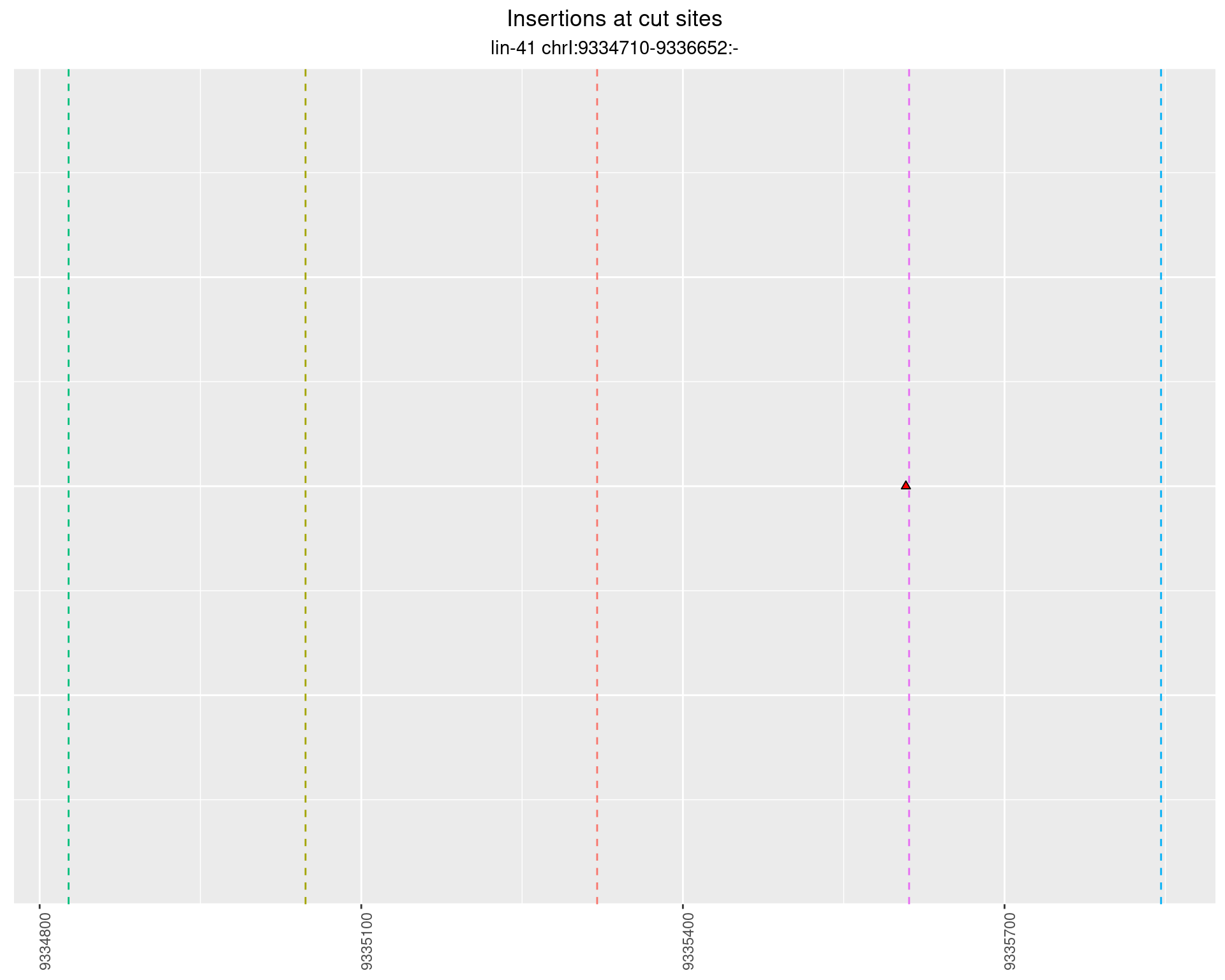
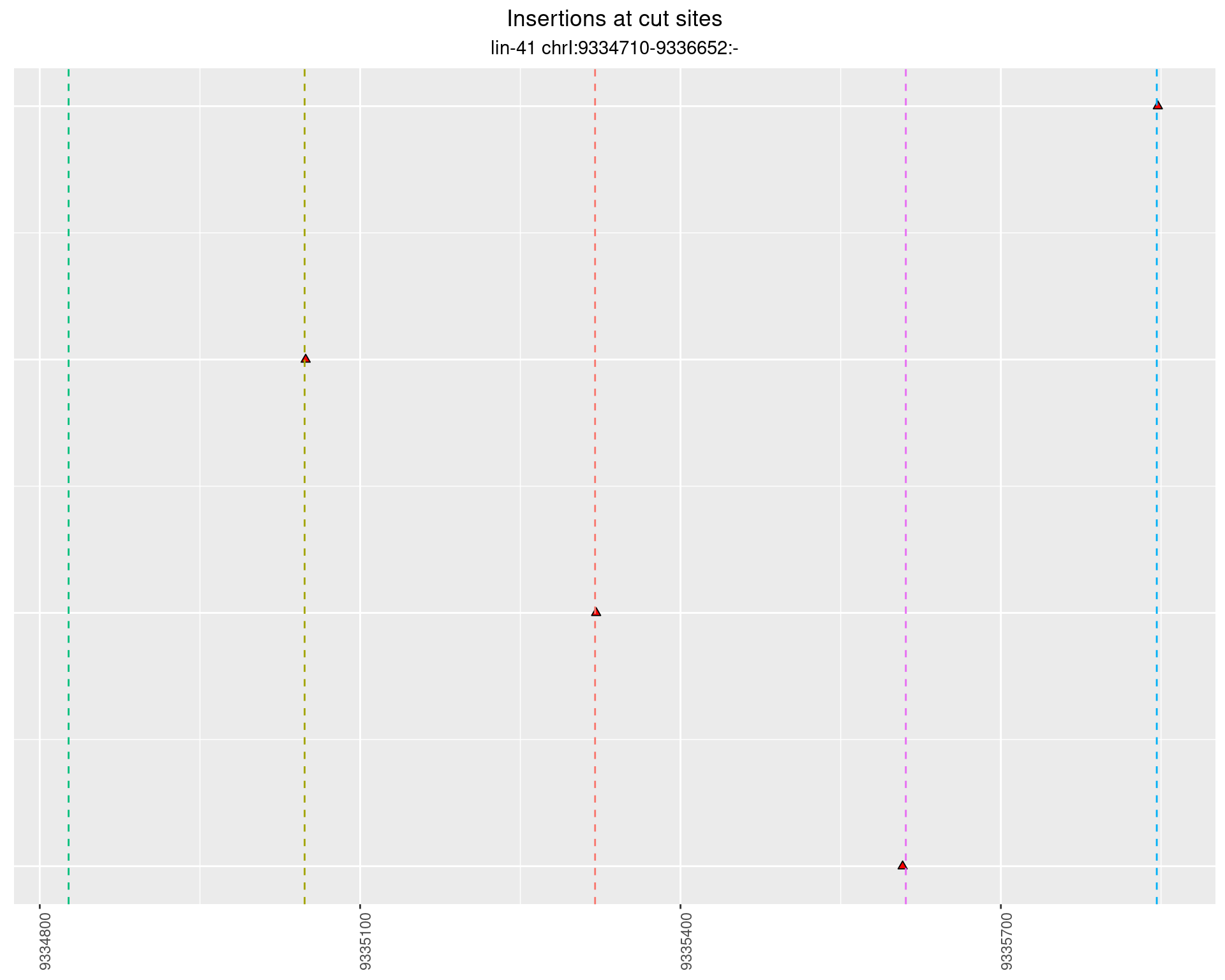
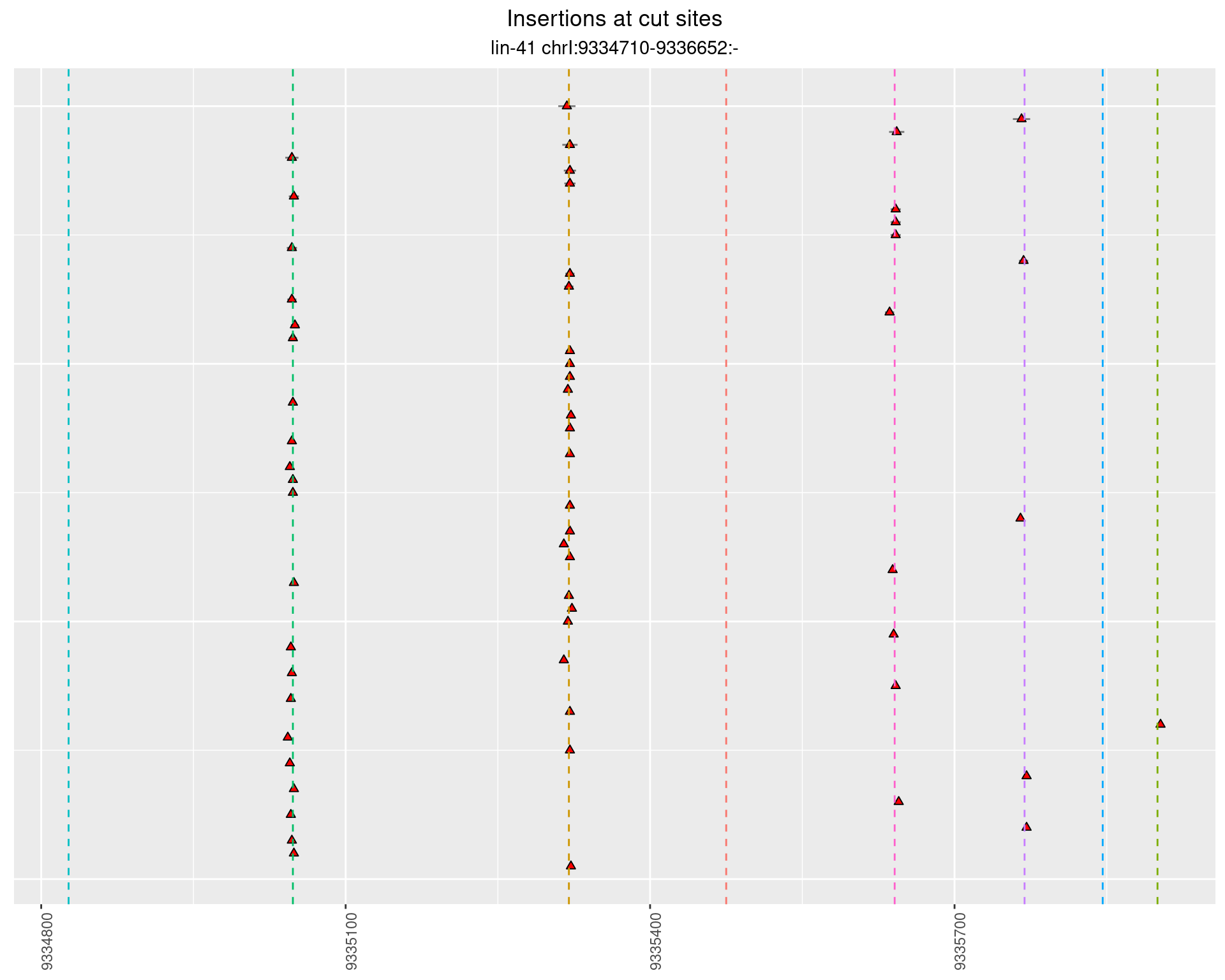
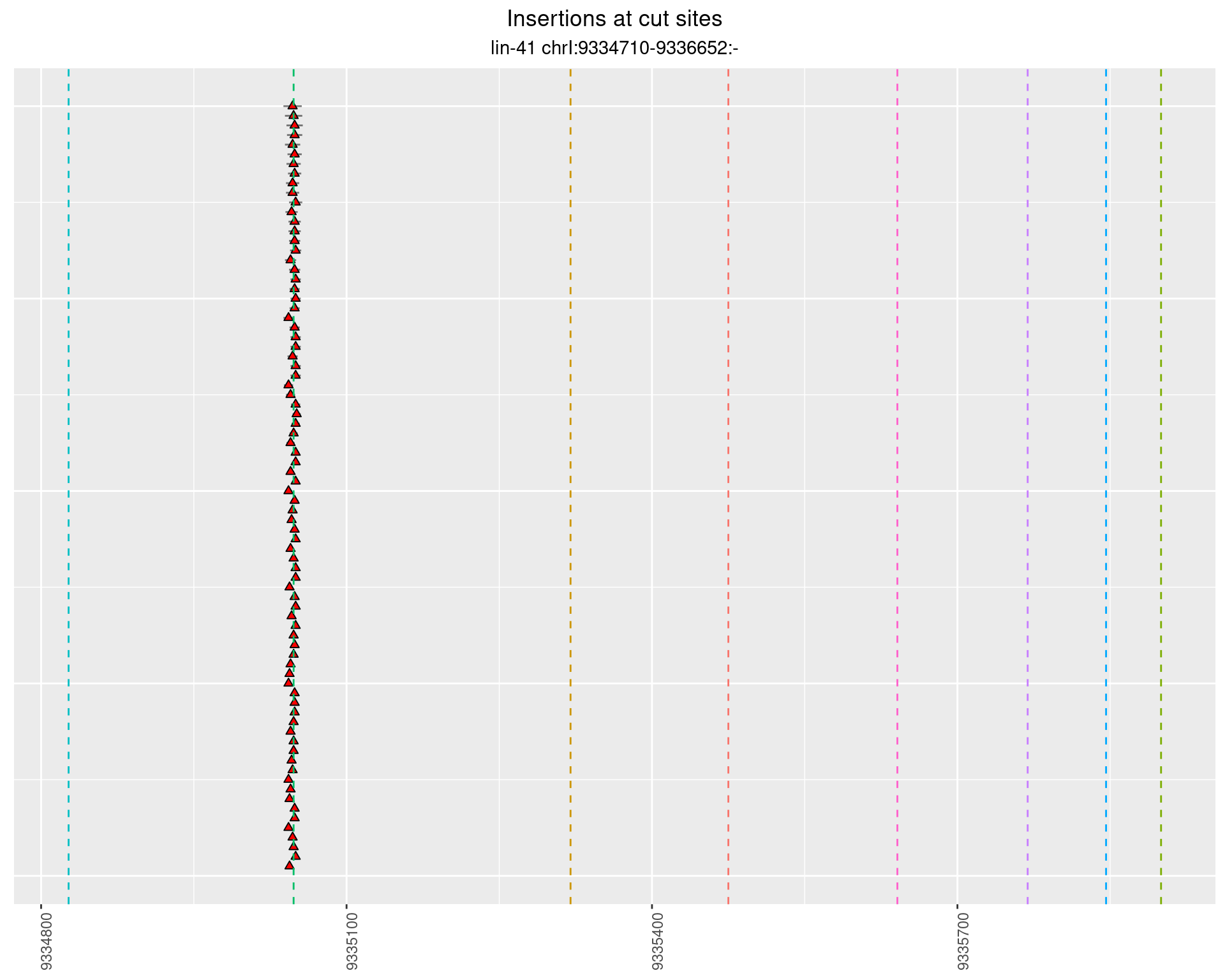
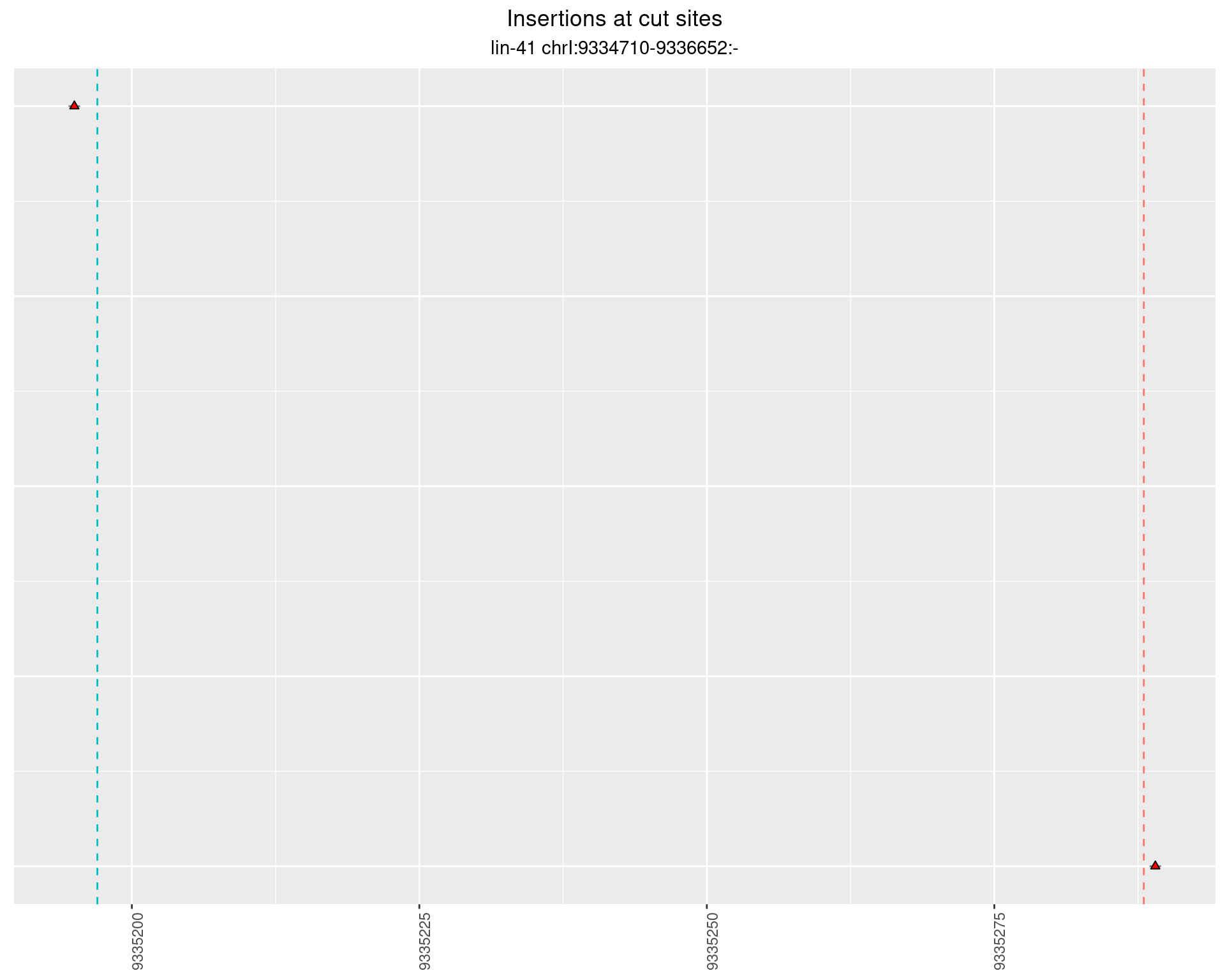
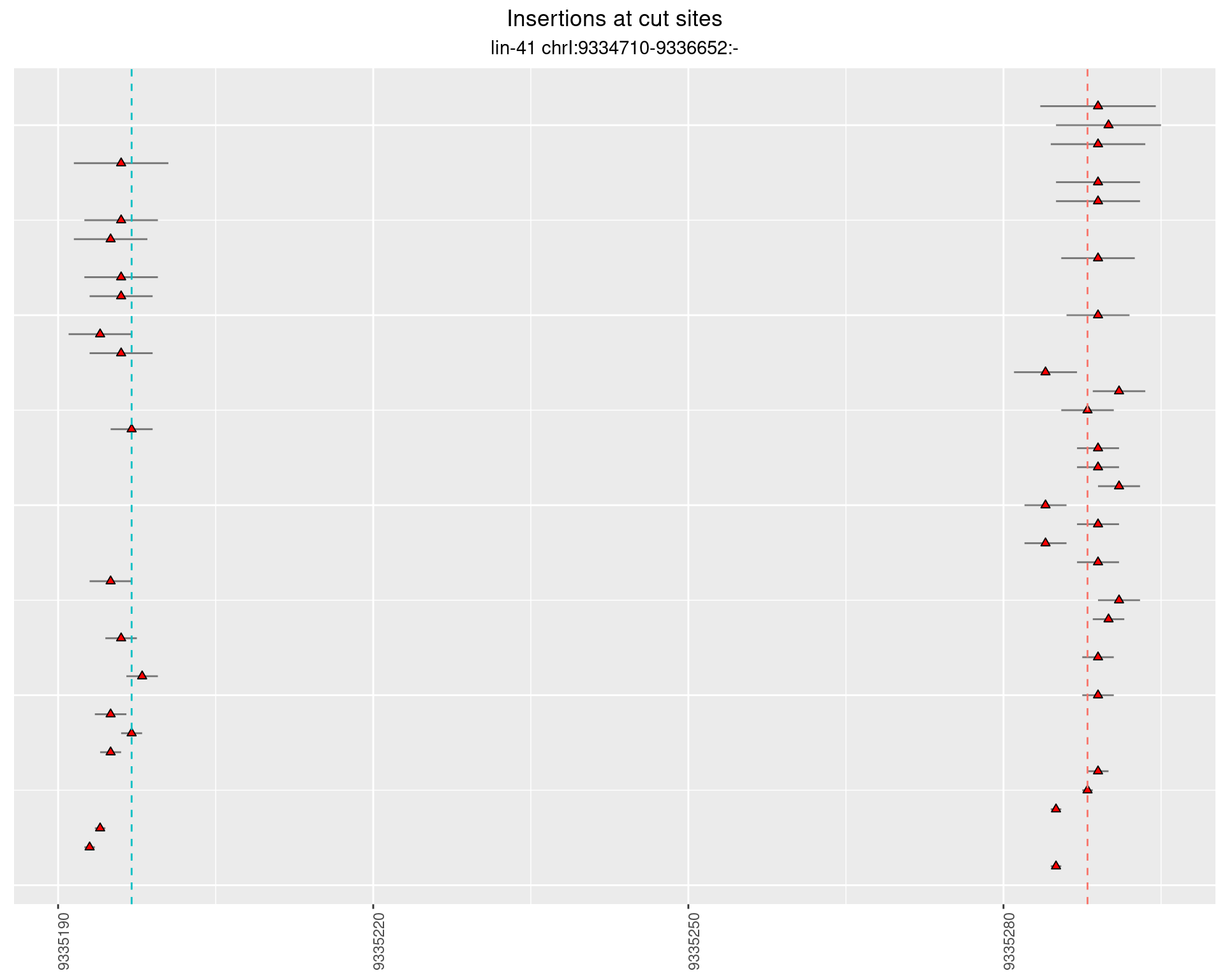
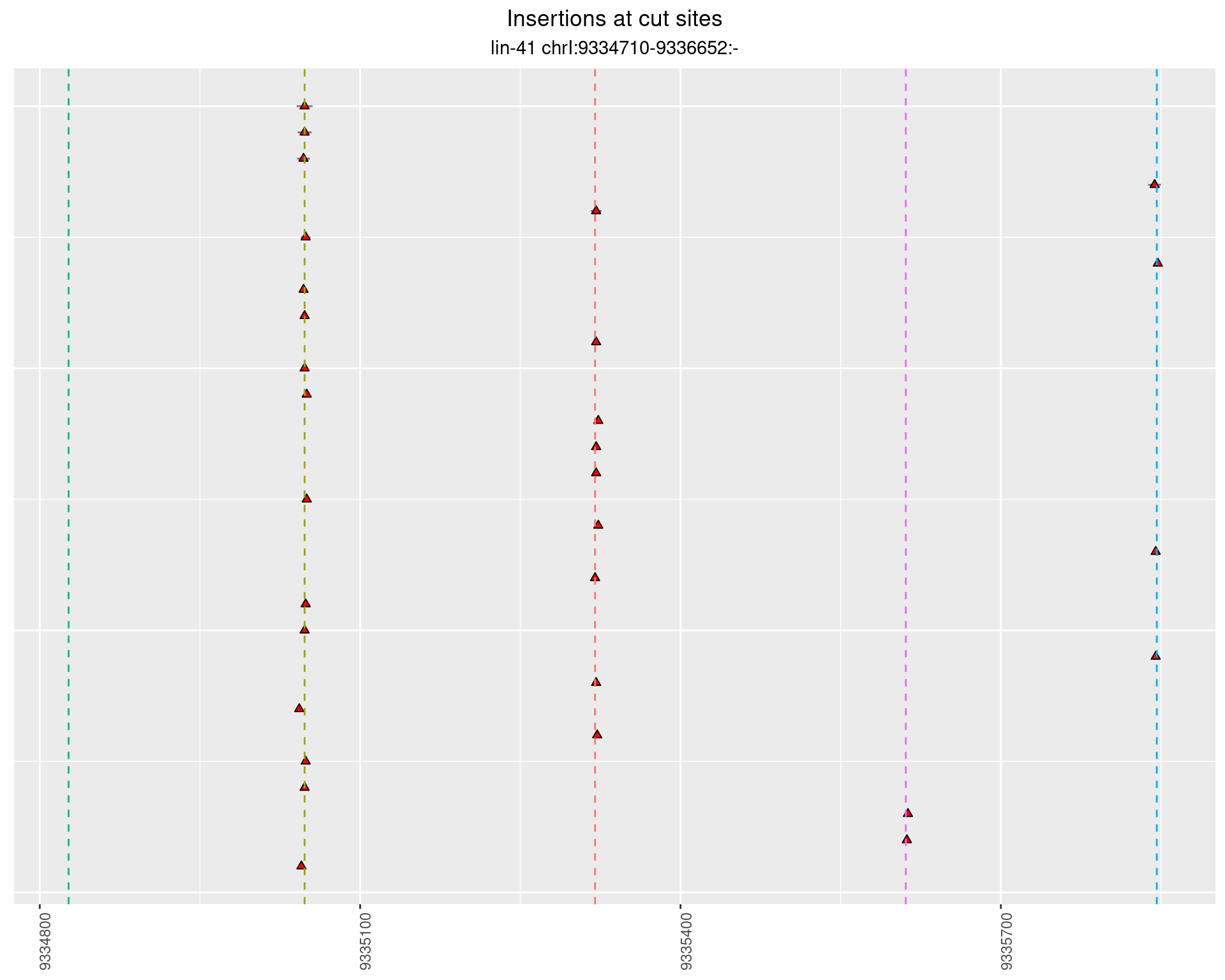
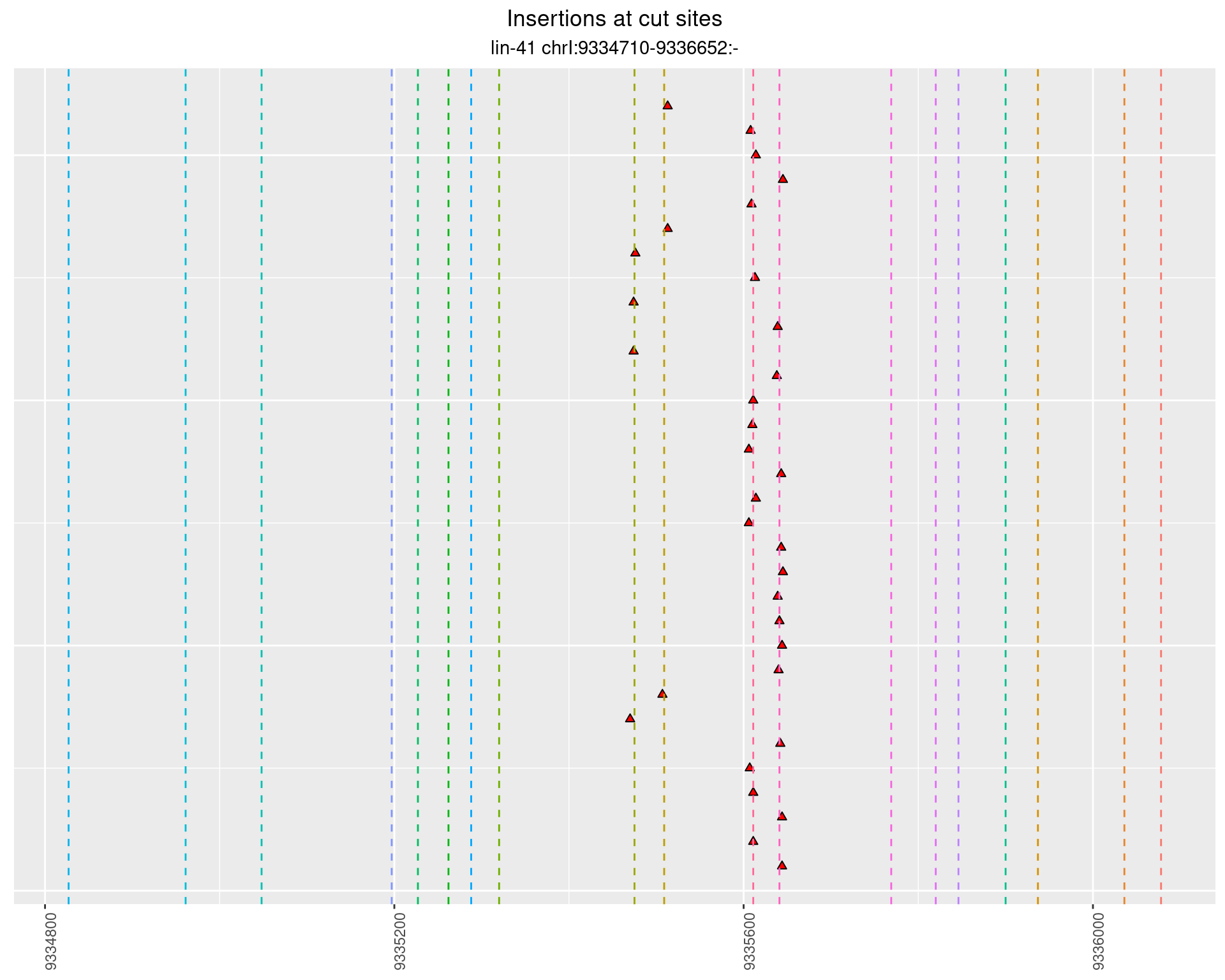
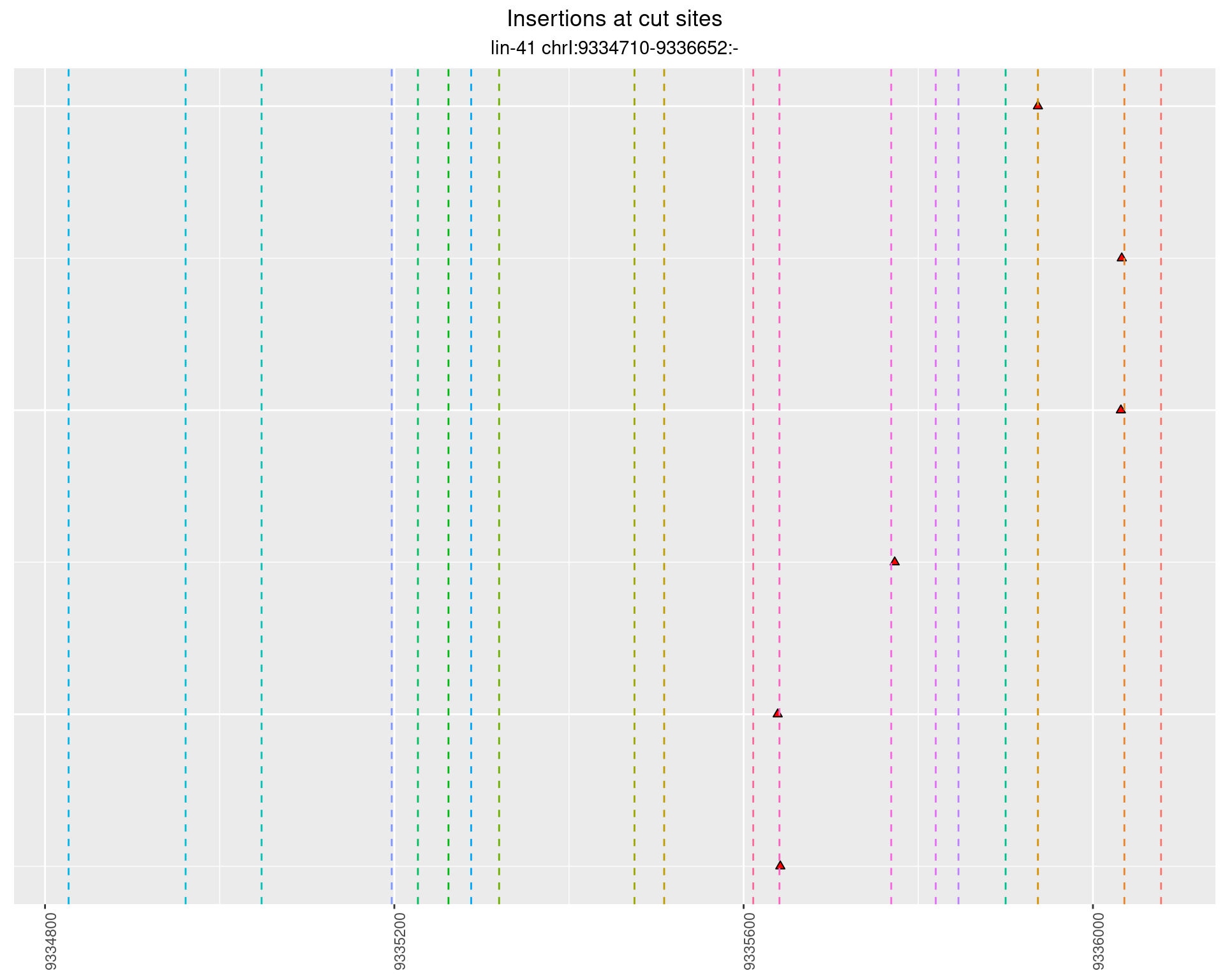
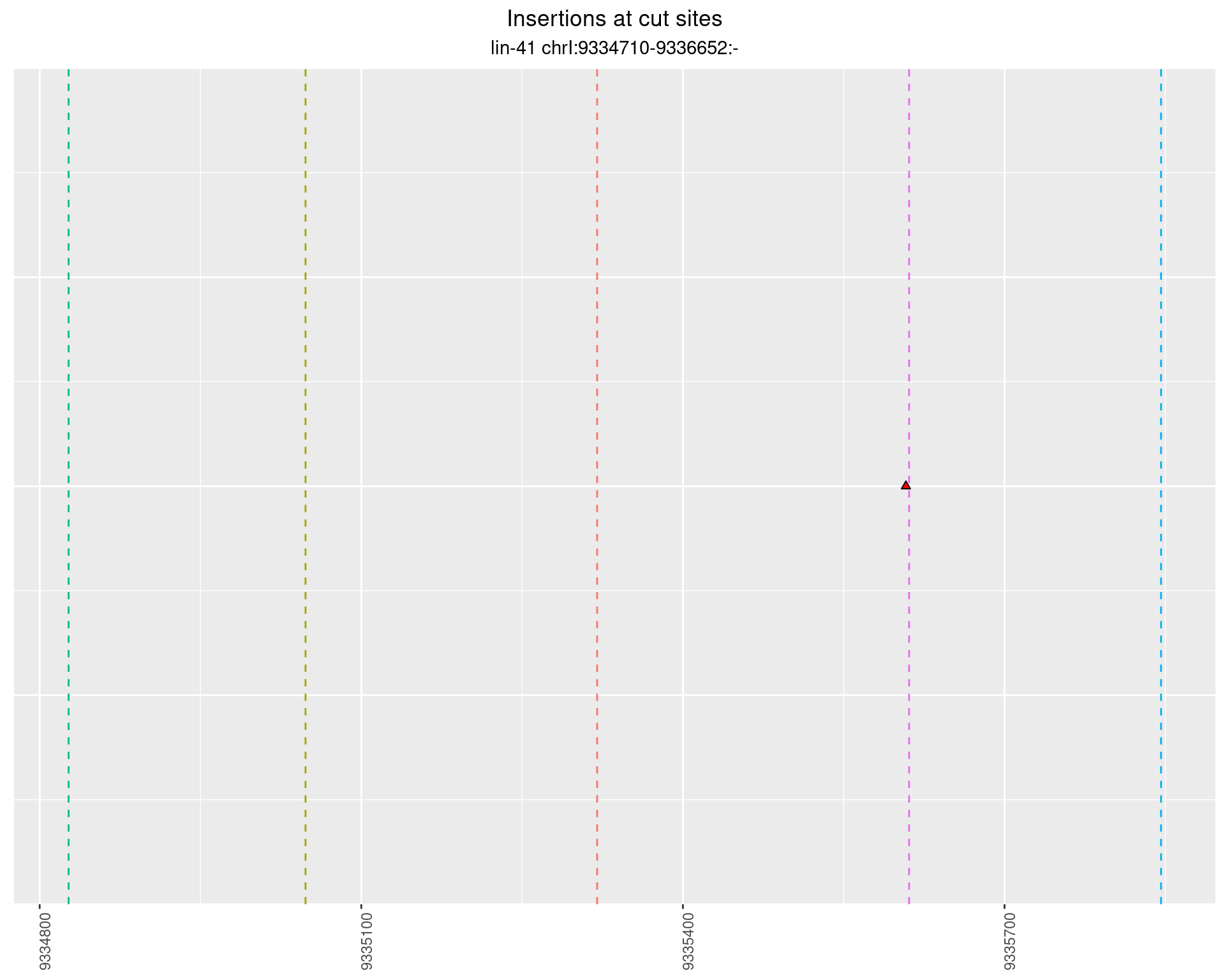
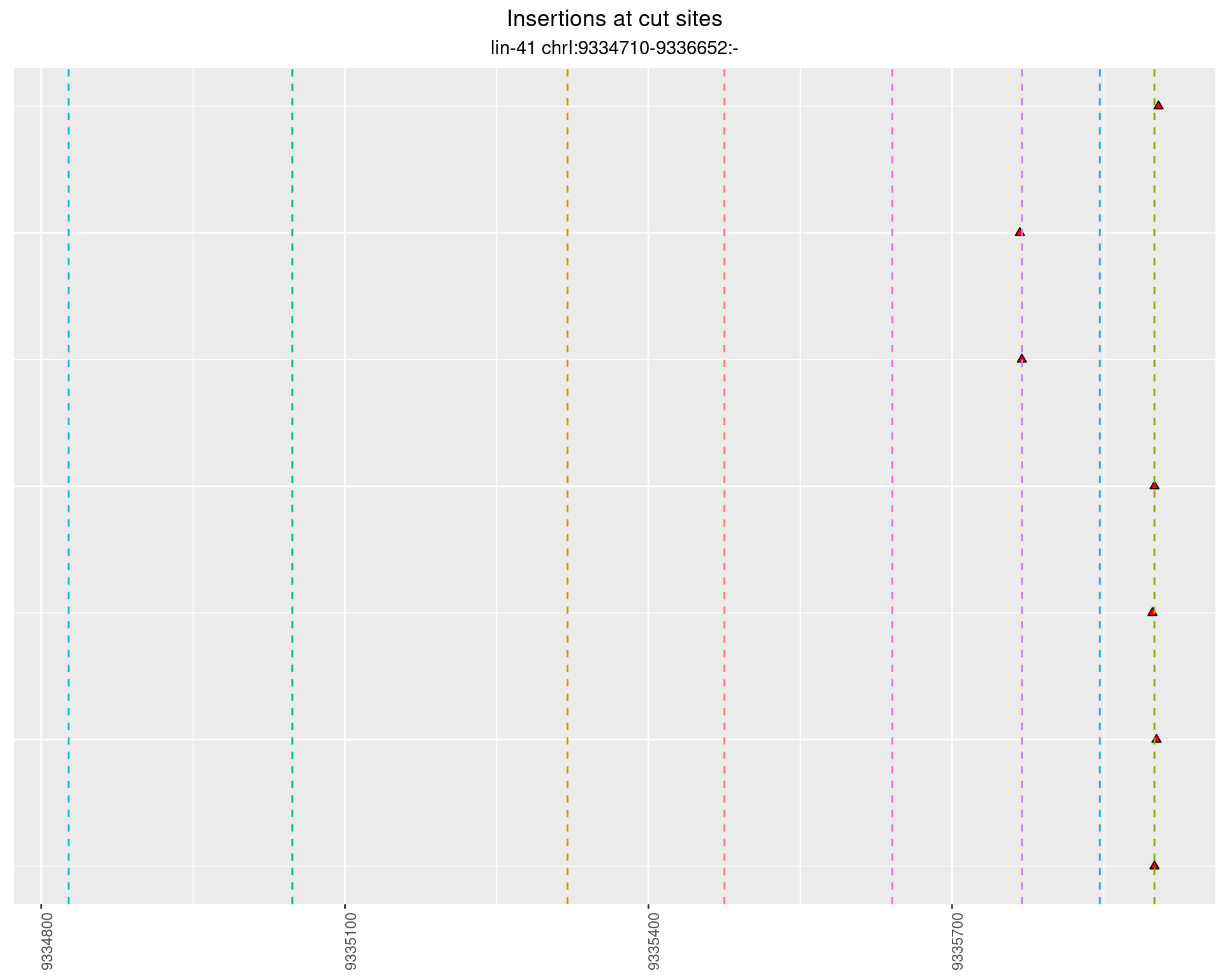
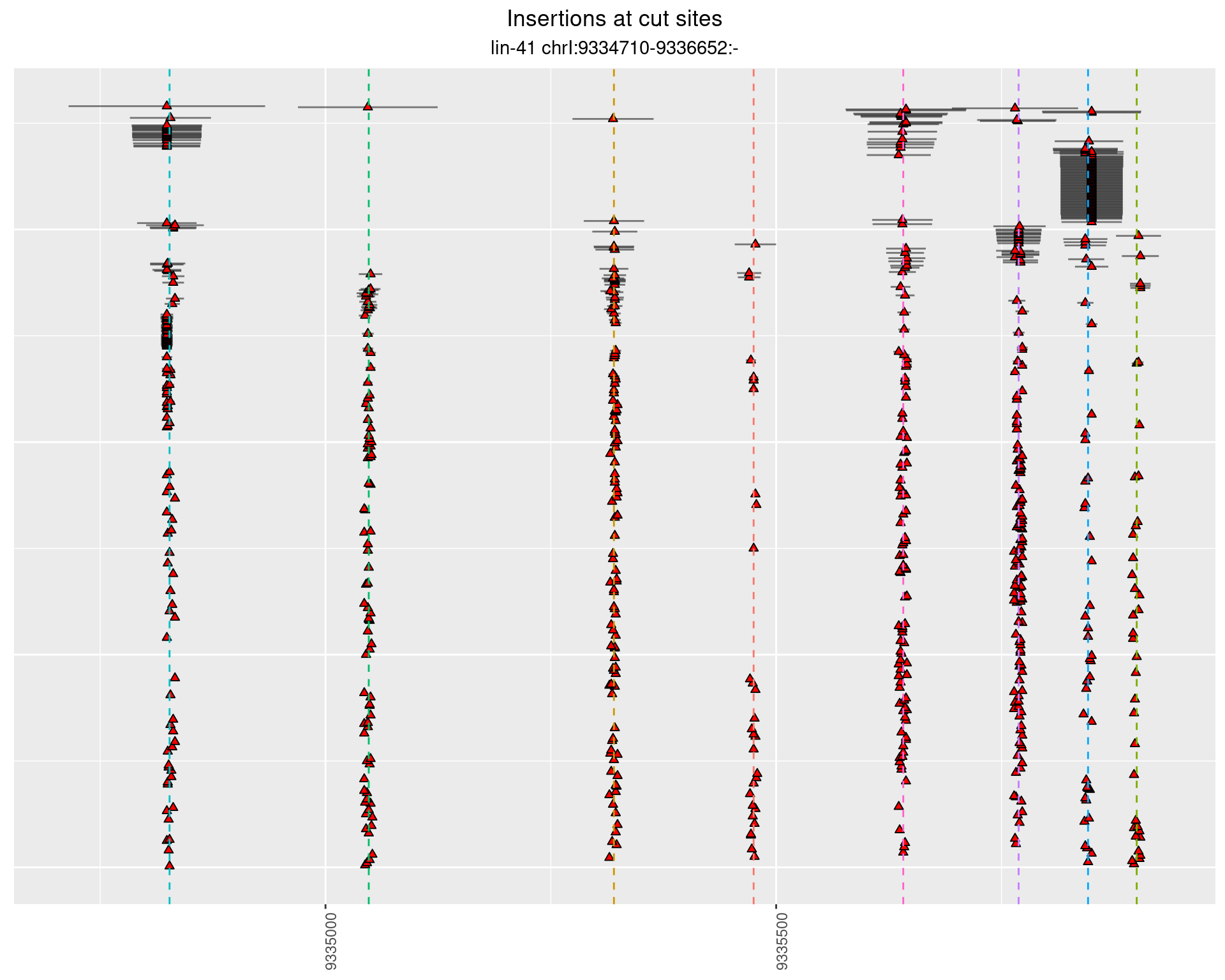
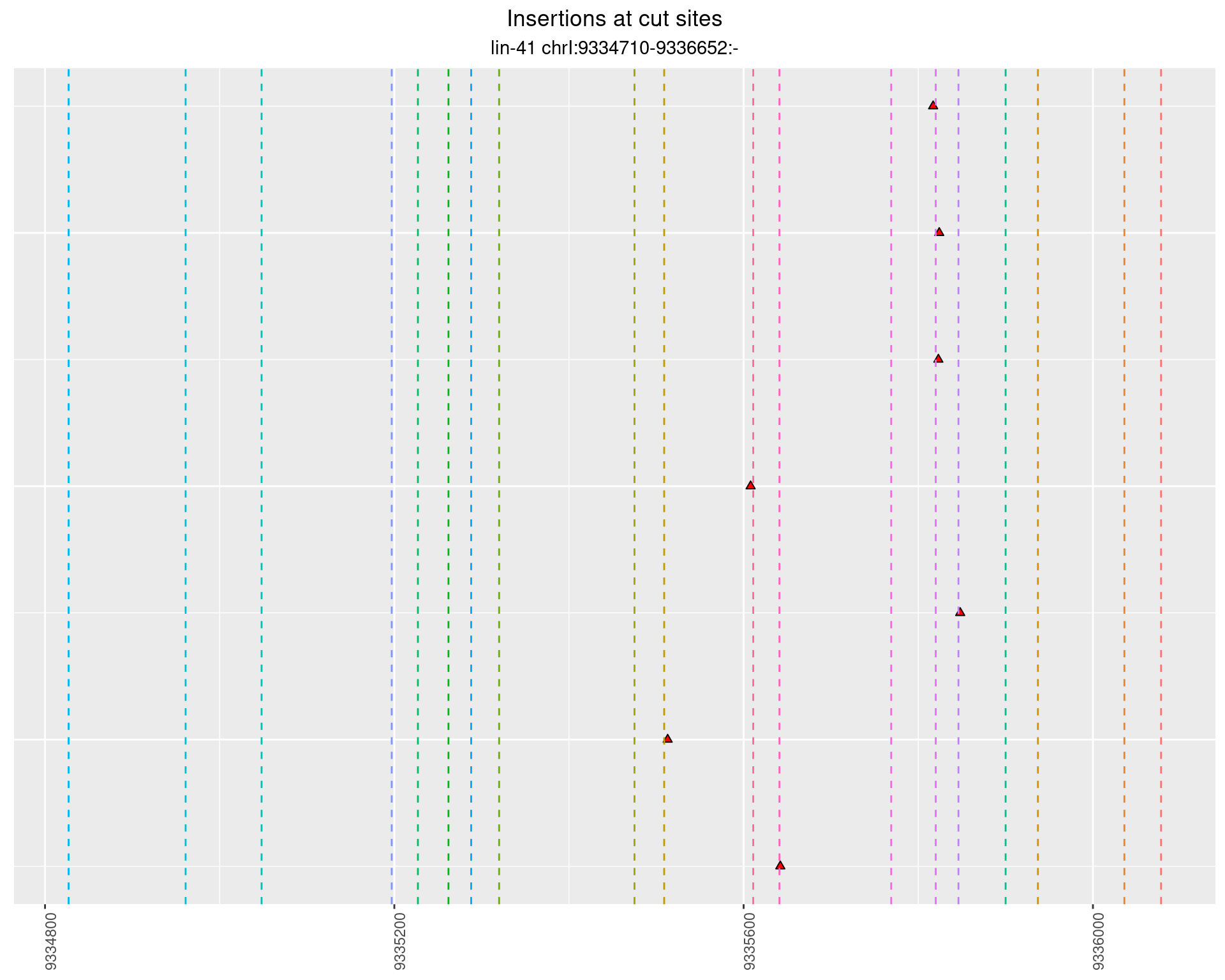
These plots show the diversity of insertions at the cut sites taking into account the actual sequence that is inserted.
## import insertions data (that contains inserted sequences, too) and make some summary plots
insertions <- as.data.table(do.call(rbind, lapply(1:nrow(sampleSheet), function(i) {
sampleName <- sampleSheet[i, 'sample_name']
f <- file.path(pipeline_output_dir, 'indels', sampleName, paste0(sampleName, '.insertedSequences.tsv'))
if(file.exists(f)) {
dt <- data.table::fread(f)
dt$sample <- sampleName
dt$end <- dt$start
return(dt)
} else {
warning("Can't open .insertedSequences.tsv file for sample ",sampleName,
" at ",f,"\n")
return(NULL)
}
})))
#collapse insertions
insertions <- insertions[,length(name),
by = c('seqname', 'sample', 'start',
'end', 'insertedSequence',
'insertionWidth')]
colnames(insertions)[7] <- 'ReadSupport'
# get alignment coverage - will need for insertion coverage
alnCoverage <- importSampleBigWig(pipeline_output_dir,
sampleSheet$sample_name, ".alnCoverage.bigwig")
insertions <- do.call(rbind, lapply(unique(insertions$sample), function(s) {
do.call(rbind, lapply(unique(insertions[sample == s]$seqname), function(chr) {
dt <- insertions[sample == s & seqname == chr]
dt$coverage <- as.vector(alnCoverage[[s]][[chr]])[dt$start]
return(dt)
}))
}))
#compute frequency value for each insertion
#(number of reads supporting the insertion divided by coverage at insertion site)
insertions$freq <- insertions$ReadSupport/insertions$coverage
#find overlaps with cut sites
insertions <- cbind(insertions, overlapCutSites(indels = insertions, cutSites = cutSites))
# for each sample, find the guides used in the sample and find out if the indels overlap cut sites
# warnings: only restrict to the cut sites relevant for the sample
# warnings: if the sample is untreated, then we check overlaps for all cut sites
# for the corresponding amplicon
insertions <- do.call(rbind, lapply(unique(insertions$sample), function(sampleName) {
dt <- insertions[sample == sampleName]
sgRNAs <- sampleGuides[[sampleName]]
dt$atCutSite <- apply(subset(dt, select = sgRNAs), 1, function(x) sum(x > 0) > 0)
return(dt)
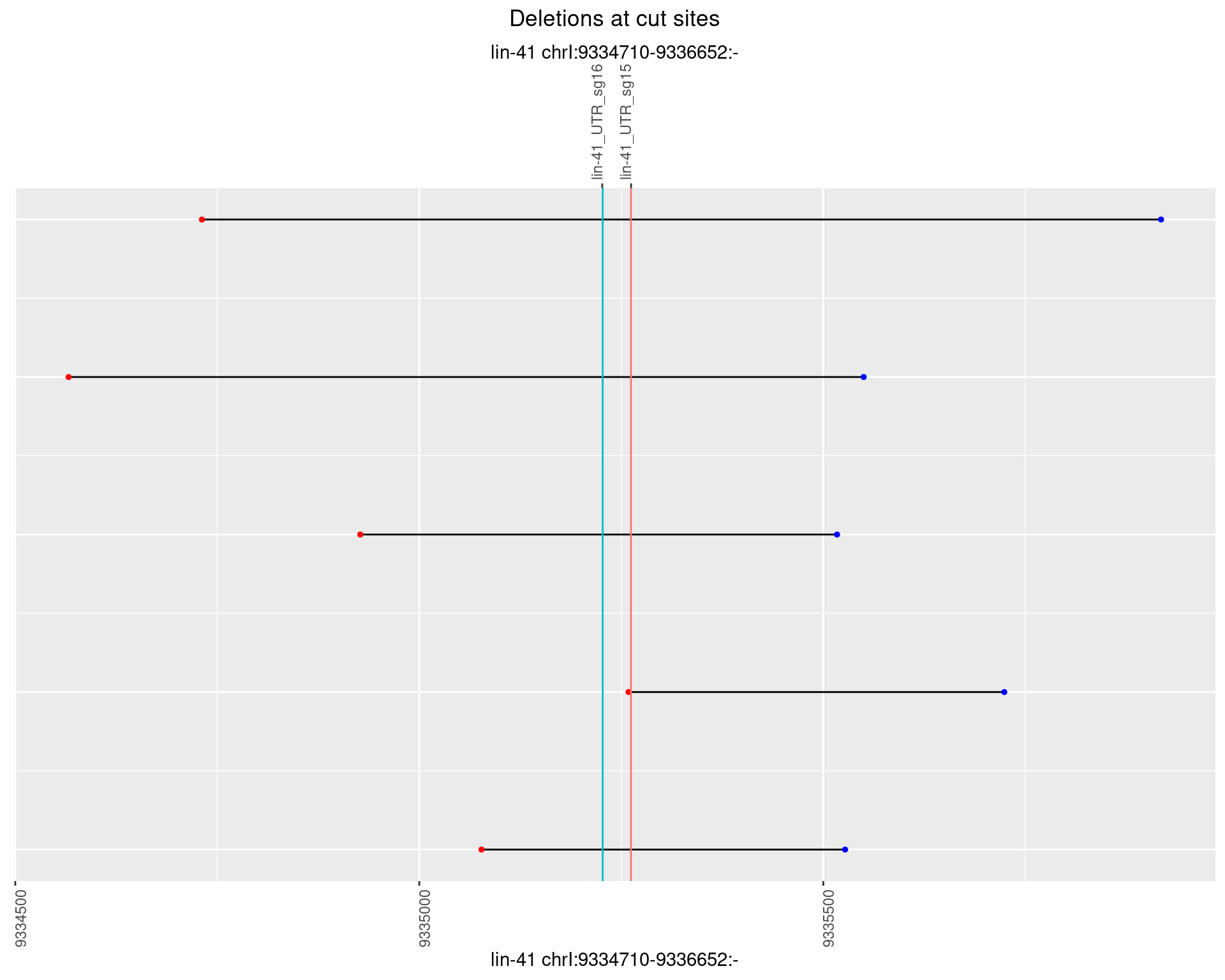
}))plotInsertions <- function(dt, cutSites, sgRNAs) {
#first randomize the order (to avoid sorting by start position)
dt <- dt[sample(1:nrow(dt), nrow(dt))]
#sort insertions by width of insertion
dt <- dt[order(insertionWidth)]
dt$linePos <- 1:nrow(dt)
ggplot2::ggplot(dt, aes(x = linePos, ymin = start - insertionWidth/2,
ymax = start + insertionWidth/2)) +
geom_linerange(size = 0.5, alpha = 0.5) +
geom_point(aes(x = linePos, y = start), shape = 24, fill = 'red') +
geom_hline(data = as.data.frame(cutSites[cutSites$name %in% sgRNAs,]),
aes(yintercept = start, color = name), linetype = 'dashed', show.legend = FALSE) +
theme(axis.text.y = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank(),
axis.ticks.y = element_blank(),
axis.text.x = element_text(angle = 90),
plot.title = element_text(hjust = 0.5),
plot.subtitle = element_text(hjust = 0.5)) +
scale_y_continuous(sec.axis = dup_axis(breaks = cutSites[cutSites$sgRNA %in% sgRNAs,]$cutSite,
labels = cutSites[cutSites$sgRNA %in% sgRNAs,]$sgRNA)) +
coord_flip()
}
insertions$freqInterval <- cut_interval(log10(insertions$freq), length = 1)
plots <- lapply(unique(insertions$sample), function(sampleName) {
dt <- insertions[sample == sampleName & atCutSite == TRUE]
if(nrow(dt) == 0) {
return(NULL)
}
#dt$freqInterval <- cut_interval(log10(dt$freq), length = 1)
#segment plots with different frequency thresholds
segmentPlots <- lapply(unique(as.character(dt$freqInterval)), function(x) {
if(nrow(dt[freqInterval == x]) > 0) {
p <- plotInsertions(dt[freqInterval == x], cutSites, sampleGuides[[sampleName]])
p <- p + labs(title = 'Insertions at cut sites',
subtitle = paste(targetName, targetRegion))
} else {
p <- NULL
}
return(p)
})
names(segmentPlots) <- lapply(unique(as.character(dt$freqInterval)), function(x) {
paste(10^(as.numeric(unlist(strsplit(sub("(\\[|\\()(.+)(\\]|\\))", "\\2", x), ',')))),
collapse = ' - ')
})
return(segmentPlots)
})
names(plots) <- unique(insertions$sample)# folder to save pdf versions of the segment plots
dirPath <- paste(targetName, 'Indel_Diversity.insertion_segment_plots', sep = '.')
if(!dir.exists(dirPath)) {
dir.create(dirPath)
}
for (sample in names(plots)) {
cat('## ',sample,'{.tabset .tabset-fade .tabset-pills}\n\n')
for(i in names(plots[[sample]])) {
cat('### Freq:',i,'\n\n')
p <- plots[[sample]][[i]]
if(!is.null(p)) {
print(p)
ggsave(filename = file.path(dirPath, paste(sample, 'freq', i, 'pdf', sep = '.')),
plot = p, width = 10, height = 8, units = 'in')
} else {
cat("No plot to show\n\n")
}
cat("\n\n")
}
cat("\n\n")
}2.1 gen_1624C_F1_lin-41_N2
2.1.1 Freq: 1e-05 - 1e-04
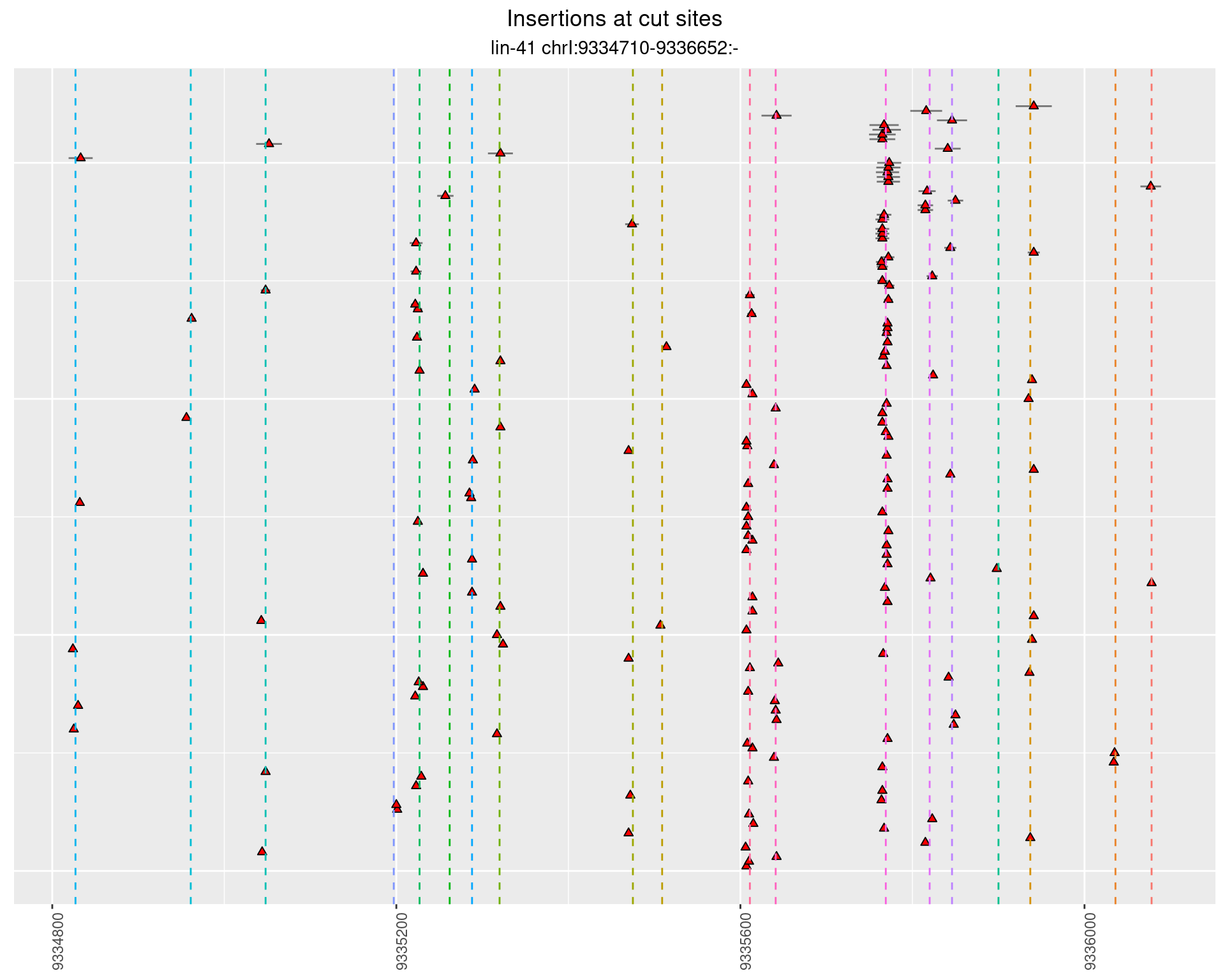
2.1.2 Freq: 1e-04 - 0.001
2.1.3 Freq: 1e-06 - 1e-05
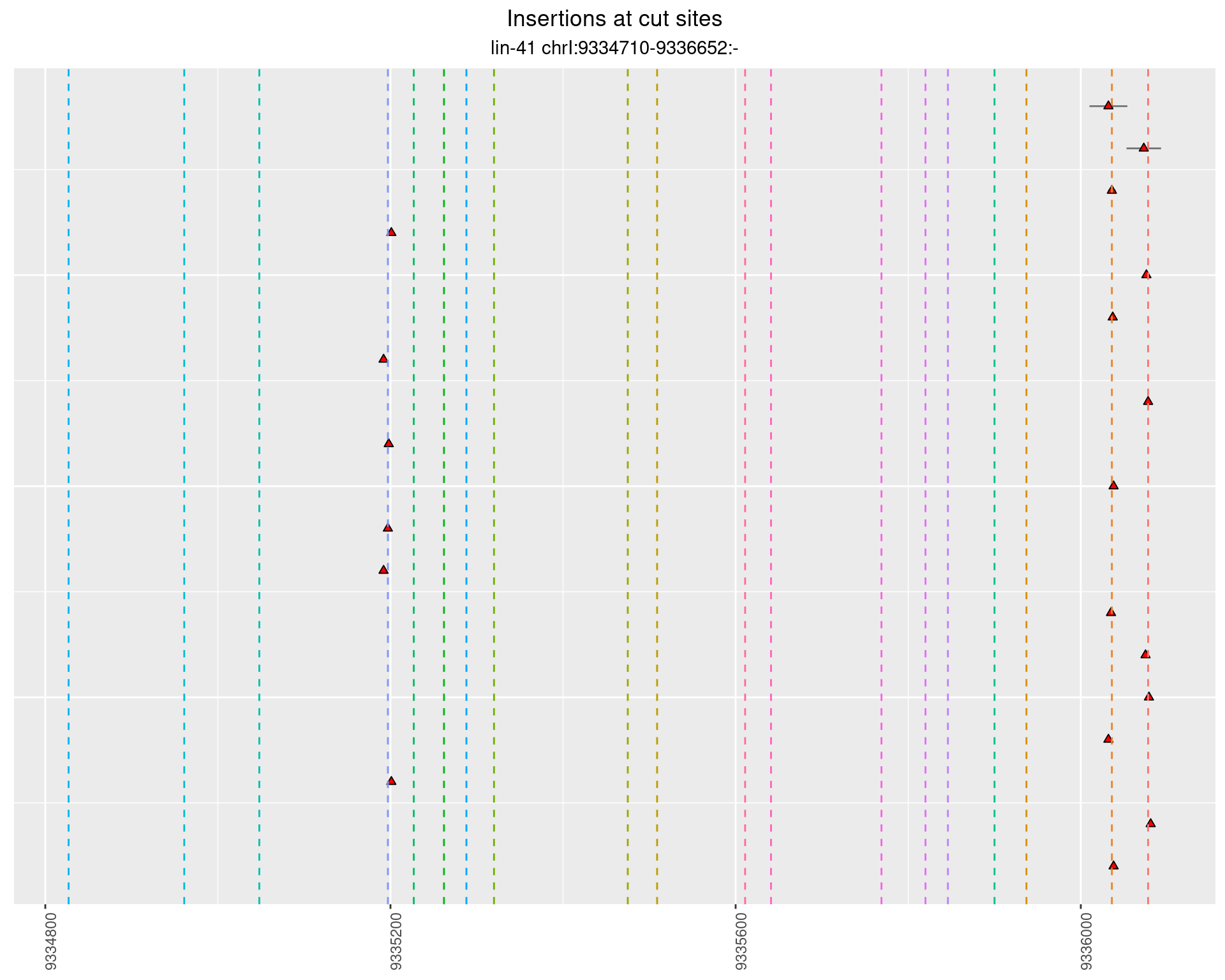
2.2 gen_1624C_F1_lin-41_sg15sg16
2.2.1 Freq: 1e-04 - 0.001
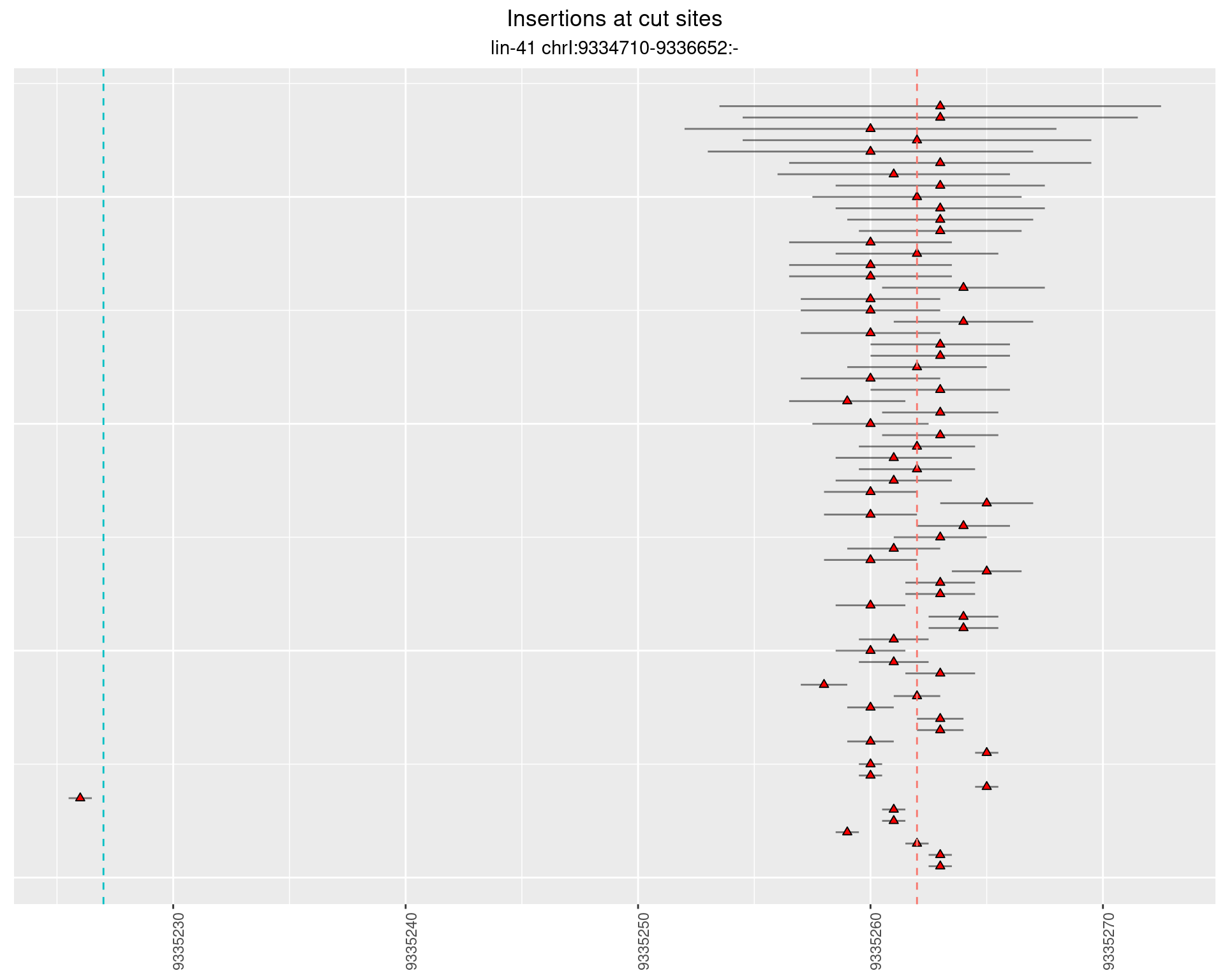
2.2.2 Freq: 1e-05 - 1e-04
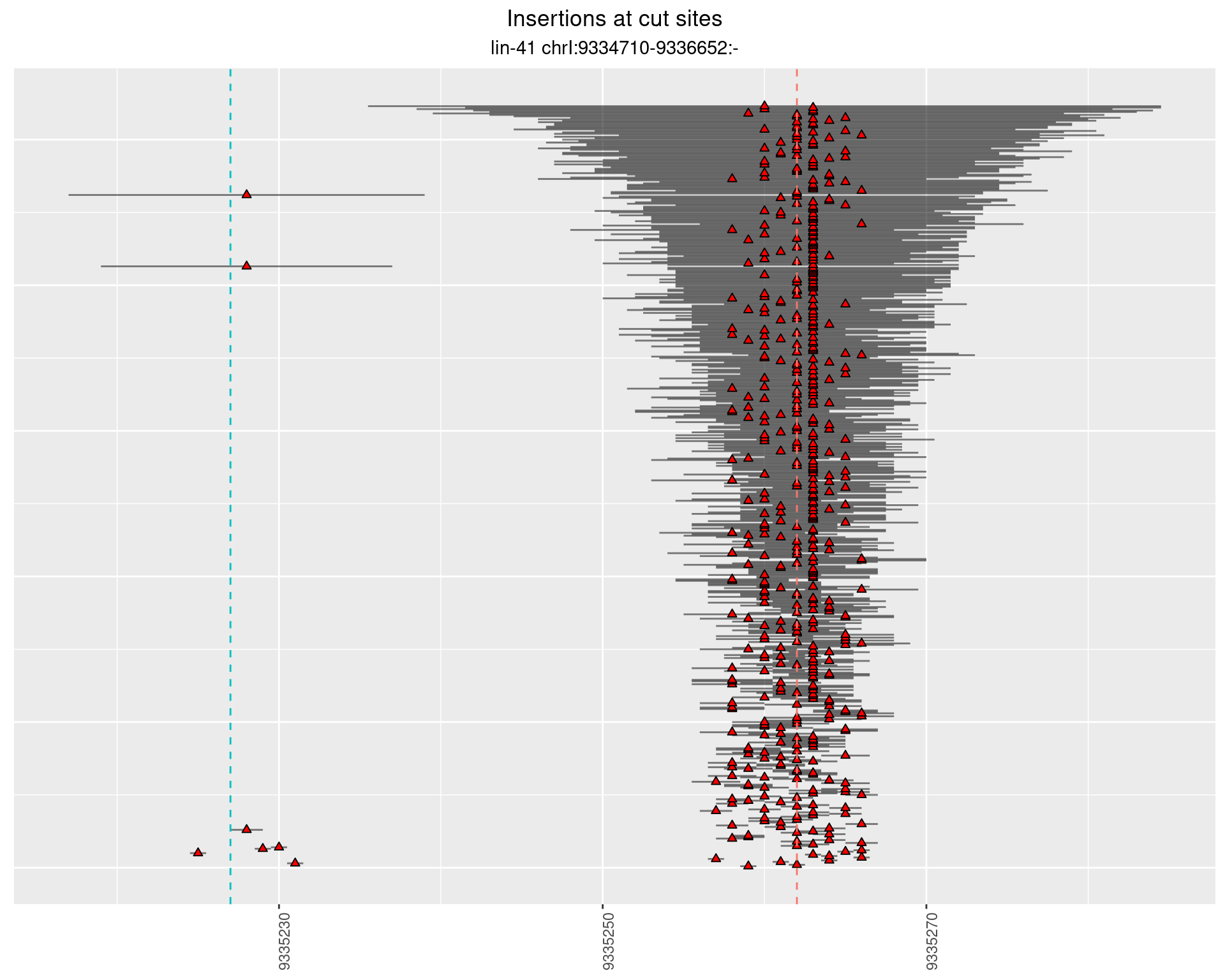
2.2.3 Freq: 1e-06 - 1e-05
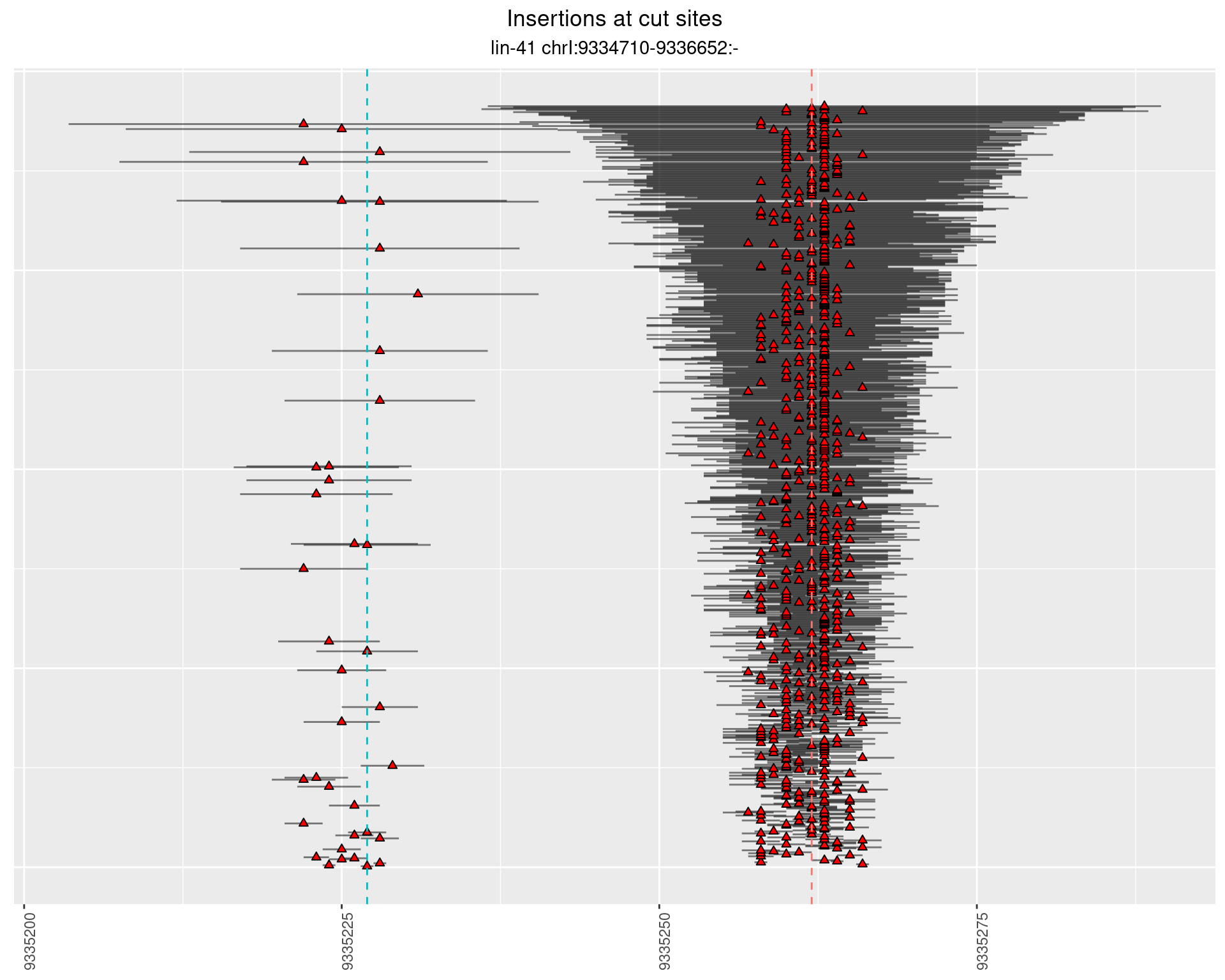
2.3 gen_1624C_F1_lin-41_pool
2.3.1 Freq: 1e-05 - 1e-04
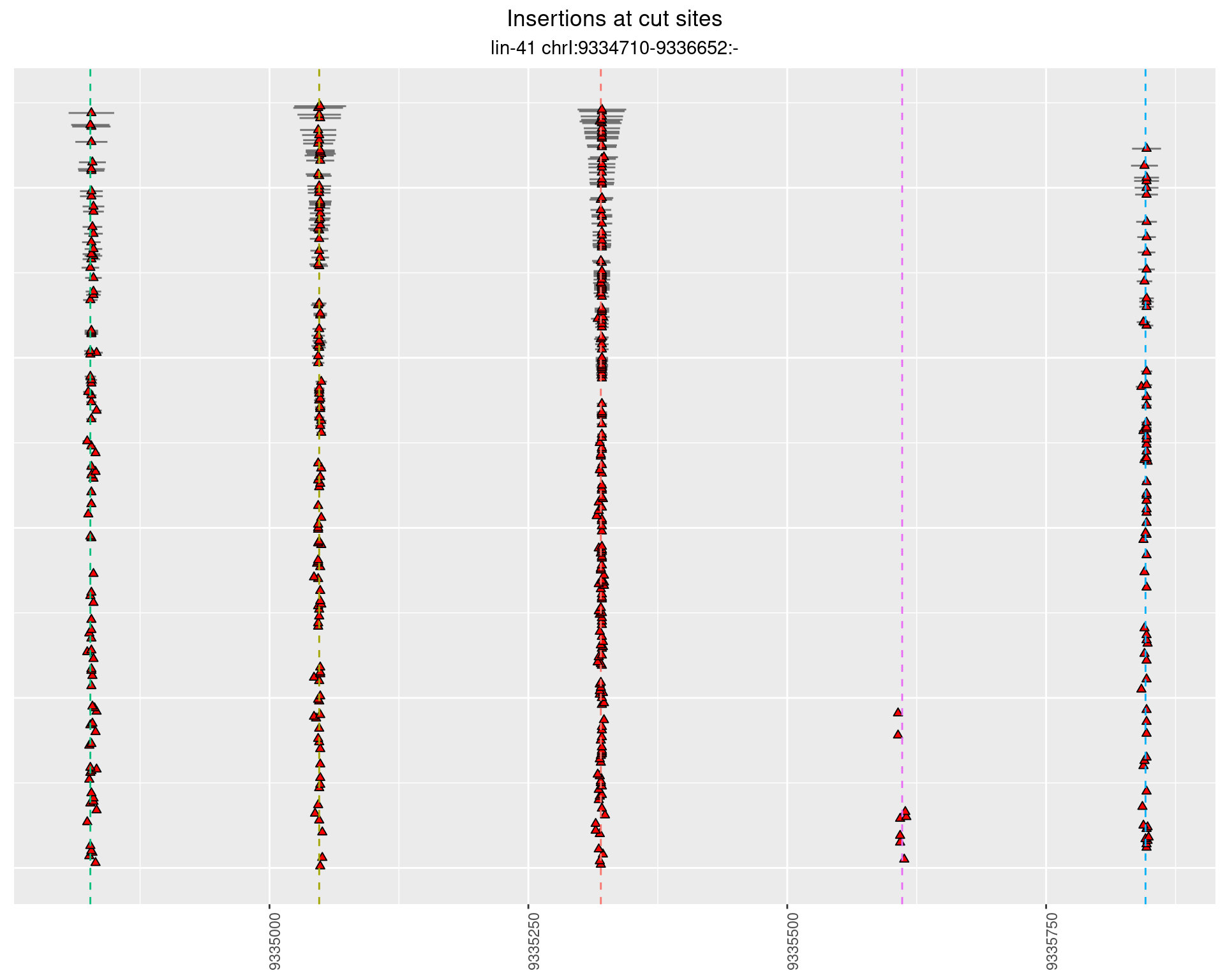
2.3.2 Freq: 1e-04 - 0.001
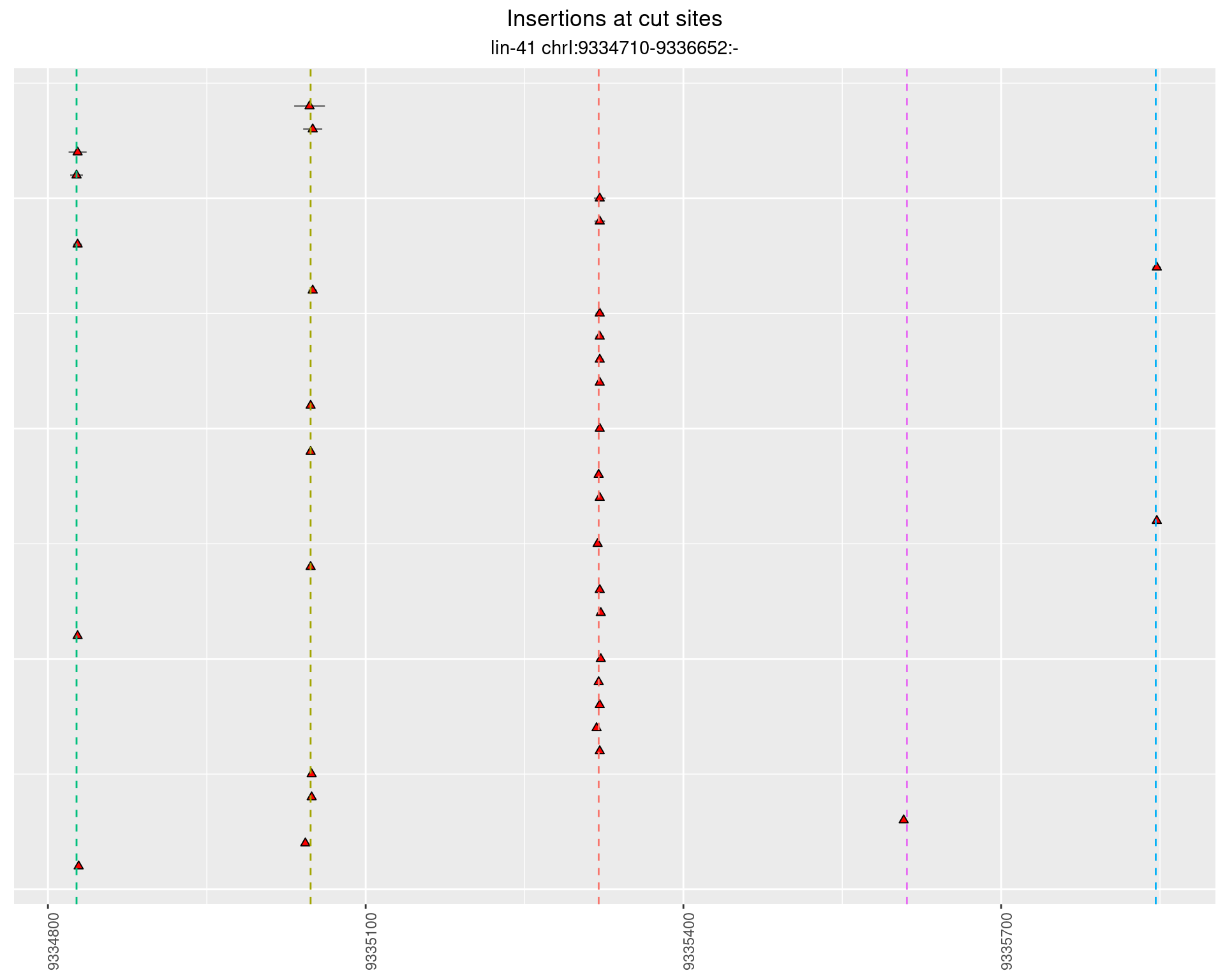
2.3.3 Freq: 1e-06 - 1e-05
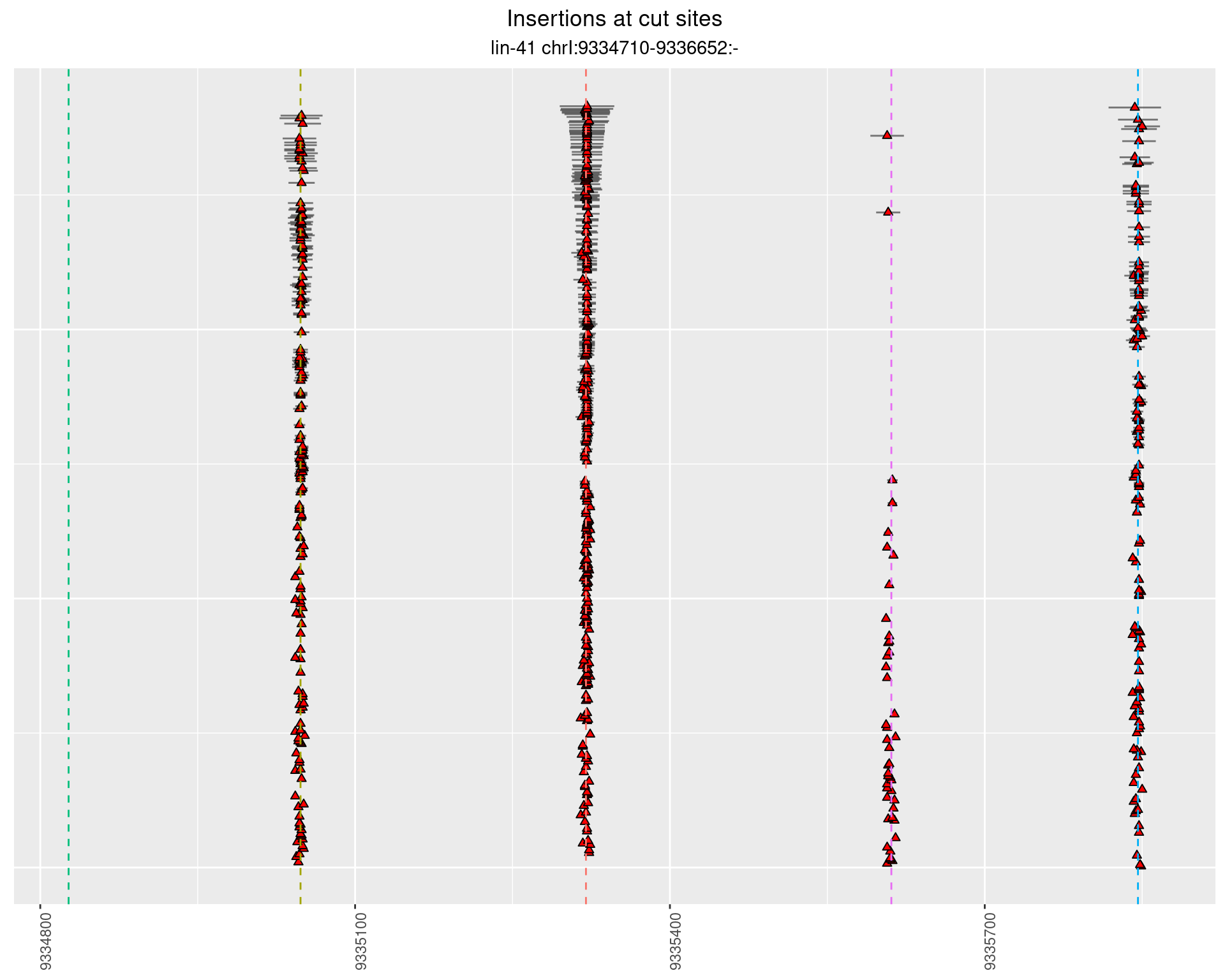
2.3.4 Freq: 0.001 - 0.01
2.4 gen_1624C_F1_lin-41_sg26sg27
2.4.1 Freq: 1e-04 - 0.001
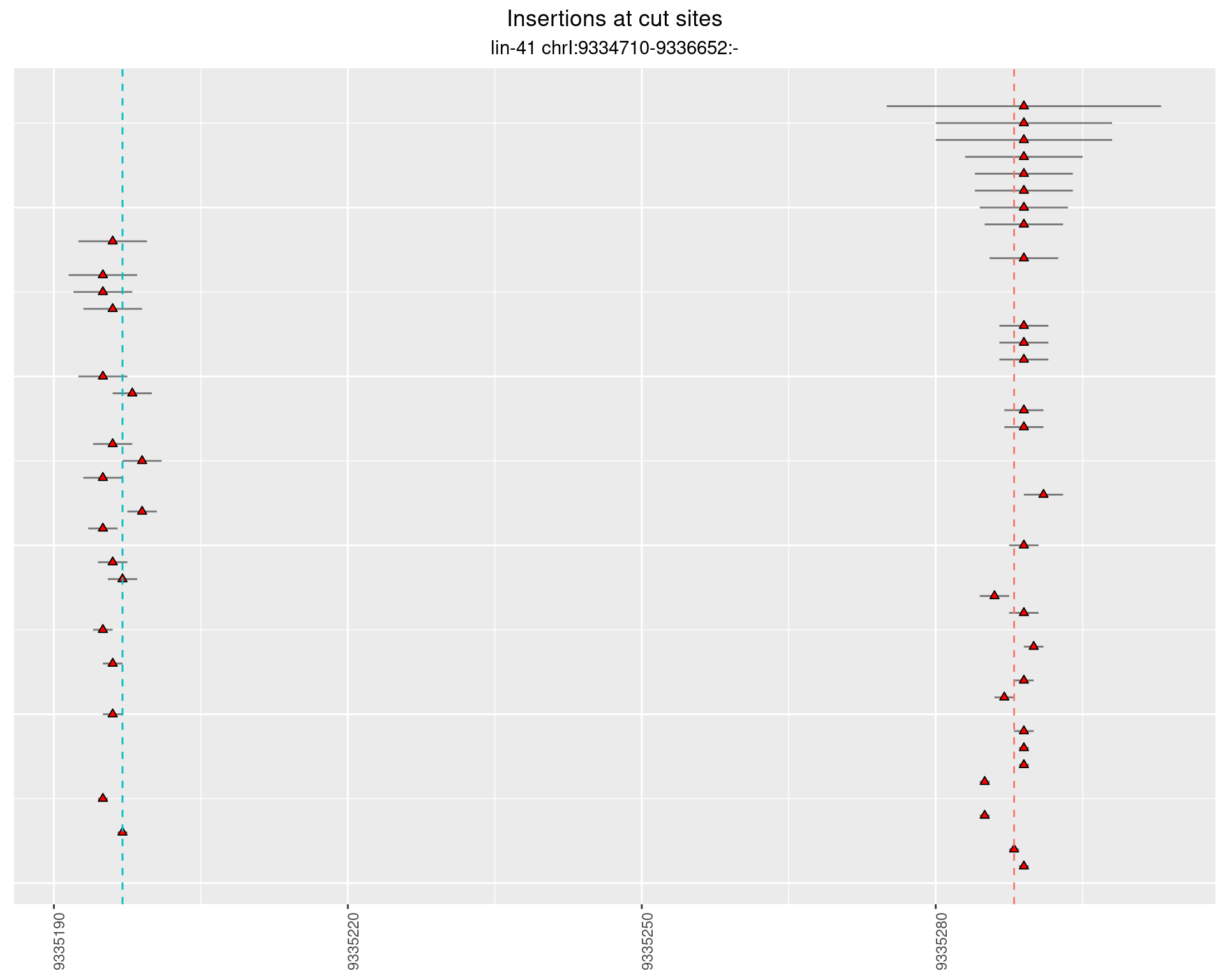
2.4.2 Freq: 1e-05 - 1e-04
2.4.3 Freq: 1e-06 - 1e-05
2.5 gen_24C_F2_lin-41_N2
2.5.1 Freq: 1e-05 - 1e-04
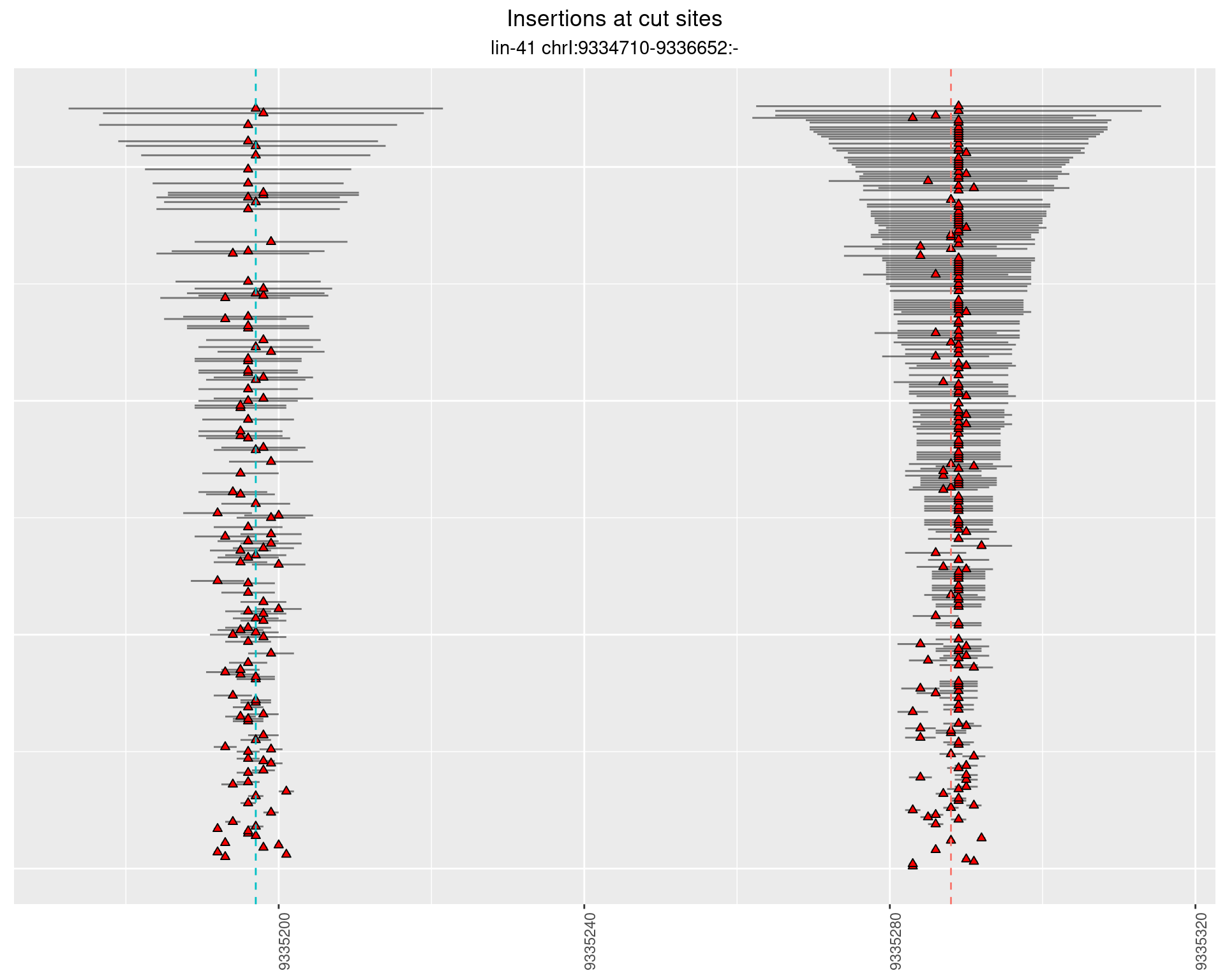
2.5.2 Freq: 1e-06 - 1e-05
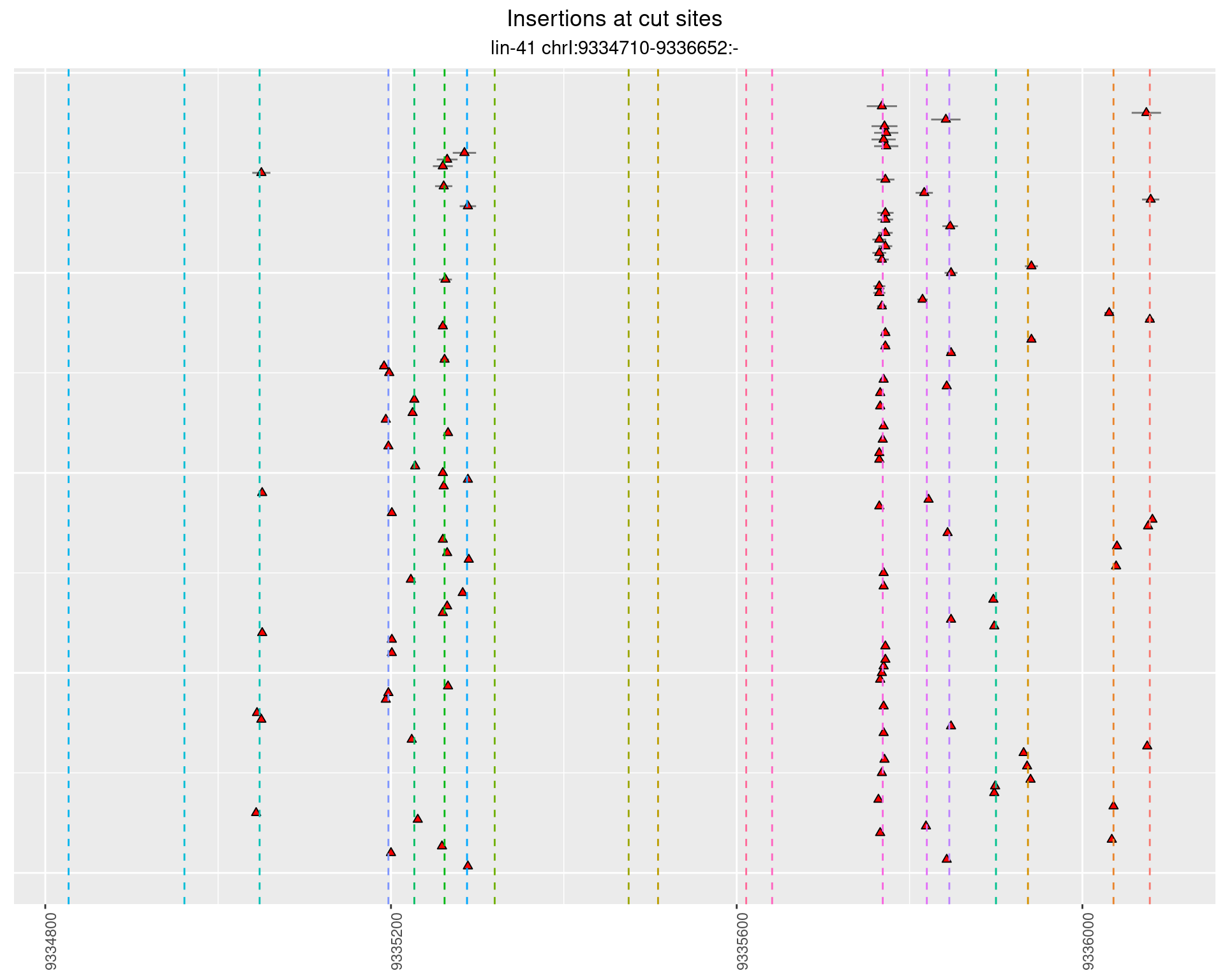
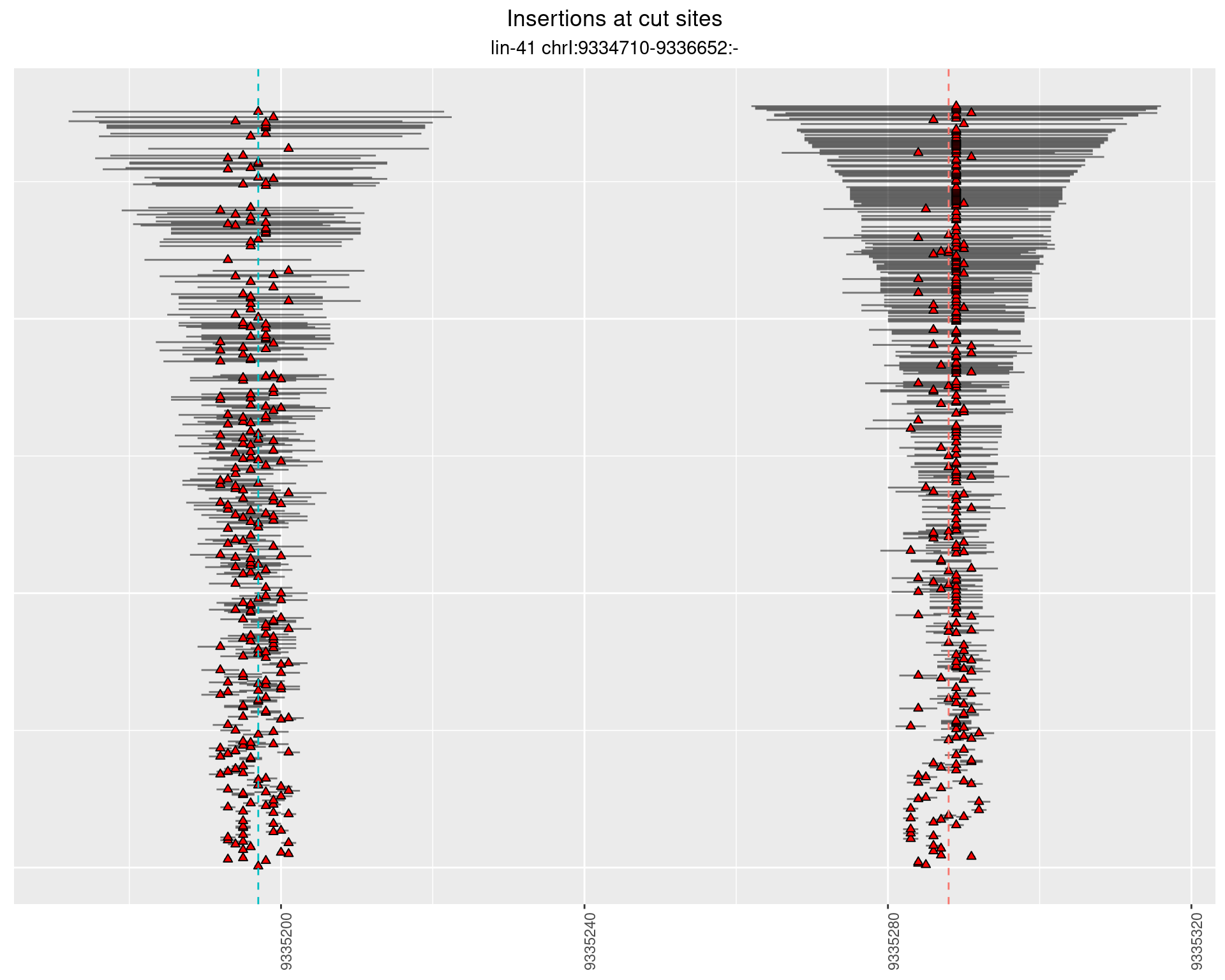
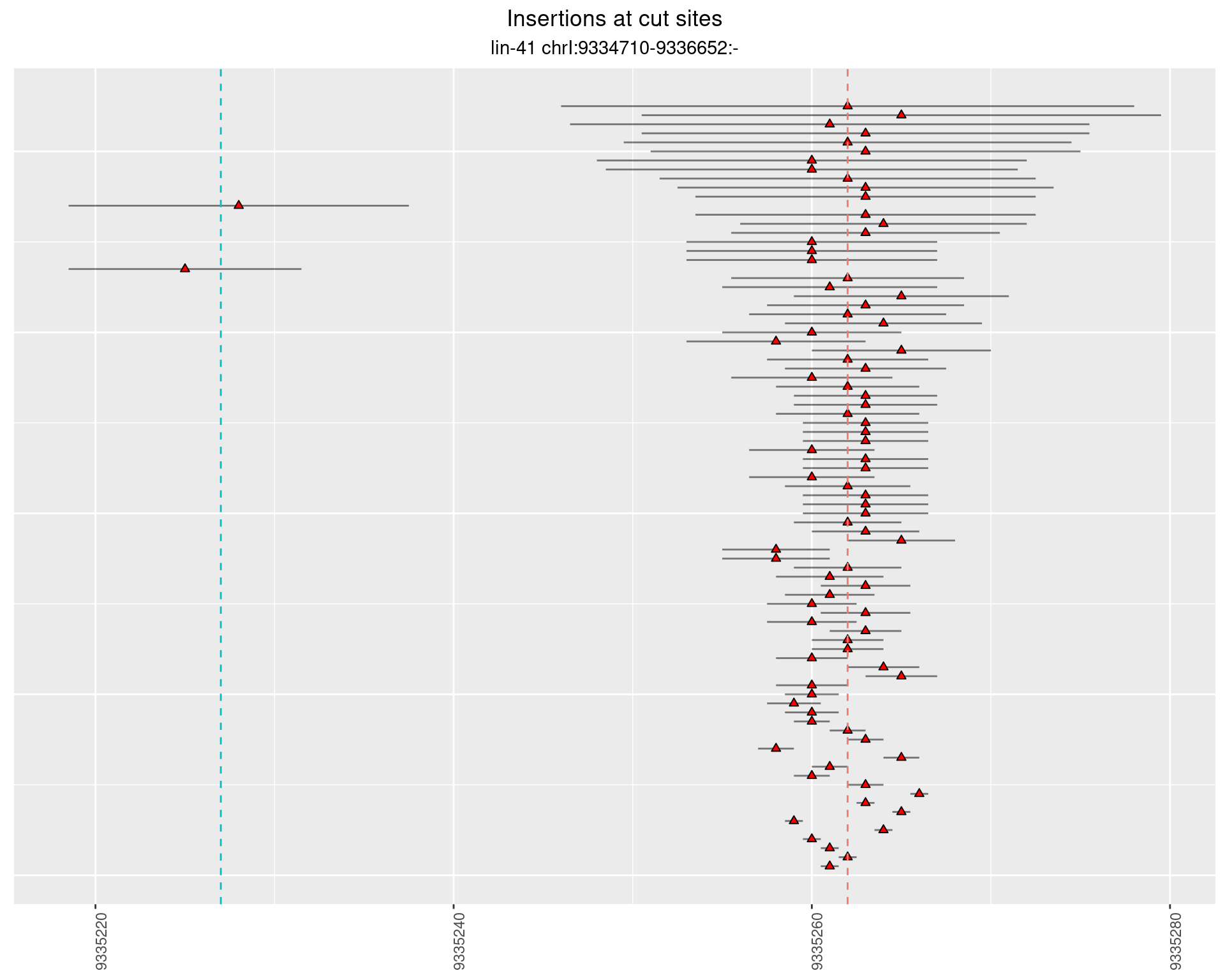
2.5.3 Freq: 1e-04 - 0.001
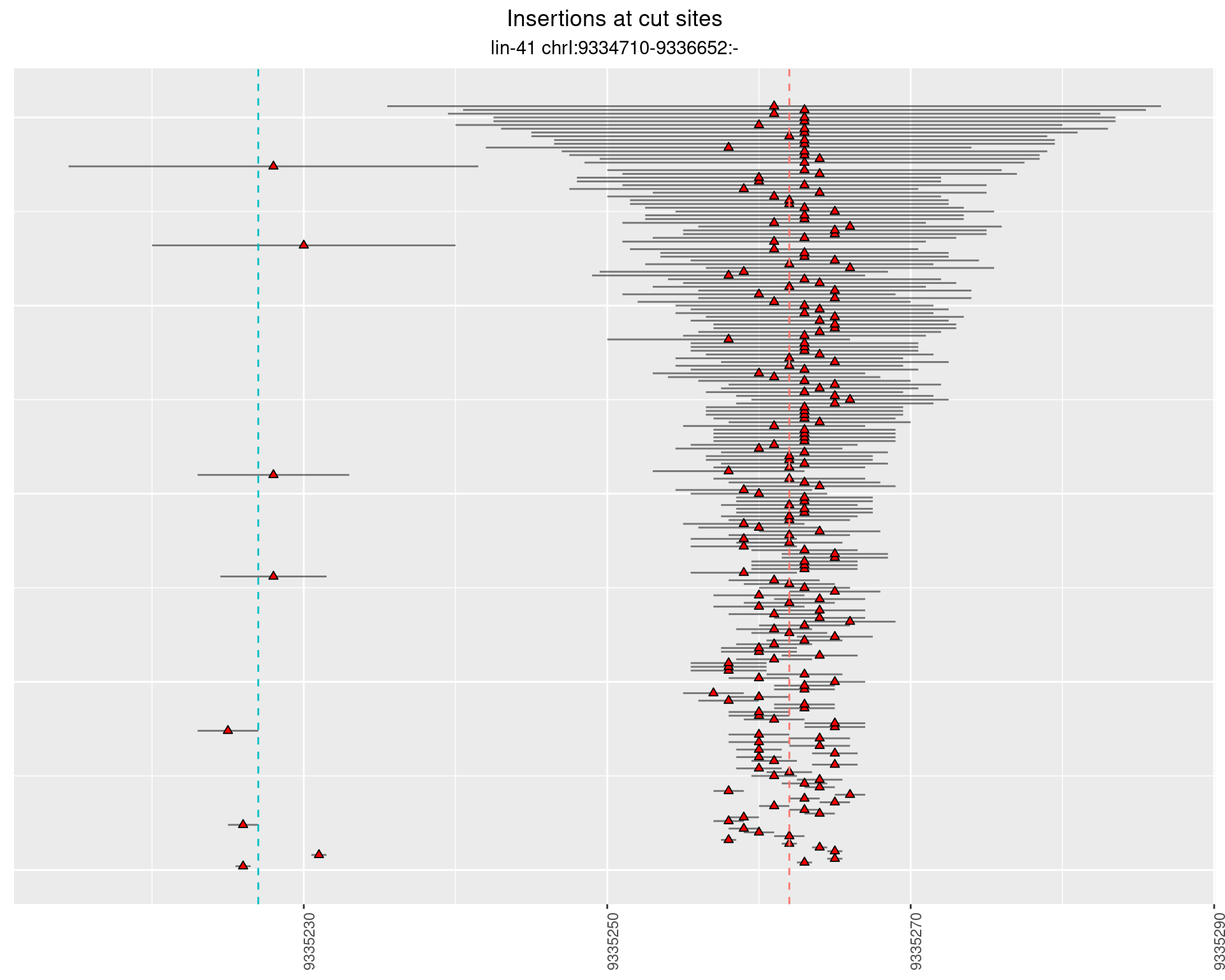
2.6 gen_24C_F2_lin-41_sg15sg16
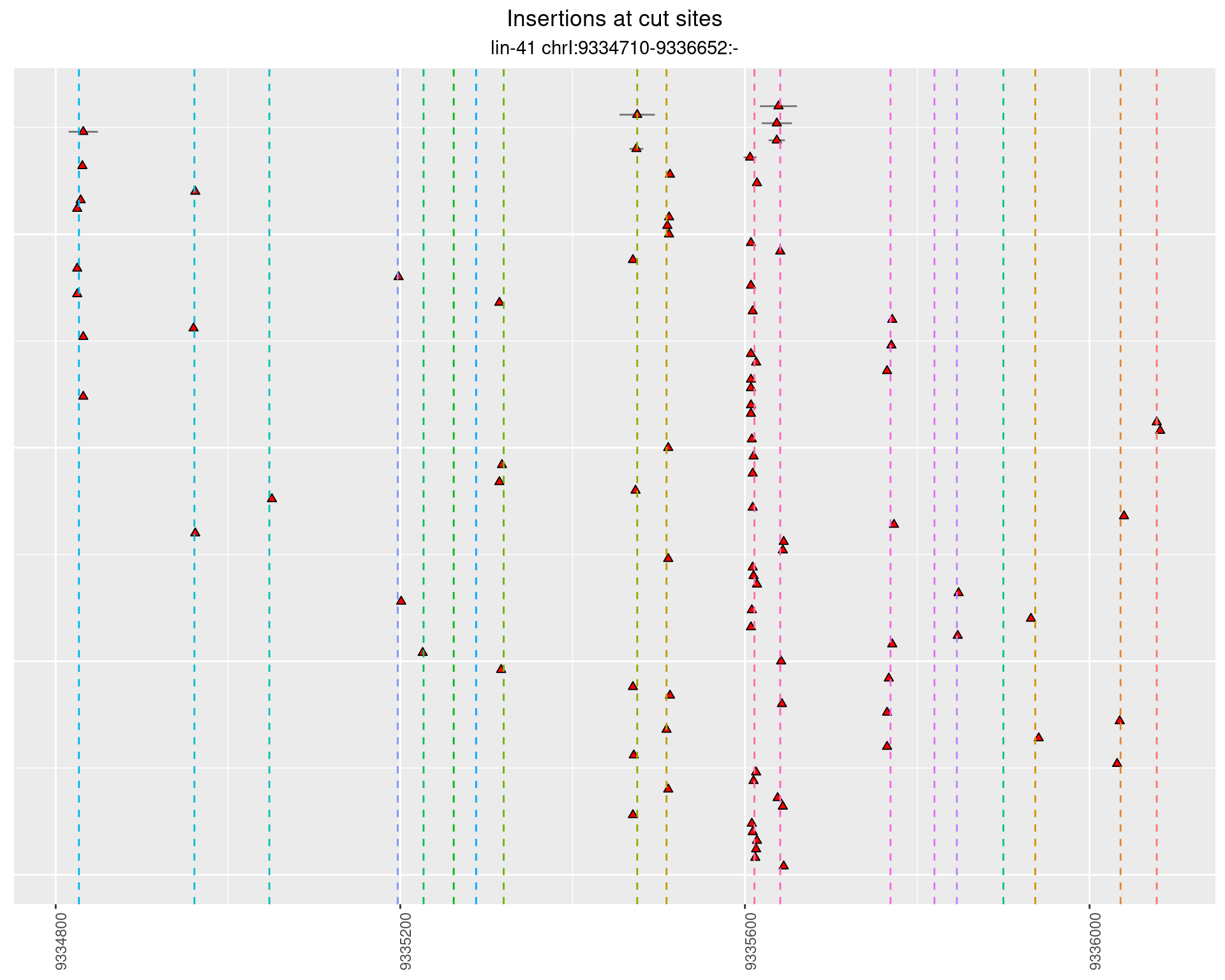
2.6.1 Freq: 1e-05 - 1e-04
2.6.2 Freq: 1e-04 - 0.001
2.6.3 Freq: 1e-06 - 1e-05
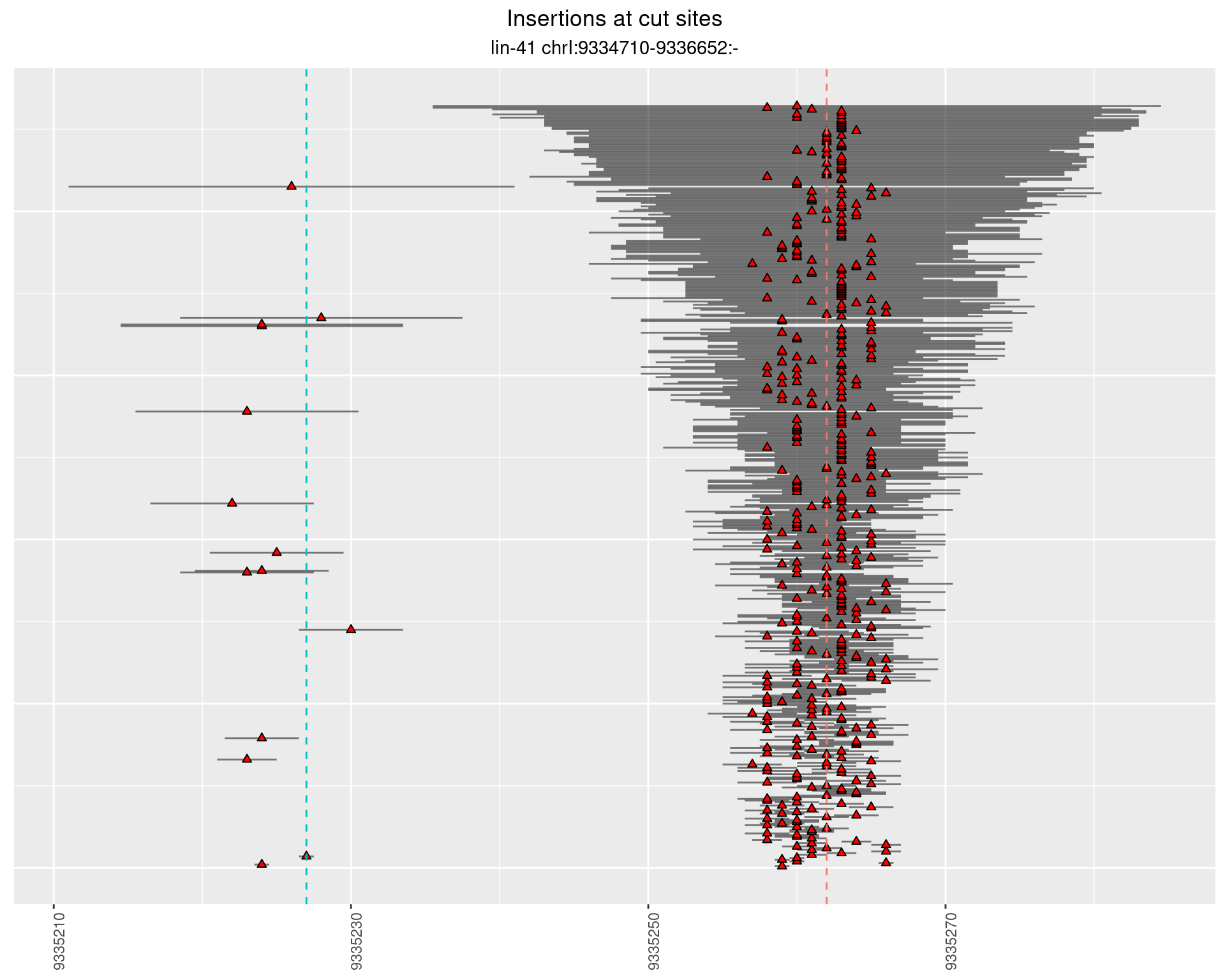
2.7 gen_24C_F2_lin-41_pool
2.7.1 Freq: 1e-05 - 1e-04
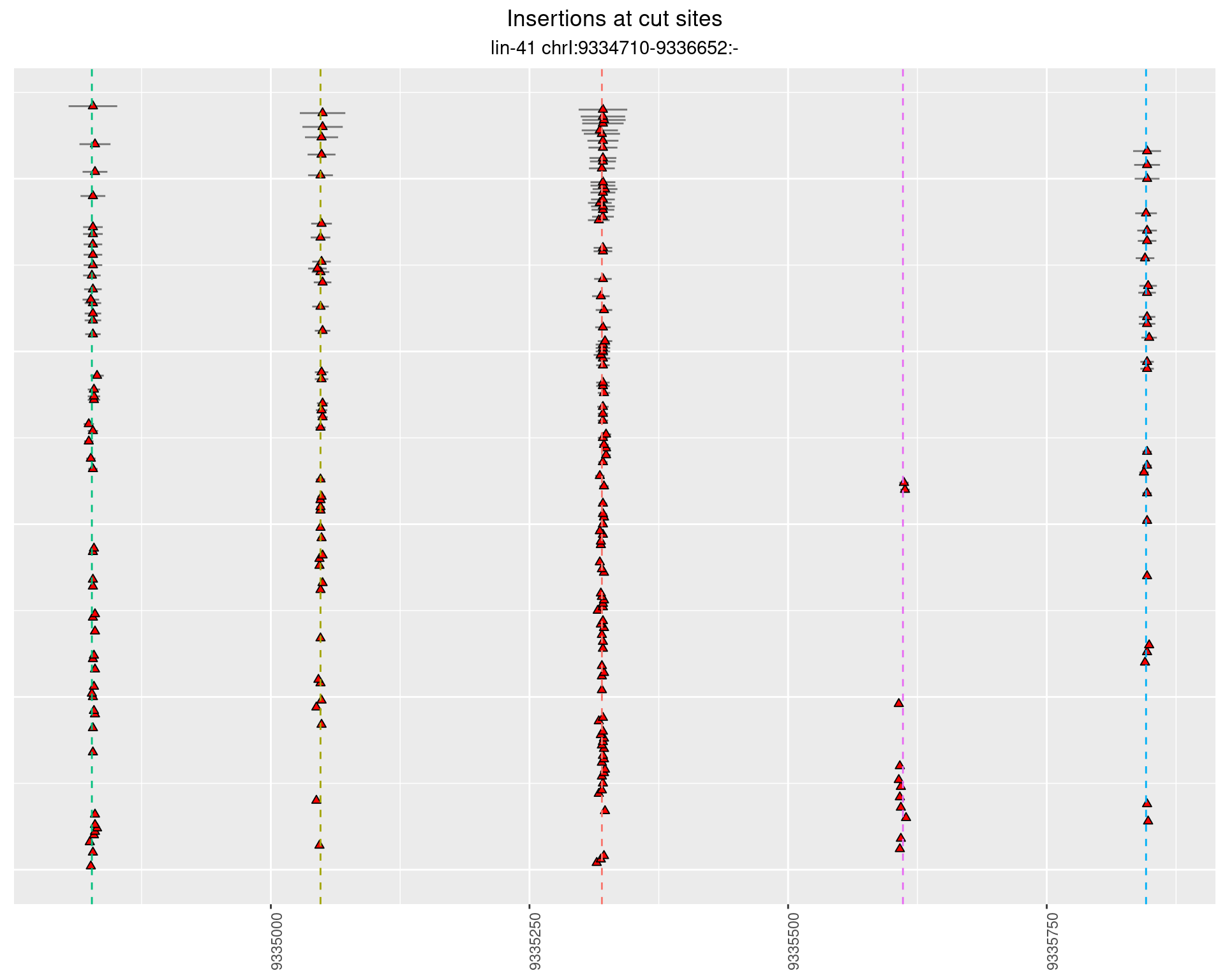
2.7.2 Freq: 1e-04 - 0.001
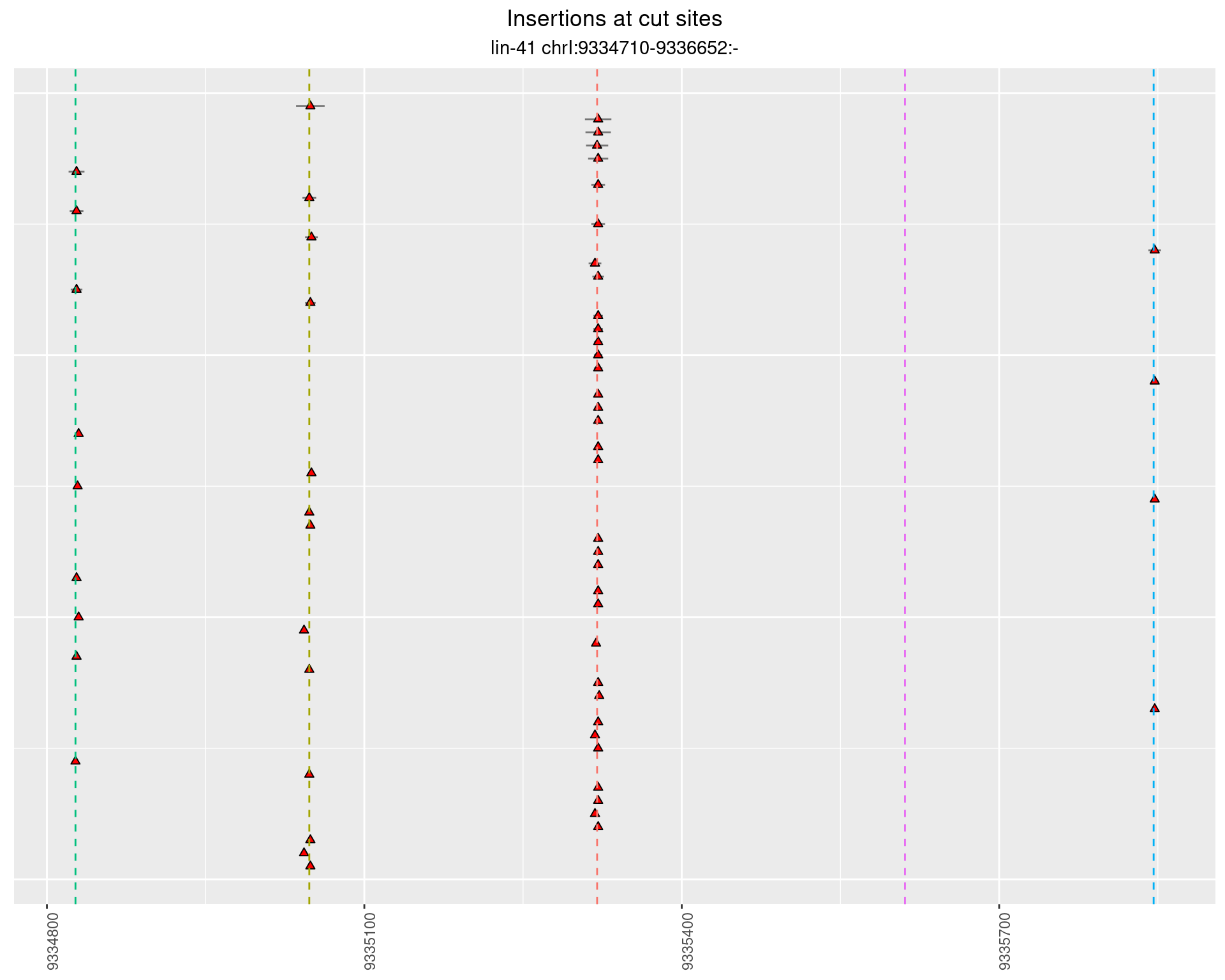
2.7.3 Freq: 1e-06 - 1e-05
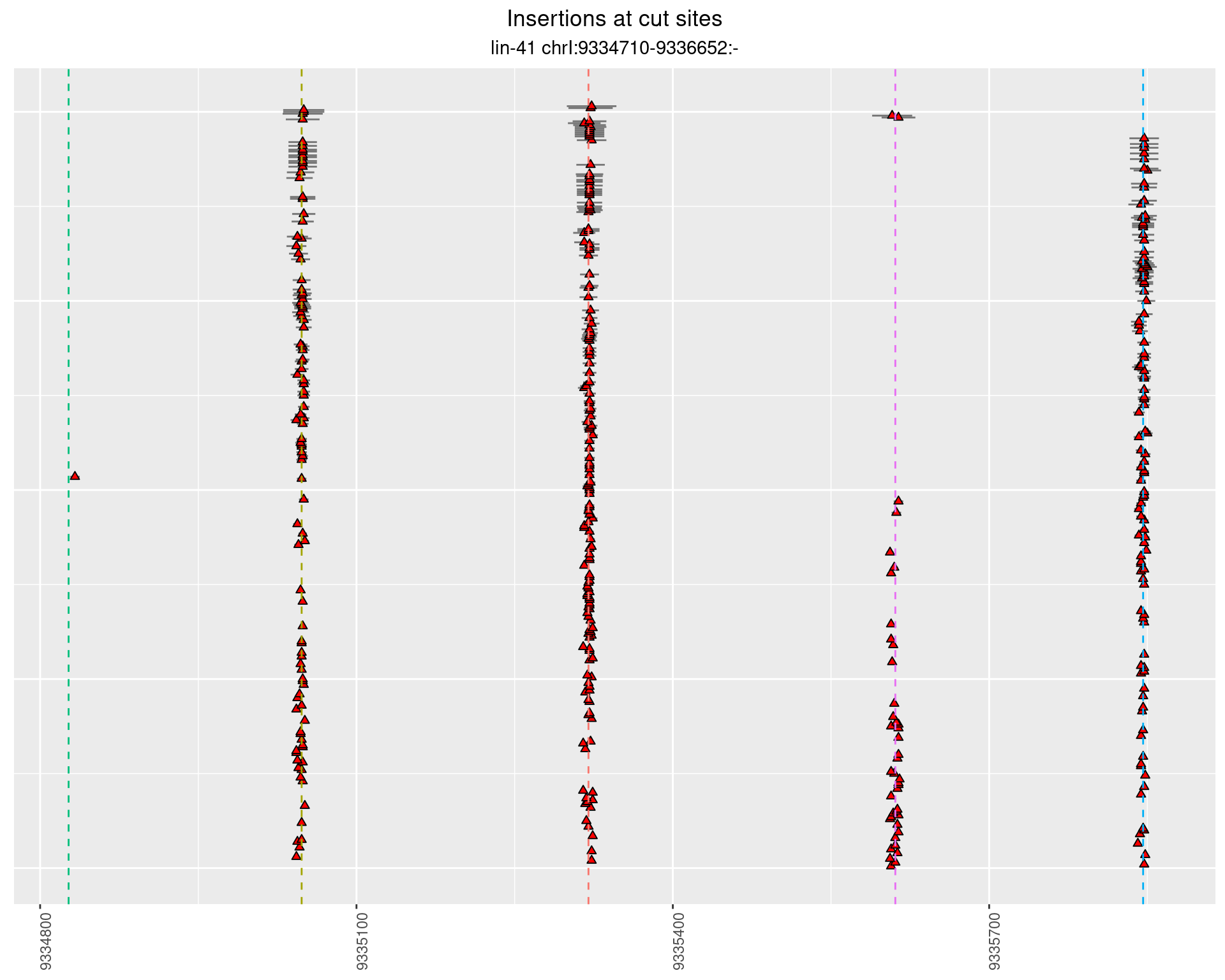
2.7.4 Freq: 0.001 - 0.01
2.8 gen_24C_F2_lin-41_sg26sg27
2.8.1 Freq: 1e-04 - 0.001
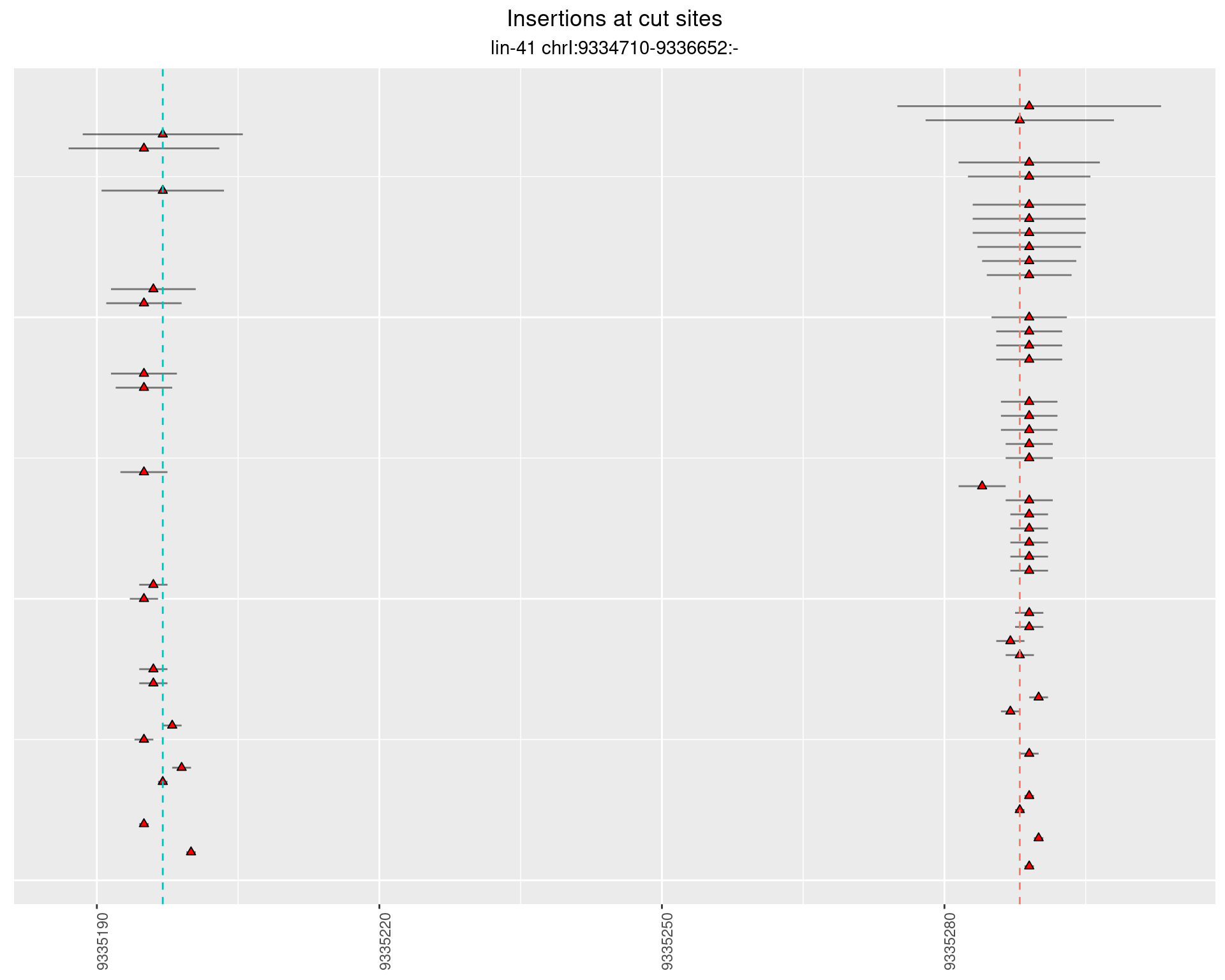
2.8.2 Freq: 1e-05 - 1e-04
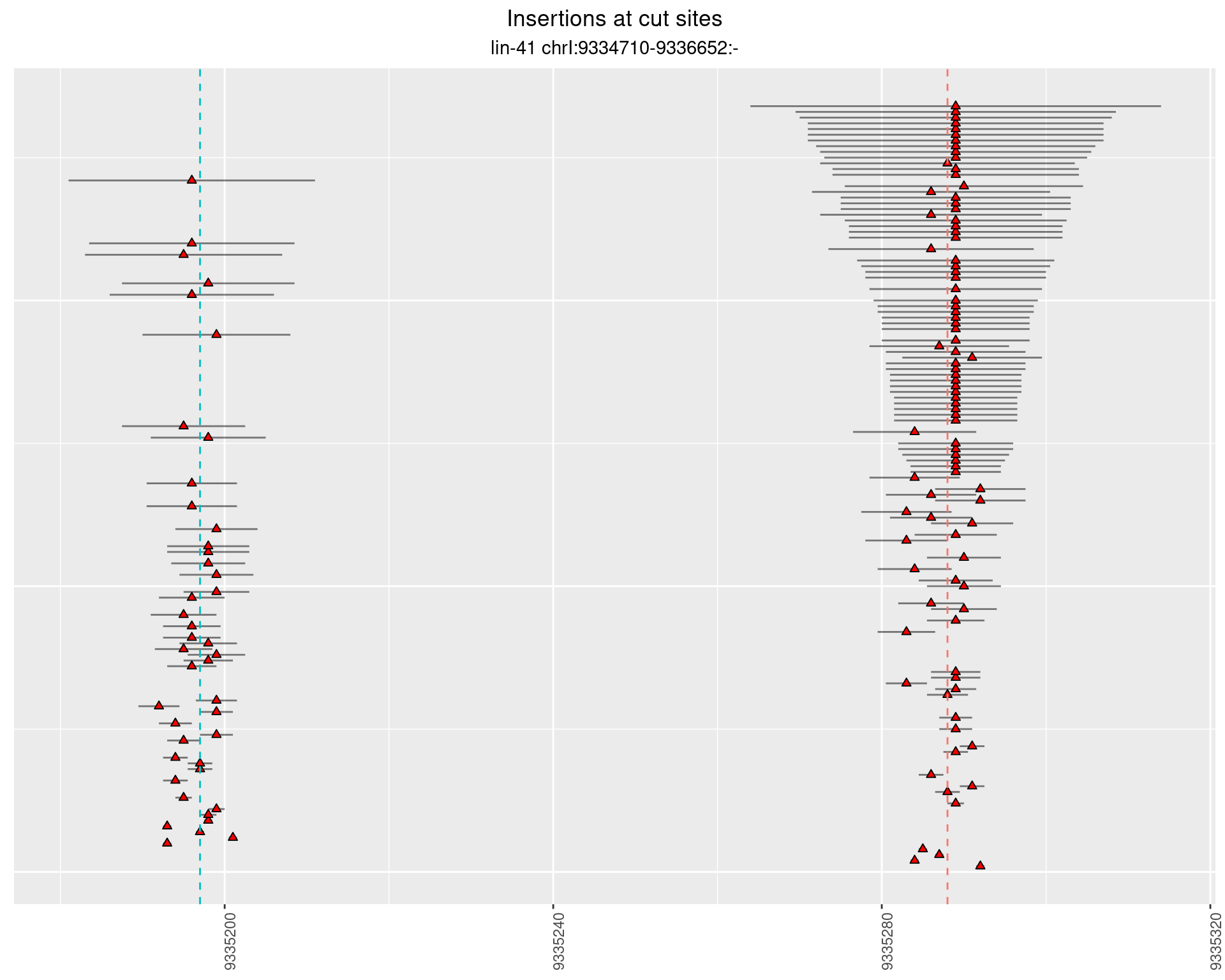
2.8.3 Freq: 1e-06 - 1e-05
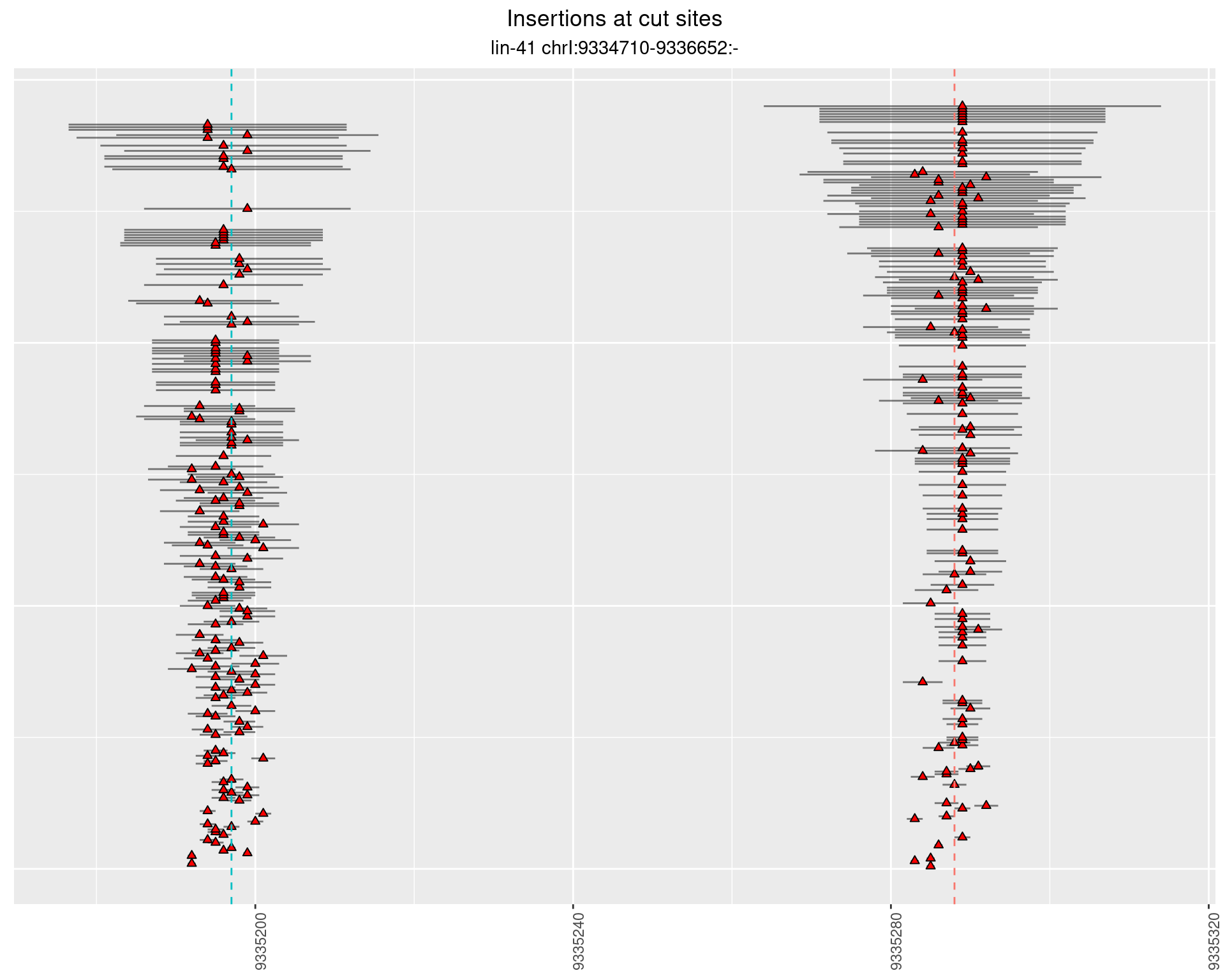
2.8.4 Freq: 0.001 - 0.01
2.9 gen_24C_F3_lin-41_N2
2.9.1 Freq: 1e-06 - 1e-05
2.9.2 Freq: 1e-05 - 1e-04
2.9.3 Freq: 1e-04 - 0.001
2.10 gen_24C_F3_lin-41_sg15sg16
2.10.1 Freq: 1e-04 - 0.001
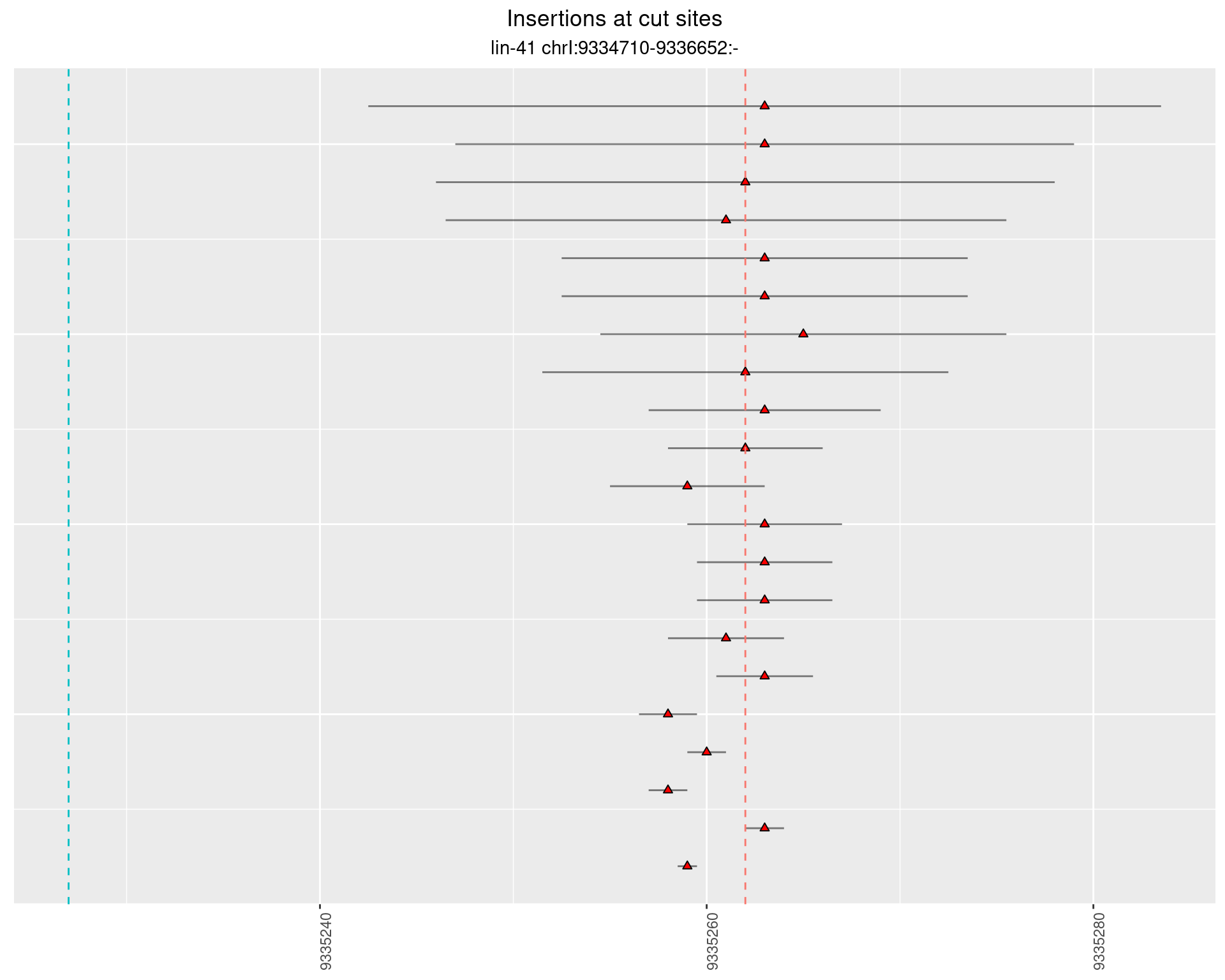
2.10.2 Freq: 0.001 - 0.01
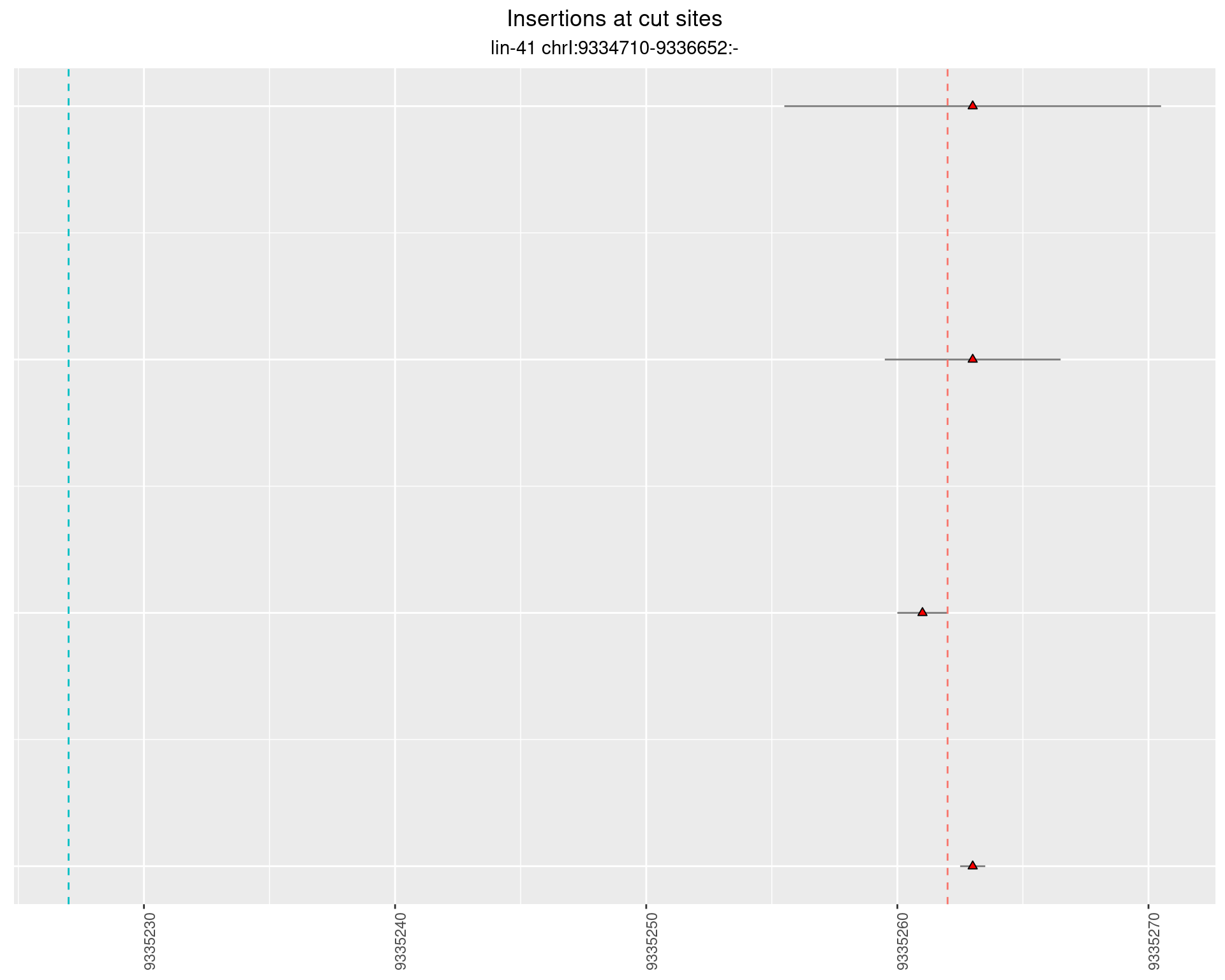
2.10.3 Freq: 1e-06 - 1e-05
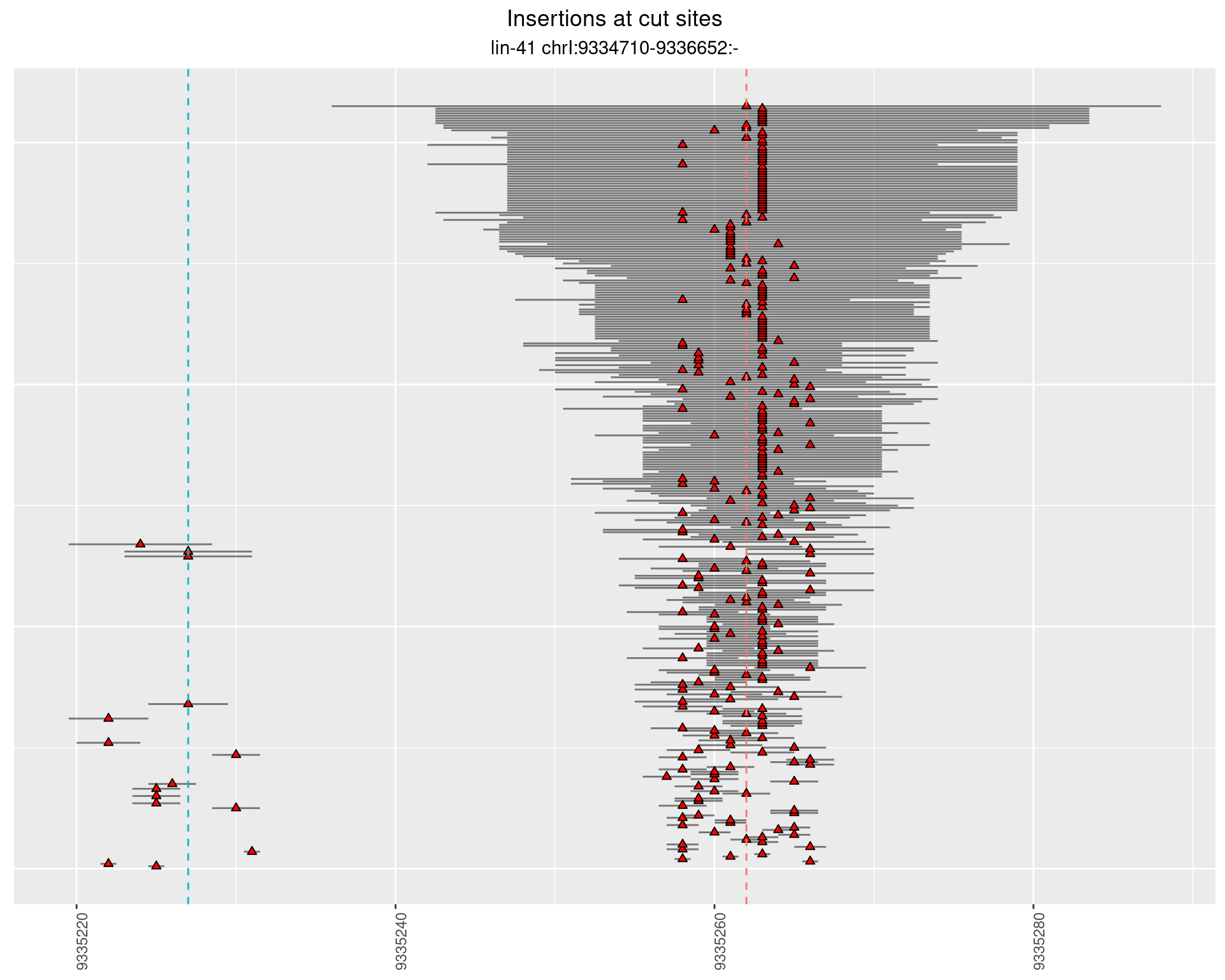
2.10.4 Freq: 1e-05 - 1e-04
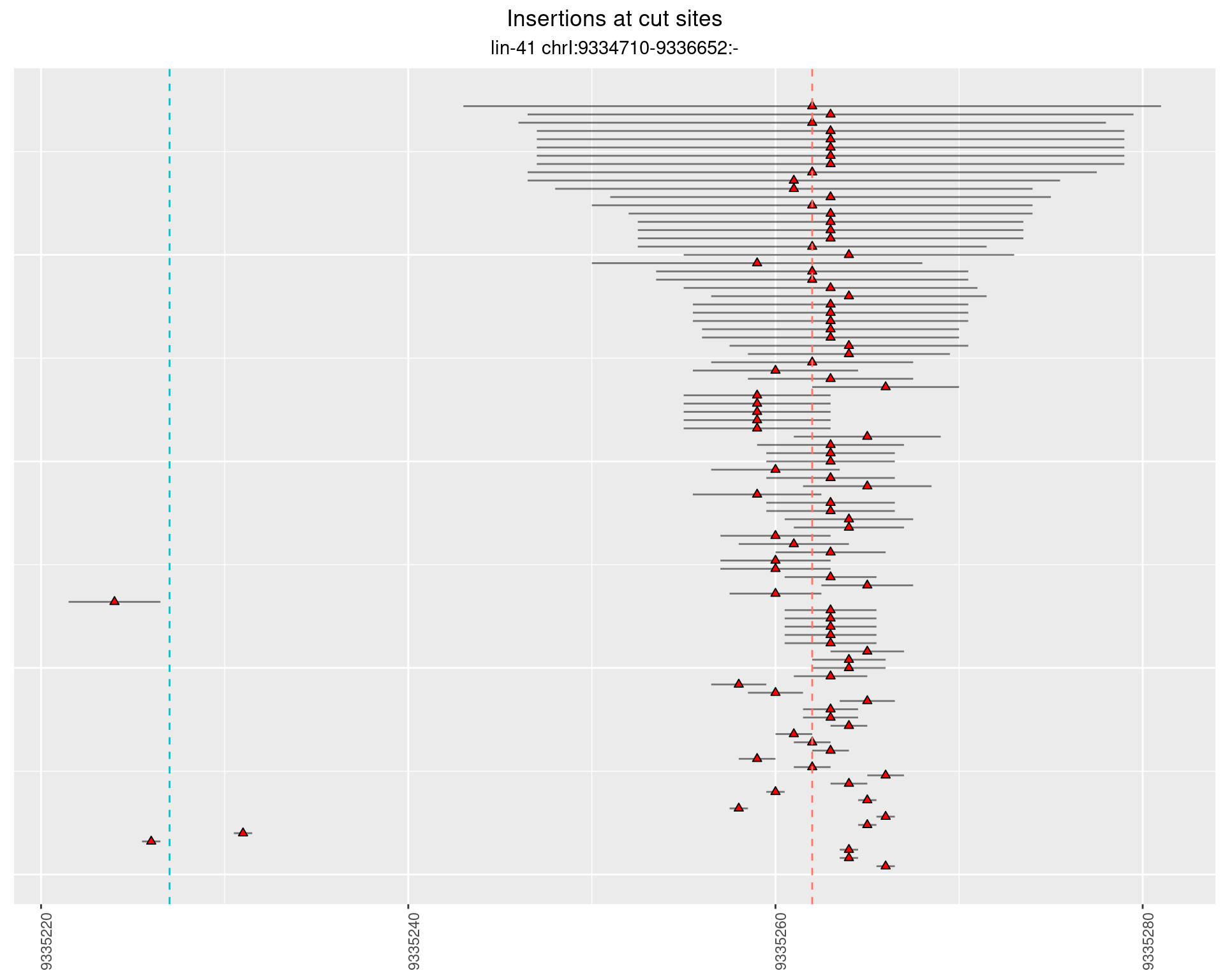
2.11 gen_24C_F3_lin-41_pool
2.11.1 Freq: 1e-05 - 1e-04
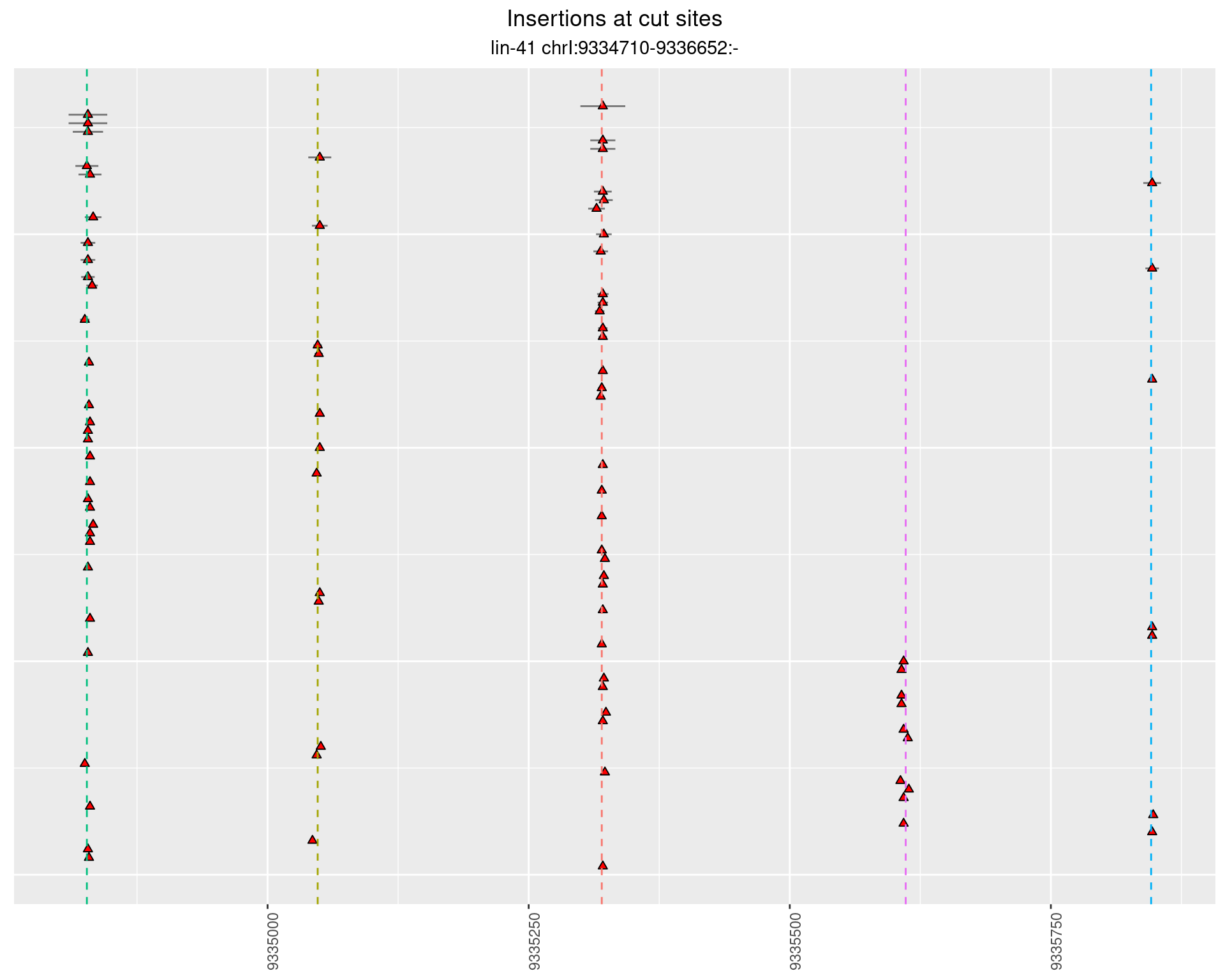
2.11.2 Freq: 1e-04 - 0.001
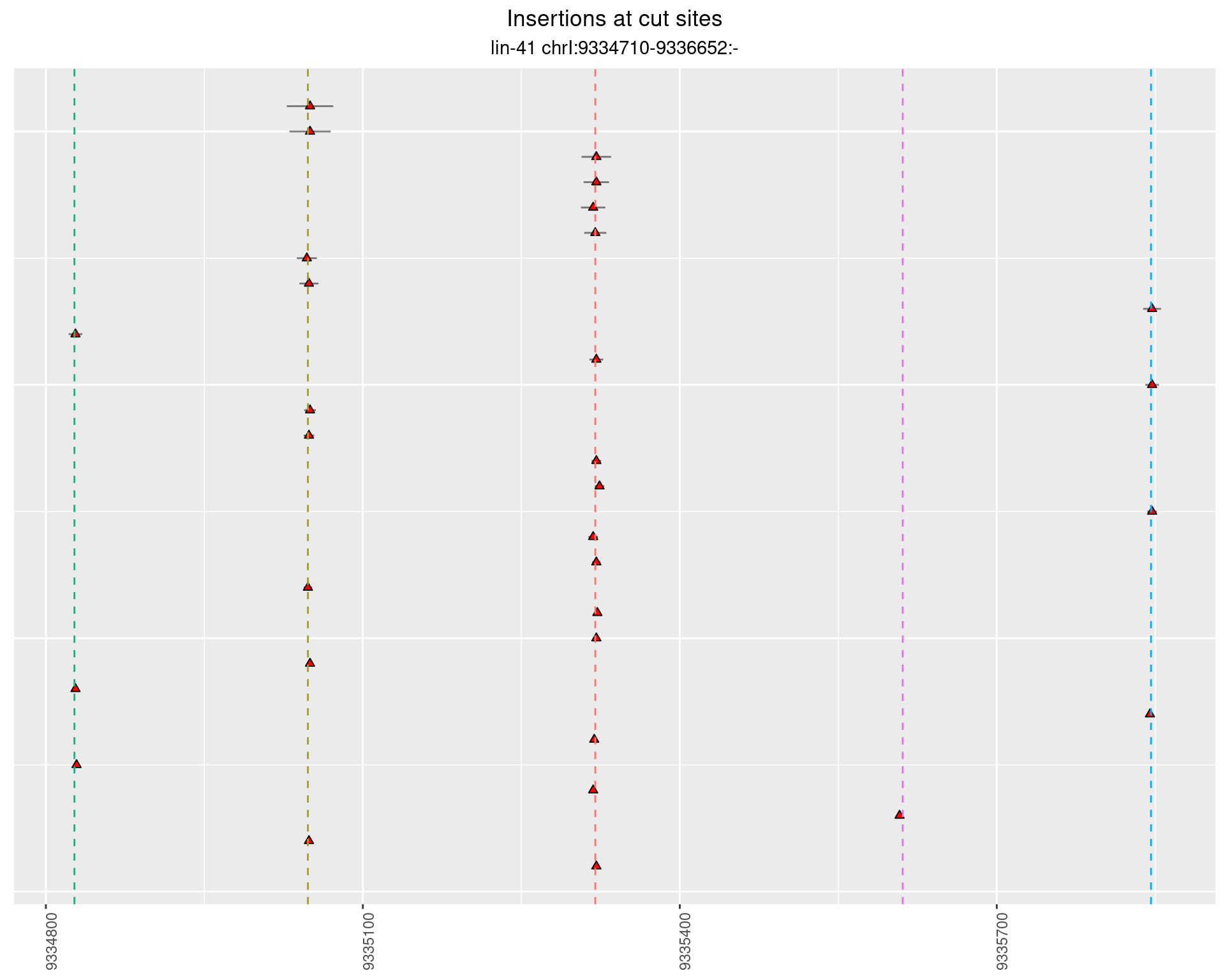
2.11.3 Freq: 0.001 - 0.01
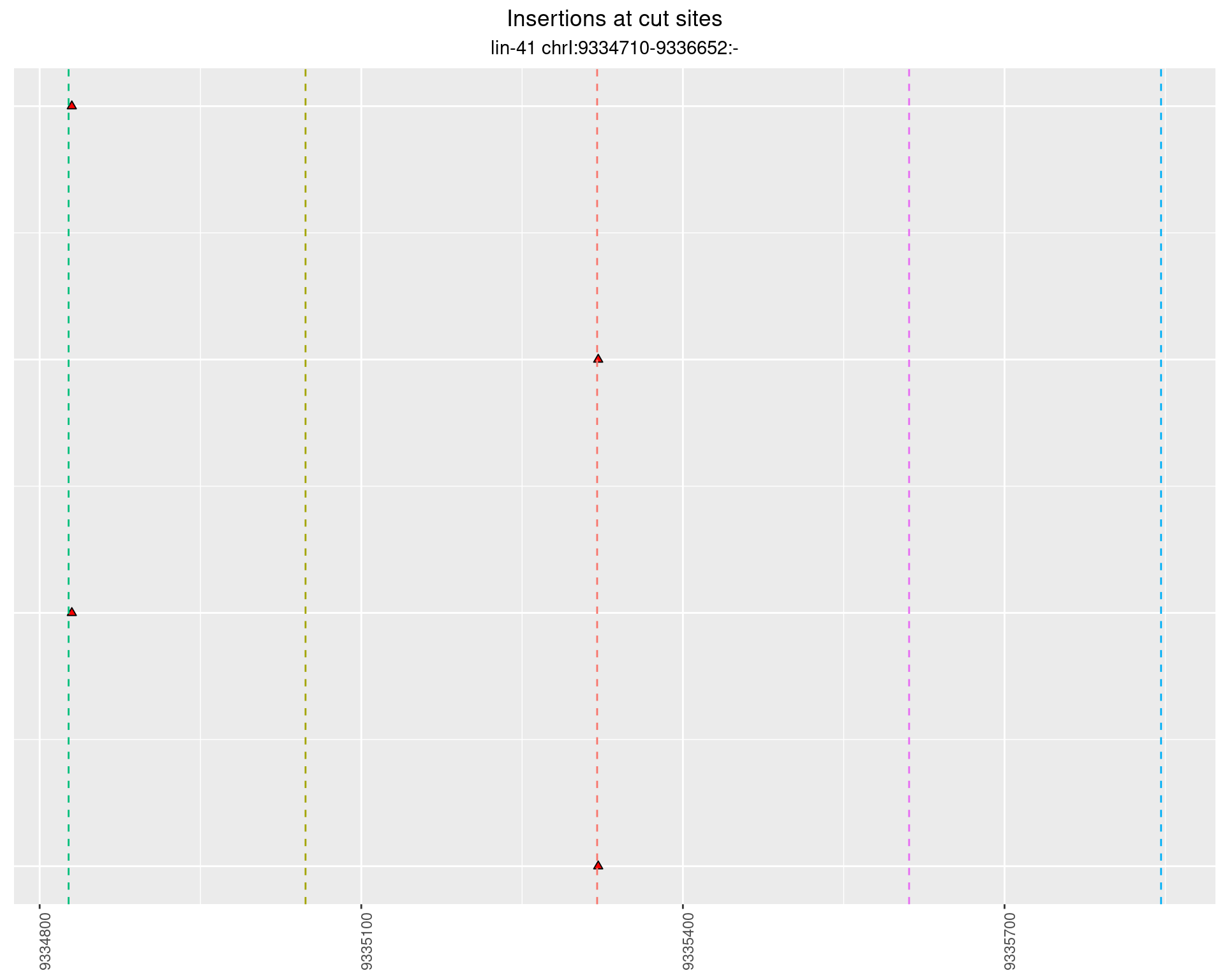
2.11.4 Freq: 1e-06 - 1e-05
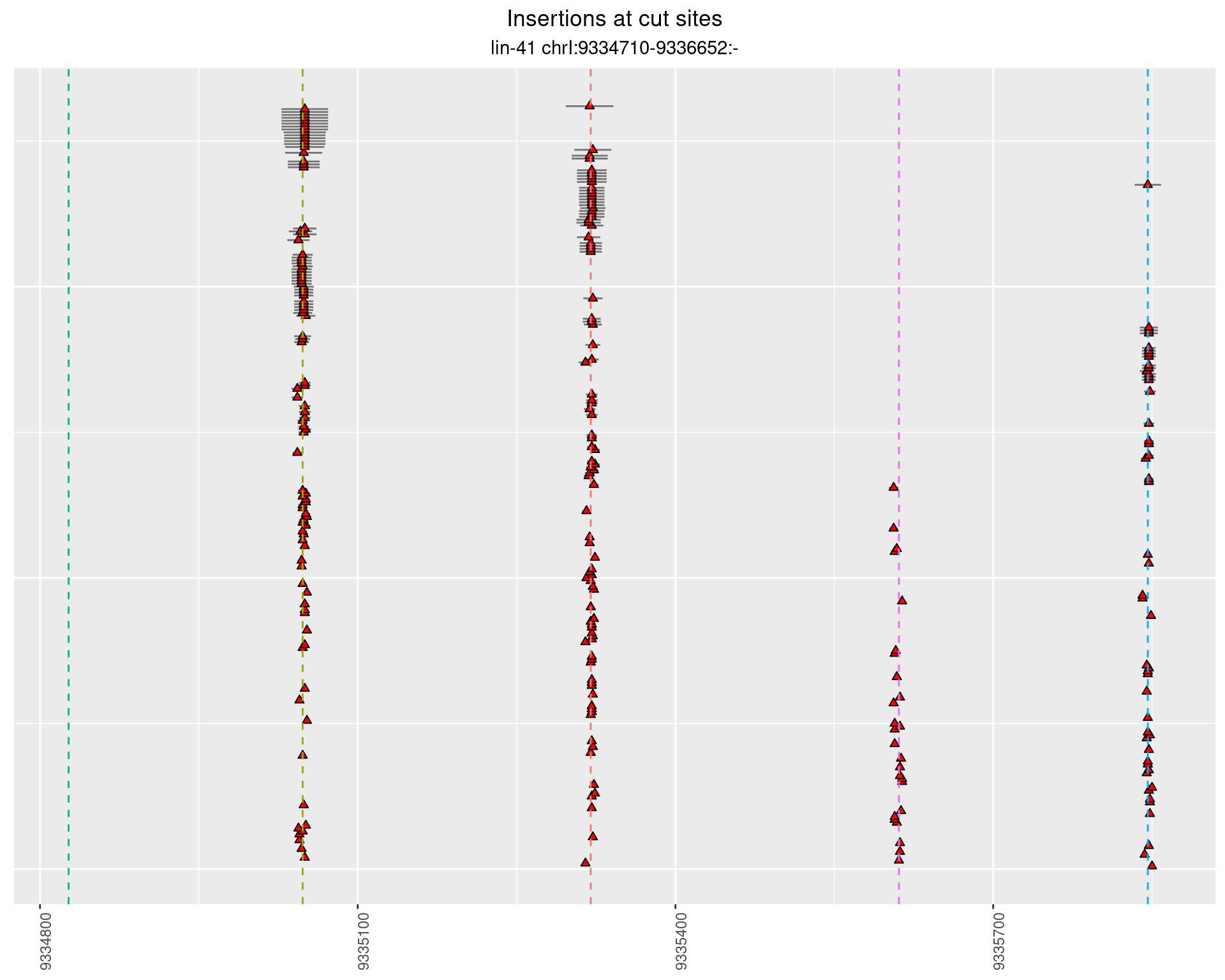
2.12 gen_24C_F3_lin-41_sg26sg27
2.12.1 Freq: 0.001 - 0.01
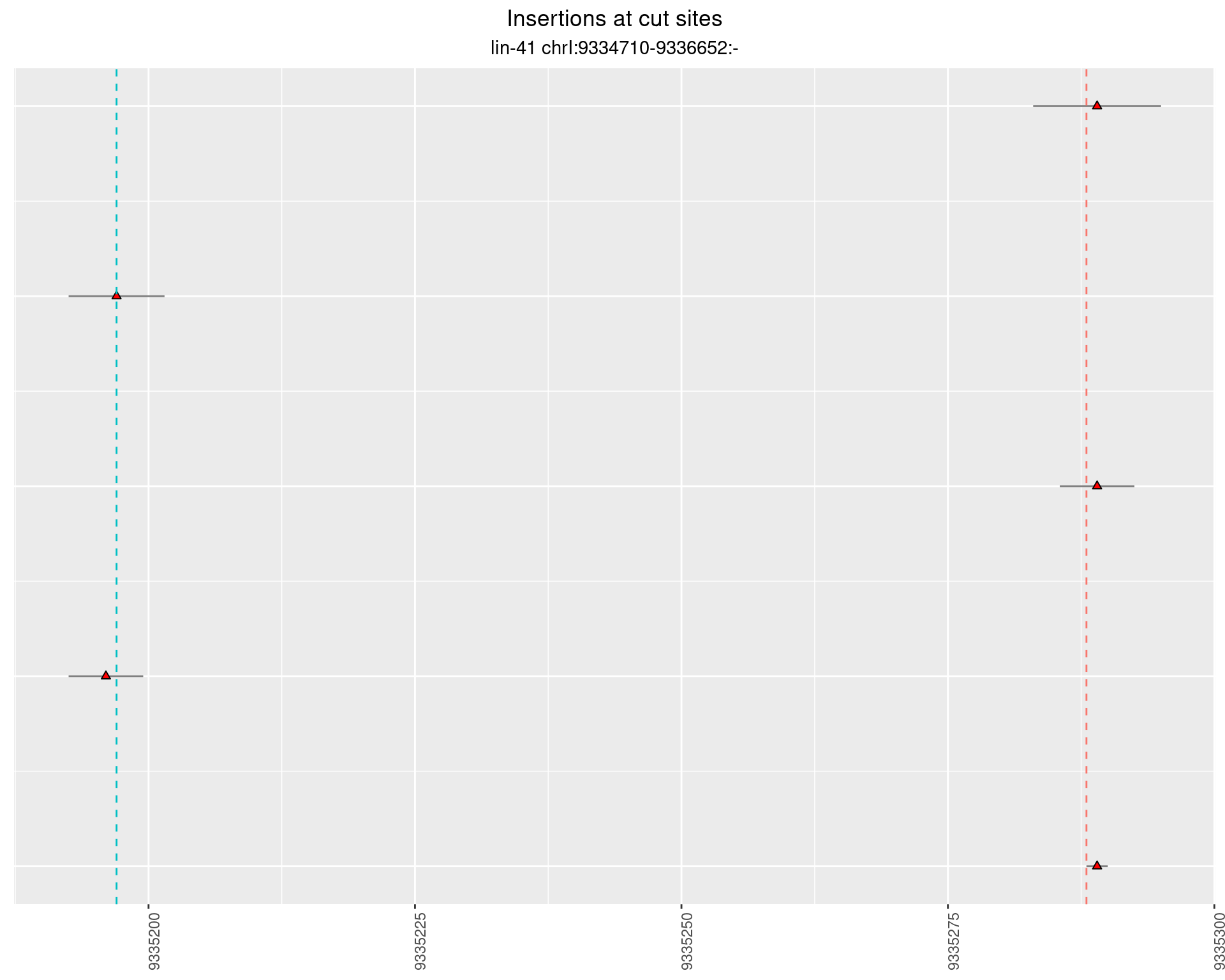
2.12.2 Freq: 1e-05 - 1e-04
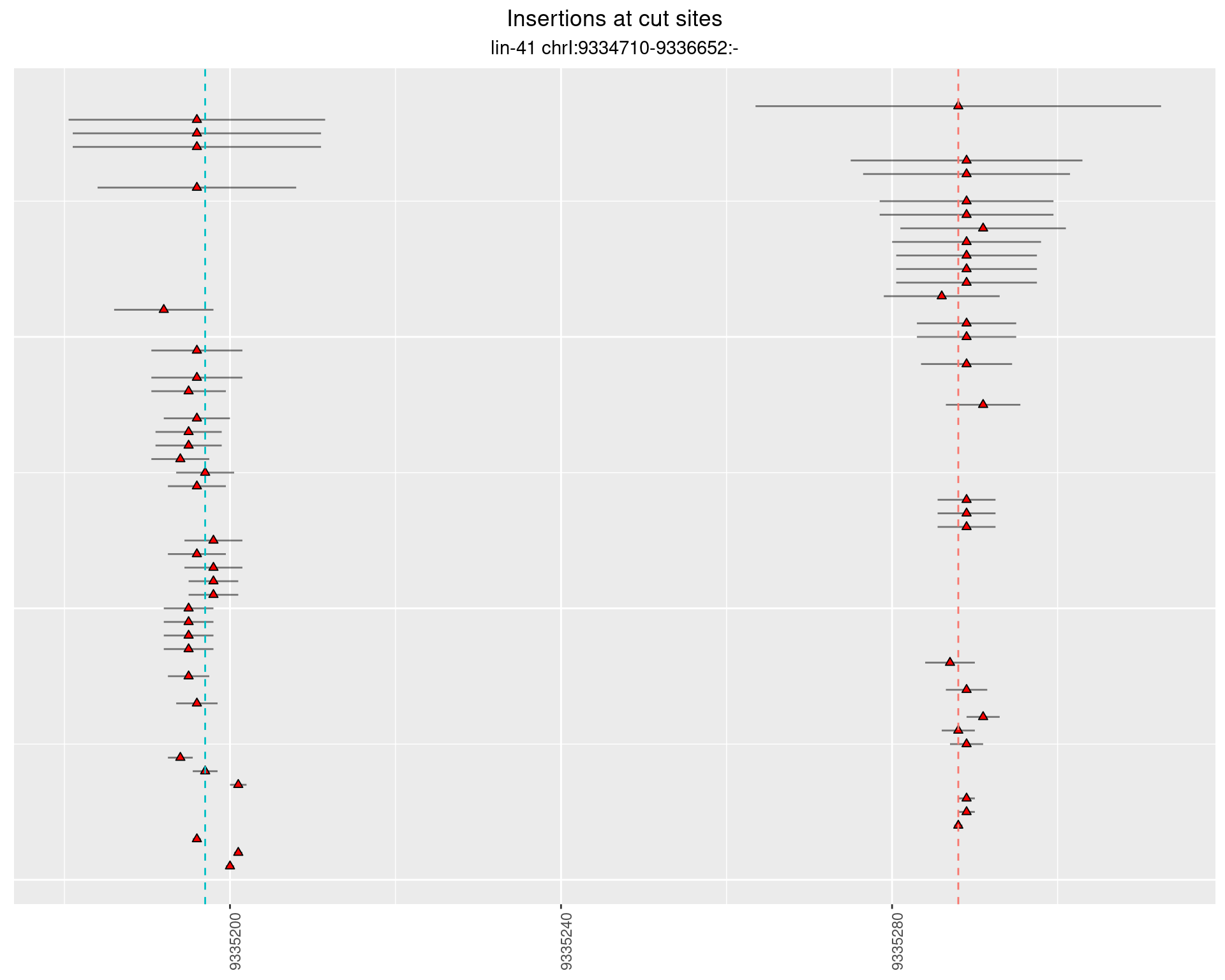
2.12.3 Freq: 1e-04 - 0.001
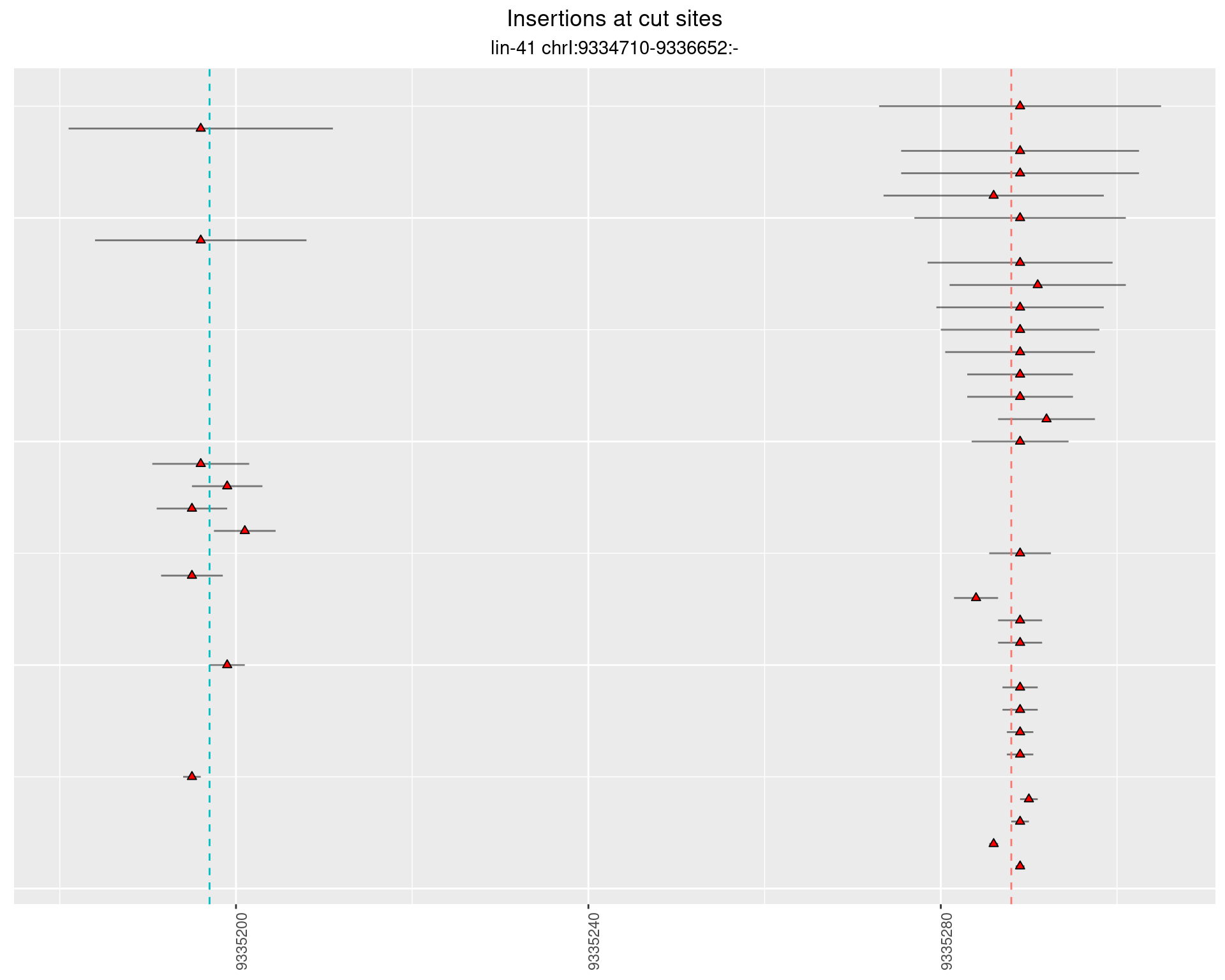
2.12.4 Freq: 1e-06 - 1e-05
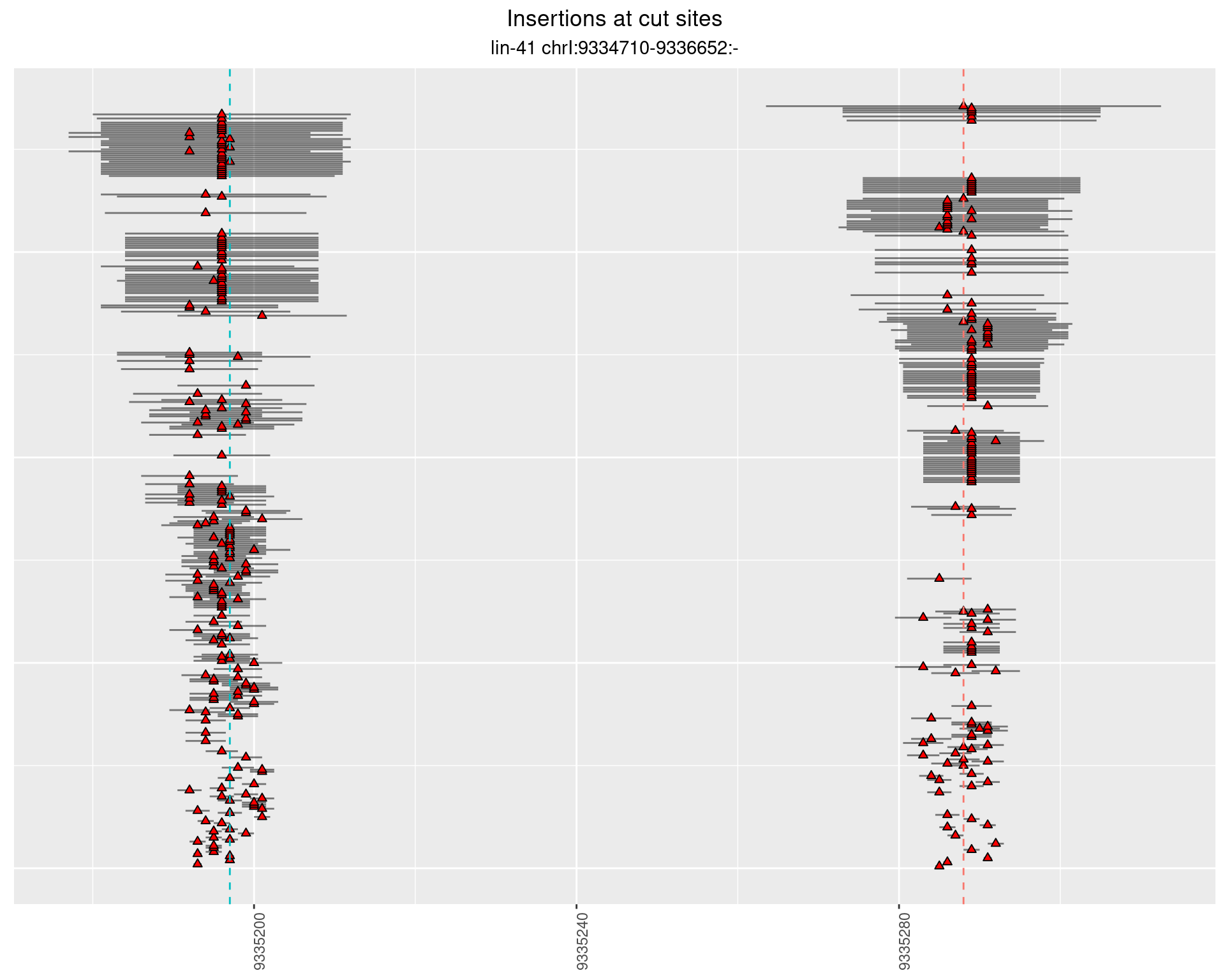
2.13 gen_24C_F4_lin-41_N2
2.13.1 Freq: 1e-06 - 1e-05
2.13.2 Freq: 1e-04 - 0.001
2.13.3 Freq: 1e-05 - 1e-04
2.14 gen_24C_F4_lin-41_sg15sg16
2.14.1 Freq: 0.001 - 0.01
2.14.2 Freq: 1e-04 - 0.001
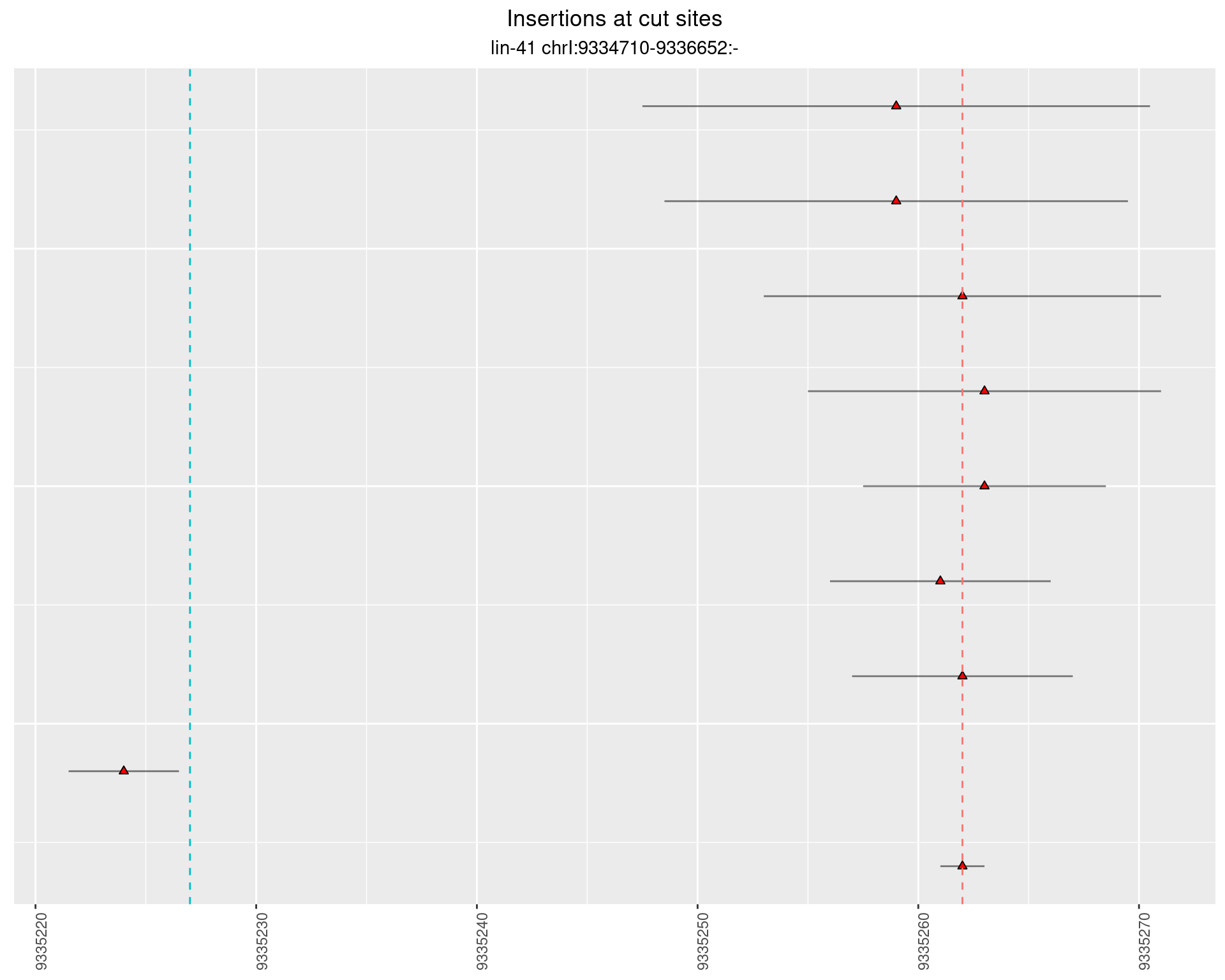
2.14.3 Freq: 1e-06 - 1e-05
2.14.4 Freq: 1e-05 - 1e-04
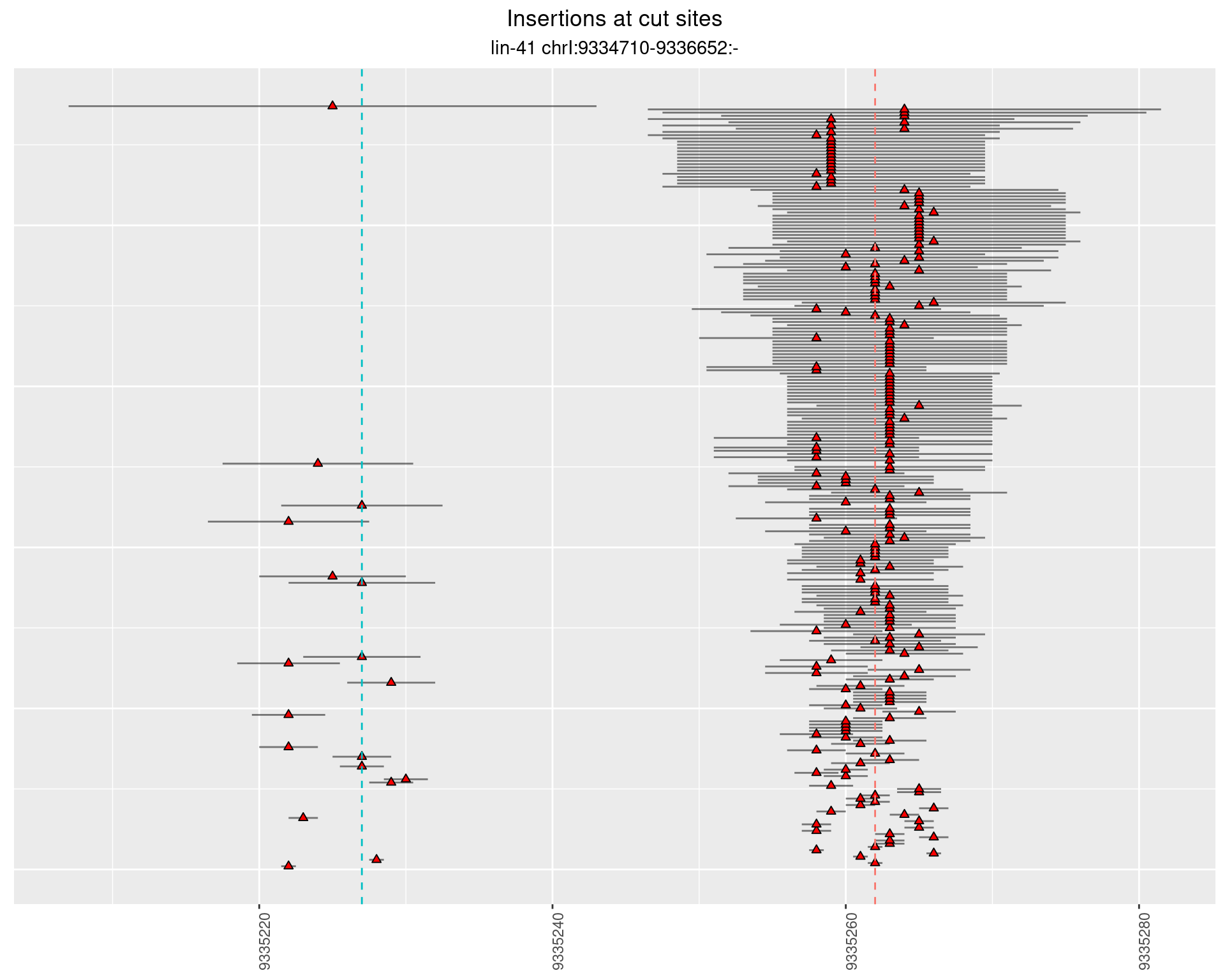
2.15 gen_24C_F4_lin-41_pool
2.15.1 Freq: 1e-04 - 0.001
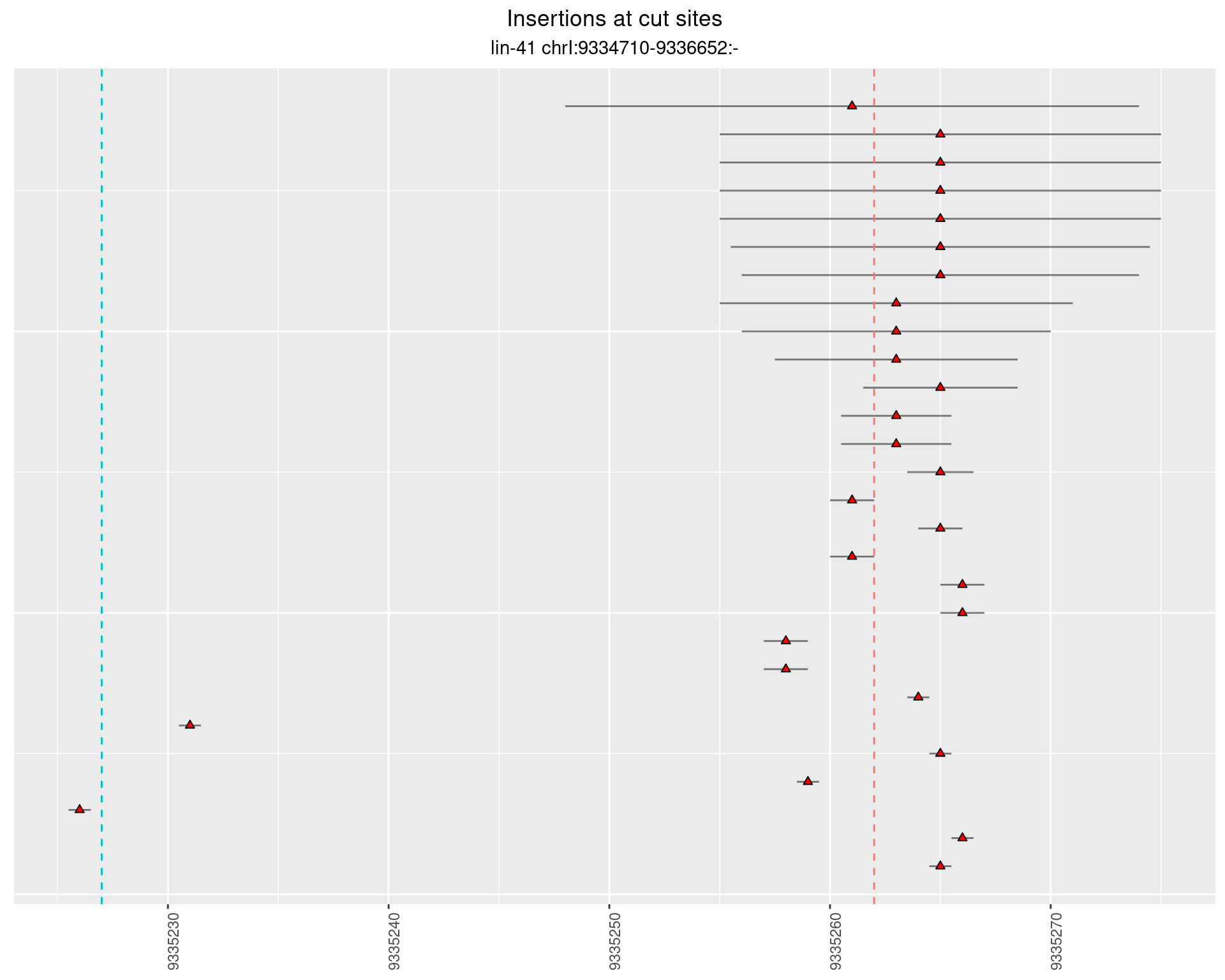
2.15.2 Freq: 0.001 - 0.01
2.15.3 Freq: 1e-05 - 1e-04
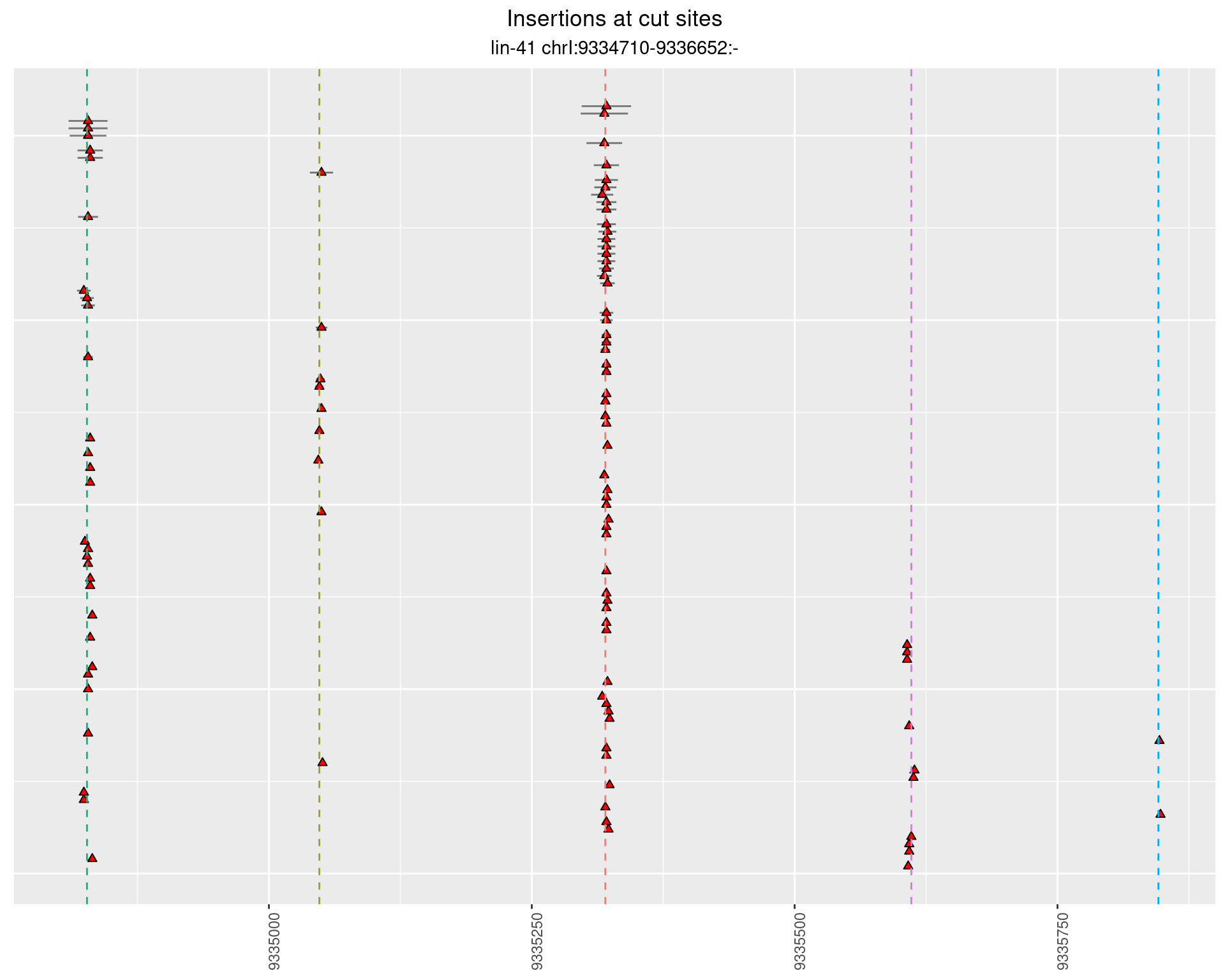
2.15.4 Freq: 1e-06 - 1e-05
2.16 gen_24C_F4_lin-41_sg26sg27
2.16.1 Freq: 1e-04 - 0.001
2.16.2 Freq: 1e-06 - 1e-05
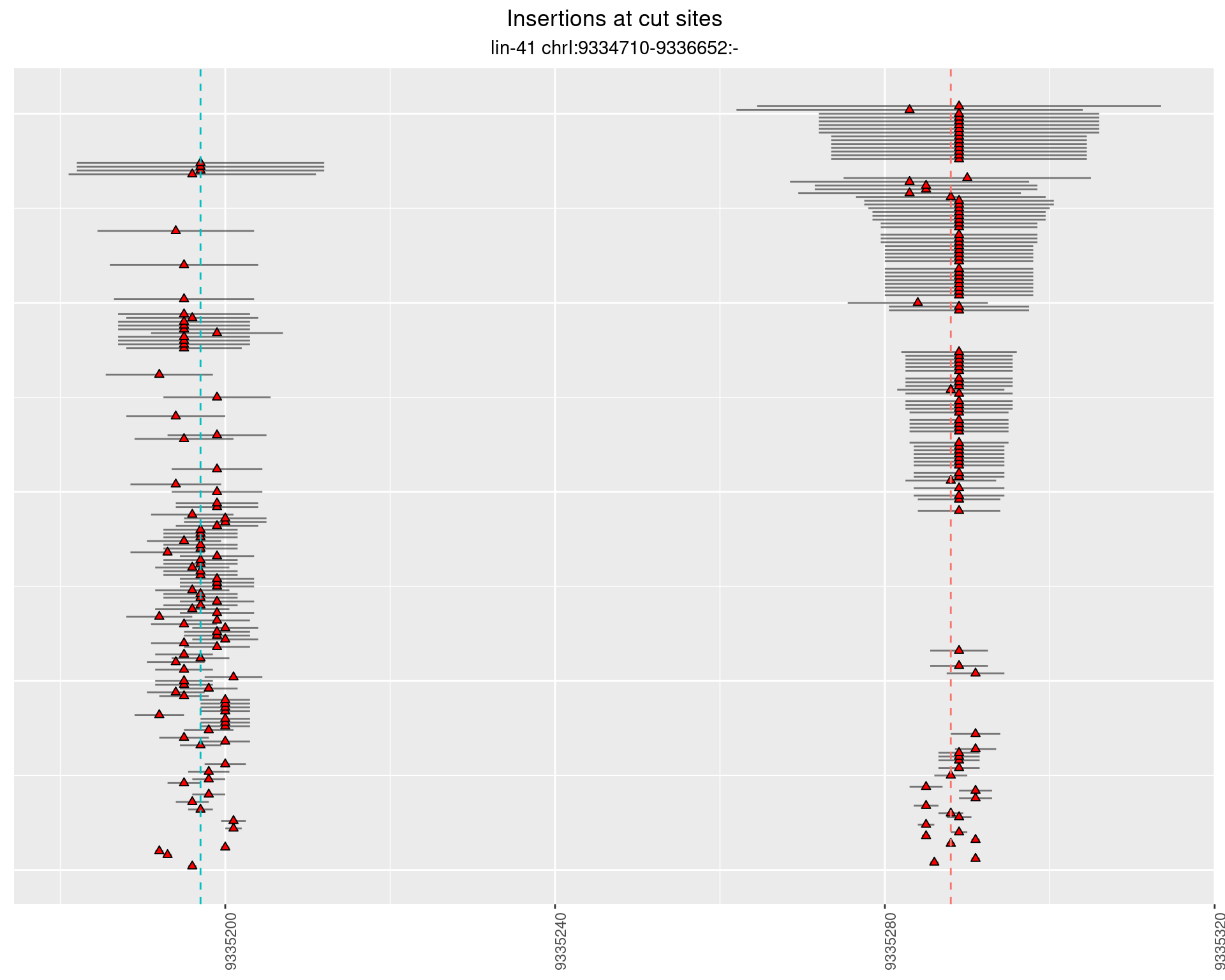
2.16.3 Freq: 0.001 - 0.01
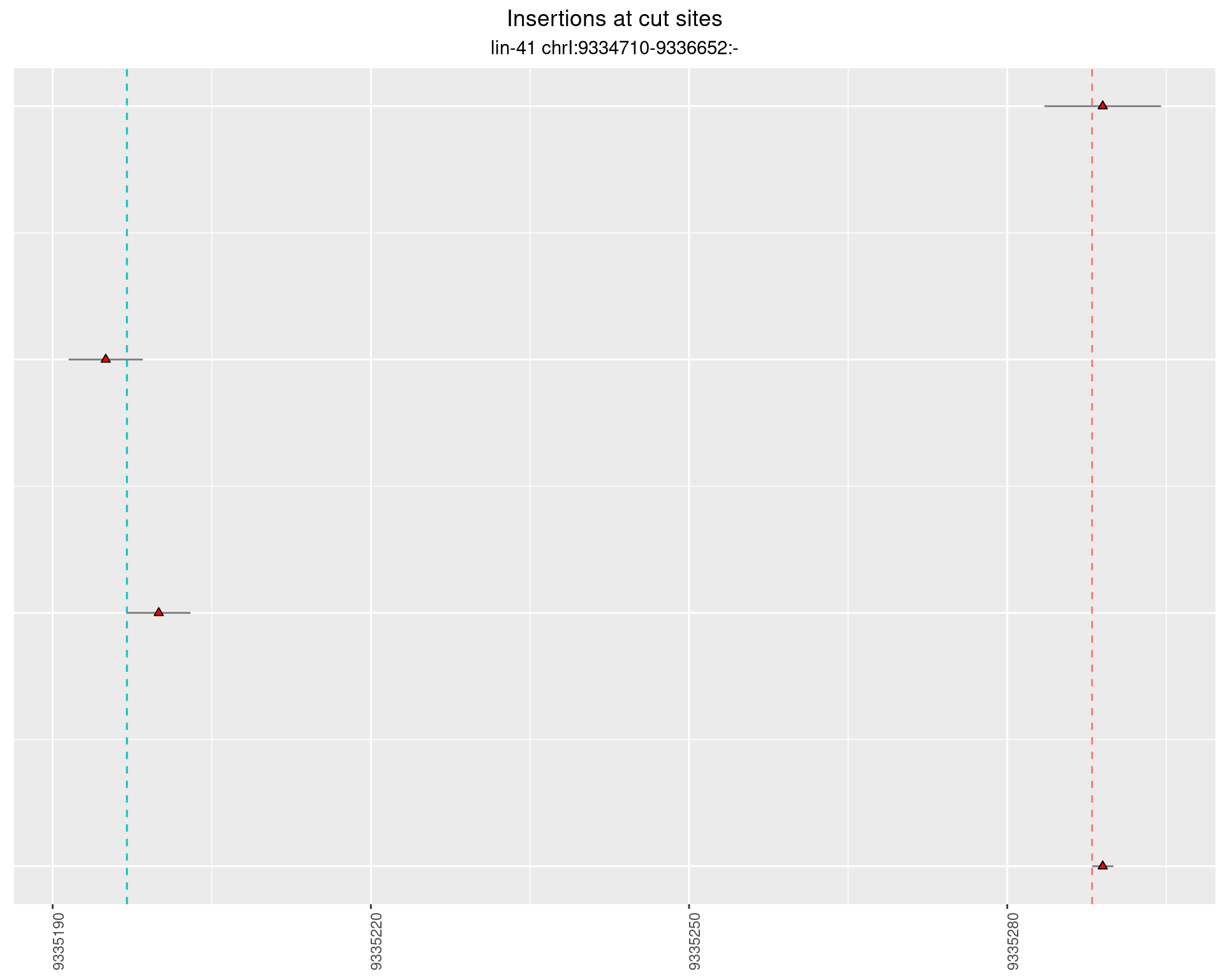
2.16.4 Freq: 1e-05 - 1e-04
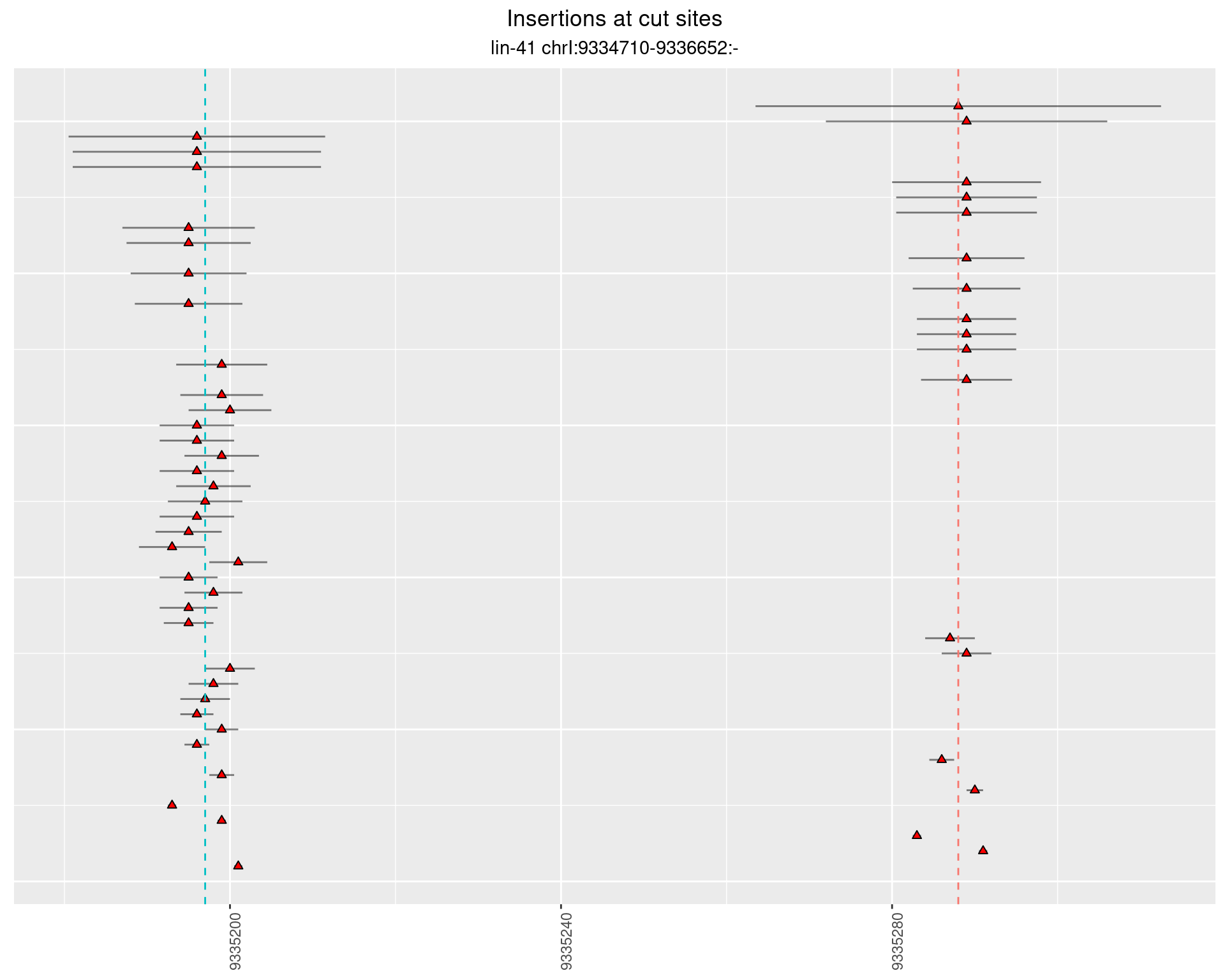
2.17 gen_24C_F5_lin-41_N2
2.17.1 Freq: 1e-04 - 0.001
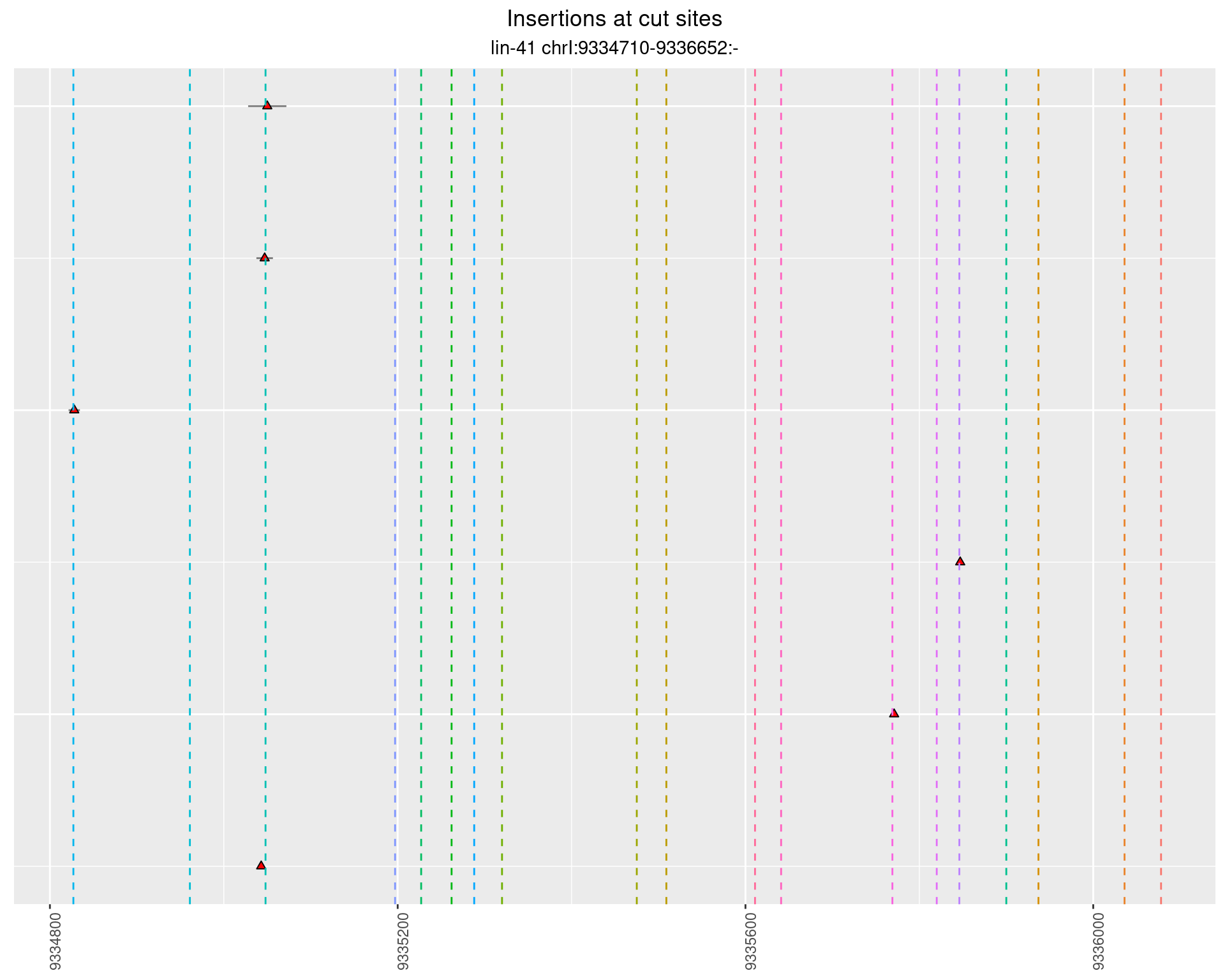
2.17.2 Freq: 1e-05 - 1e-04
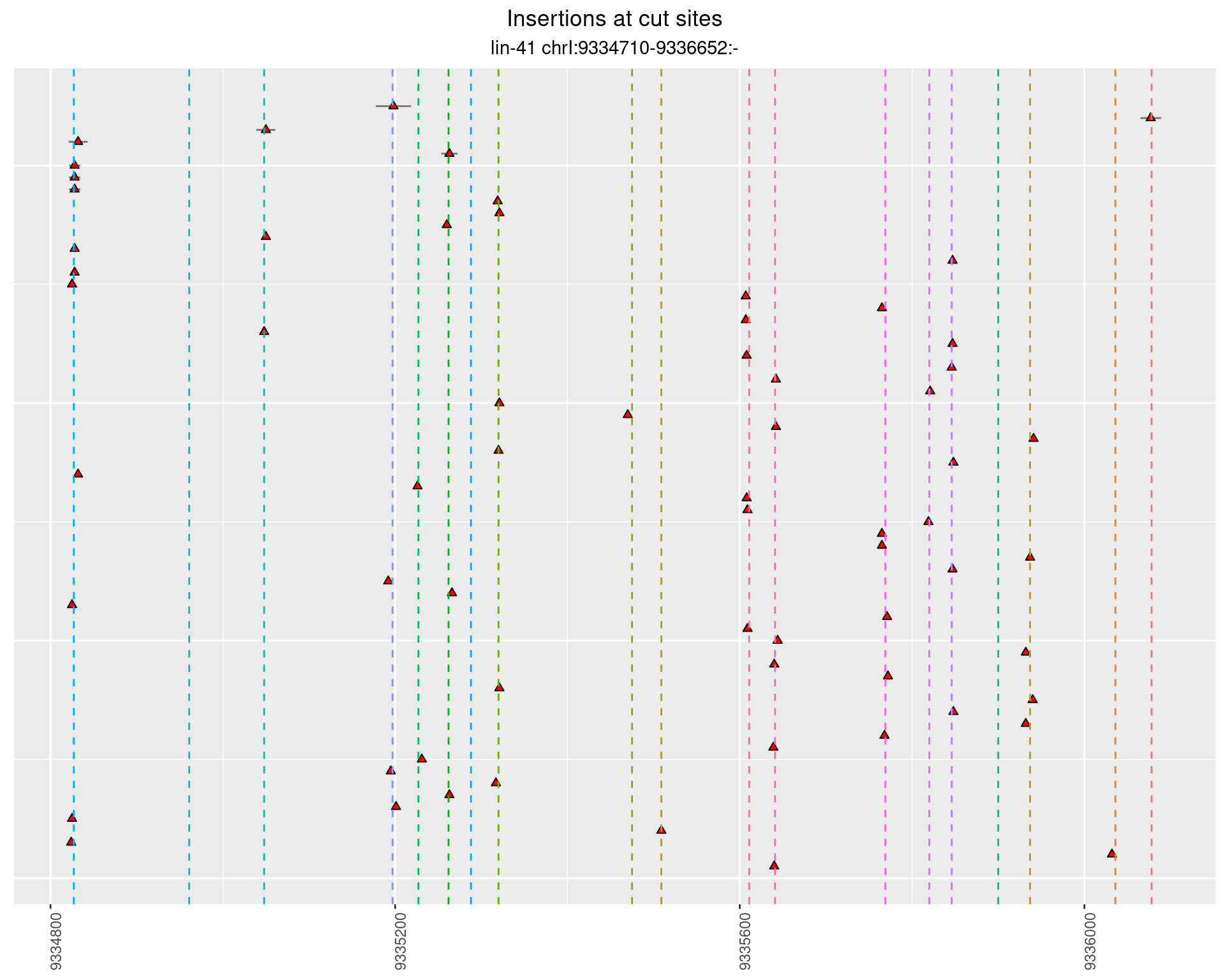
2.17.3 Freq: 1e-06 - 1e-05
2.17.4 Freq: 0.001 - 0.01
2.18 gen_24C_F5_lin-41_sg15sg16
2.18.1 Freq: 0.001 - 0.01

2.18.2 Freq: 1e-06 - 1e-05
2.18.3 Freq: 1e-05 - 1e-04
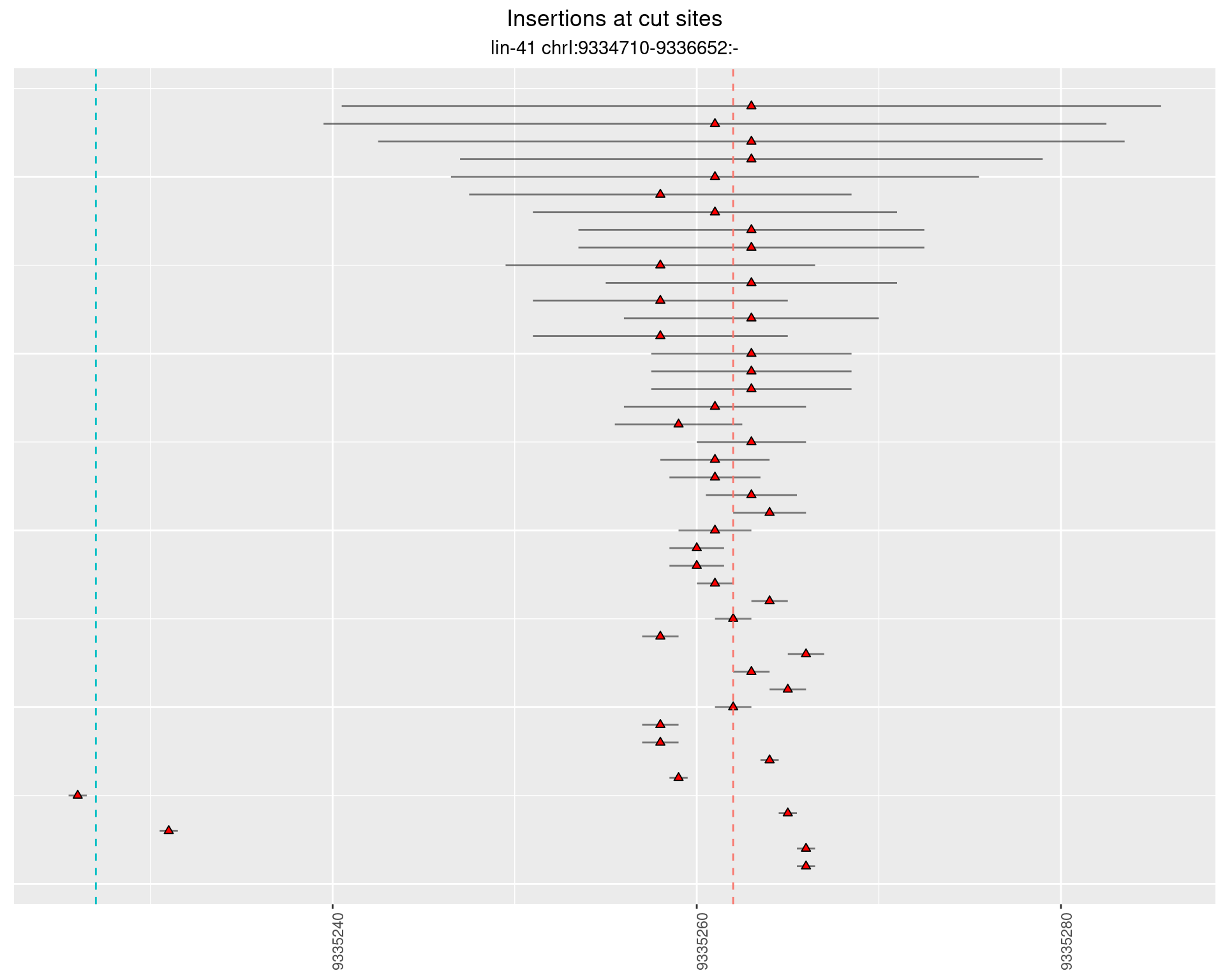
2.18.4 Freq: 1e-04 - 0.001
2.19 gen_24C_F5_lin-41_pool
2.19.1 Freq: 1e-05 - 1e-04
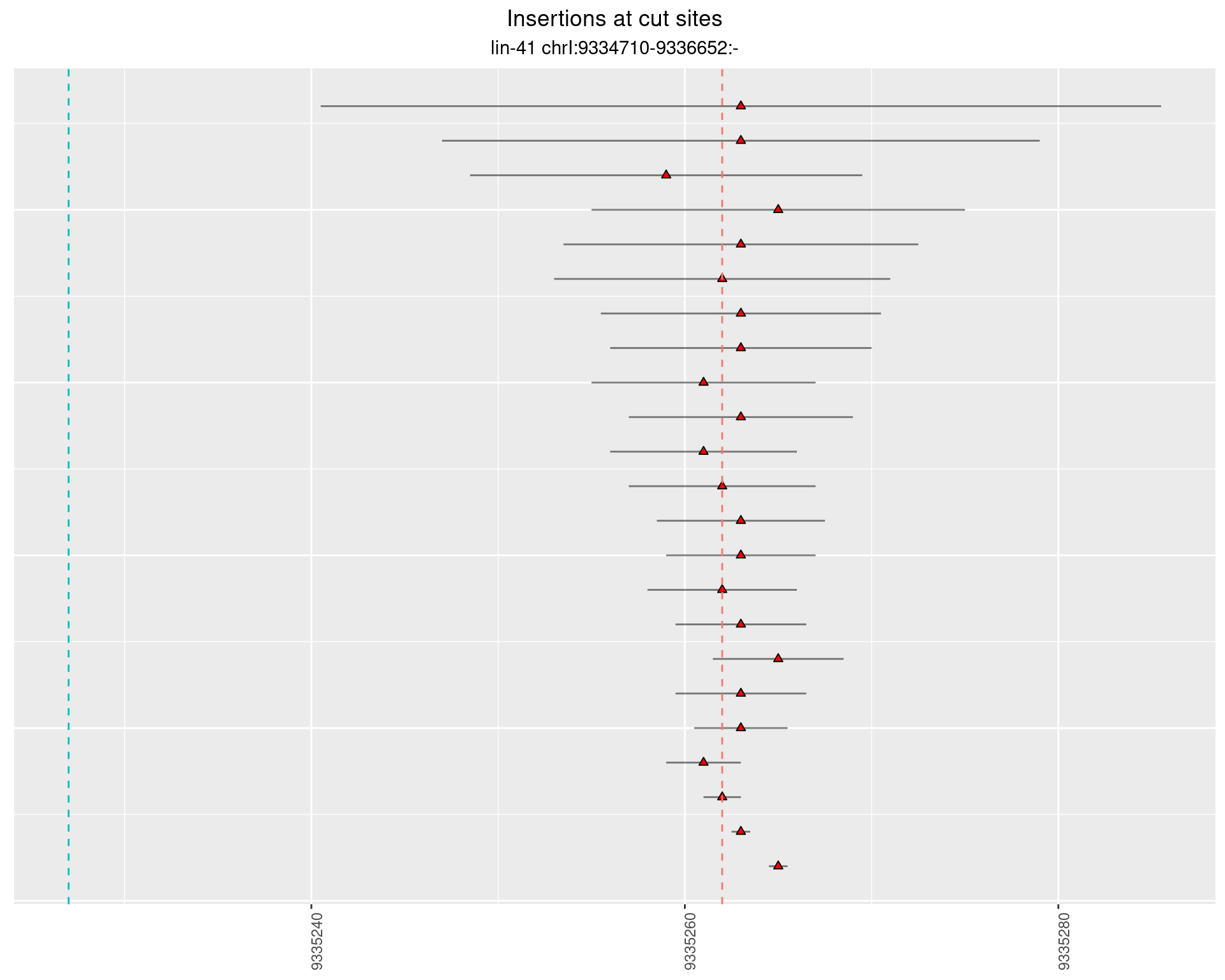
2.19.2 Freq: 1e-04 - 0.001
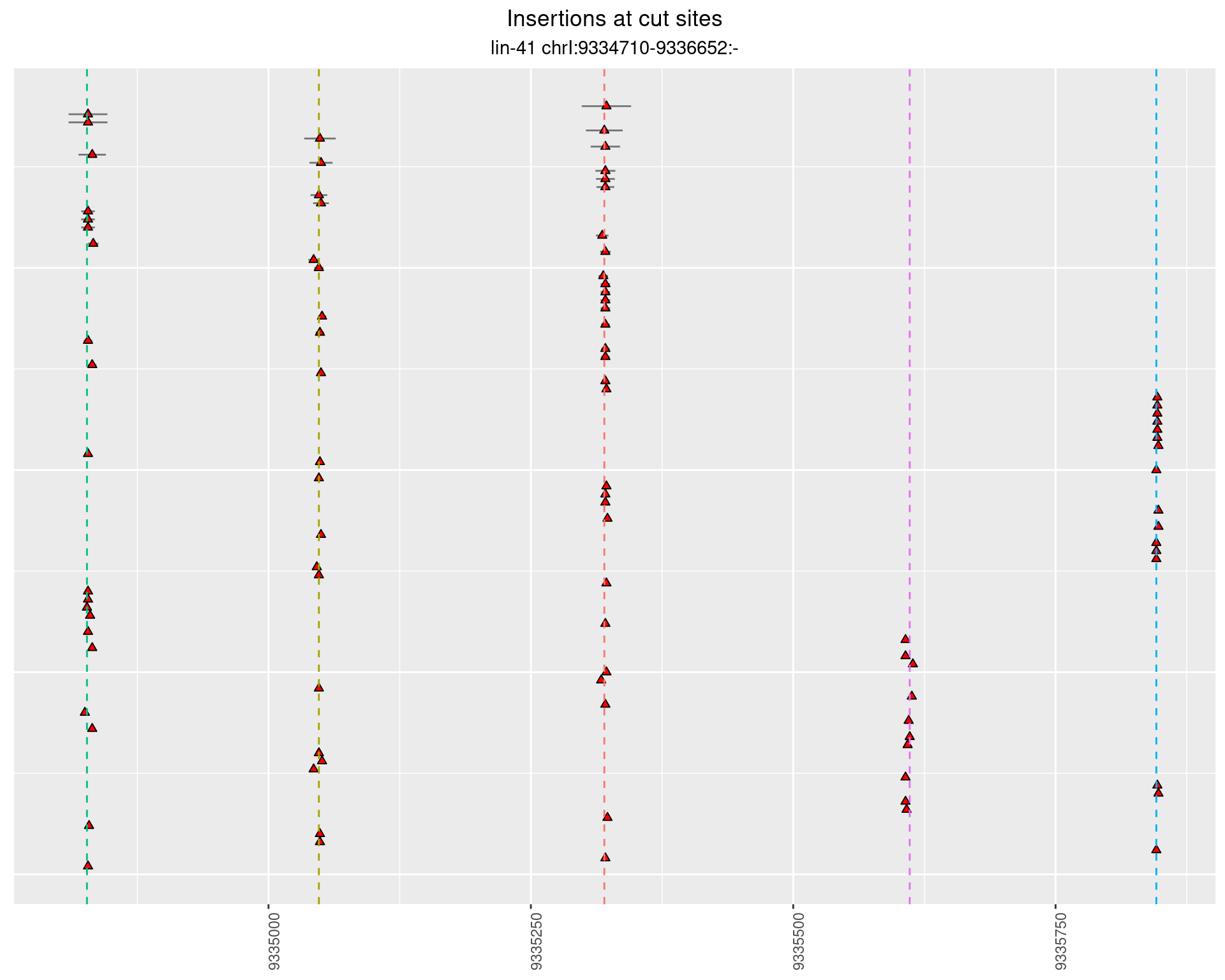
2.19.3 Freq: 0.001 - 0.01
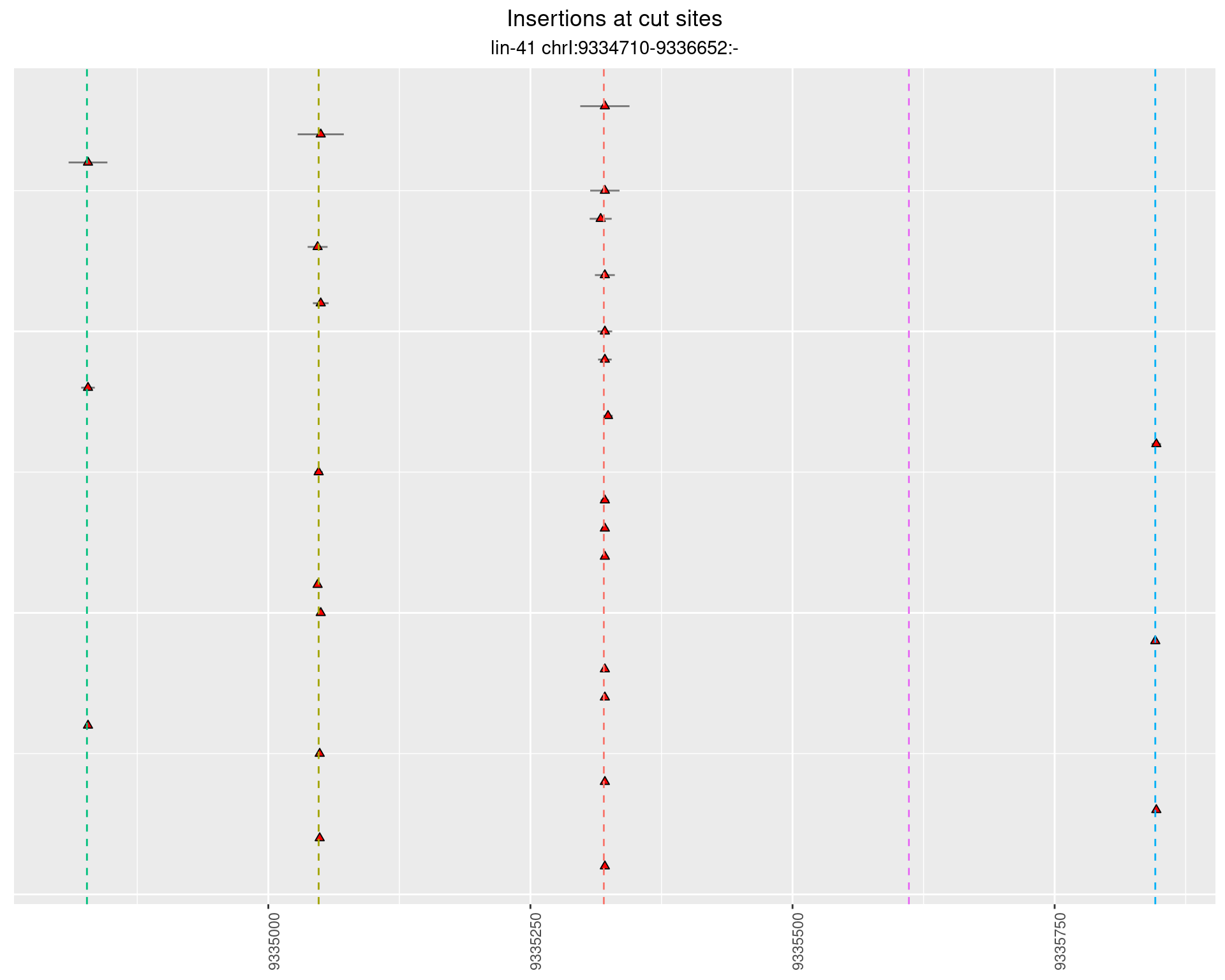
2.19.4 Freq: 1e-06 - 1e-05
2.20 gen_24C_F5_lin-41_sg26sg27
2.20.1 Freq: 1e-04 - 0.001
2.20.2 Freq: 1e-06 - 1e-05
2.20.3 Freq: 0.001 - 0.01
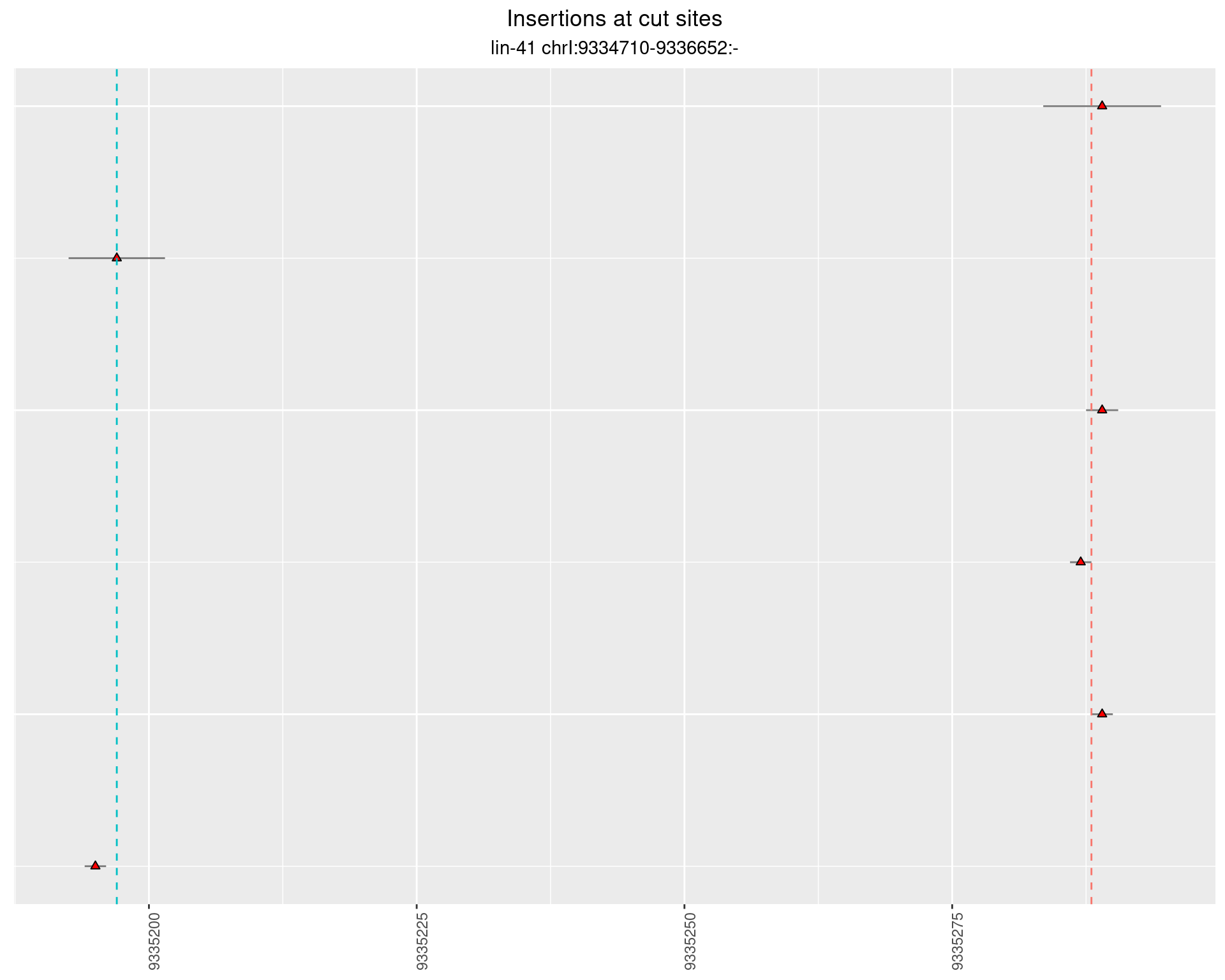
2.20.4 Freq: 1e-05 - 1e-04
2.21 gen_16C_F2_lin-41_N2
2.21.1 Freq: 1e-06 - 1e-05
2.21.2 Freq: 1e-05 - 1e-04
2.21.3 Freq: 1e-04 - 0.001
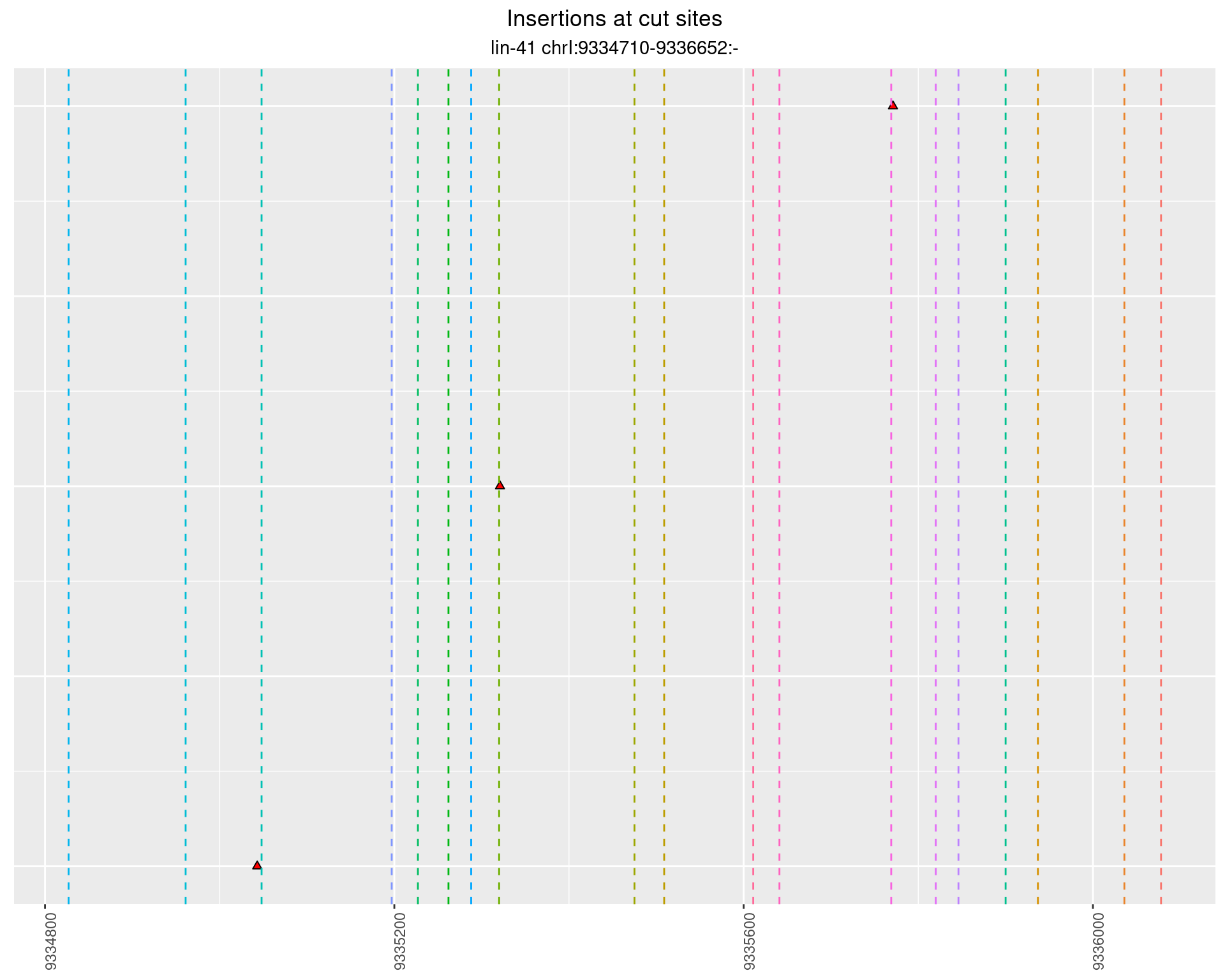
2.22 gen_16C_F2_lin-41_sg15sg16
2.22.1 Freq: 1e-04 - 0.001
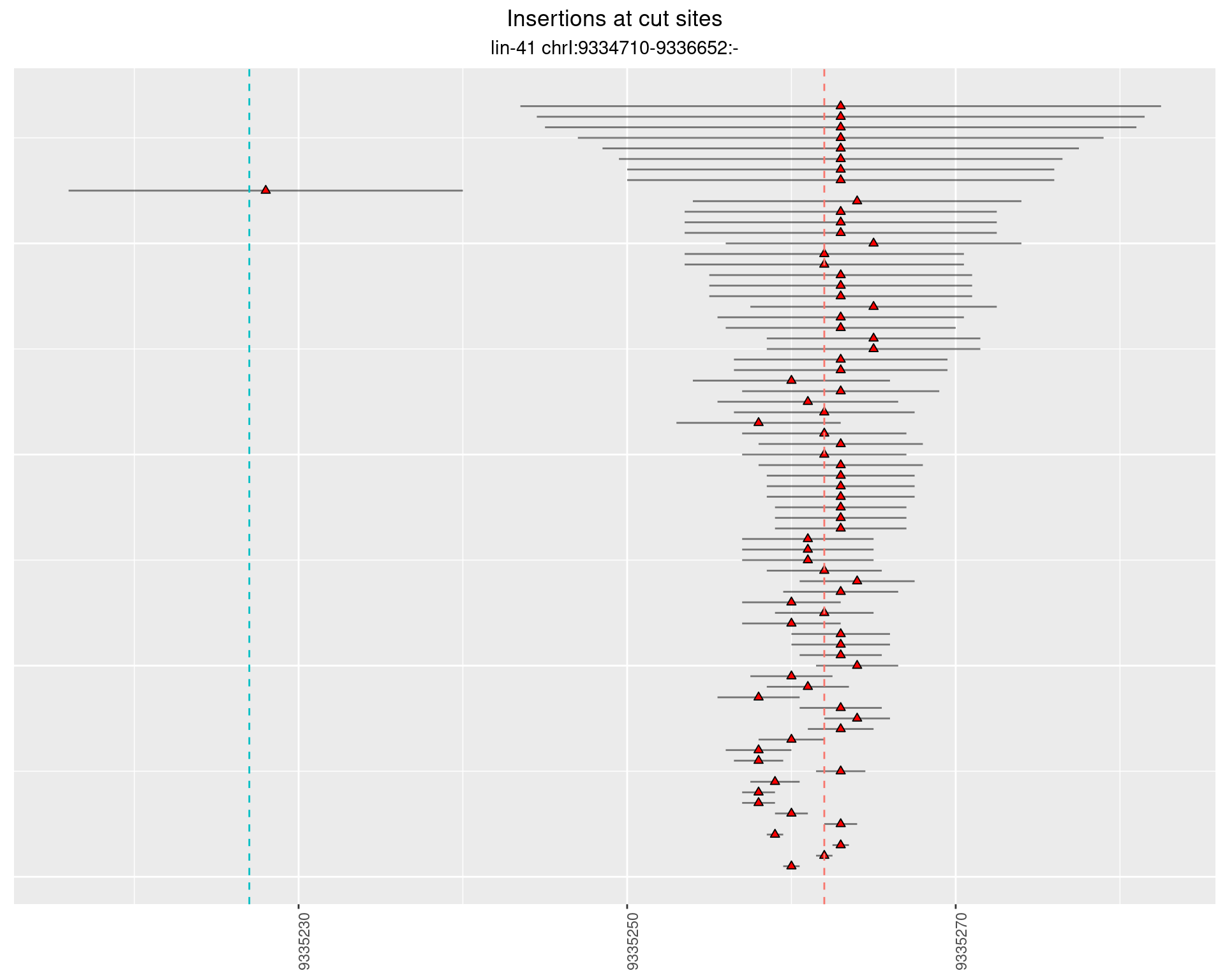
2.22.2 Freq: 1e-05 - 1e-04
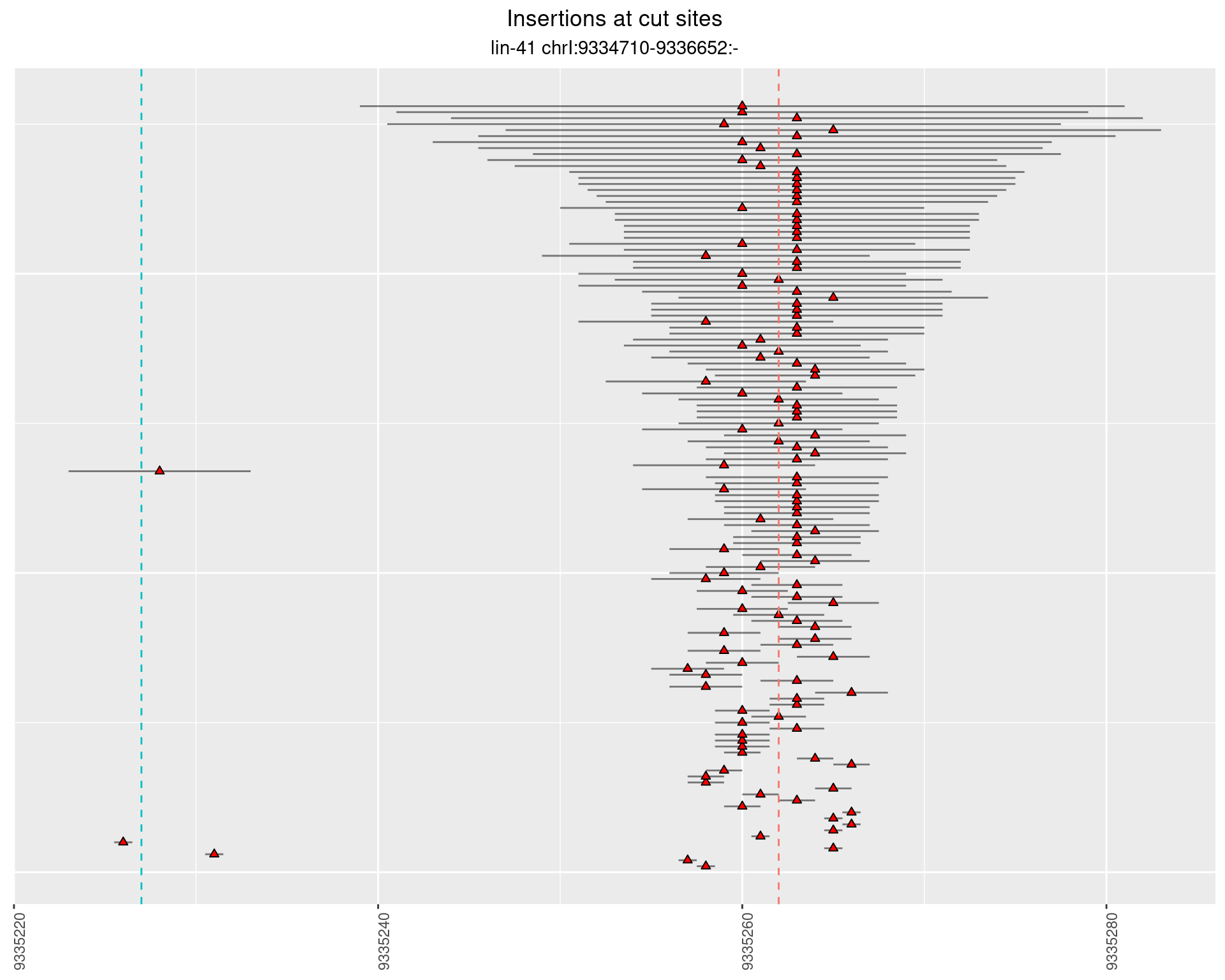
2.22.3 Freq: 1e-06 - 1e-05
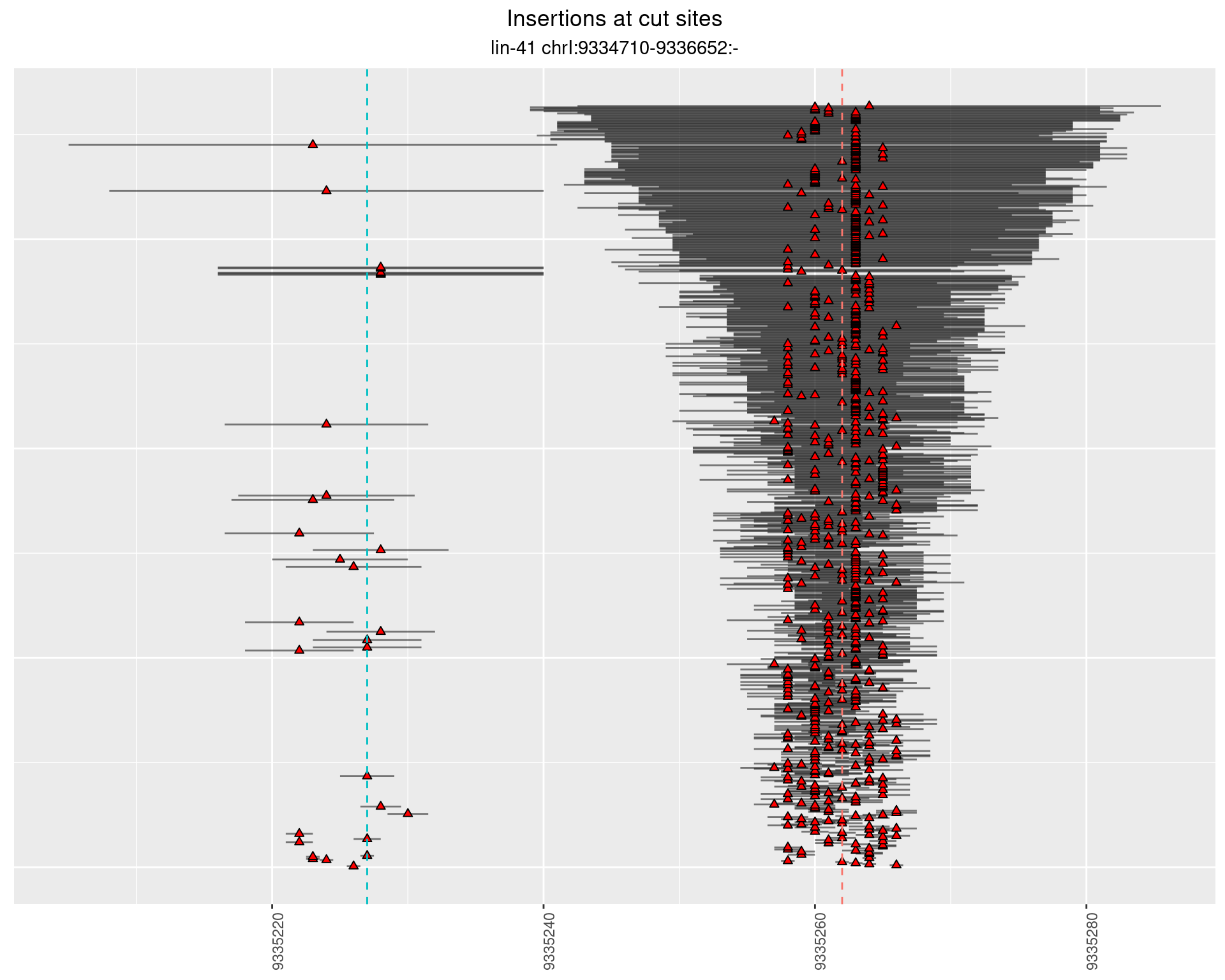
2.22.4 Freq: 0.001 - 0.01
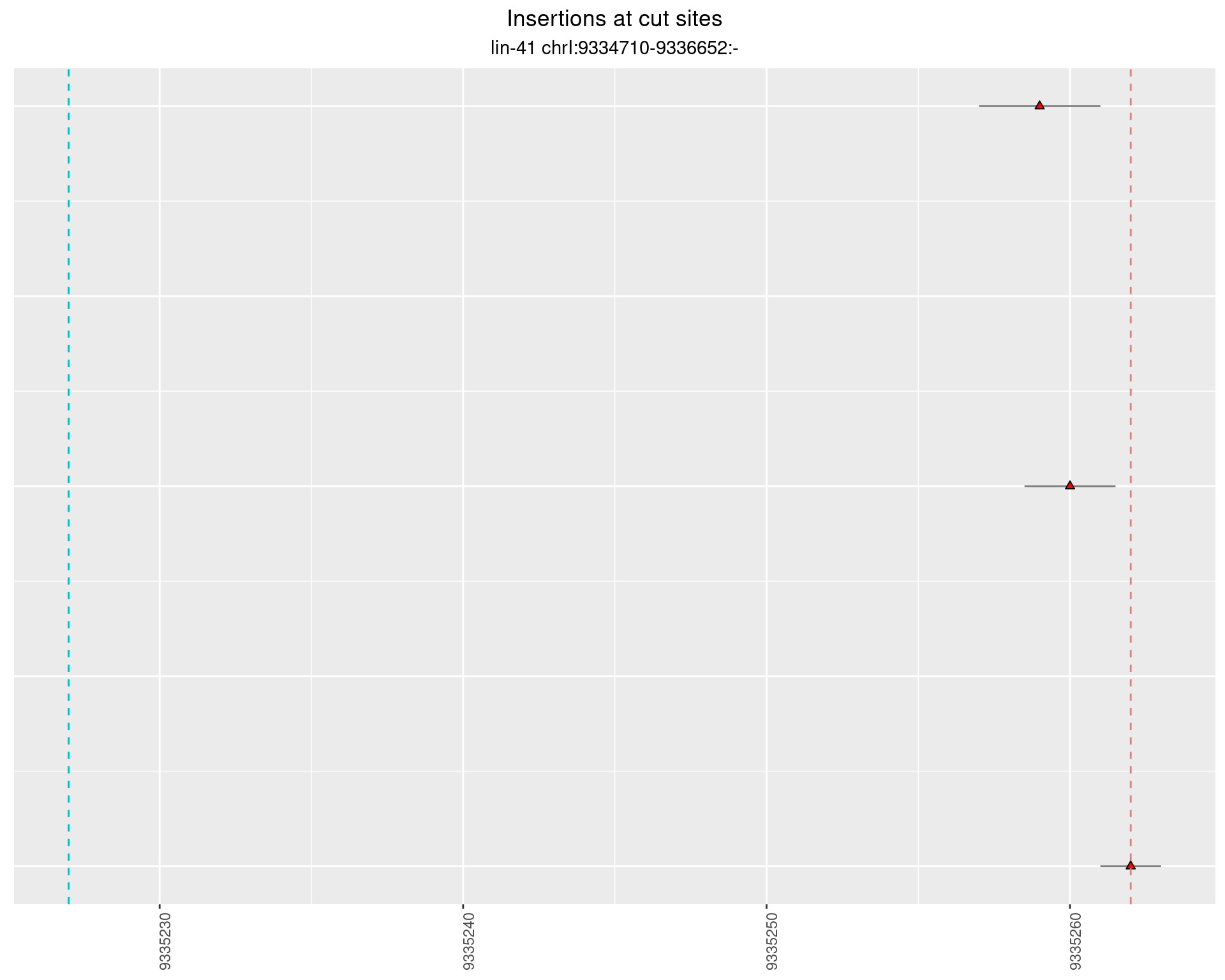
2.23 gen_16C_F2_lin-41_pool
2.23.1 Freq: 1e-05 - 1e-04
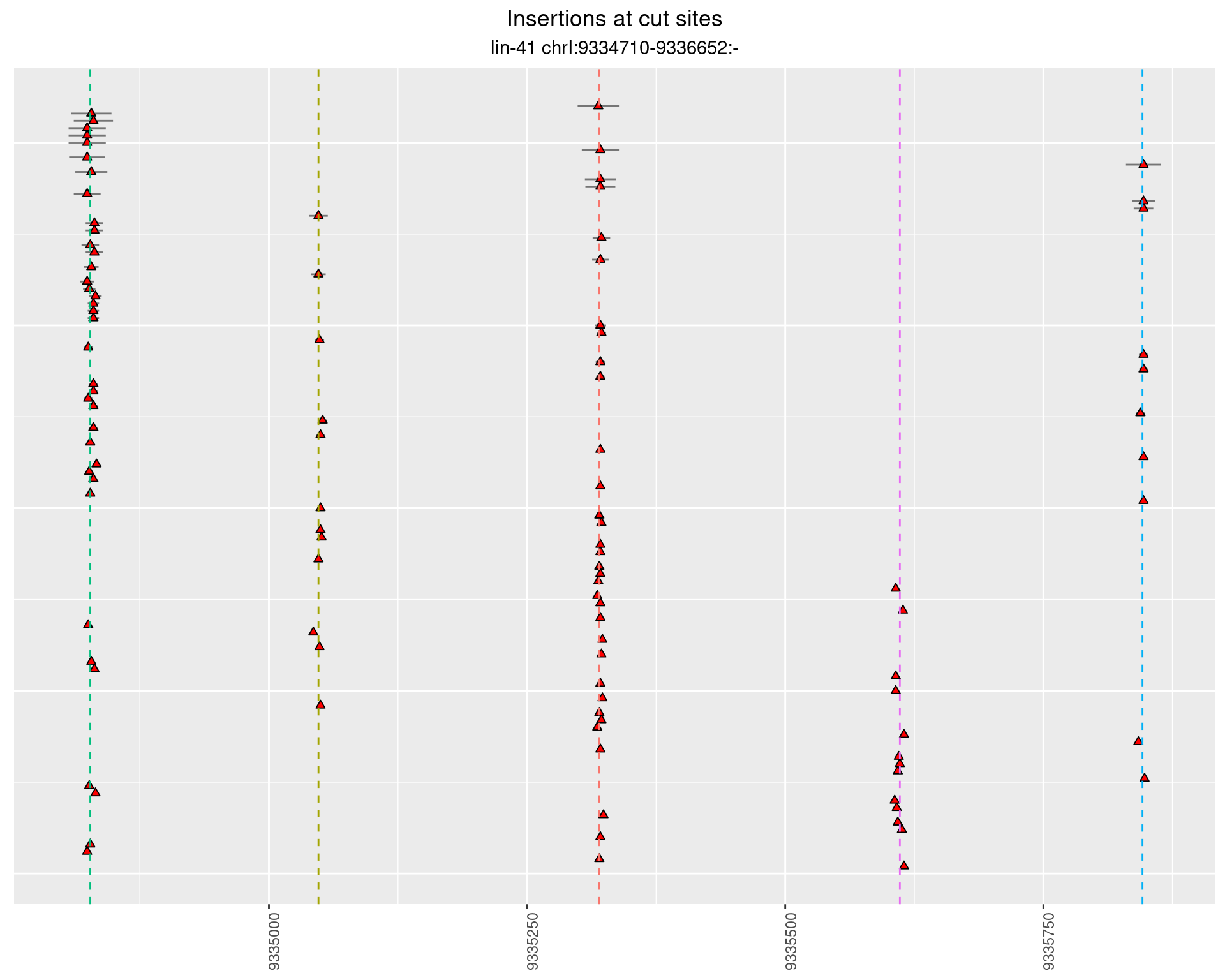
2.23.2 Freq: 1e-04 - 0.001
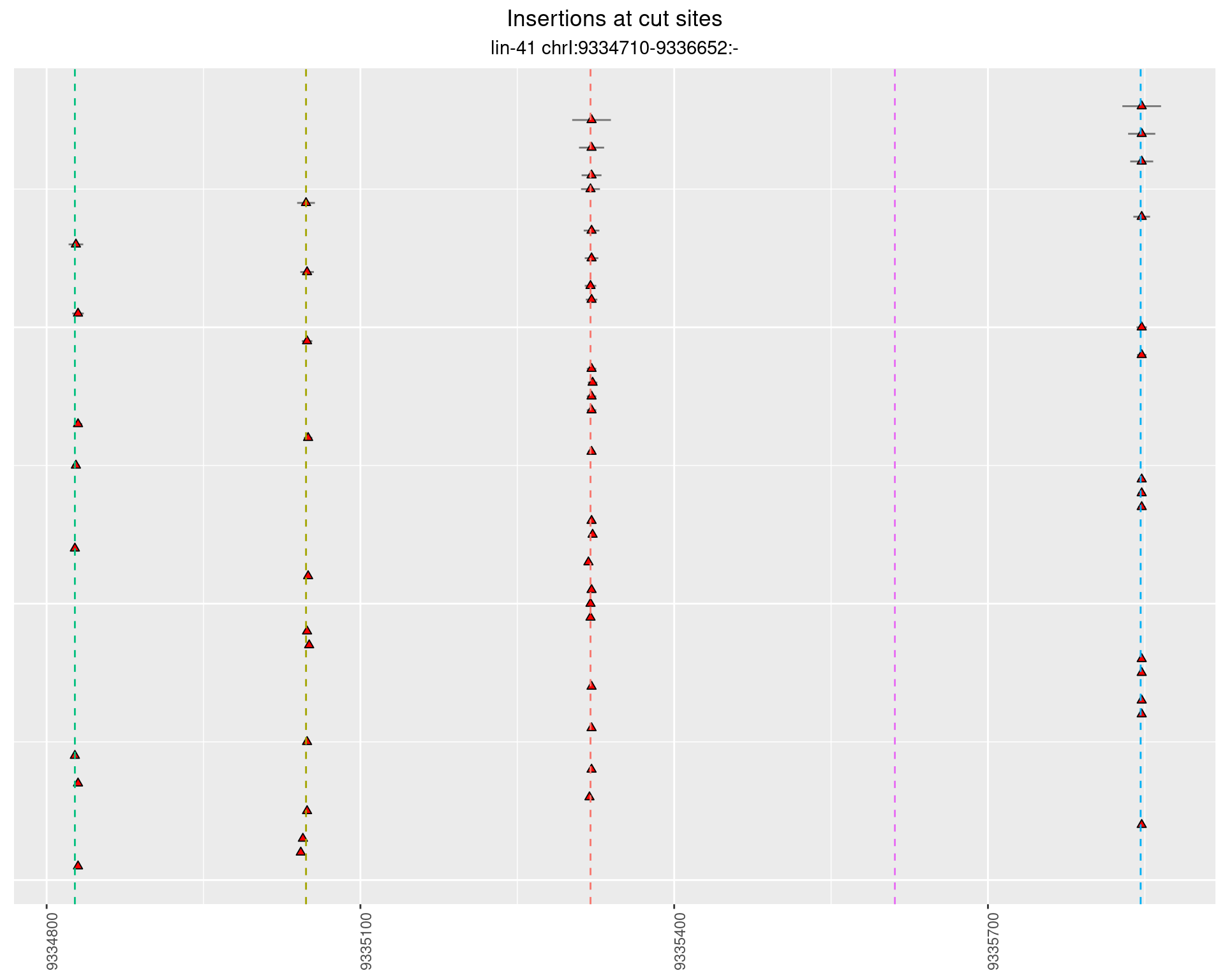
2.23.3 Freq: 0.001 - 0.01
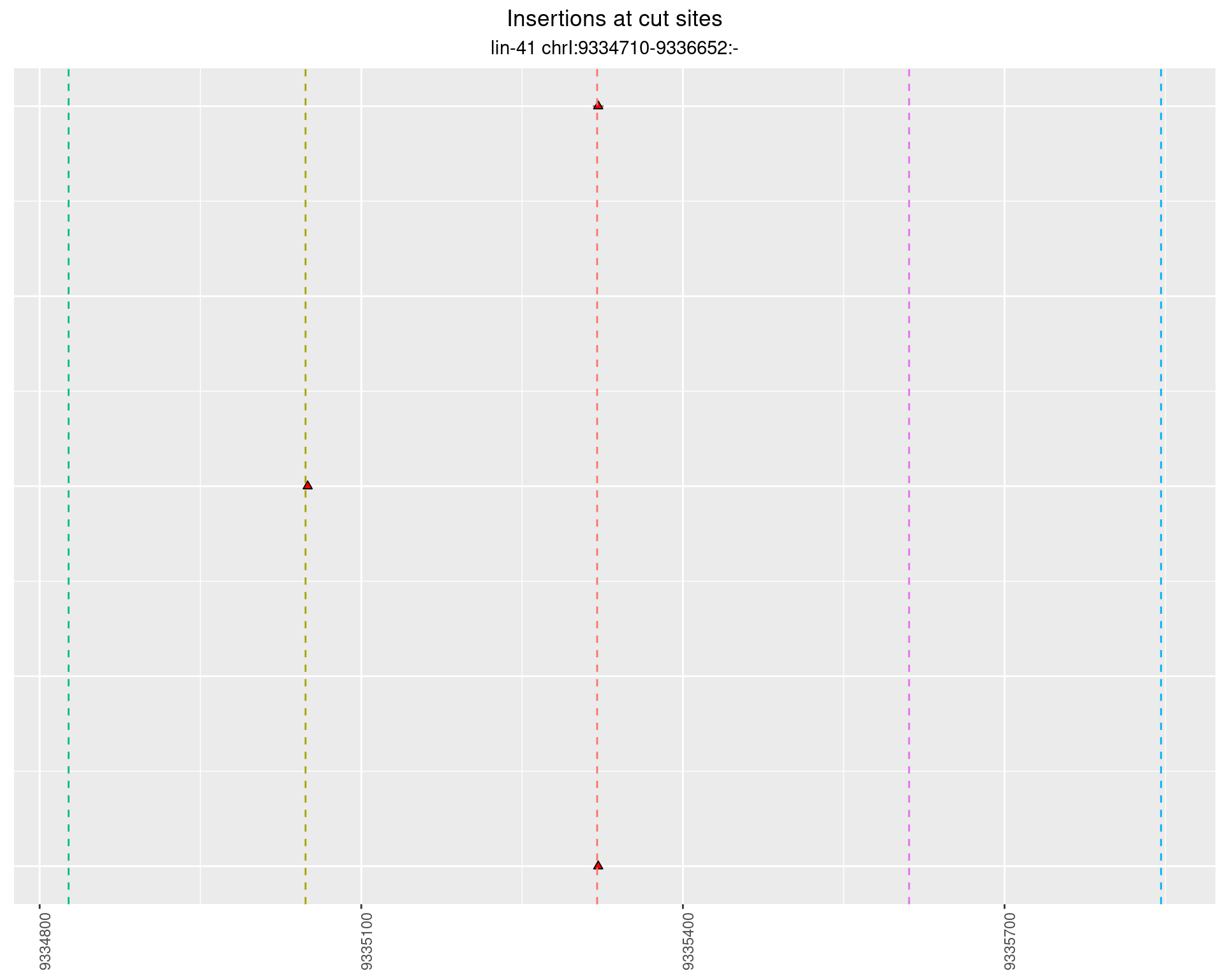
2.23.4 Freq: 1e-06 - 1e-05
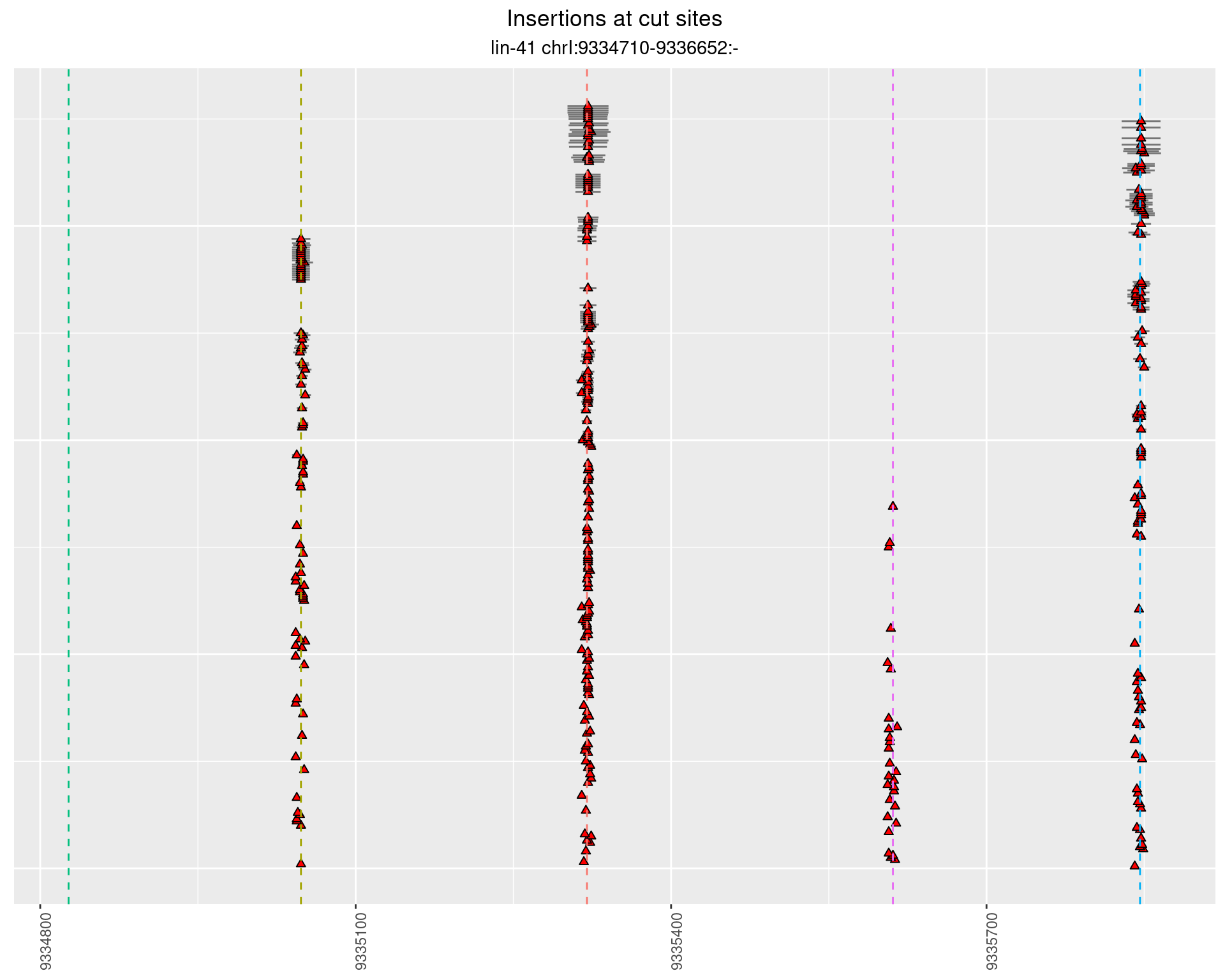
2.24 gen_16C_F2_lin-41_sg26sg27
2.24.1 Freq: 1e-04 - 0.001
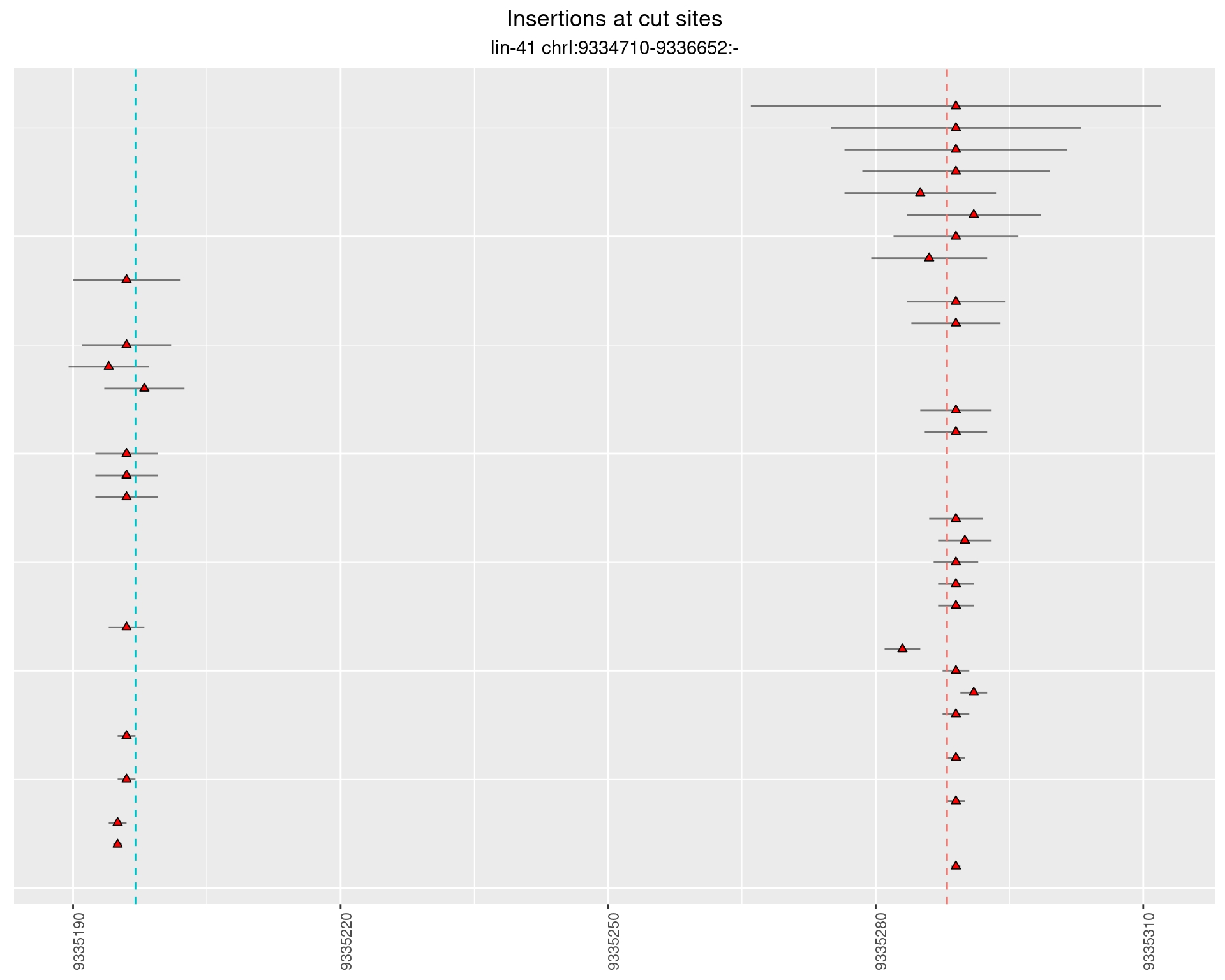
2.24.2 Freq: 1e-06 - 1e-05
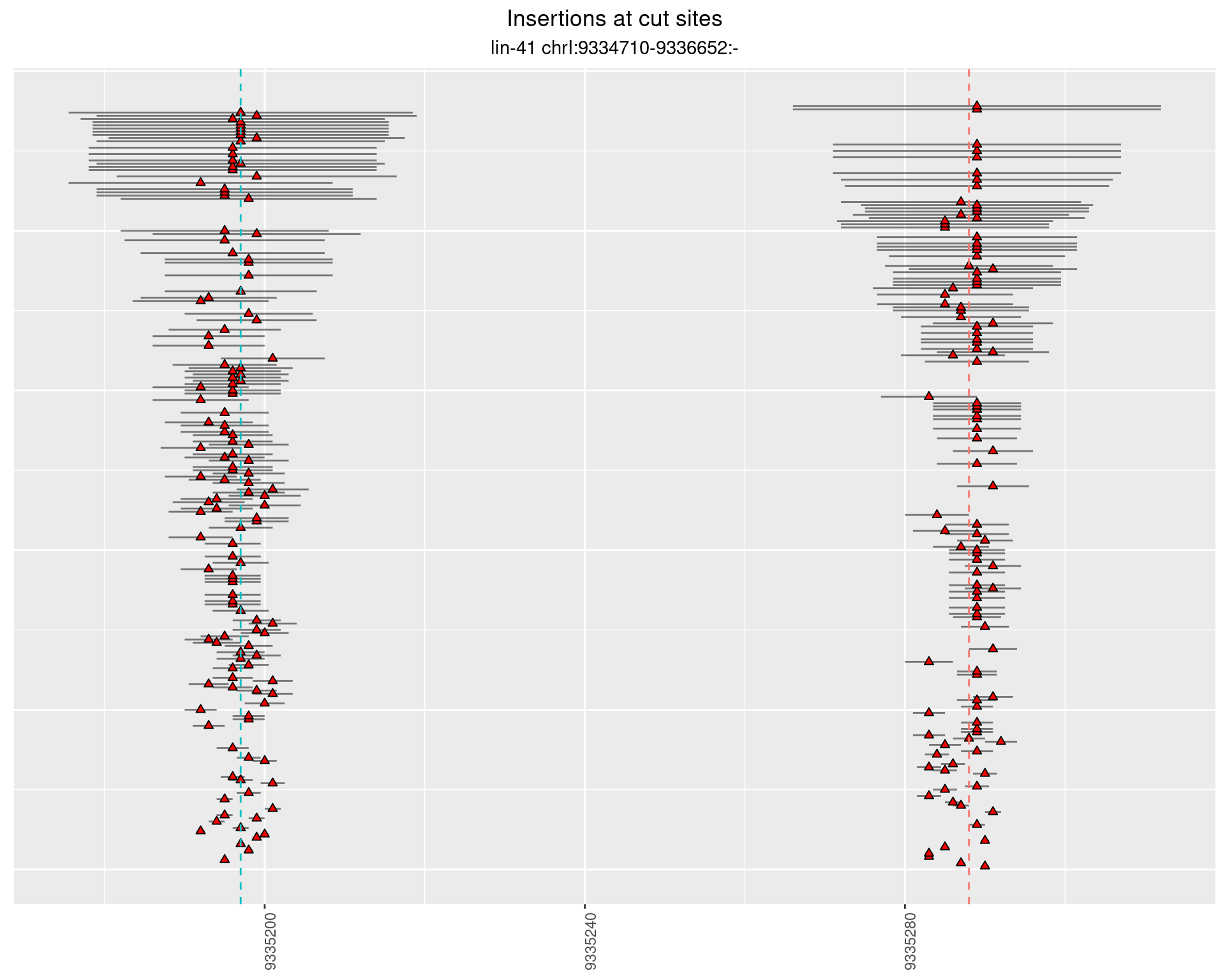
2.24.3 Freq: 1e-05 - 1e-04
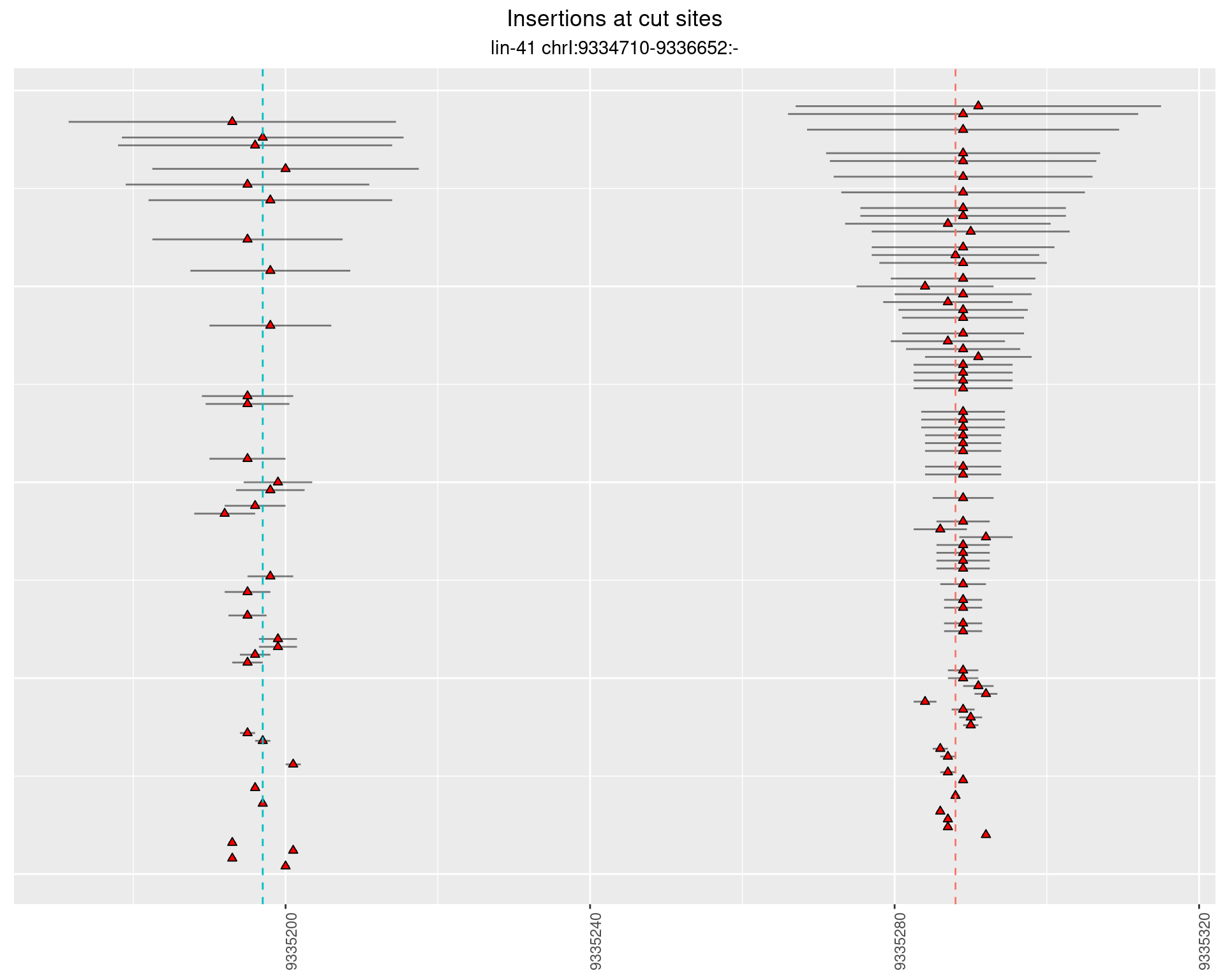
2.24.4 Freq: 0.001 - 0.01
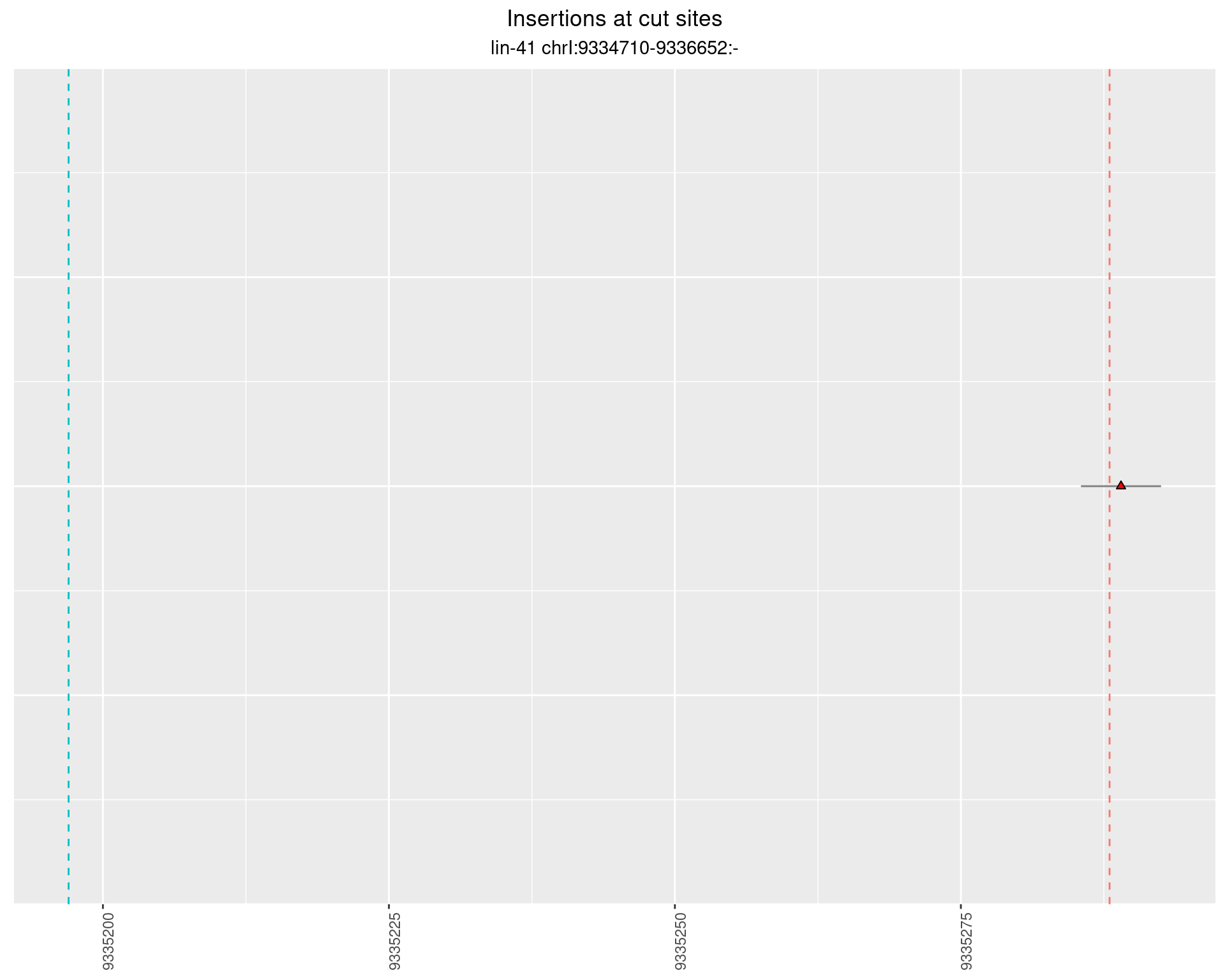
2.25 gen_16C_F3_lin-41_N2
2.25.1 Freq: 1e-06 - 1e-05
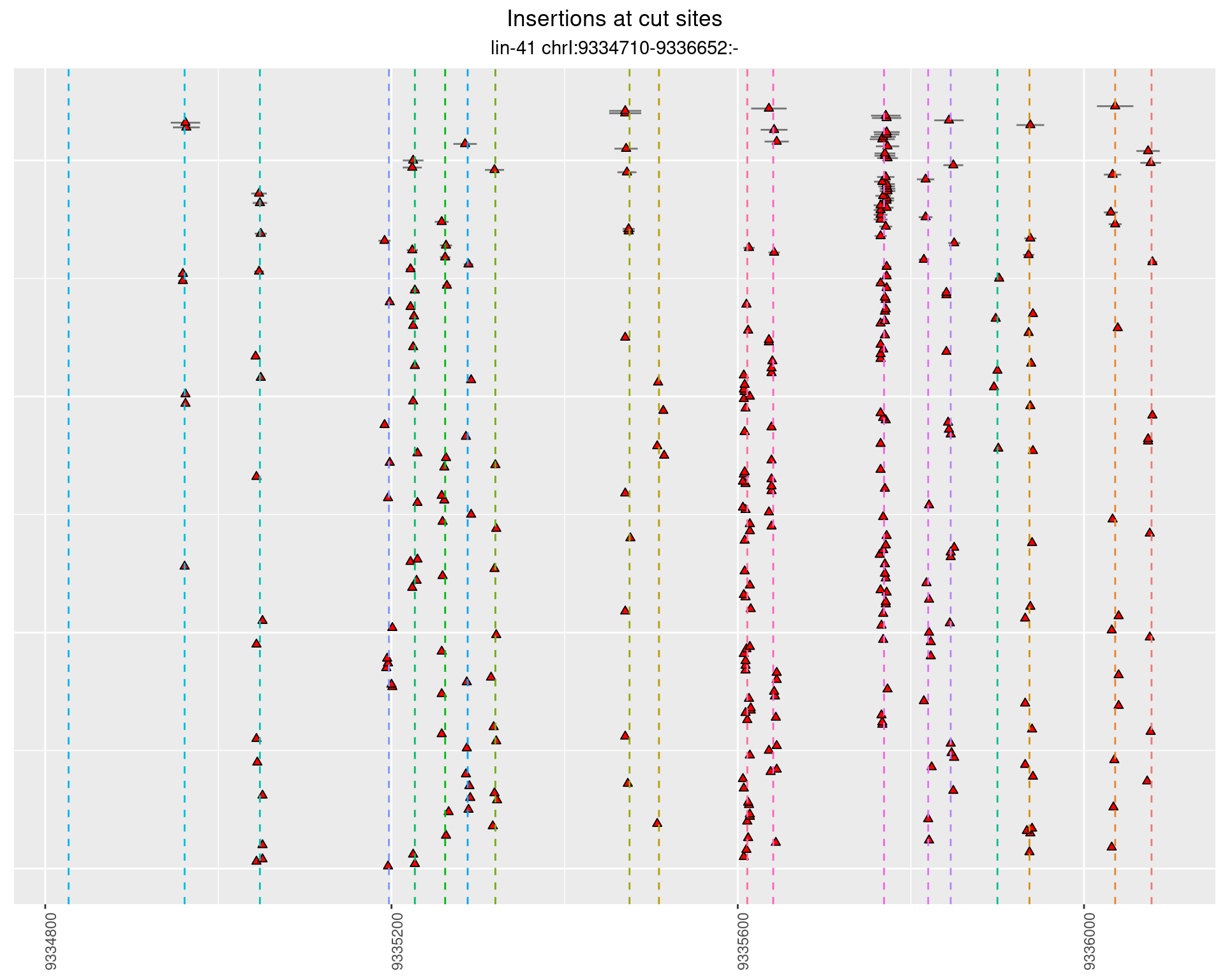
2.25.2 Freq: 1e-05 - 1e-04
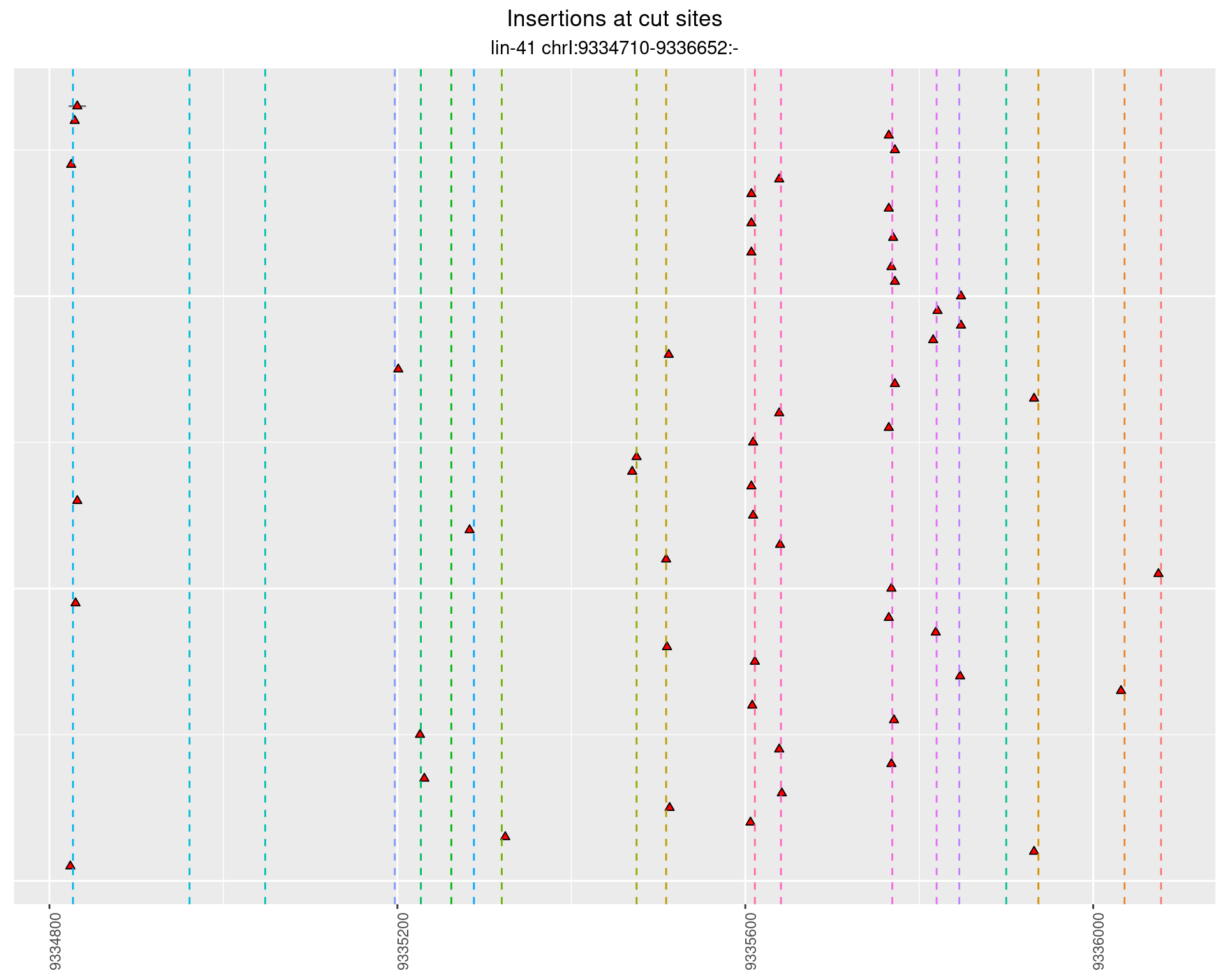
2.25.3 Freq: 1e-04 - 0.001
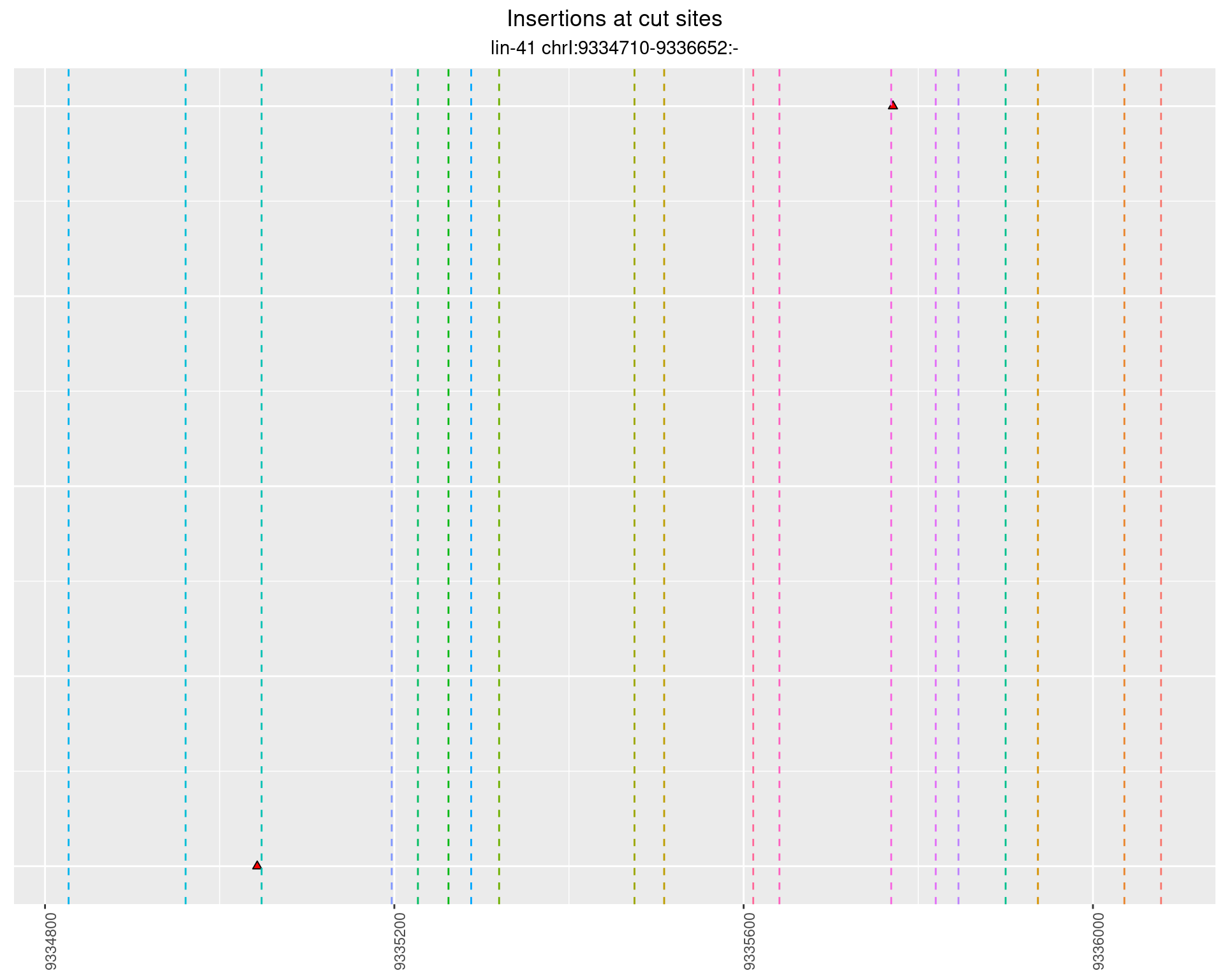
2.26 gen_16C_F3_lin-41_sg15sg16
2.26.1 Freq: 1e-04 - 0.001
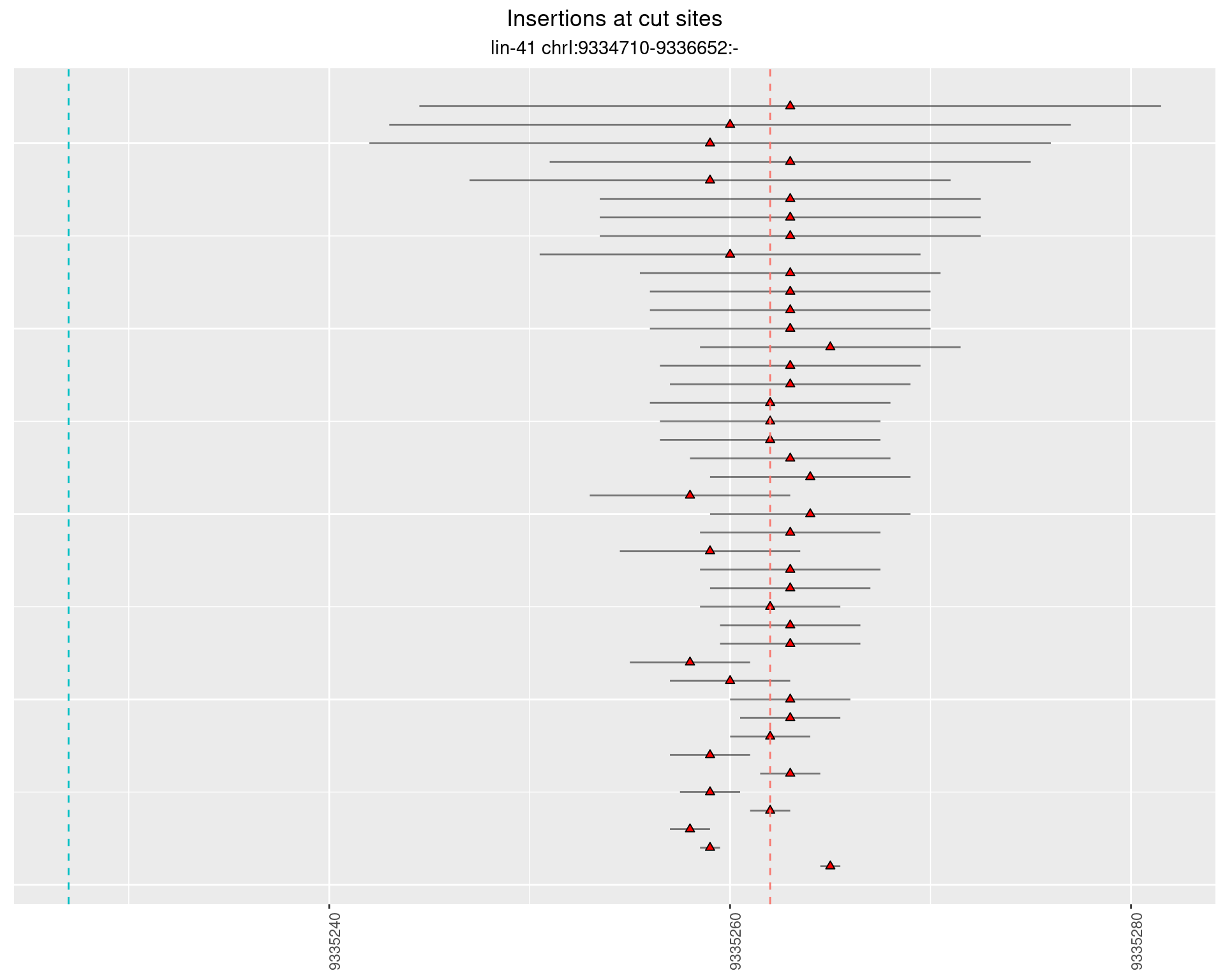
2.26.2 Freq: 1e-06 - 1e-05
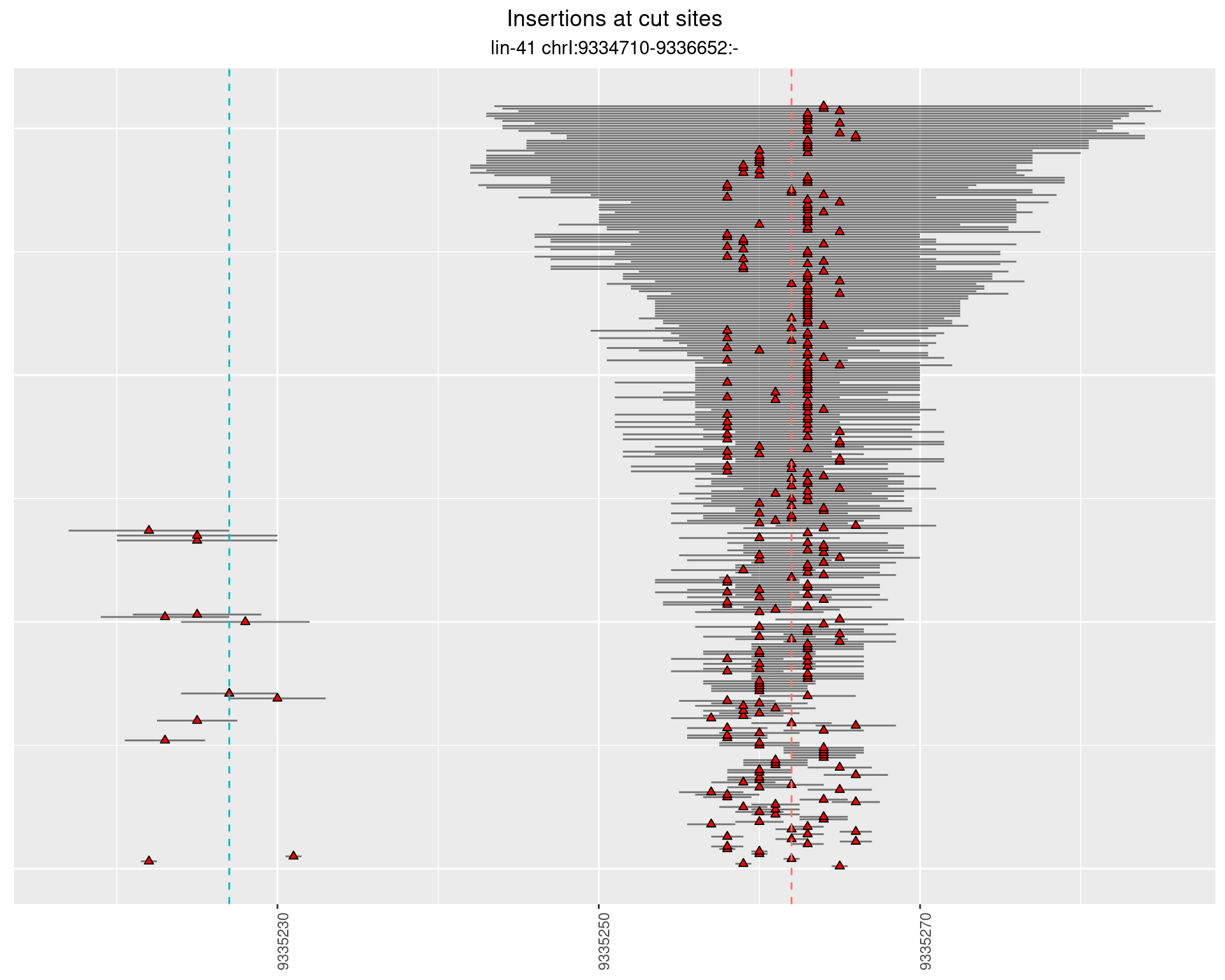
2.26.3 Freq: 0.001 - 0.01
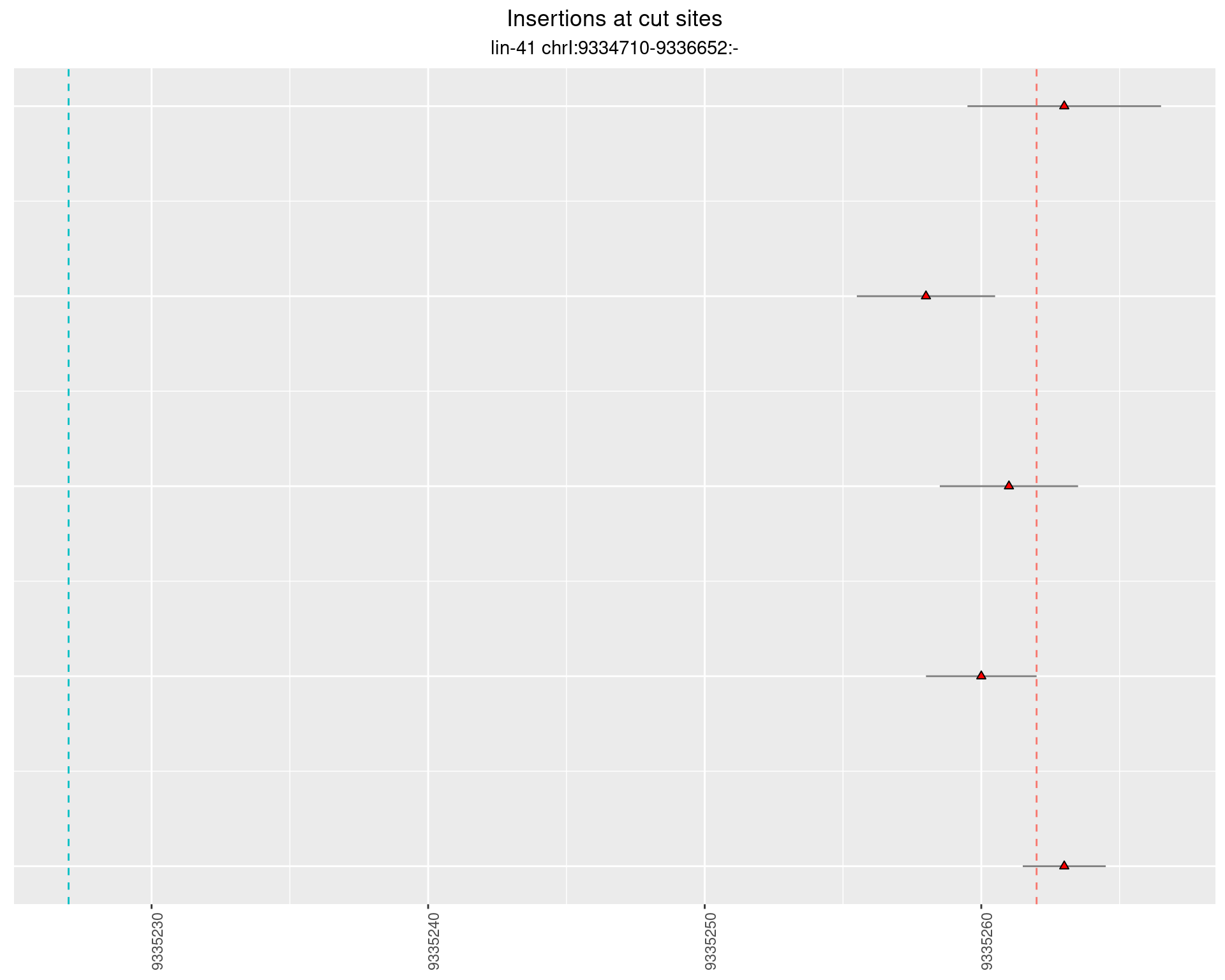
2.26.4 Freq: 1e-05 - 1e-04
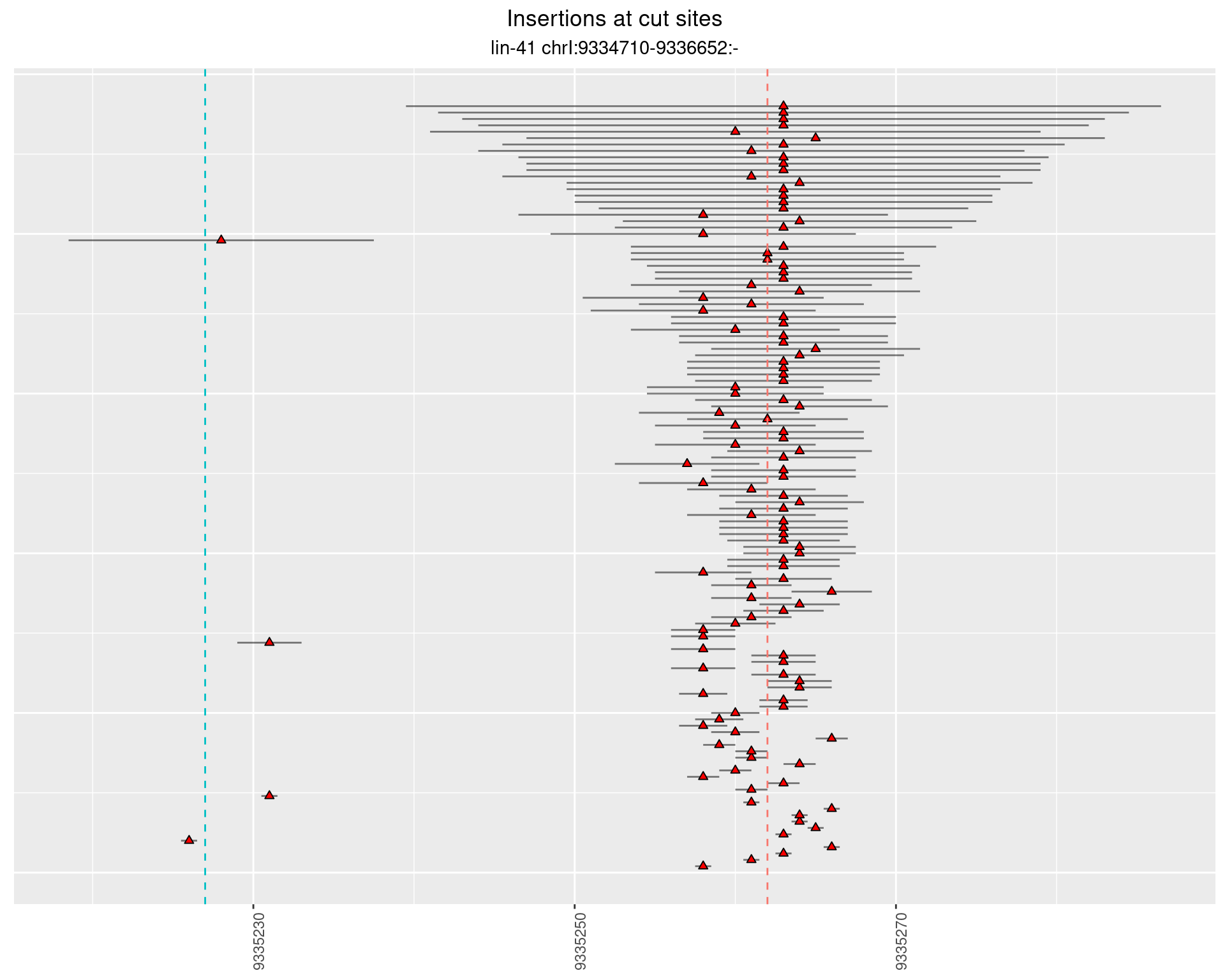
2.27 gen_16C_F3_lin-41_pool
2.27.1 Freq: 0.001 - 0.01
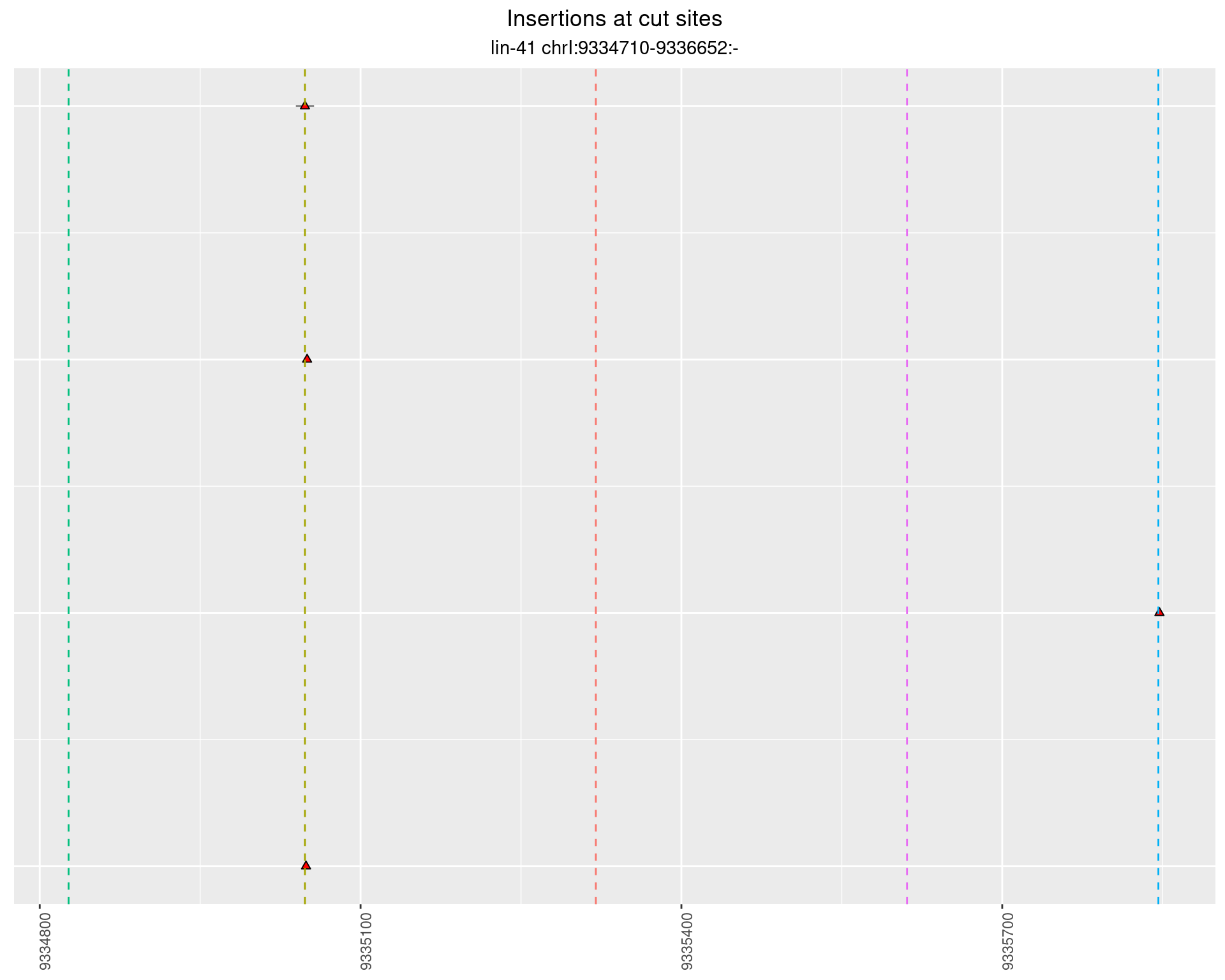
2.27.2 Freq: 1e-04 - 0.001
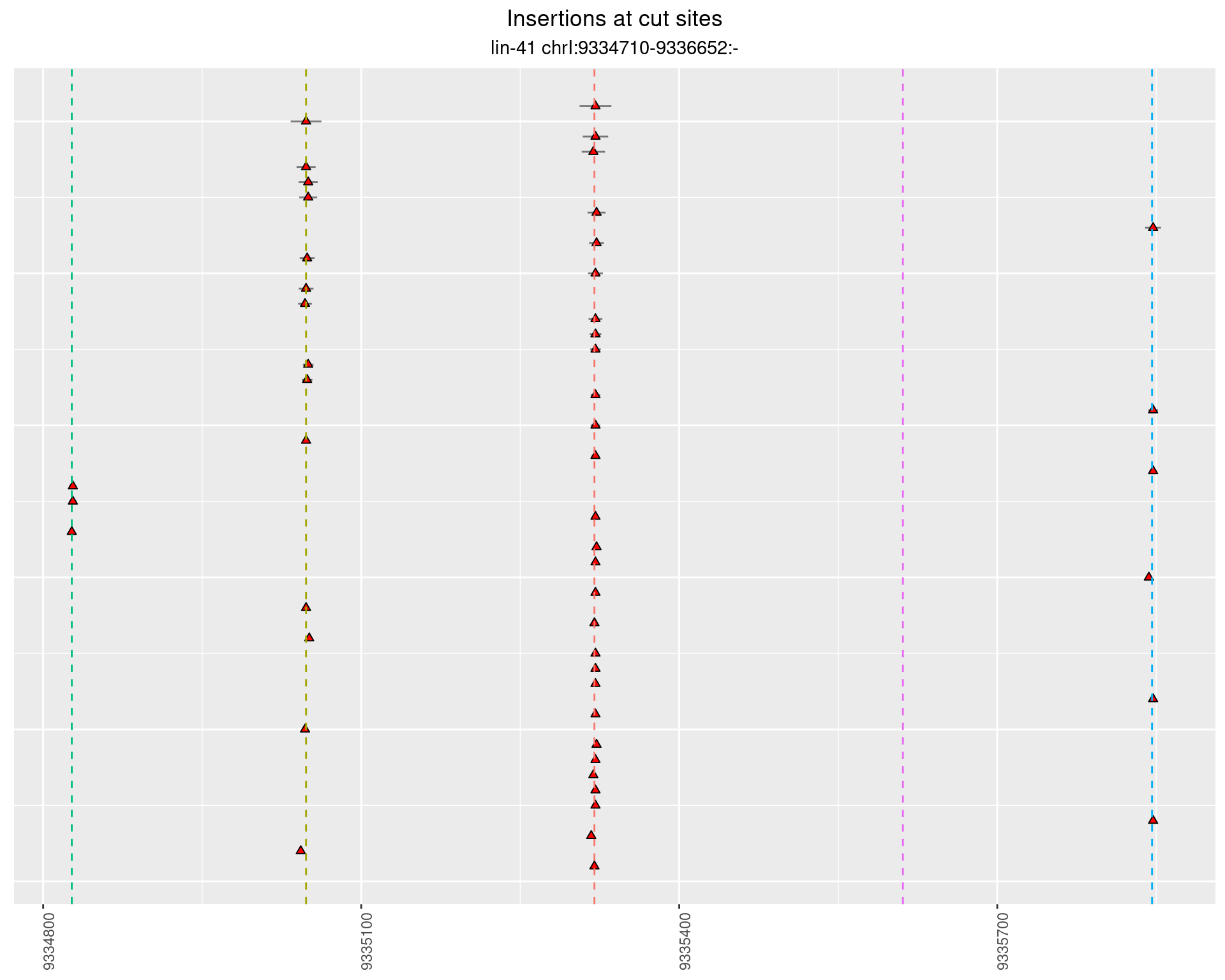
2.27.3 Freq: 1e-05 - 1e-04
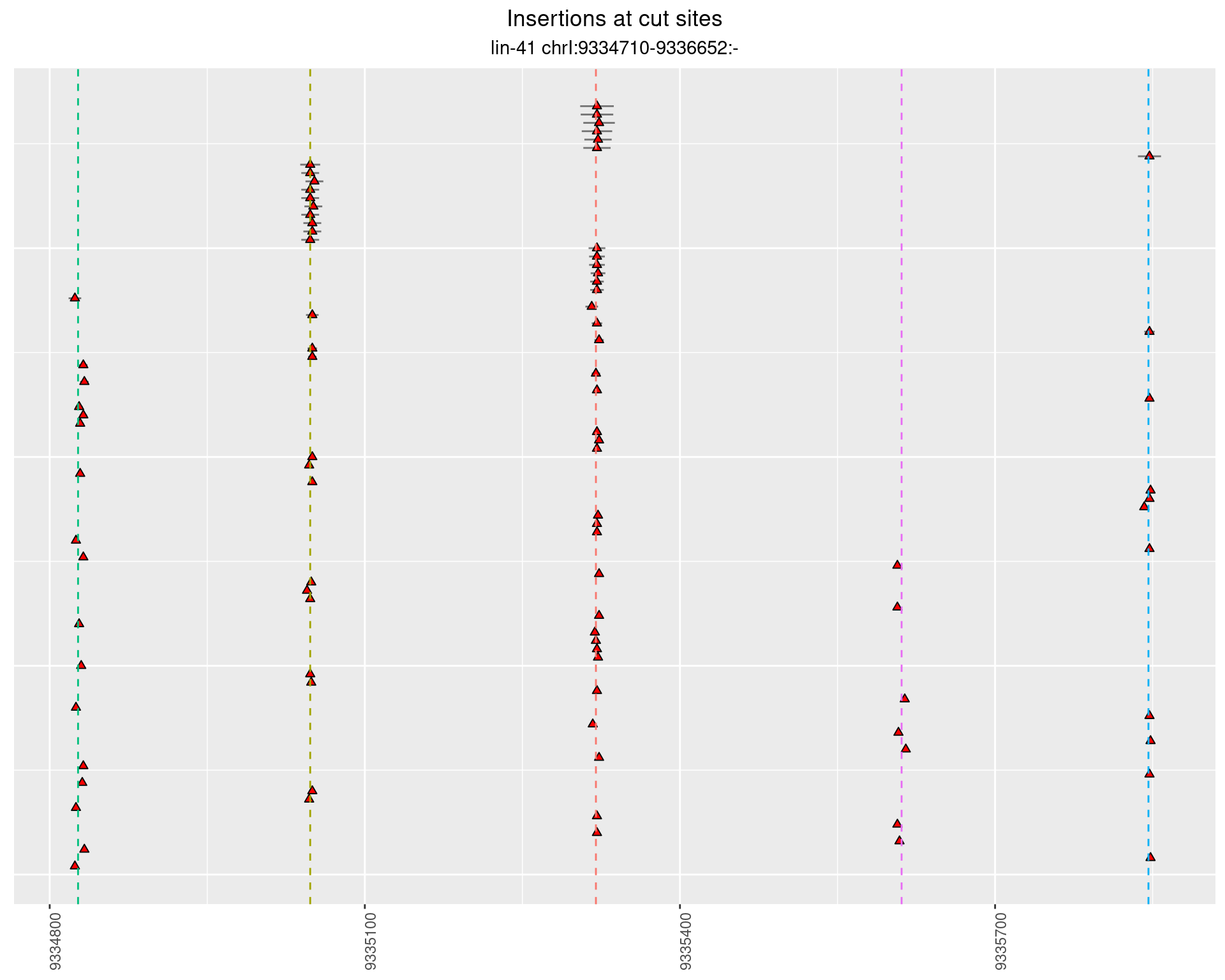
2.27.4 Freq: 1e-06 - 1e-05
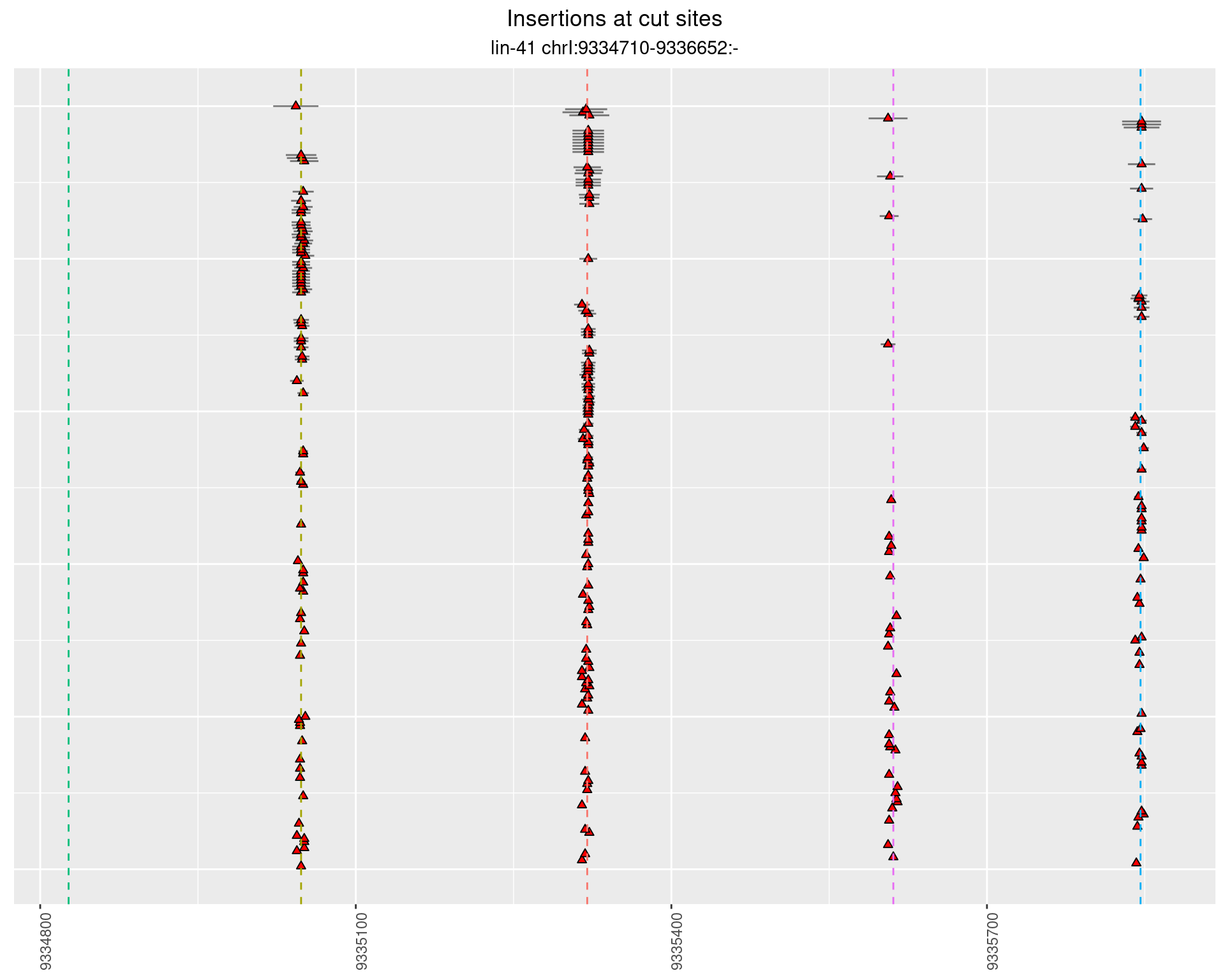
2.28 gen_16C_F3_lin-41_sg26sg27
2.28.1 Freq: 0.001 - 0.01
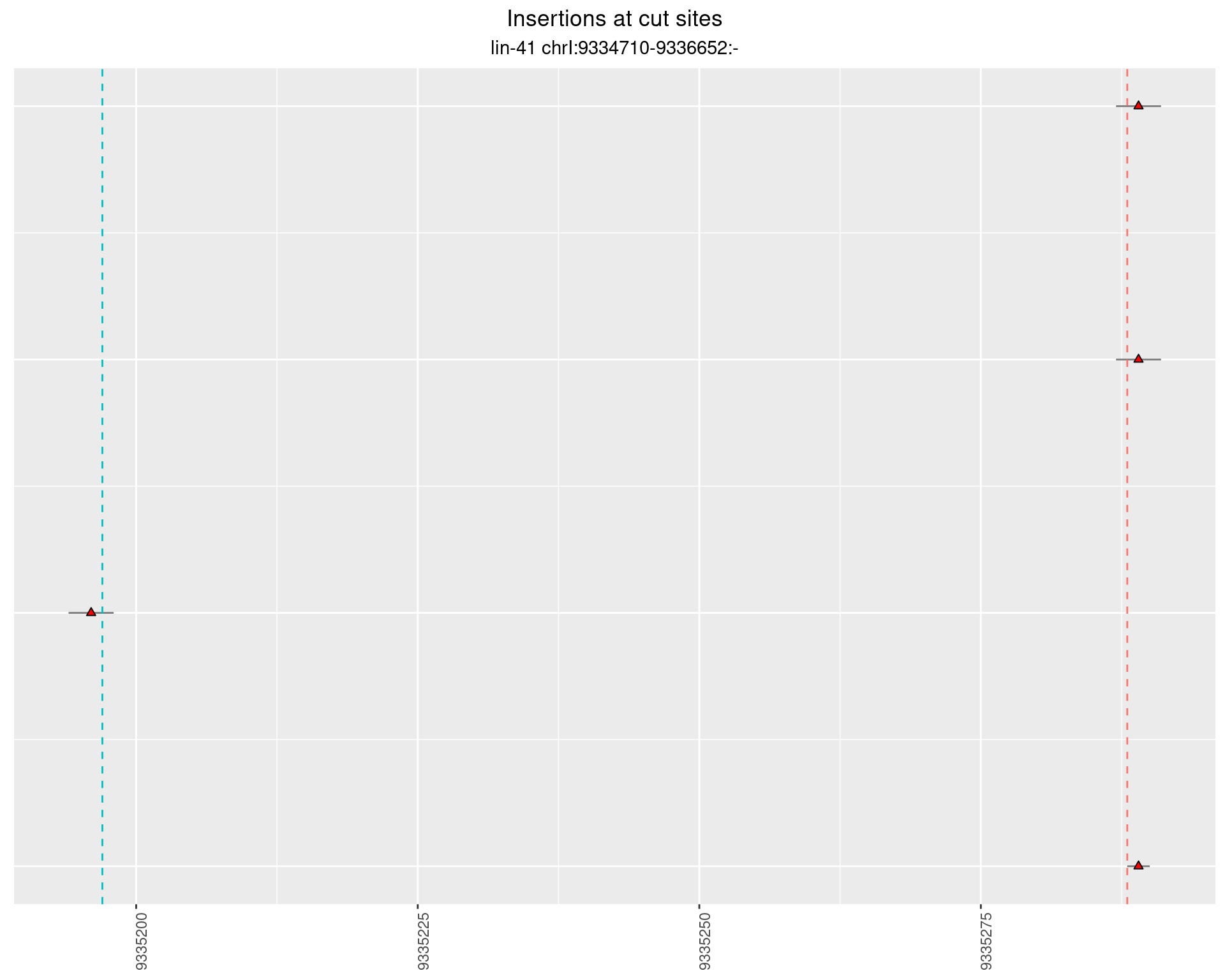
2.28.2 Freq: 1e-04 - 0.001
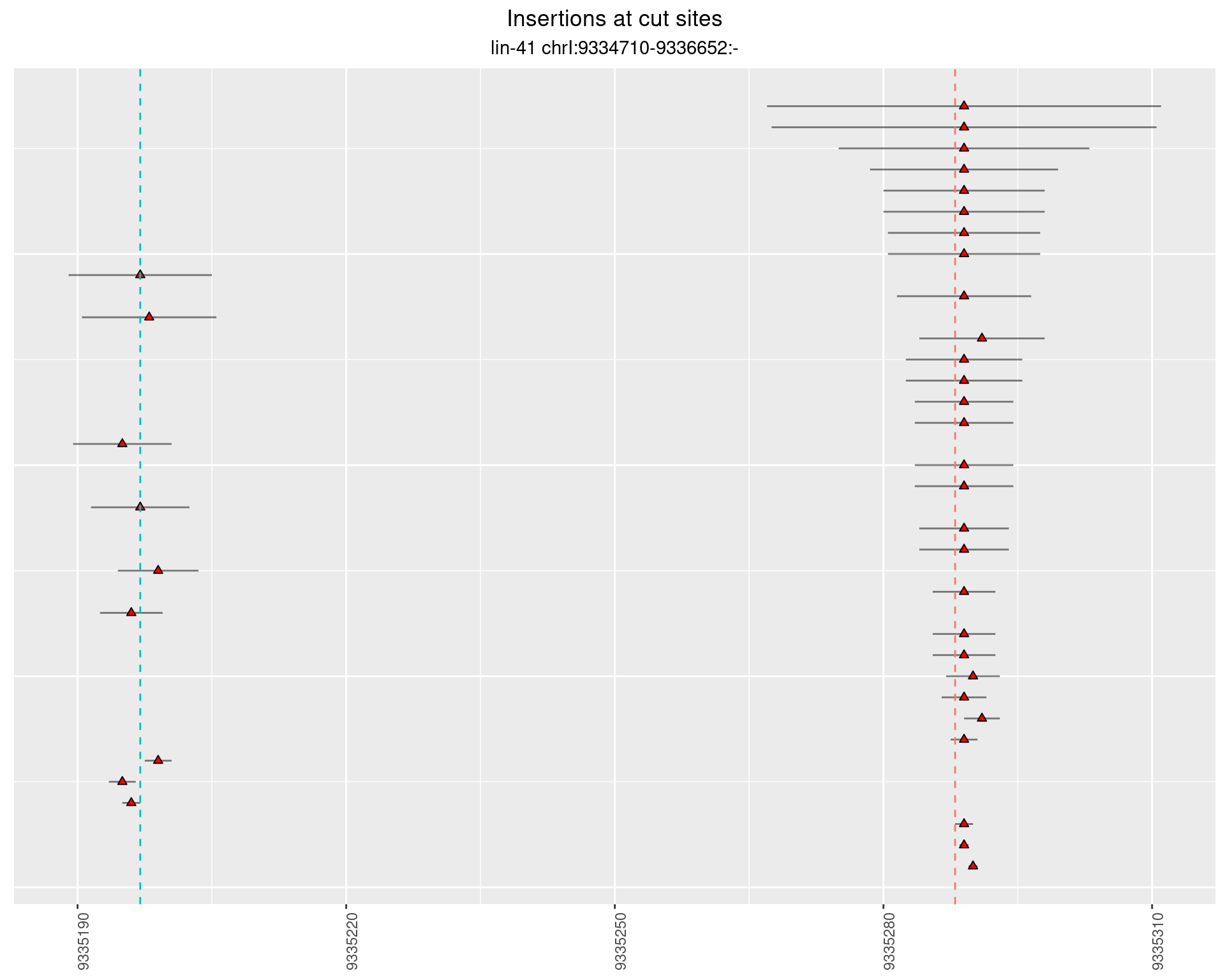
2.28.3 Freq: 1e-05 - 1e-04
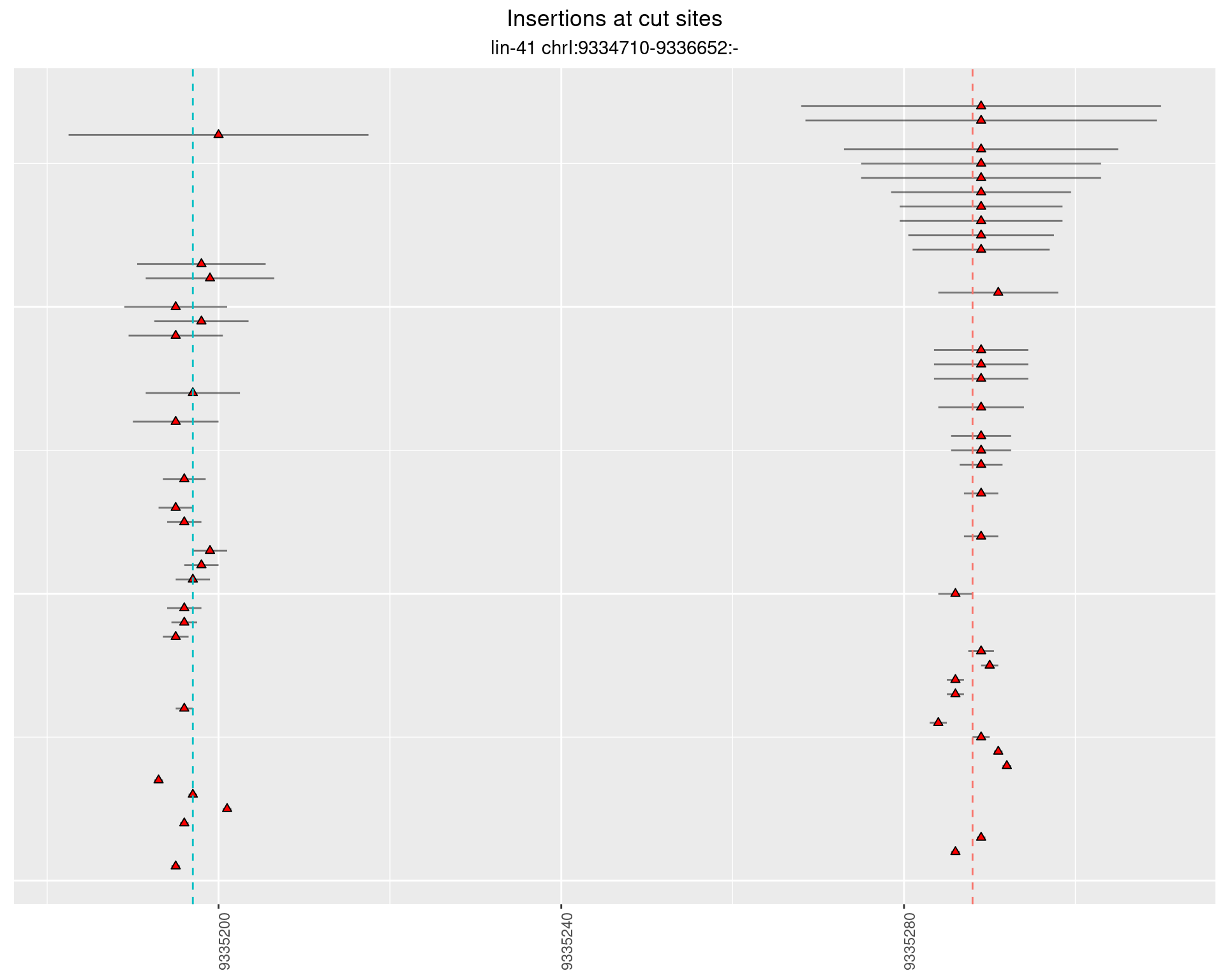
2.28.4 Freq: 1e-06 - 1e-05
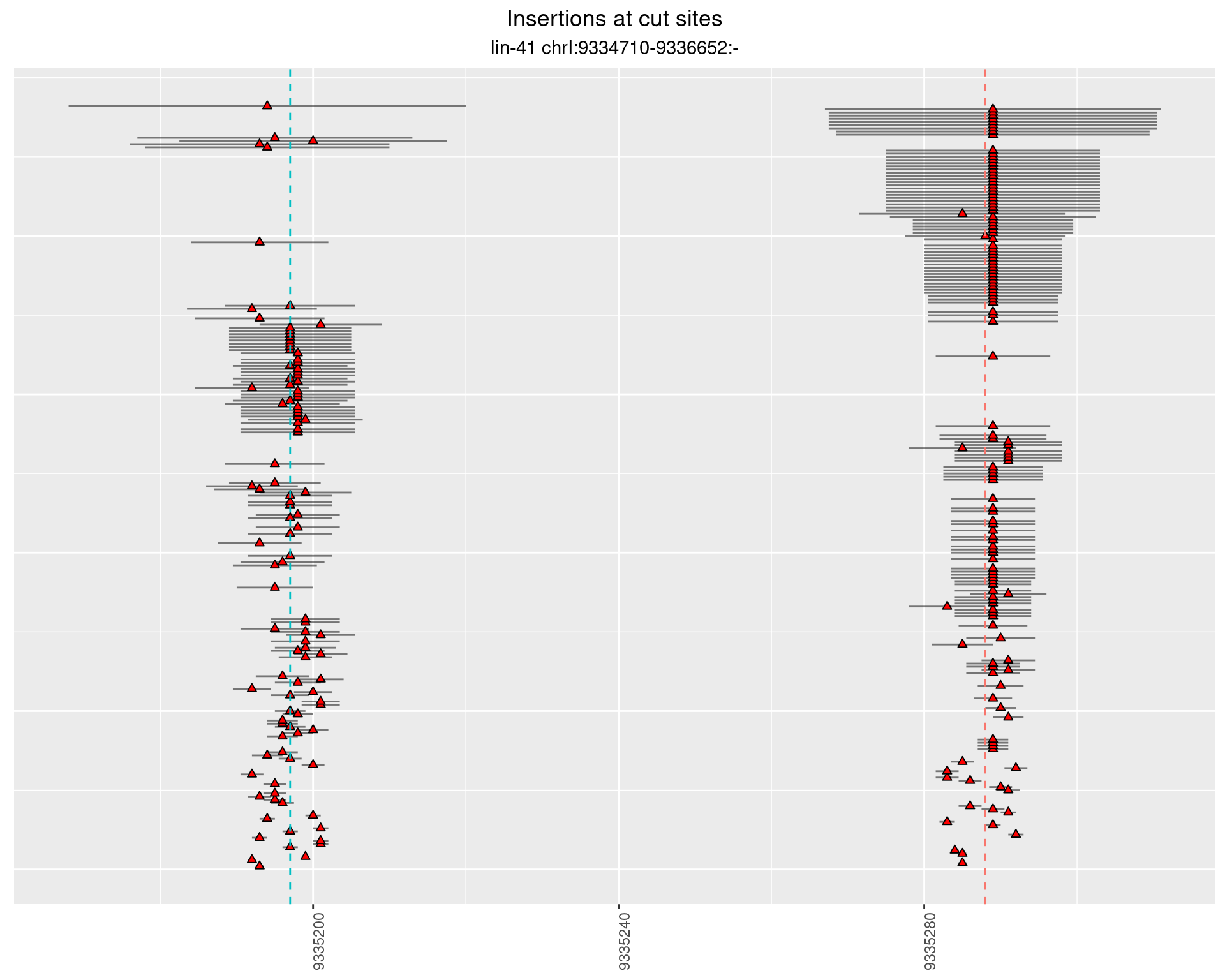
2.29 gen_16C_F4_lin-41_N2
2.29.1 Freq: 1e-05 - 1e-04
2.29.2 Freq: 1e-04 - 0.001
2.30 gen_16C_F4_lin-41_sg15sg16
2.30.1 Freq: 1e-04 - 0.001
2.30.2 Freq: 0.001 - 0.01
2.30.3 Freq: 1e-05 - 1e-04
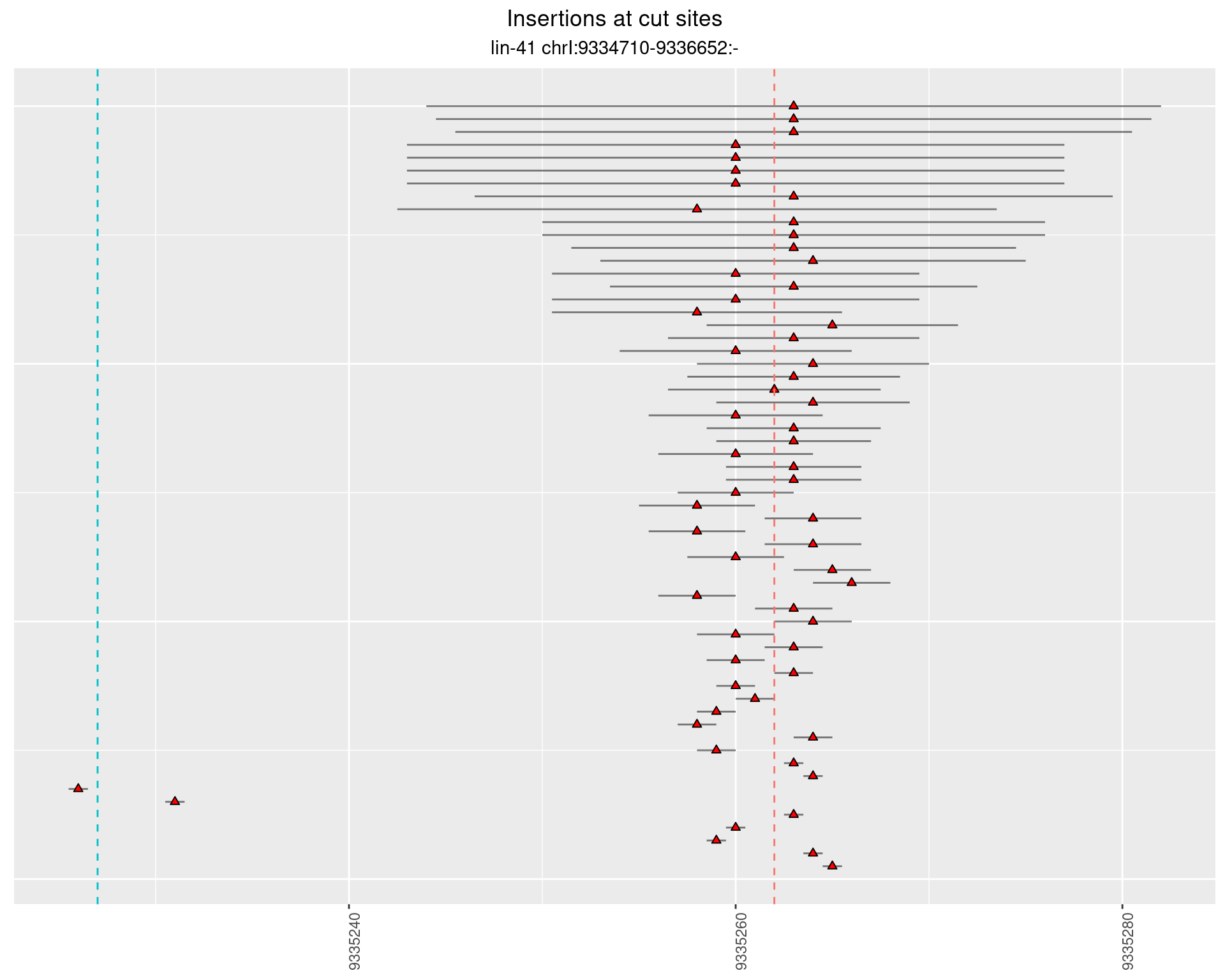
2.30.4 Freq: 1e-06 - 1e-05
2.31 gen_16C_F4_lin-41_pool
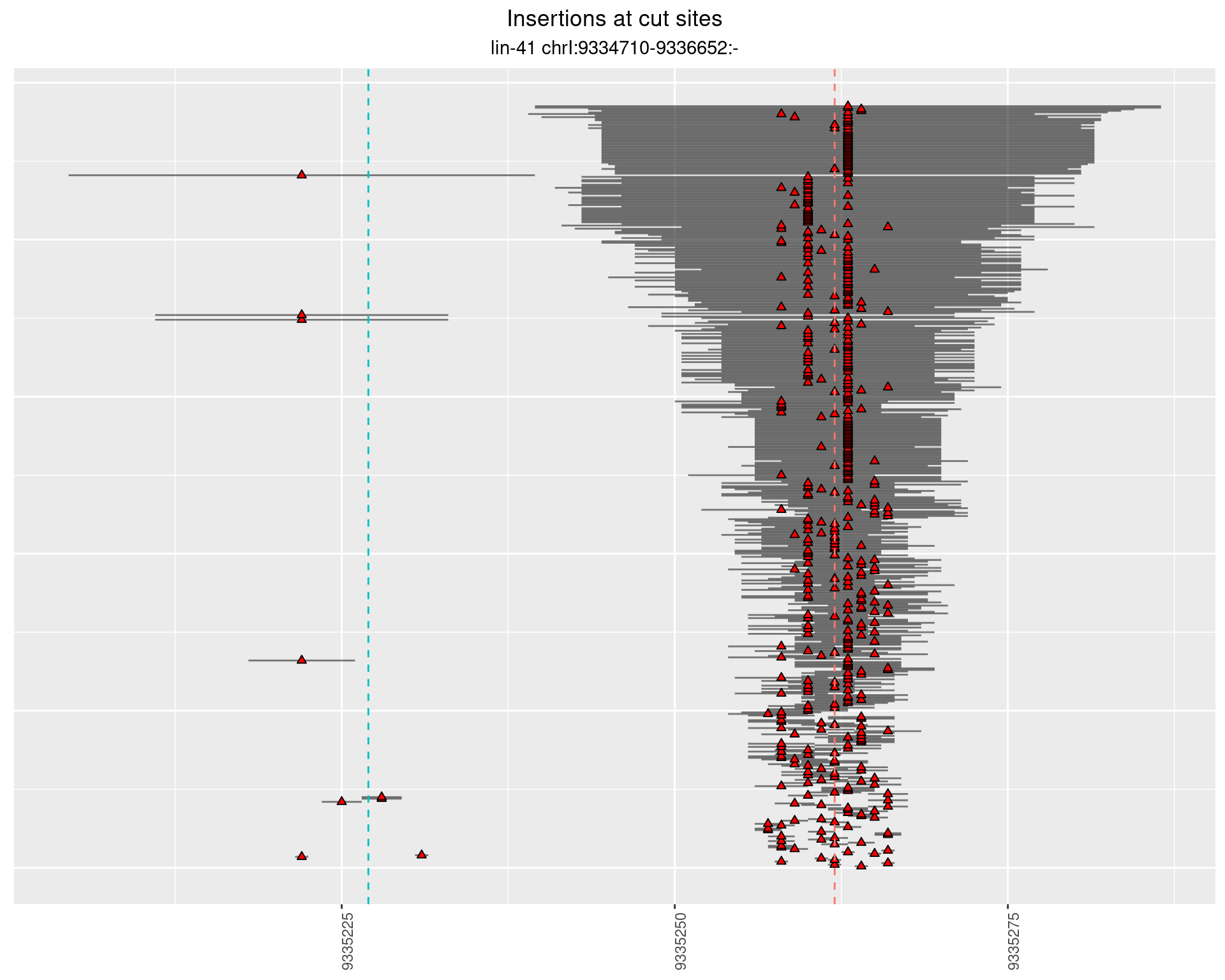
2.31.1 Freq: 1e-04 - 0.001
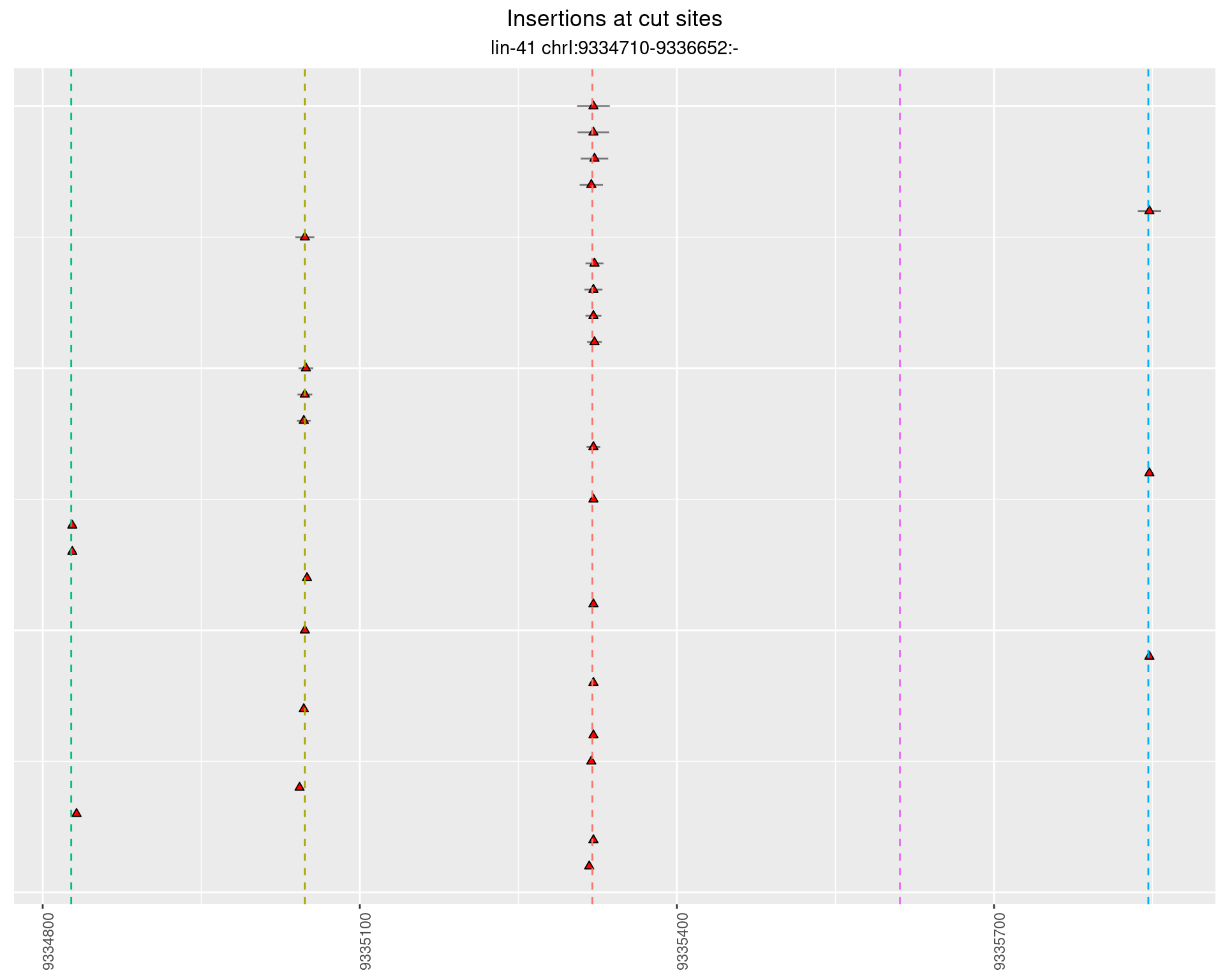
2.31.2 Freq: 0.001 - 0.01
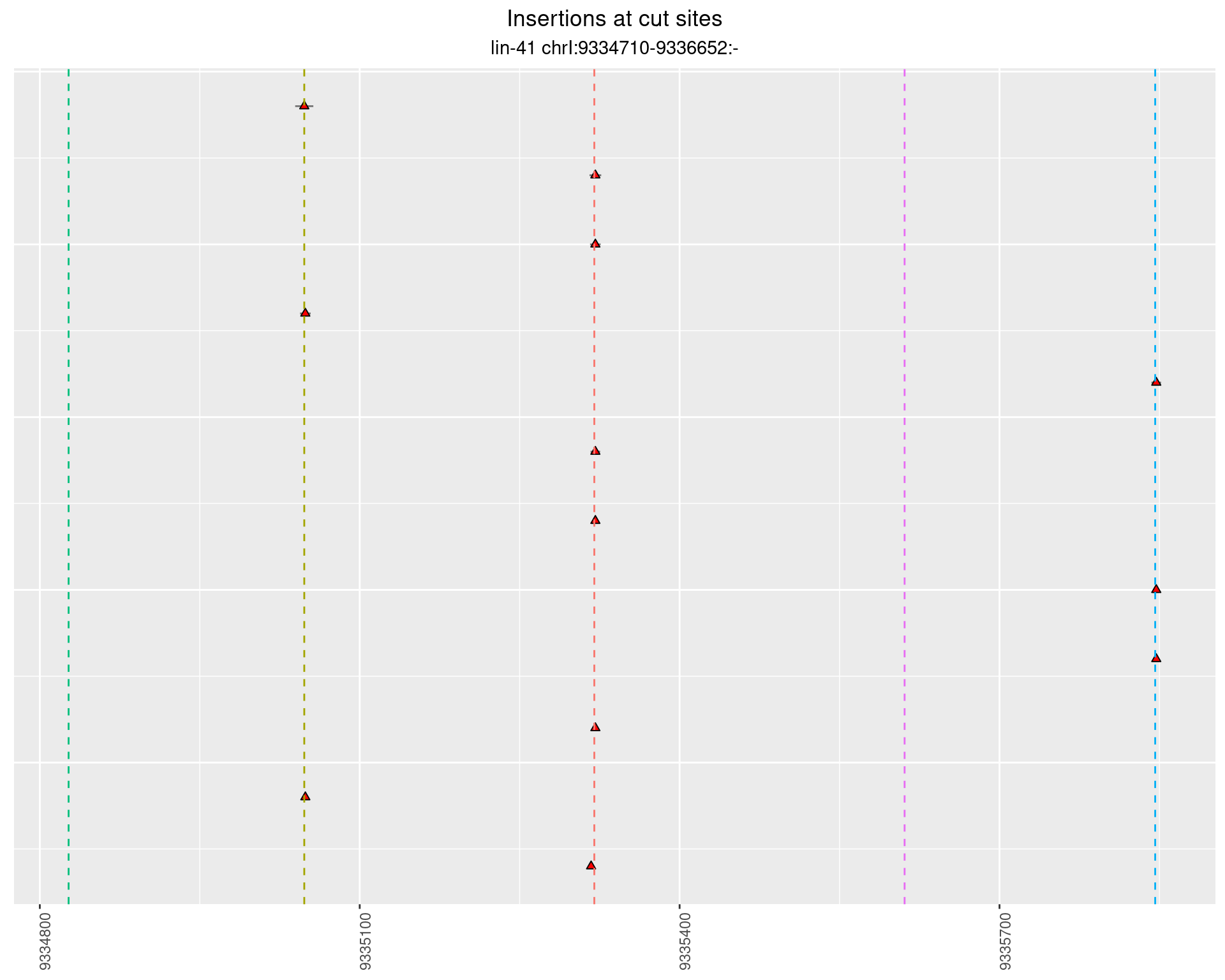
2.31.3 Freq: 1e-05 - 1e-04
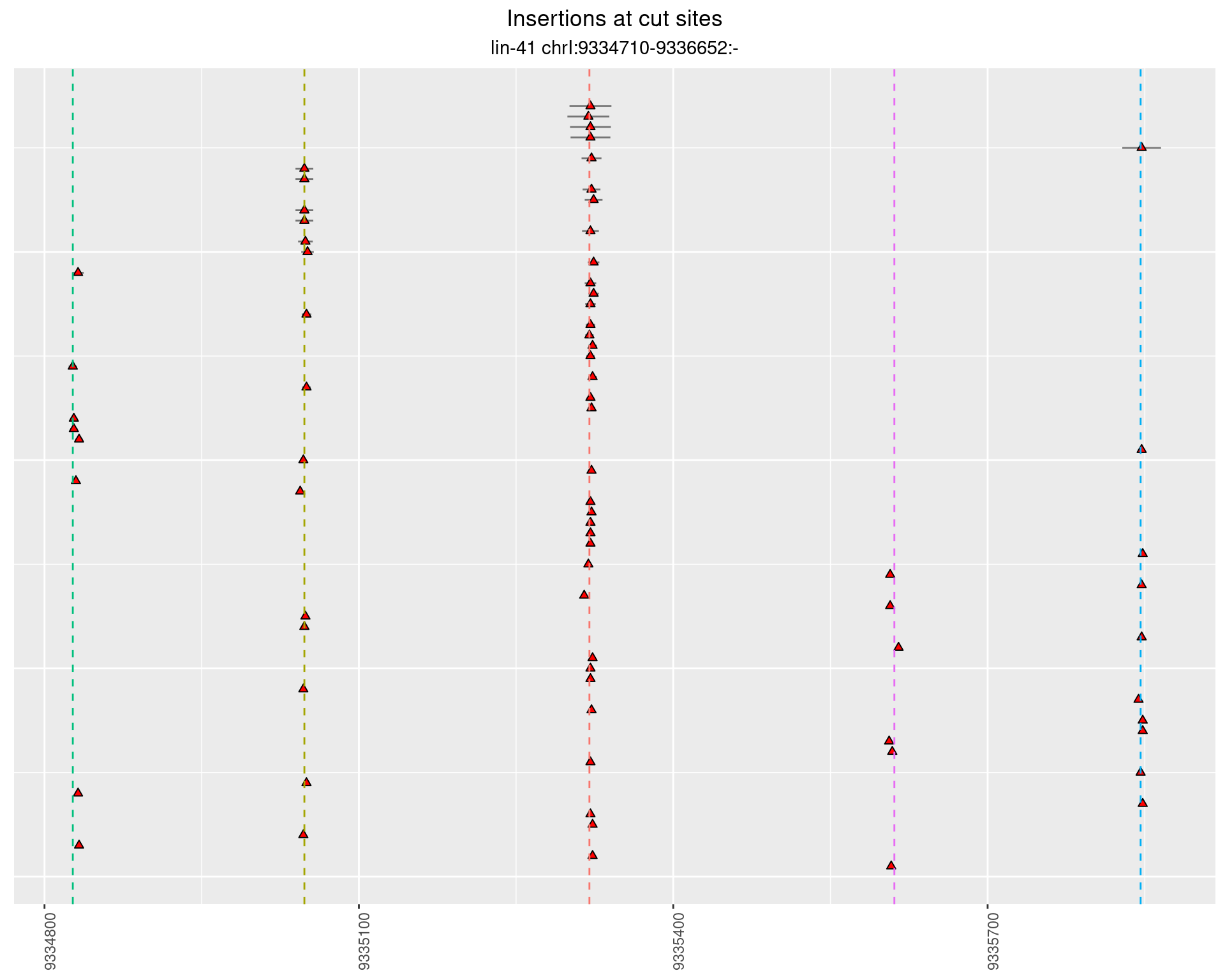
2.31.4 Freq: 1e-06 - 1e-05
2.32 gen_16C_F4_lin-41_sg26sg27
2.32.1 Freq: 0.001 - 0.01
2.32.2 Freq: 1e-04 - 0.001
2.32.3 Freq: 1e-05 - 1e-04
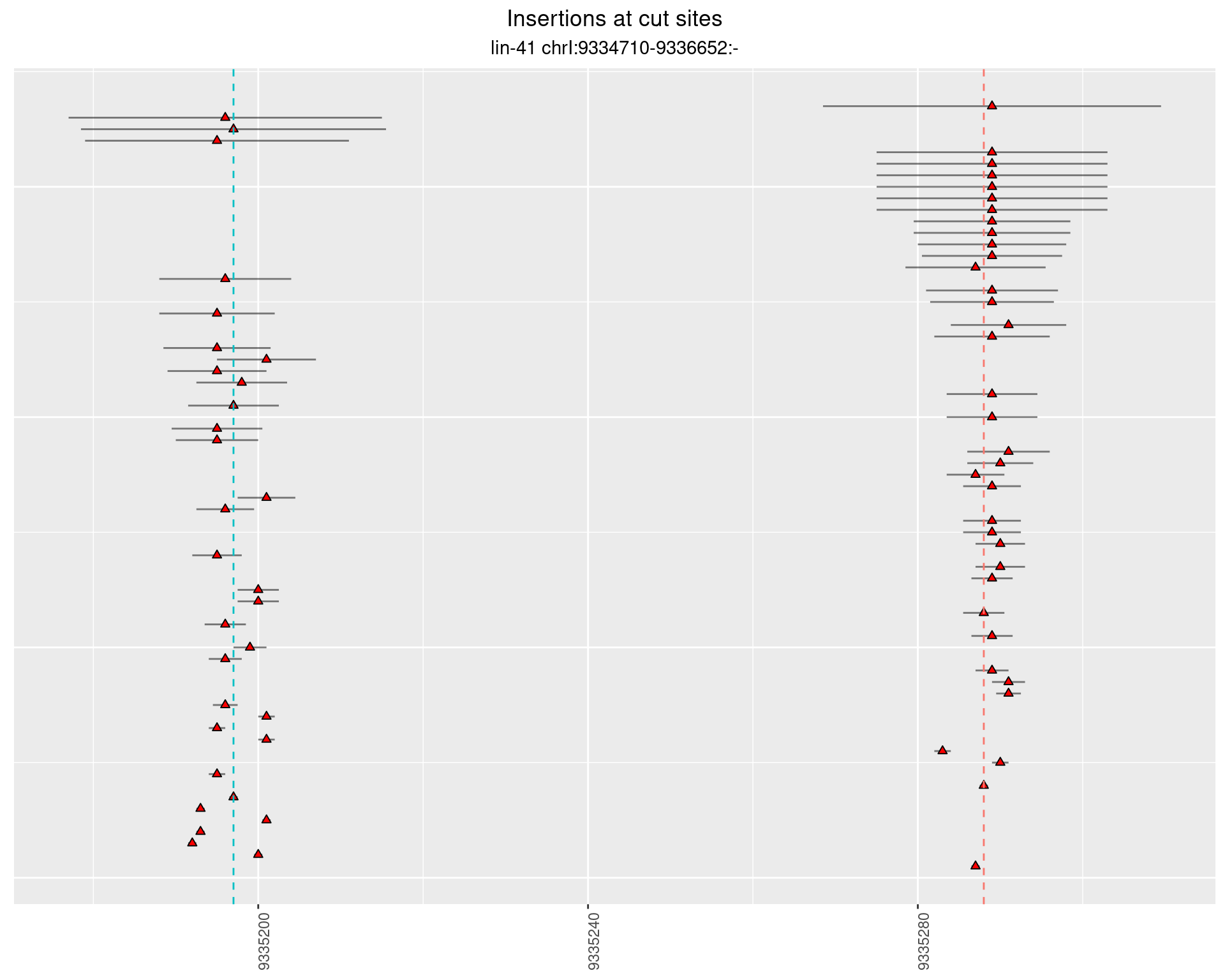
2.32.4 Freq: 1e-06 - 1e-05
2.33 gen_16C_F5_lin-41_N2
2.33.1 Freq: 1e-06 - 1e-05
2.33.2 Freq: 1e-05 - 1e-04
2.33.3 Freq: 1e-04 - 0.001
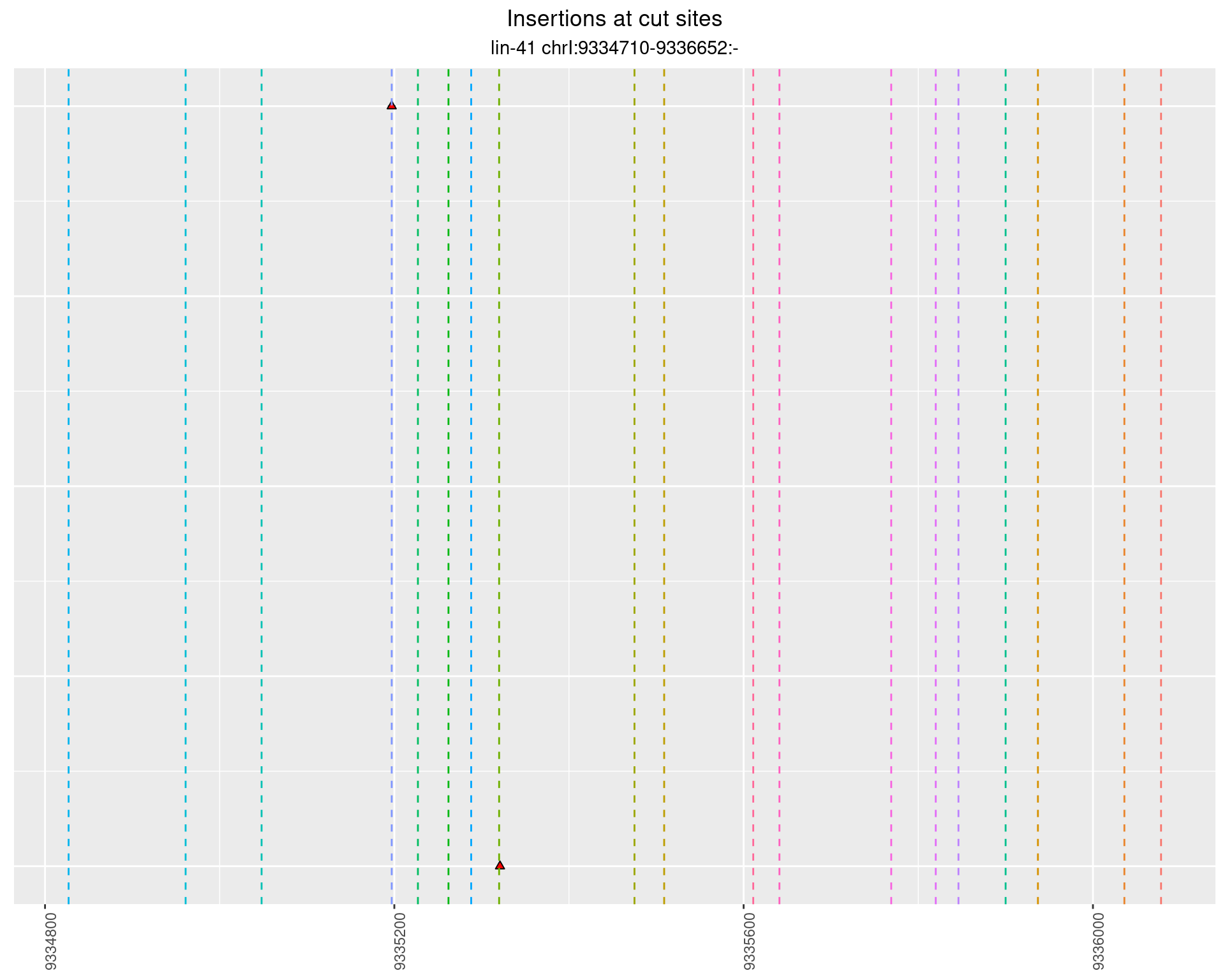
2.34 gen_16C_F5_lin-41_sg15sg16
2.34.1 Freq: 1e-04 - 0.001
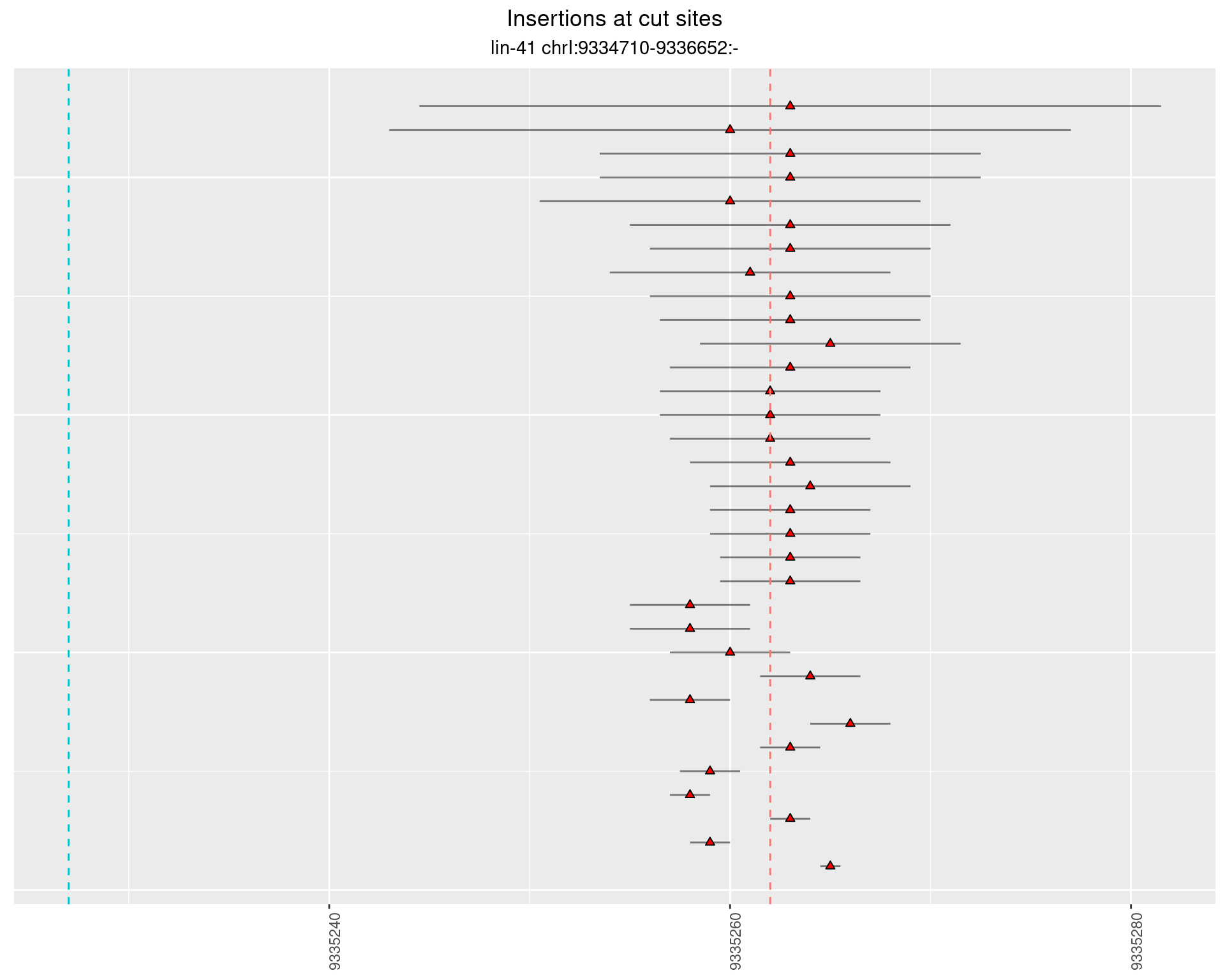
2.34.2 Freq: 0.001 - 0.01
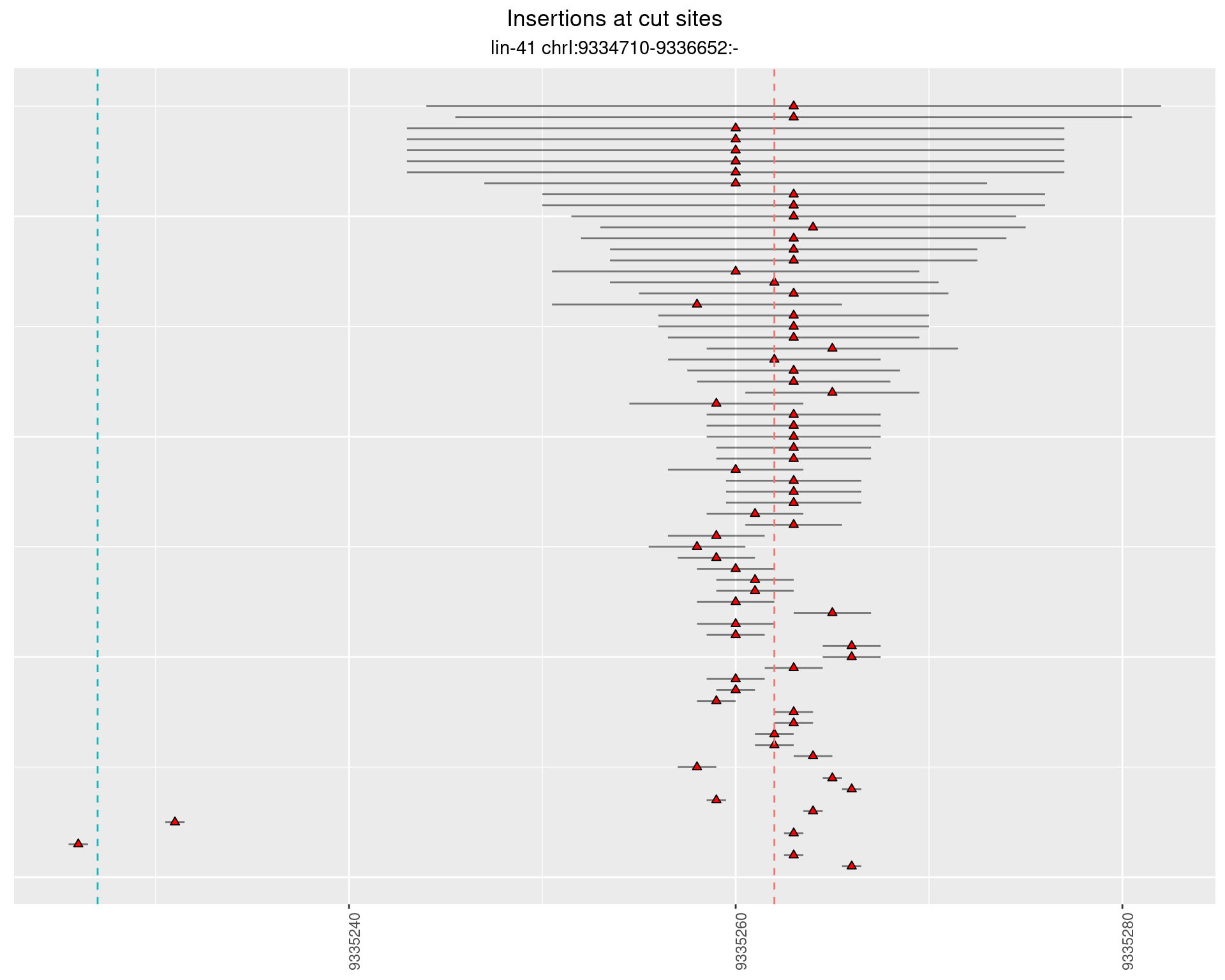
2.34.3 Freq: 1e-05 - 1e-04
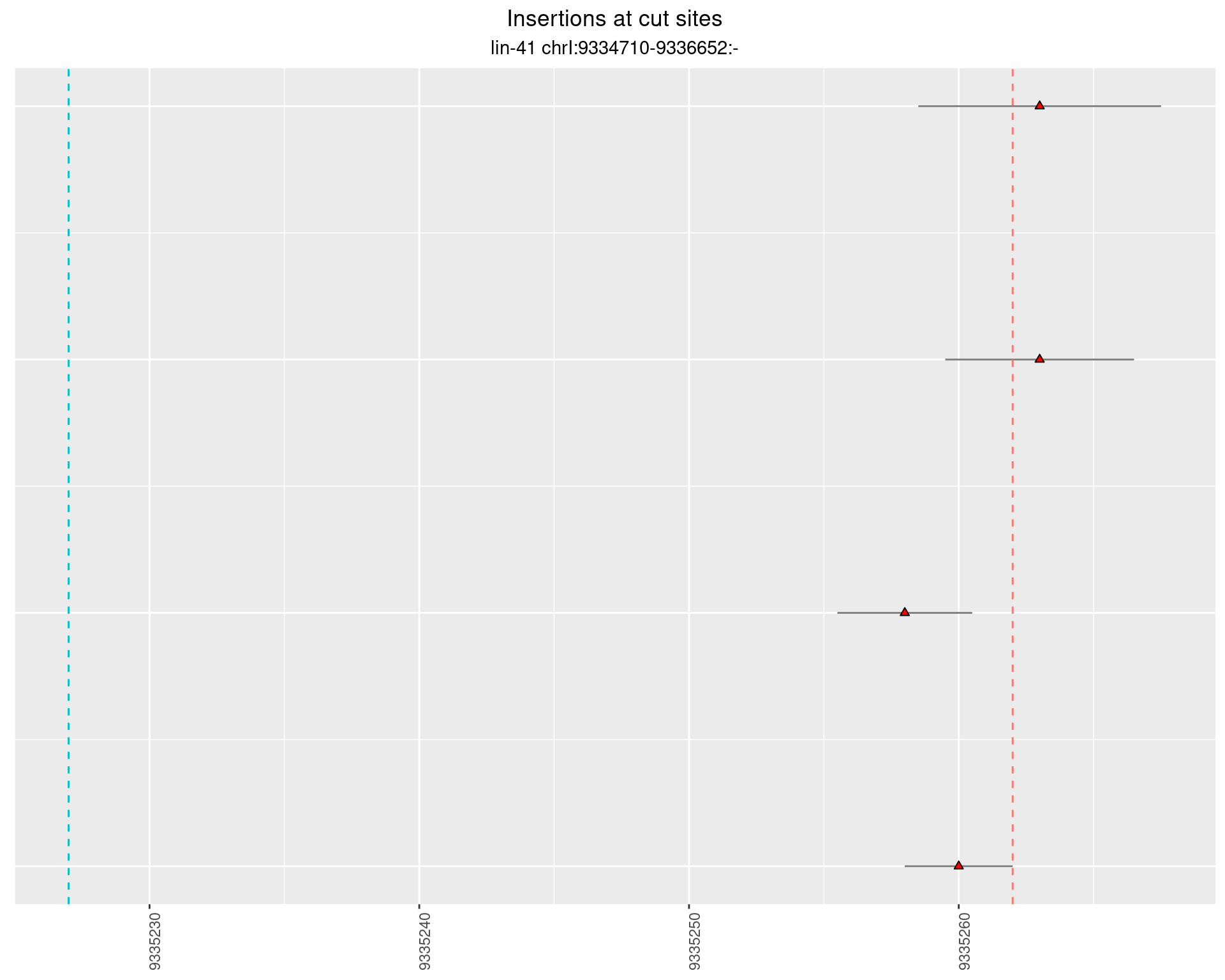
2.34.4 Freq: 1e-06 - 1e-05
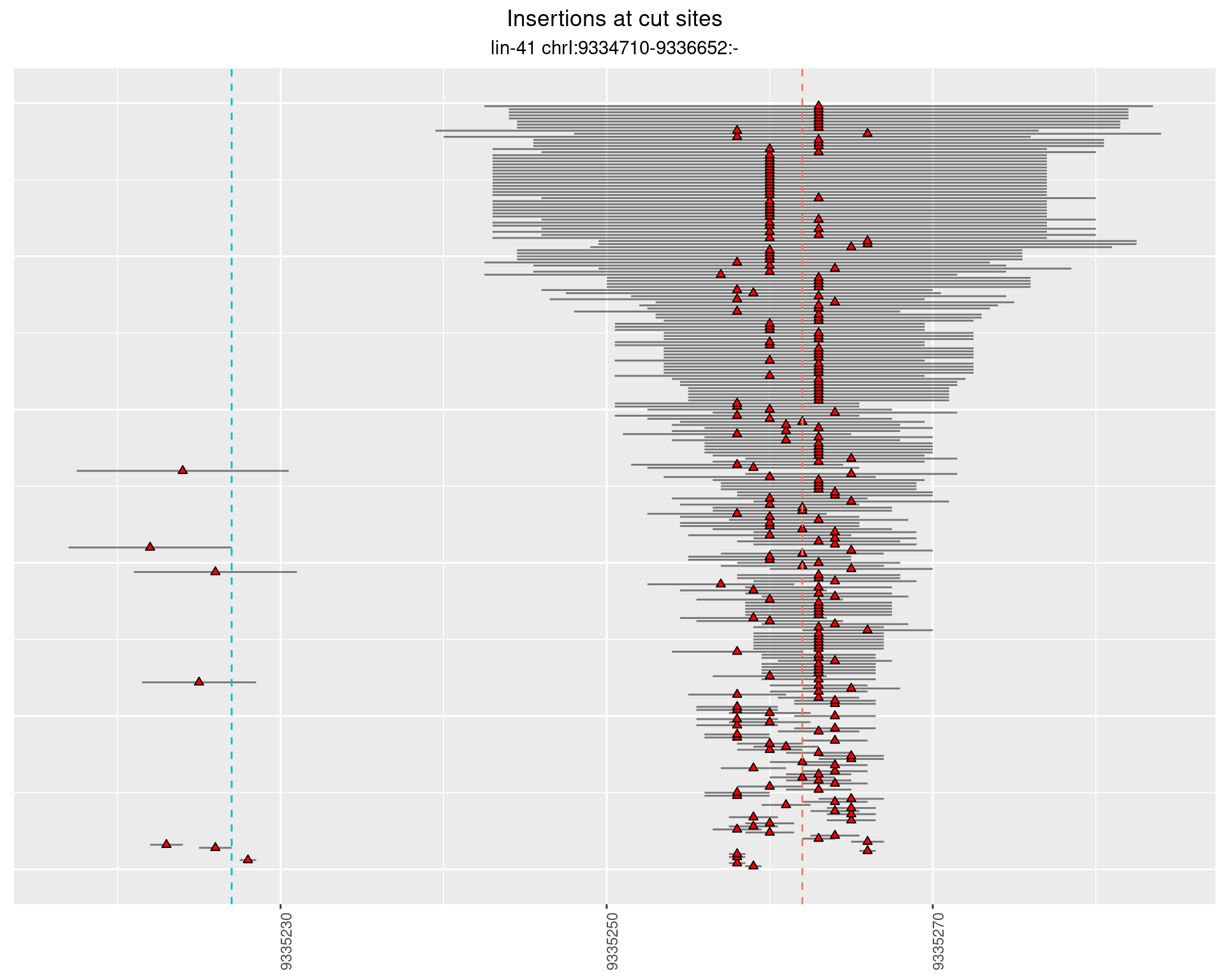
2.35 gen_16C_F5_lin-41_pool
2.35.1 Freq: 0.001 - 0.01
2.35.2 Freq: 1e-04 - 0.001
2.35.3 Freq: 1e-05 - 1e-04
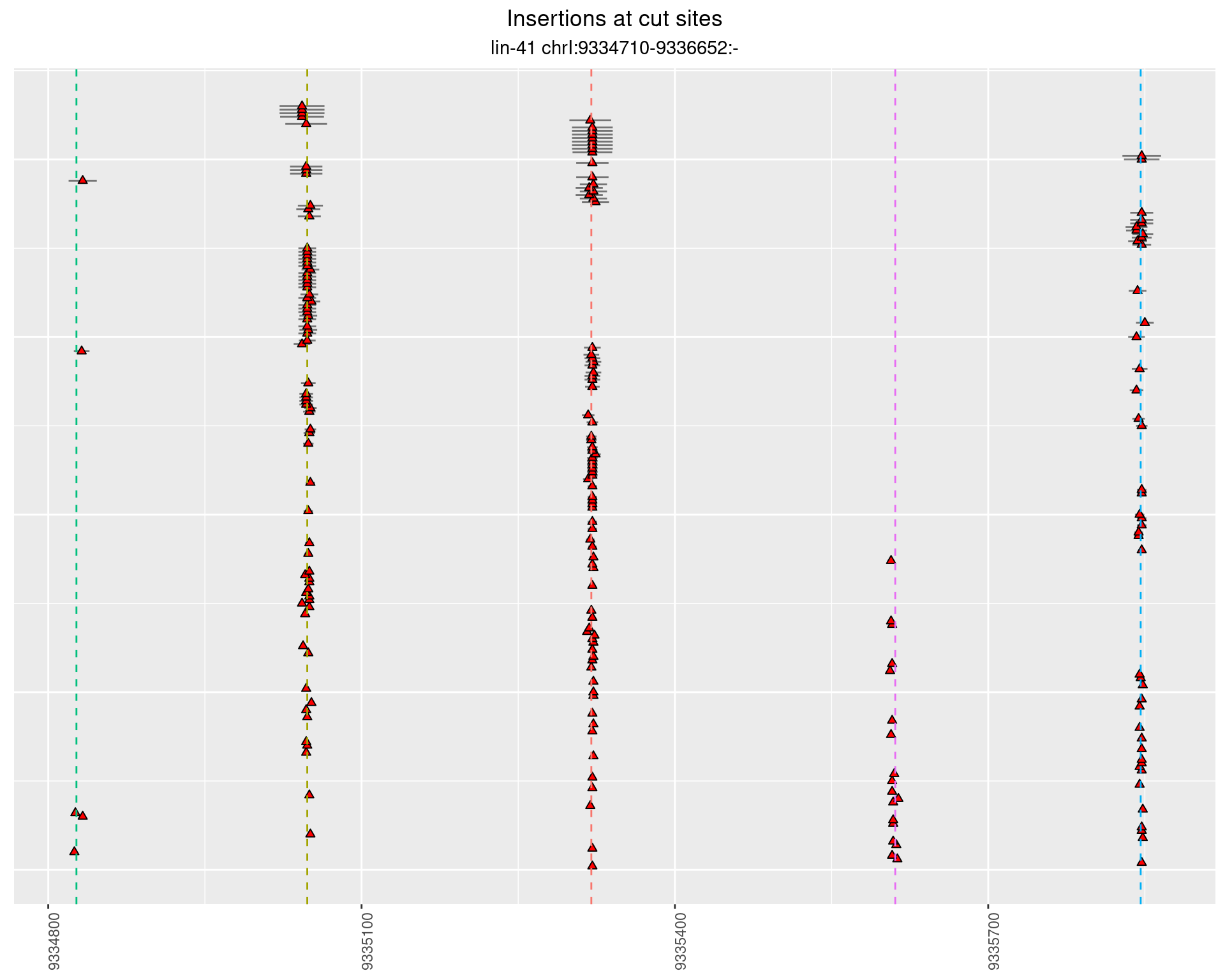
2.36 gen_16C_F5_lin-41_sg26sg27
2.36.1 Freq: 1e-04 - 0.001
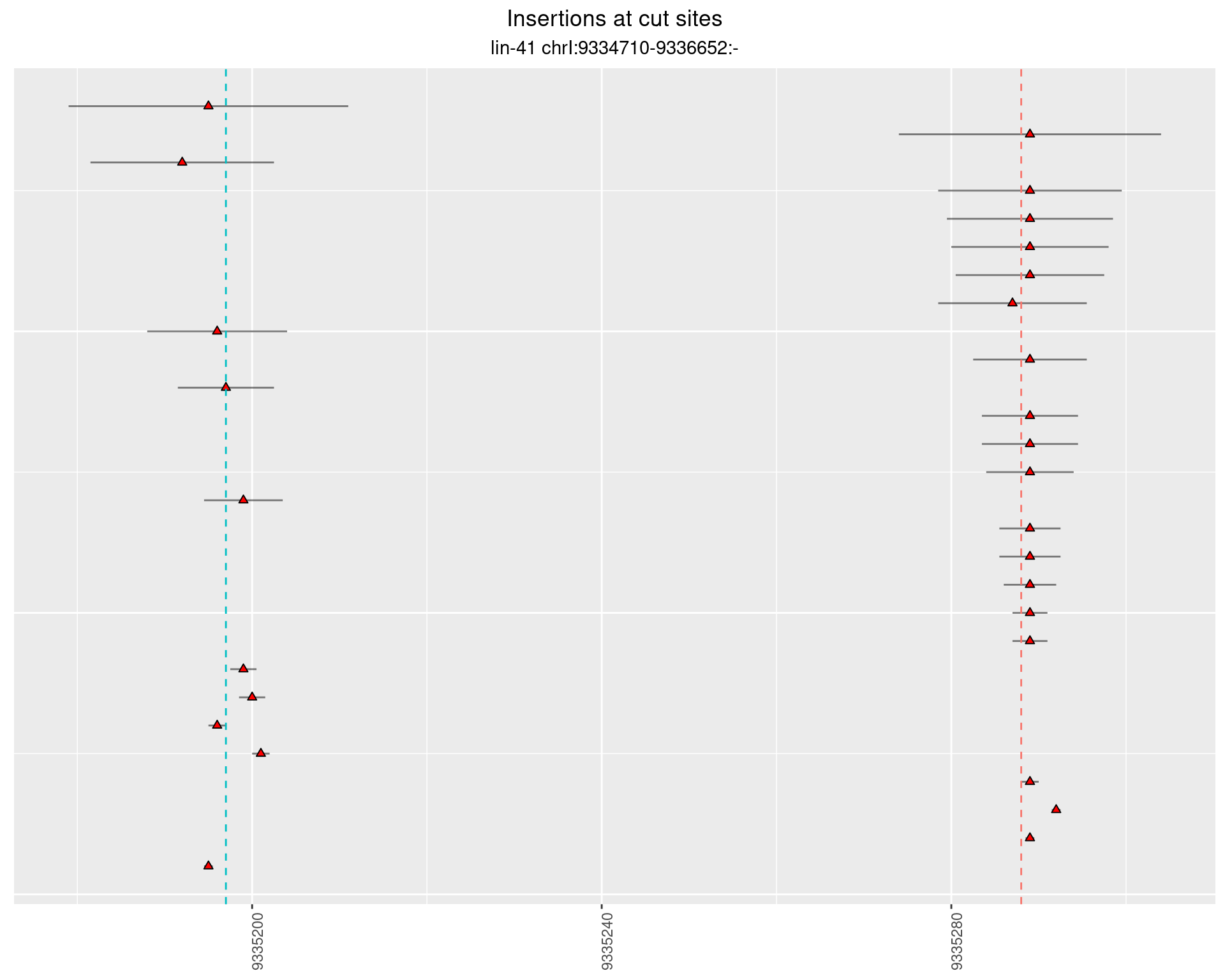
2.36.2 Freq: 1e-05 - 1e-04
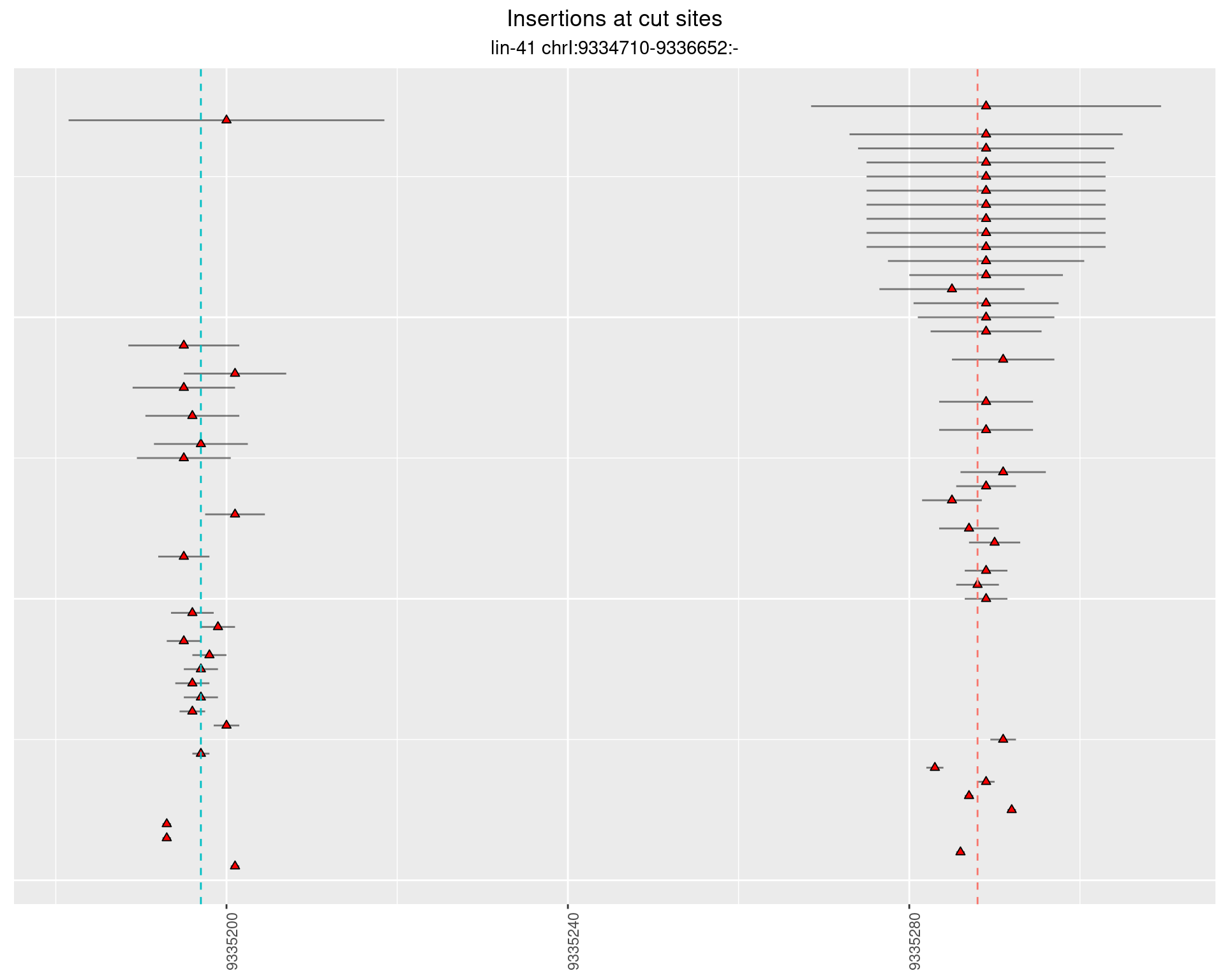
2.36.3 Freq: 0.001 - 0.01
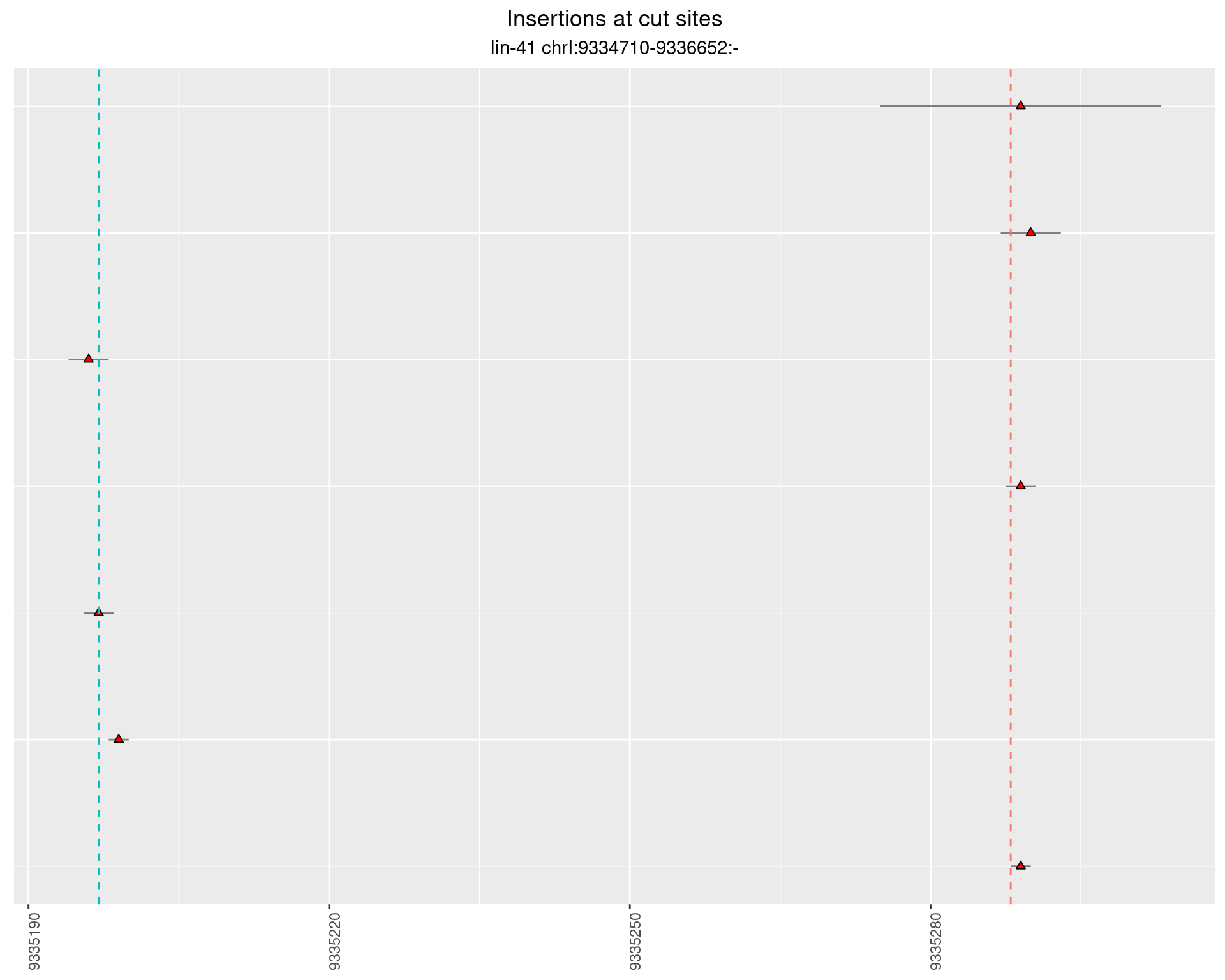
2.36.4 Freq: 1e-06 - 1e-05
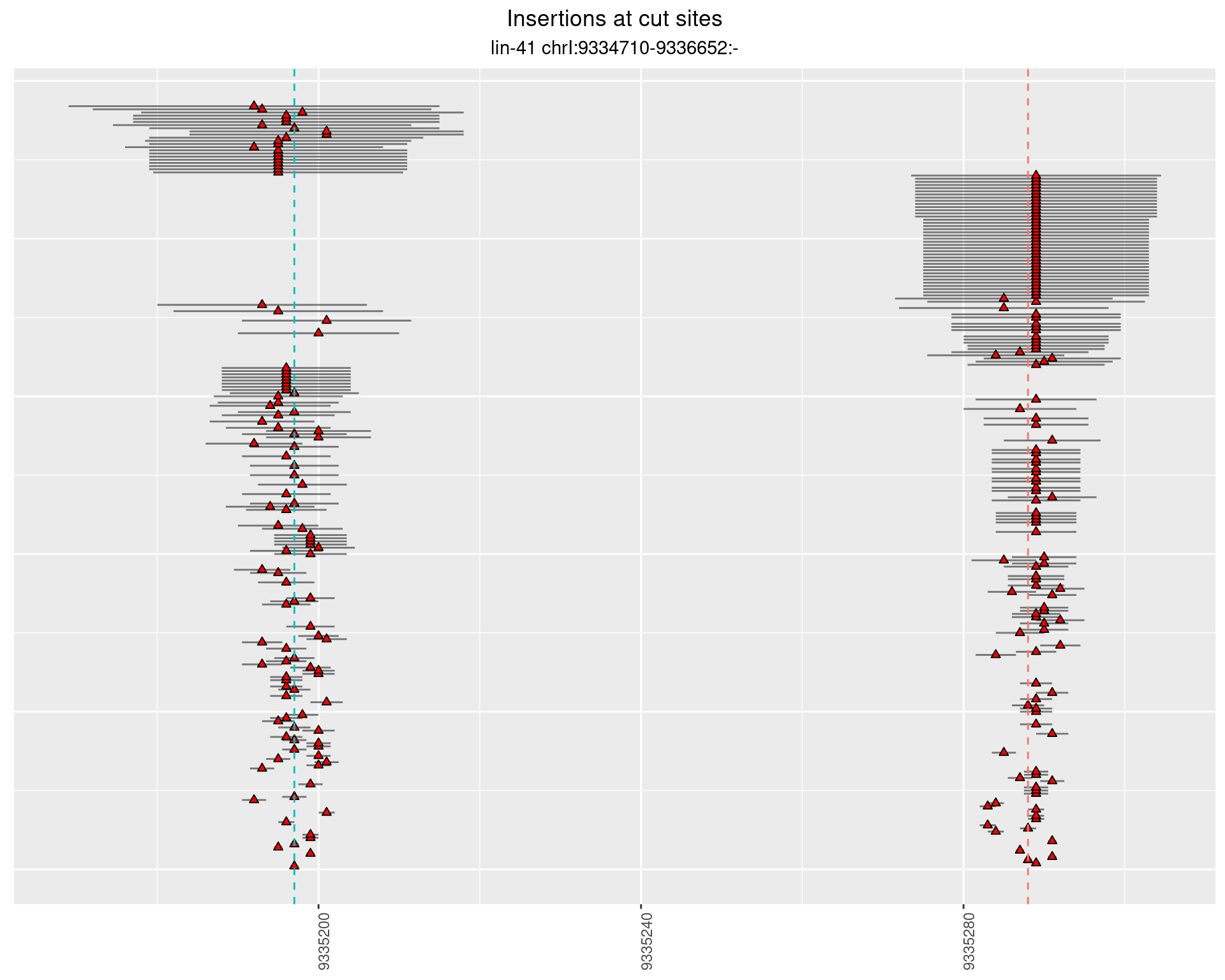
2.37 lin41_DNA_N2
2.37.1 Freq: 1e-05 - 1e-04
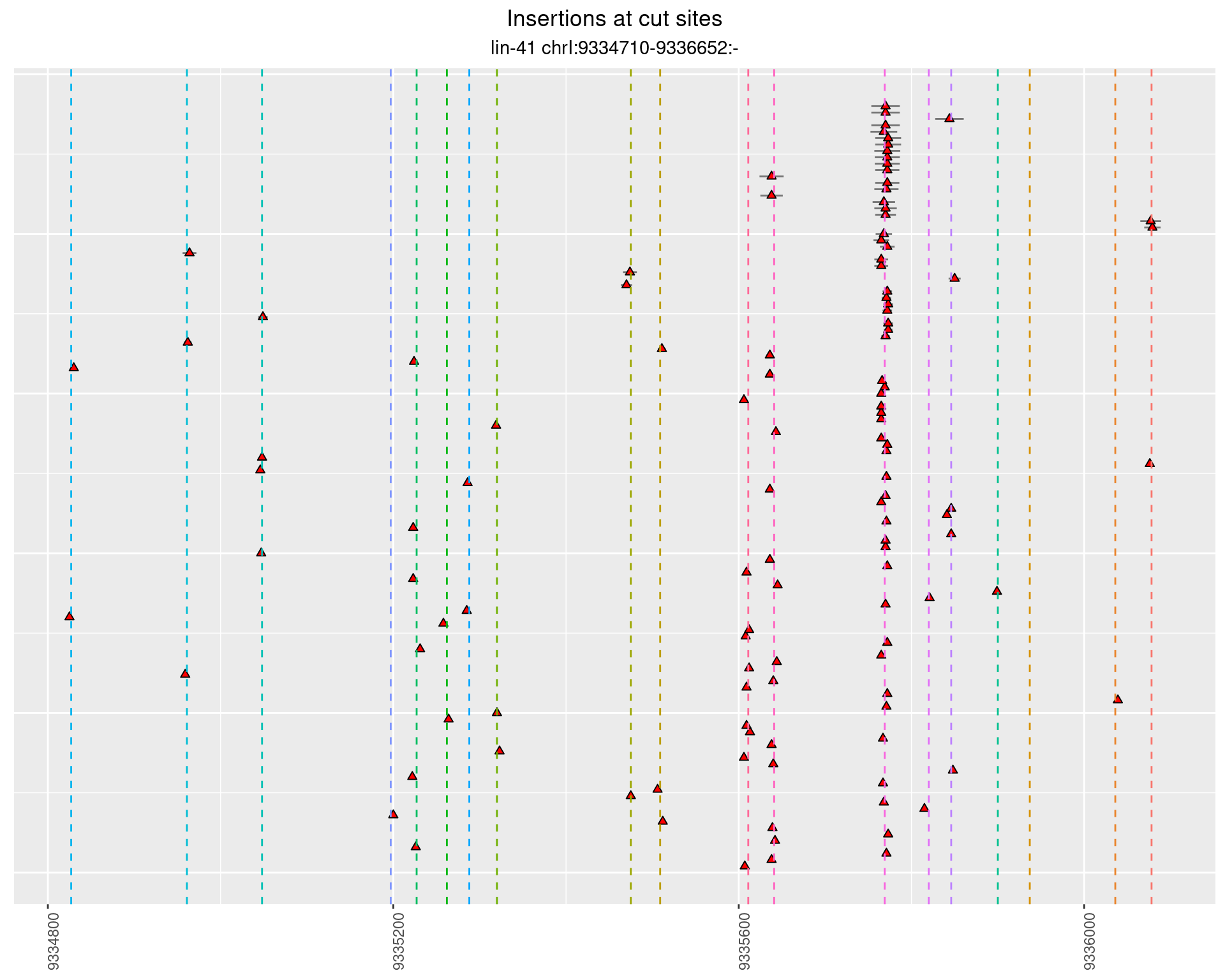
2.37.2 Freq: 1e-06 - 1e-05
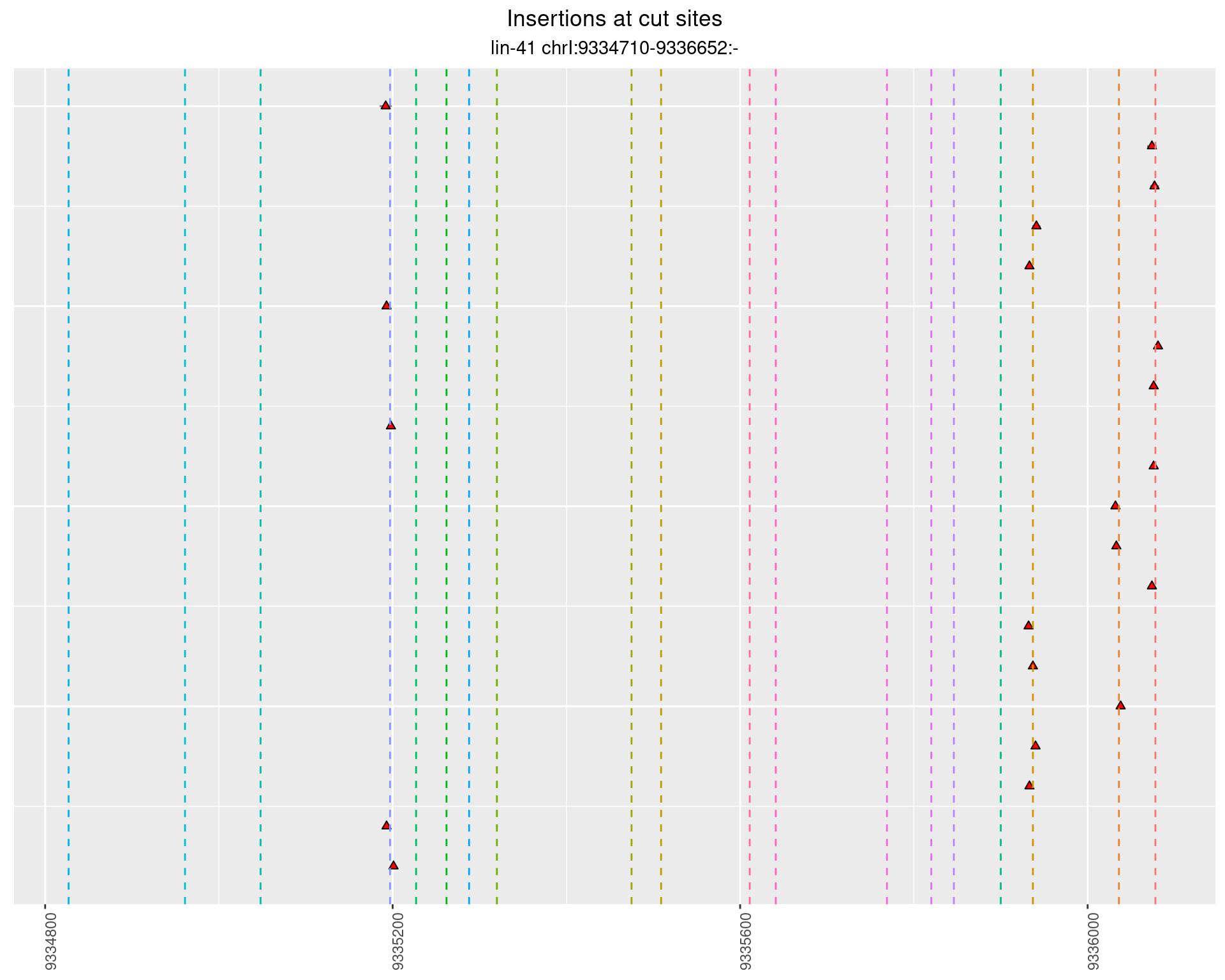
2.37.3 Freq: 1e-04 - 0.001
2.38 lin41_DNA_CDS
2.38.1 Freq: 1e-04 - 0.001
2.38.2 Freq: 1e-05 - 1e-04
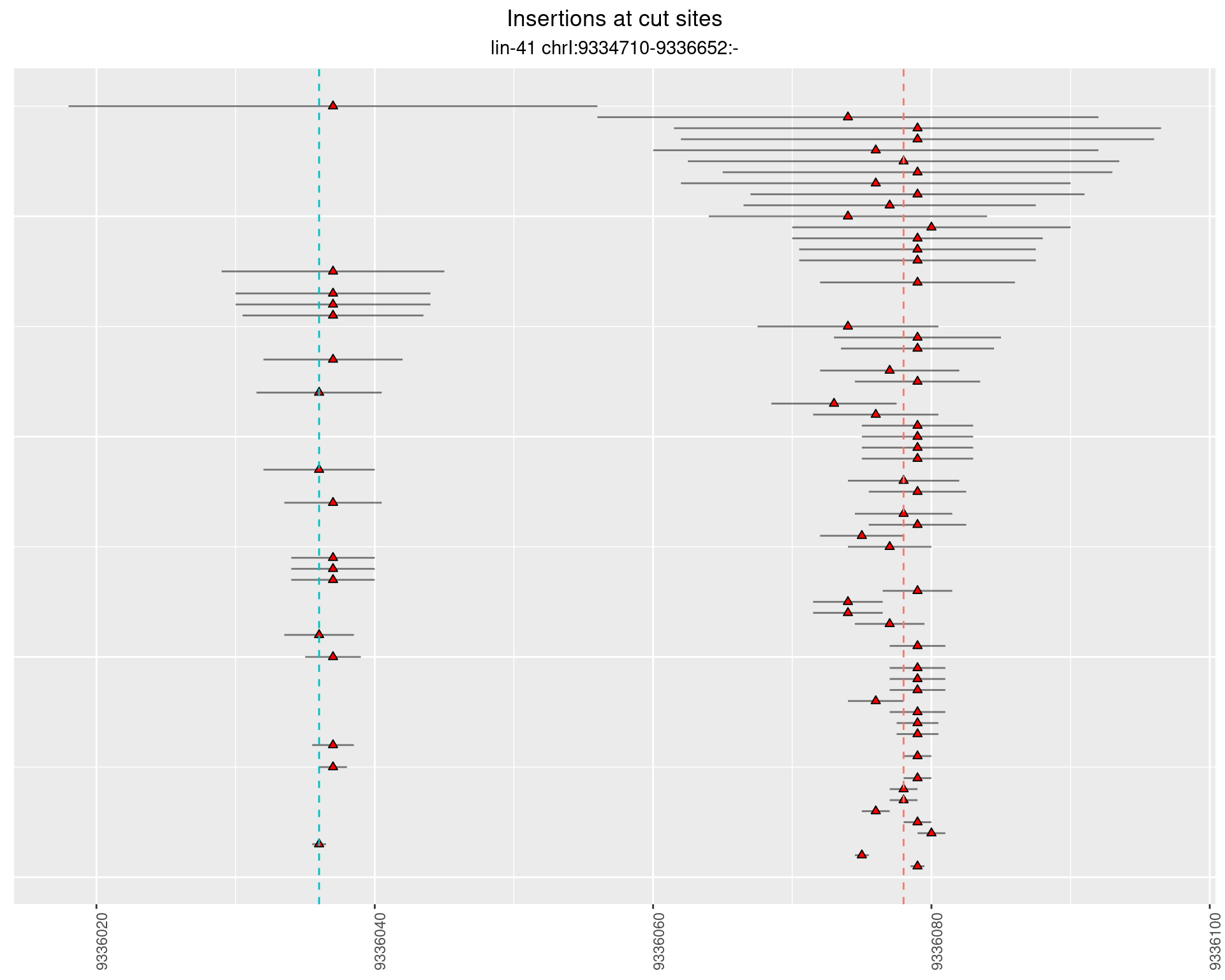
2.38.3 Freq: 1e-06 - 1e-05
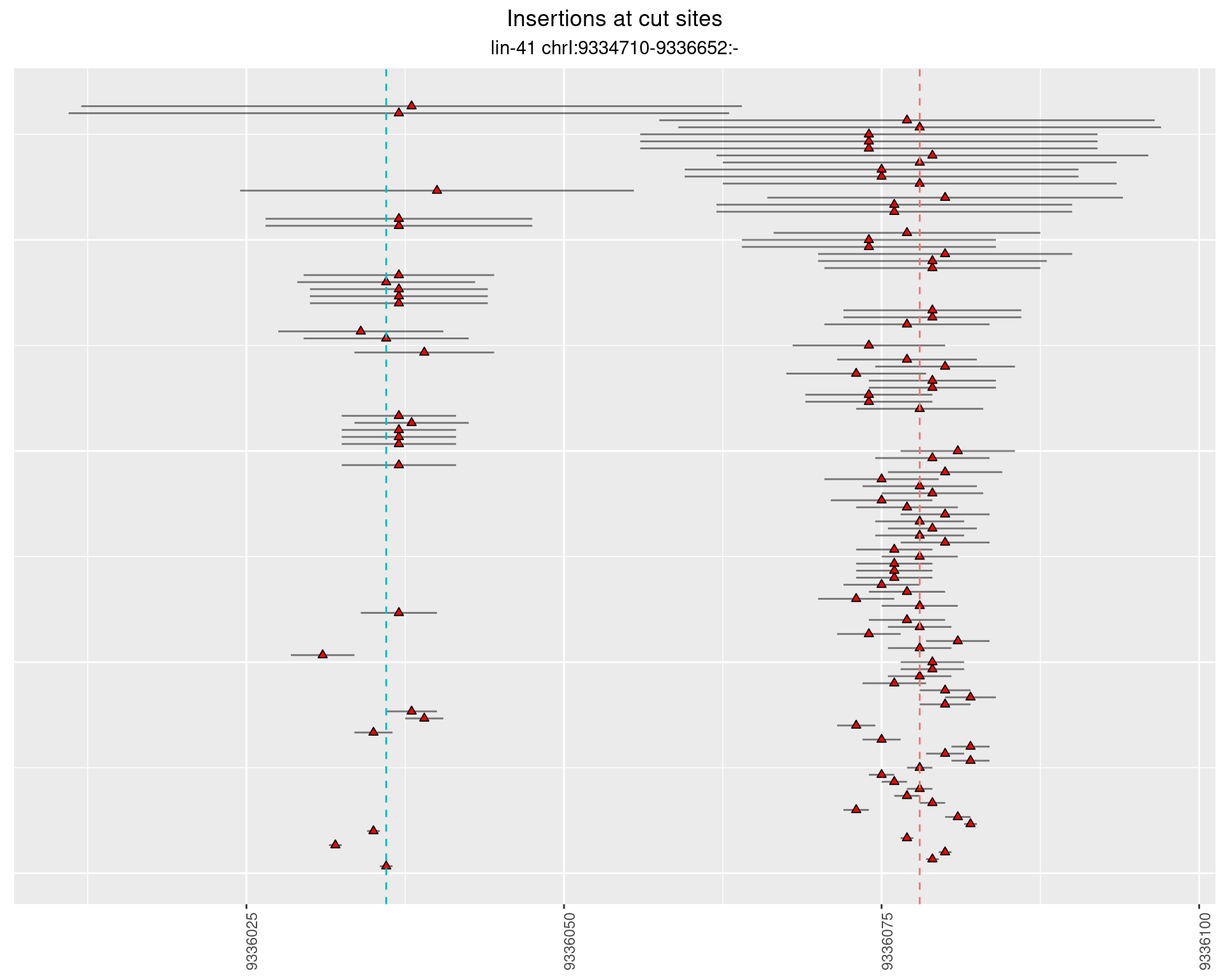
2.39 lin41_DNA_1516
2.39.1 Freq: 1e-06 - 1e-05
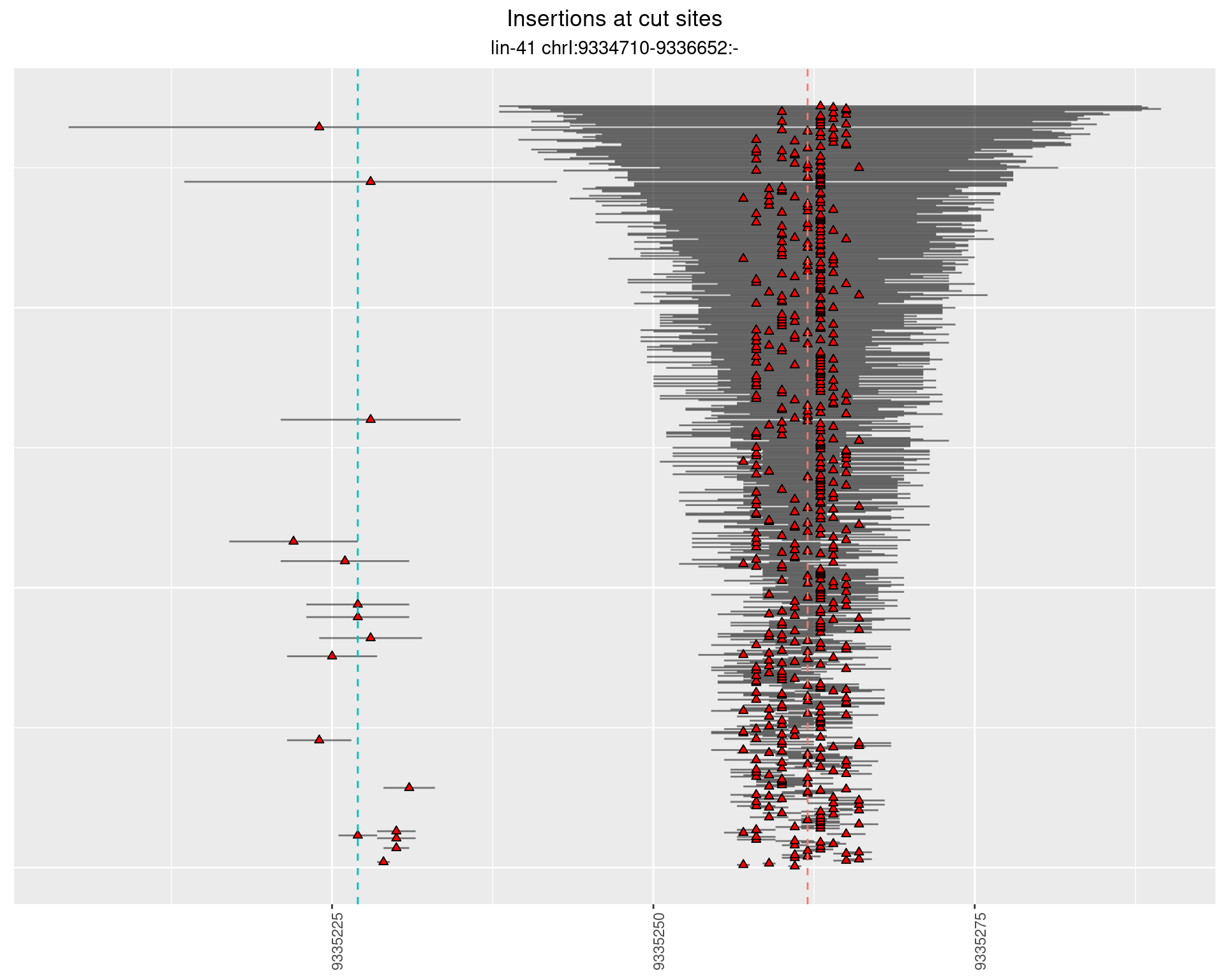
2.39.2 Freq: 1e-05 - 1e-04
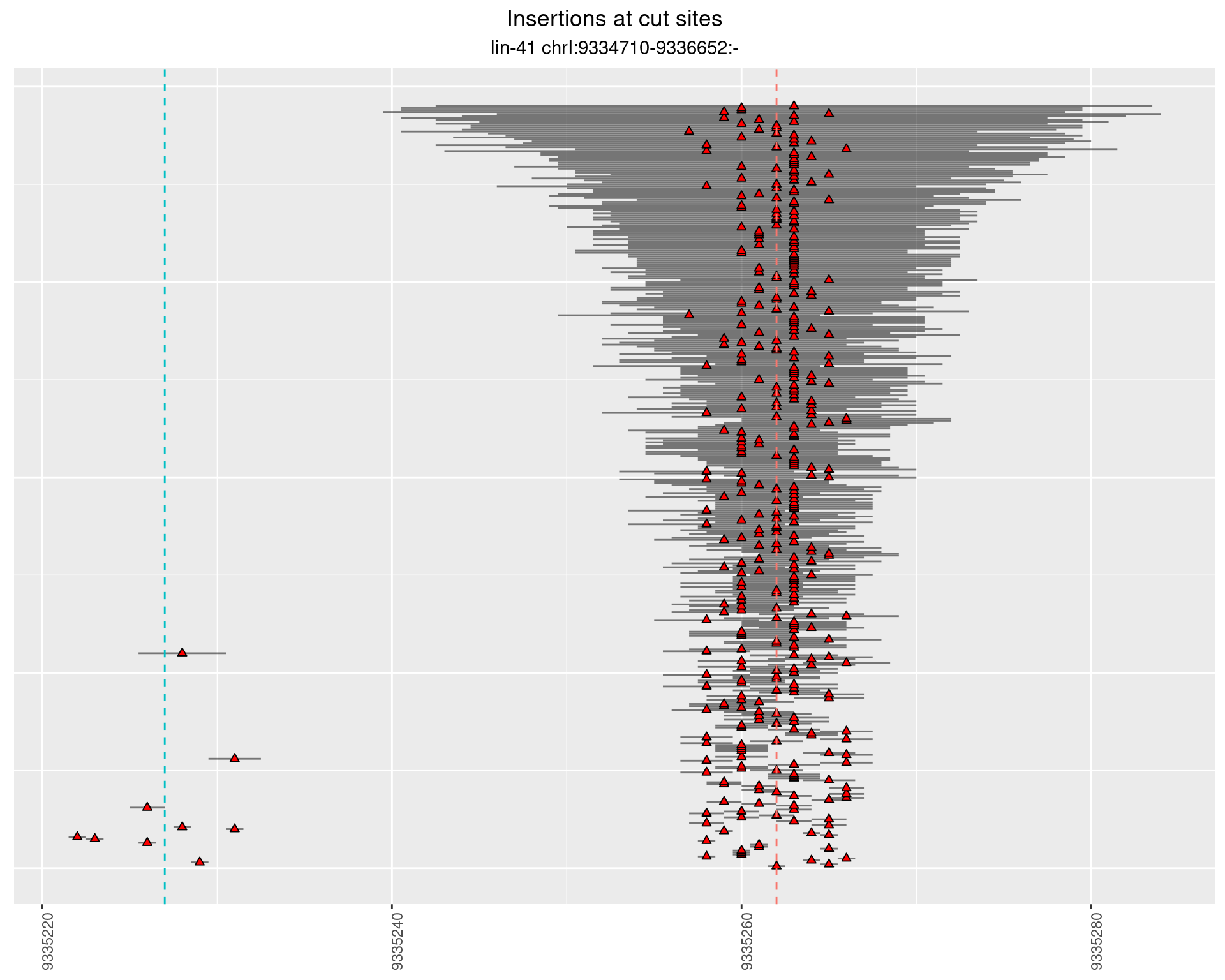
2.39.3 Freq: 1e-04 - 0.001
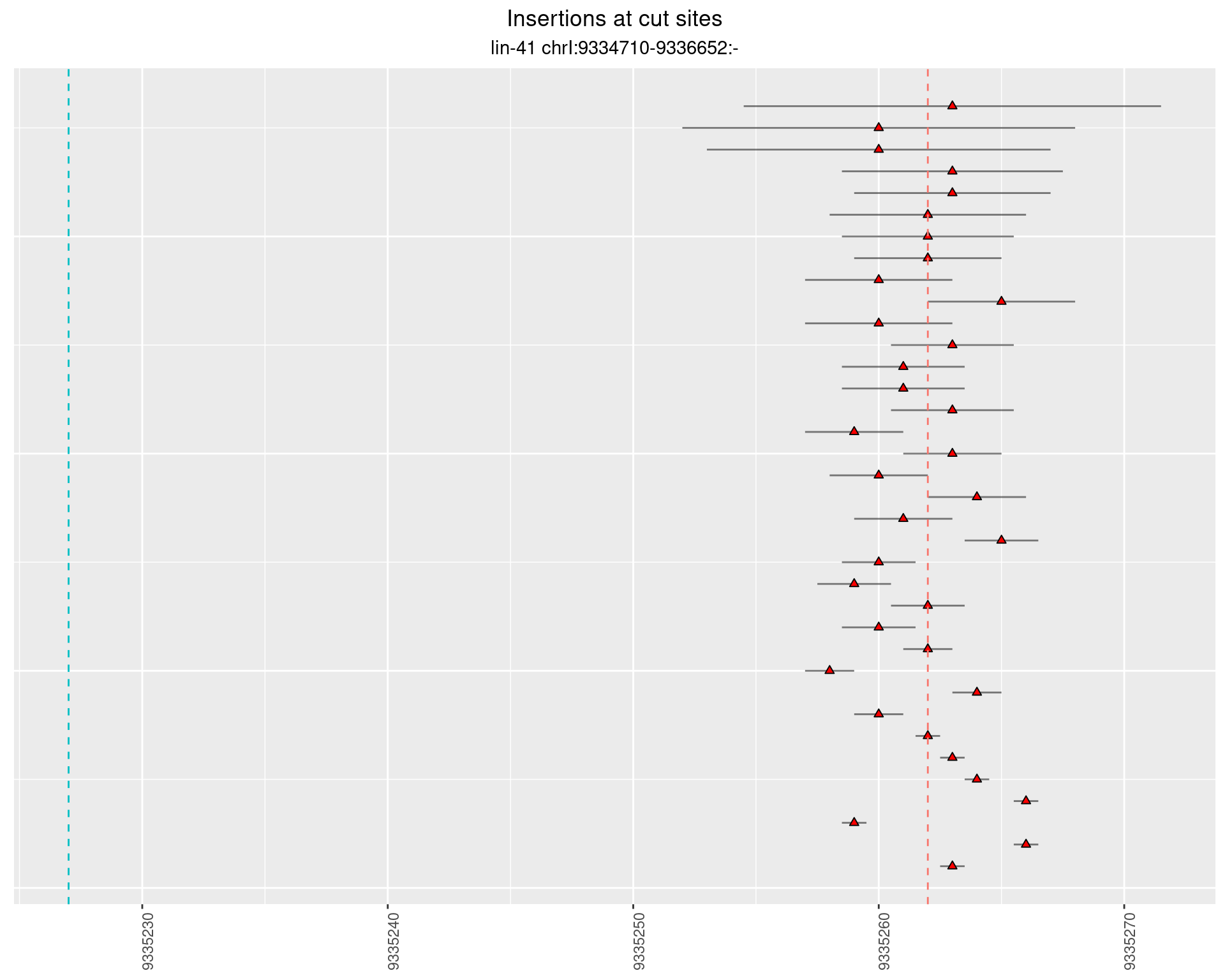
2.40 lin41_DNA_pool1
2.40.1 Freq: 1e-05 - 1e-04
2.40.2 Freq: 1e-06 - 1e-05
2.40.3 Freq: 1e-04 - 0.001
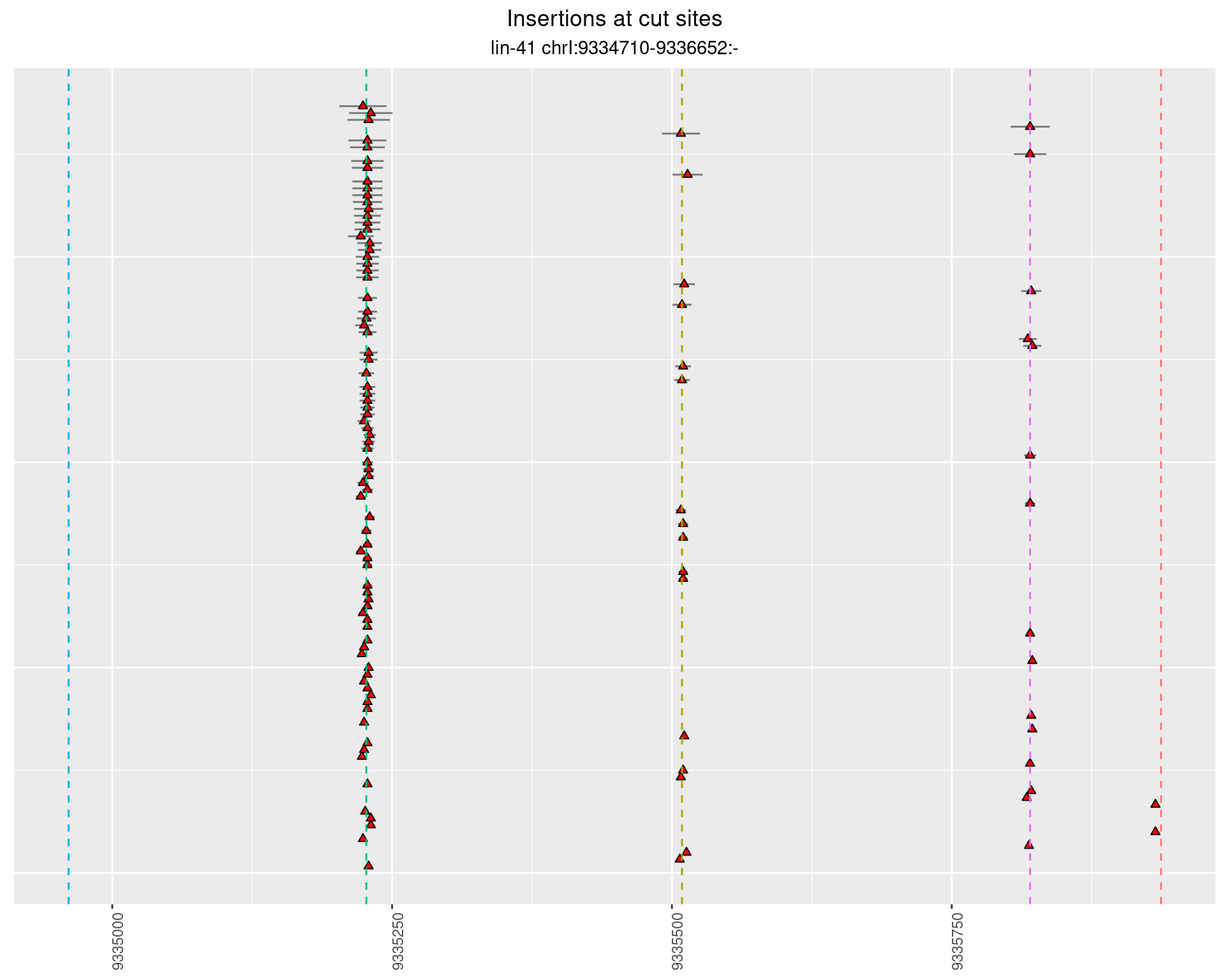
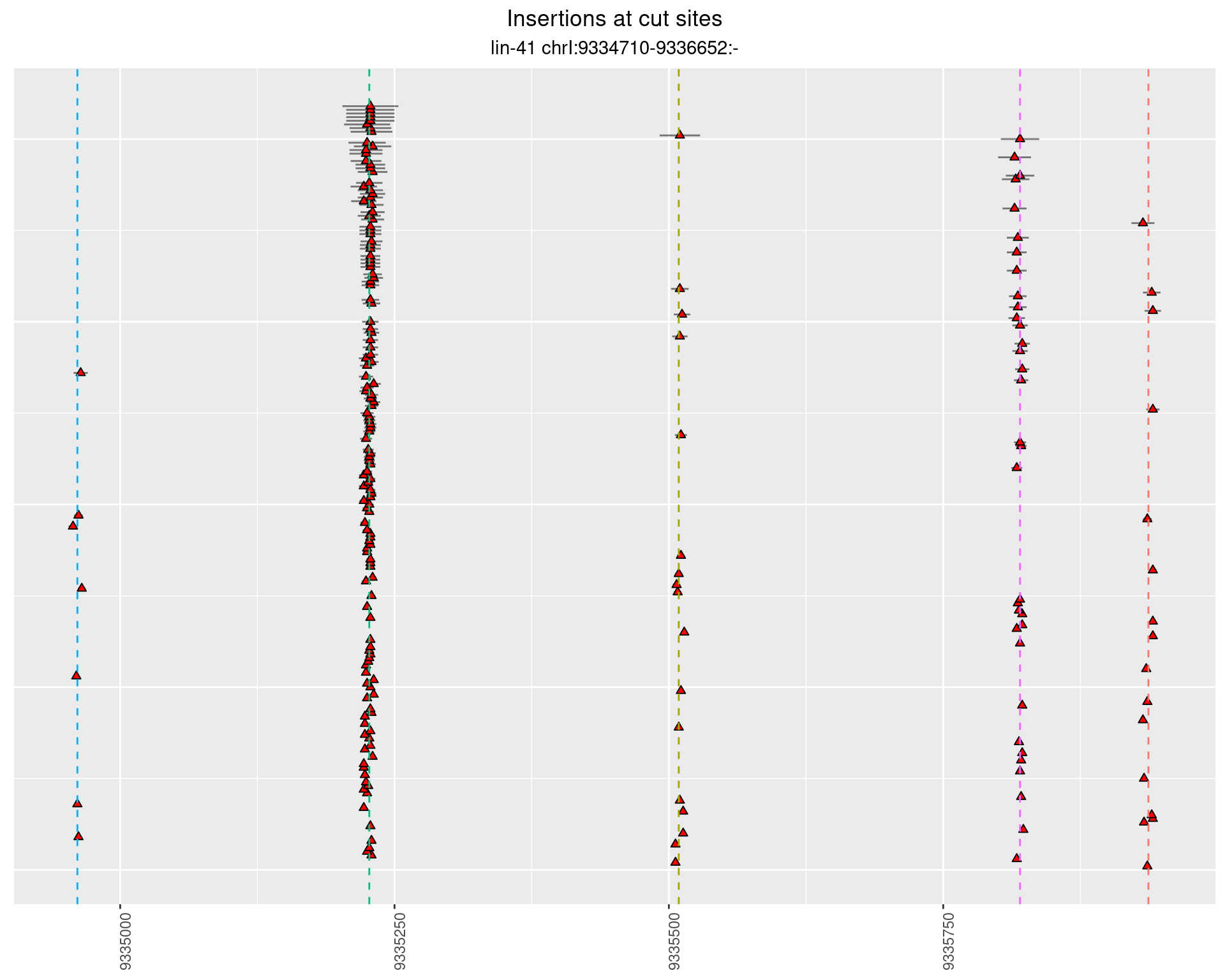
2.41 lin41_DNA_pool2
2.41.1 Freq: 1e-04 - 0.001
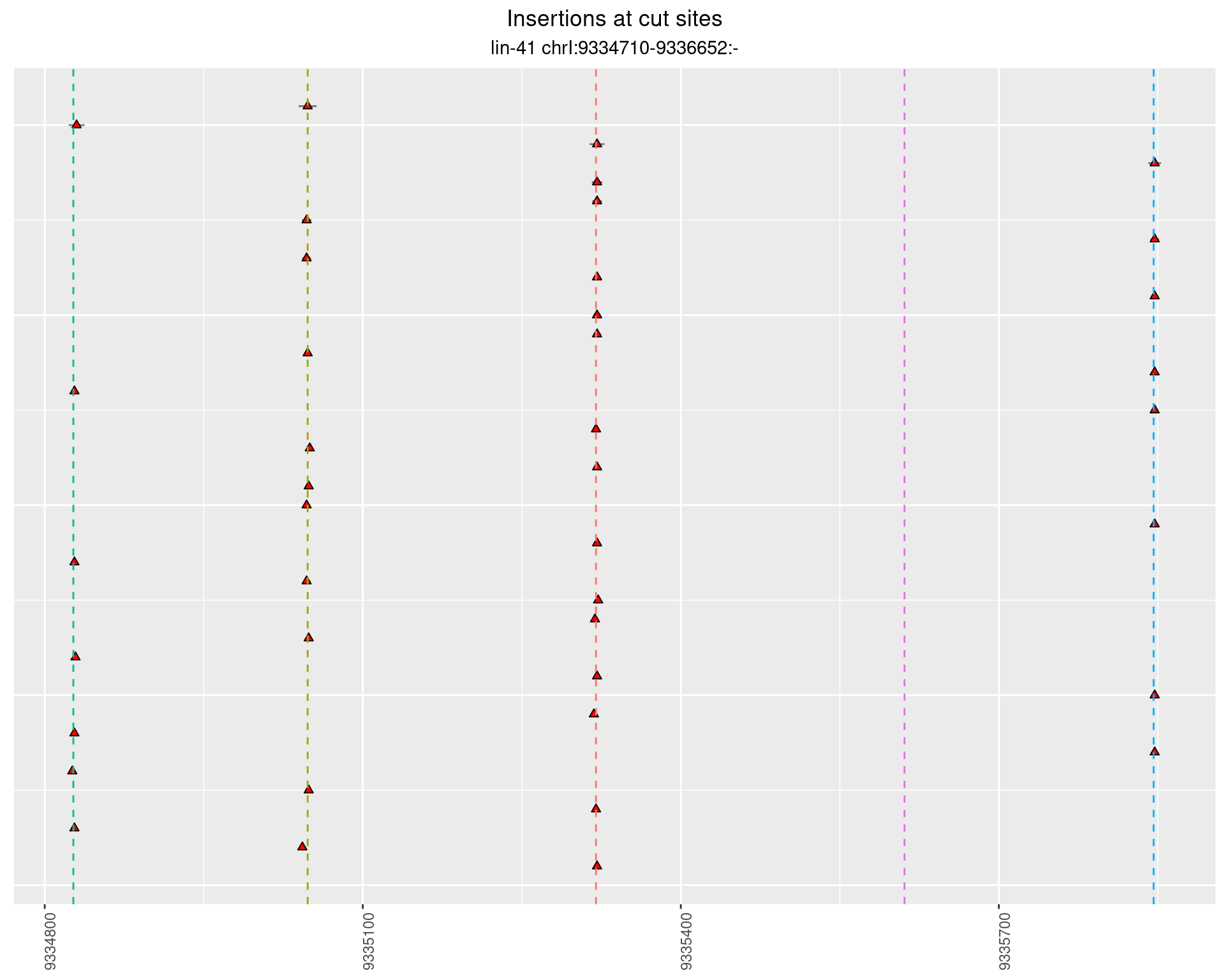
2.41.2 Freq: 1e-05 - 1e-04
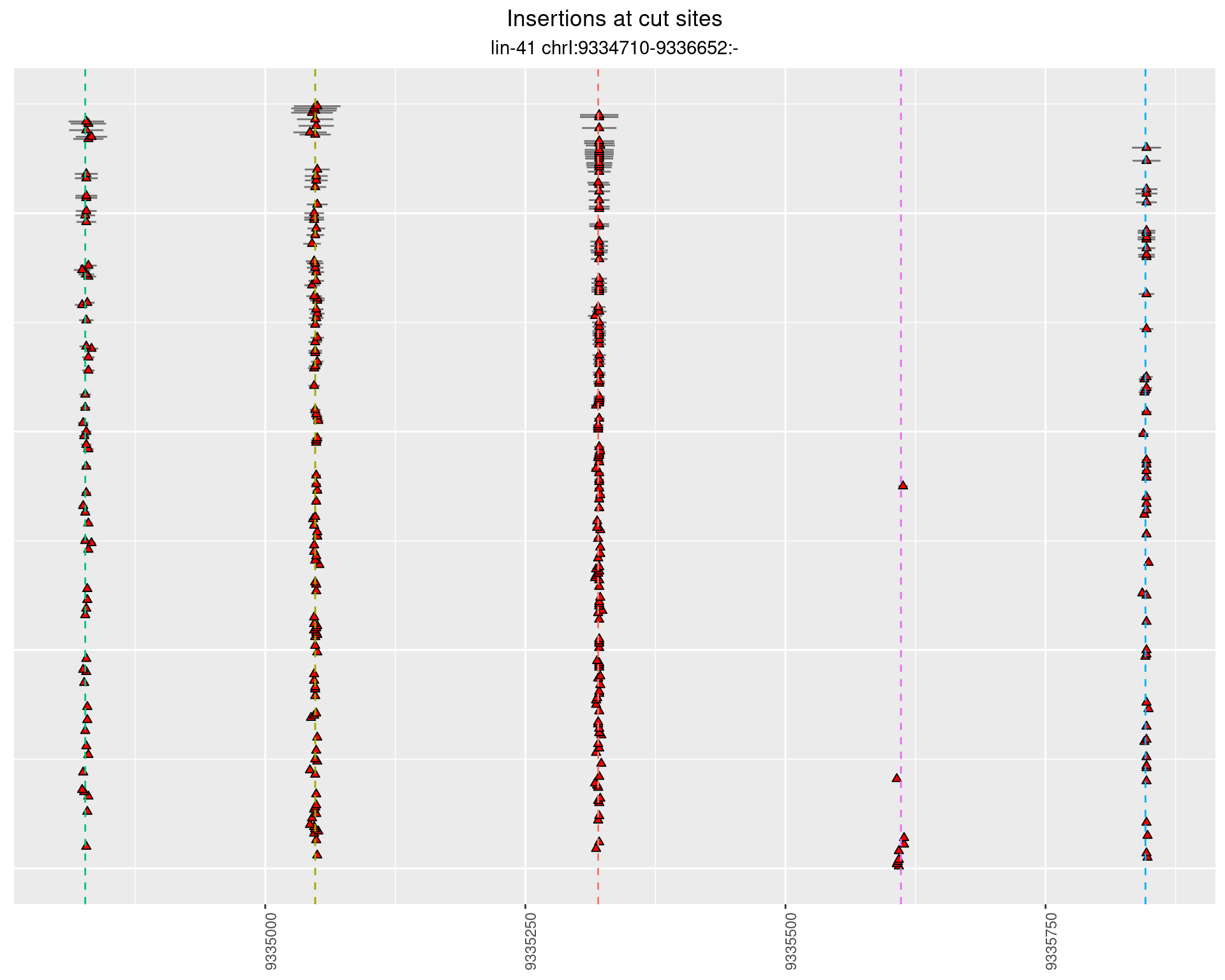
2.41.3 Freq: 1e-06 - 1e-05
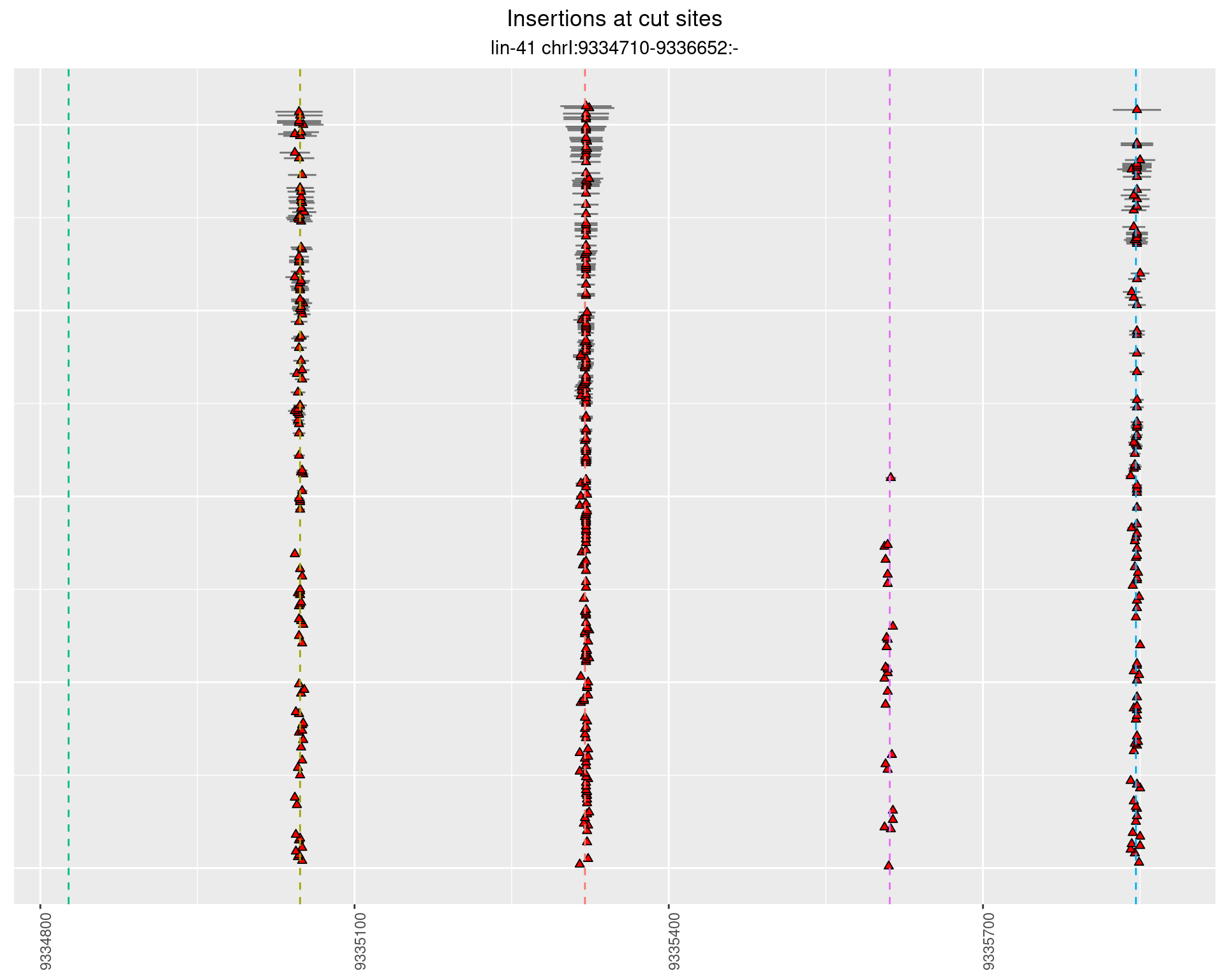
2.41.4 Freq: 0.001 - 0.01
2.42 lin41_DNA_pool3
2.42.1 Freq: 1e-05 - 1e-04
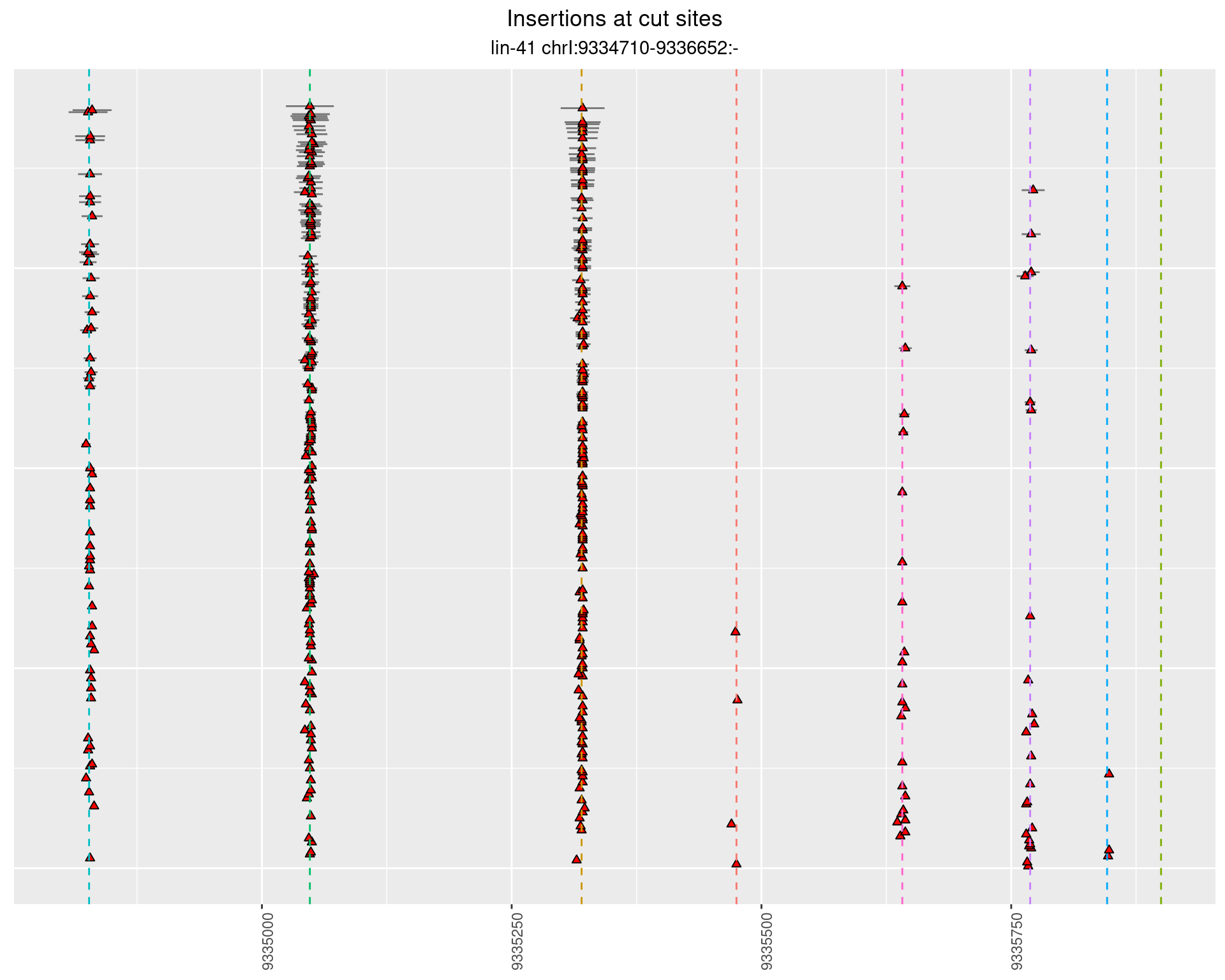
2.42.2 Freq: 1e-06 - 1e-05
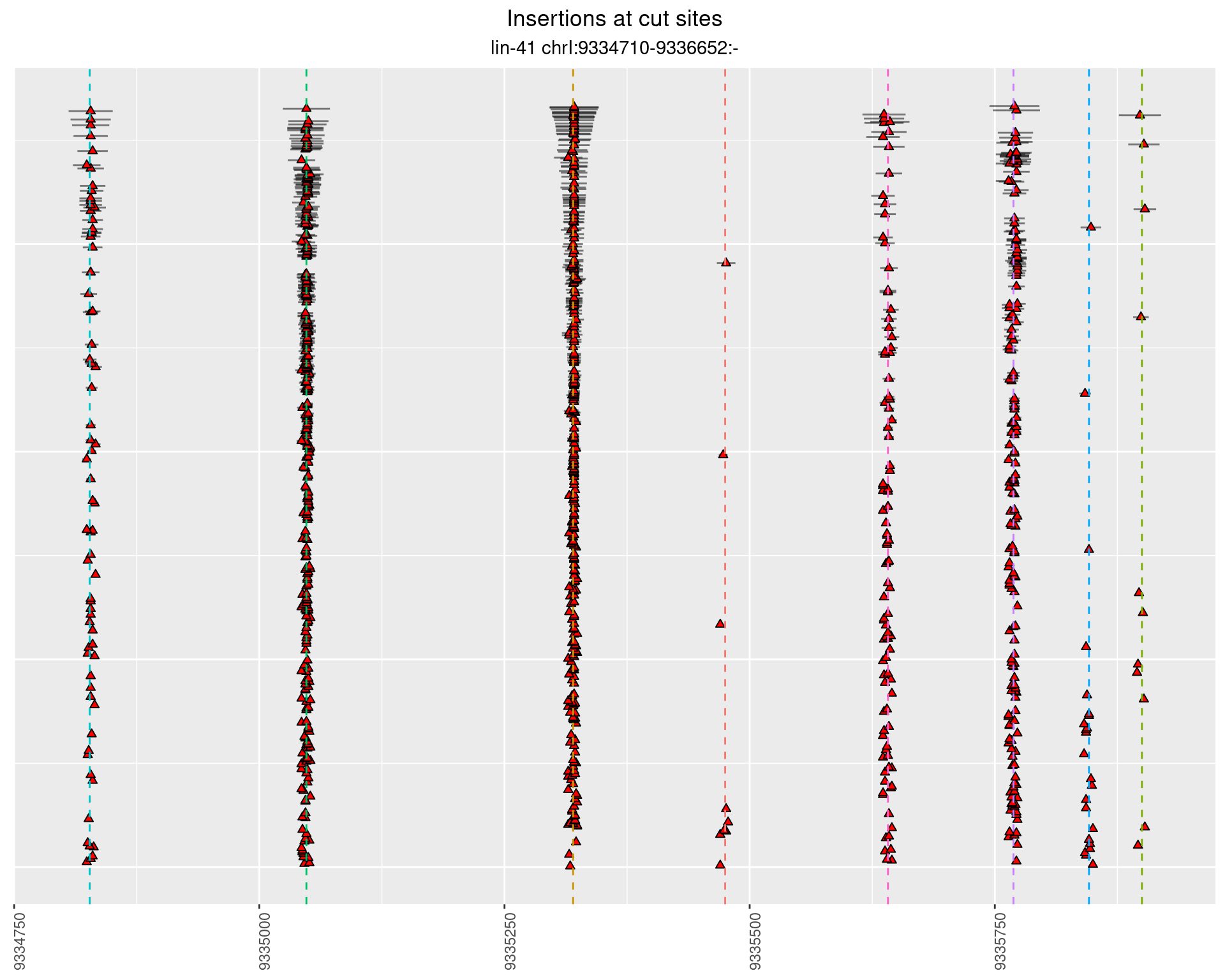
2.42.3 Freq: 1e-04 - 0.001
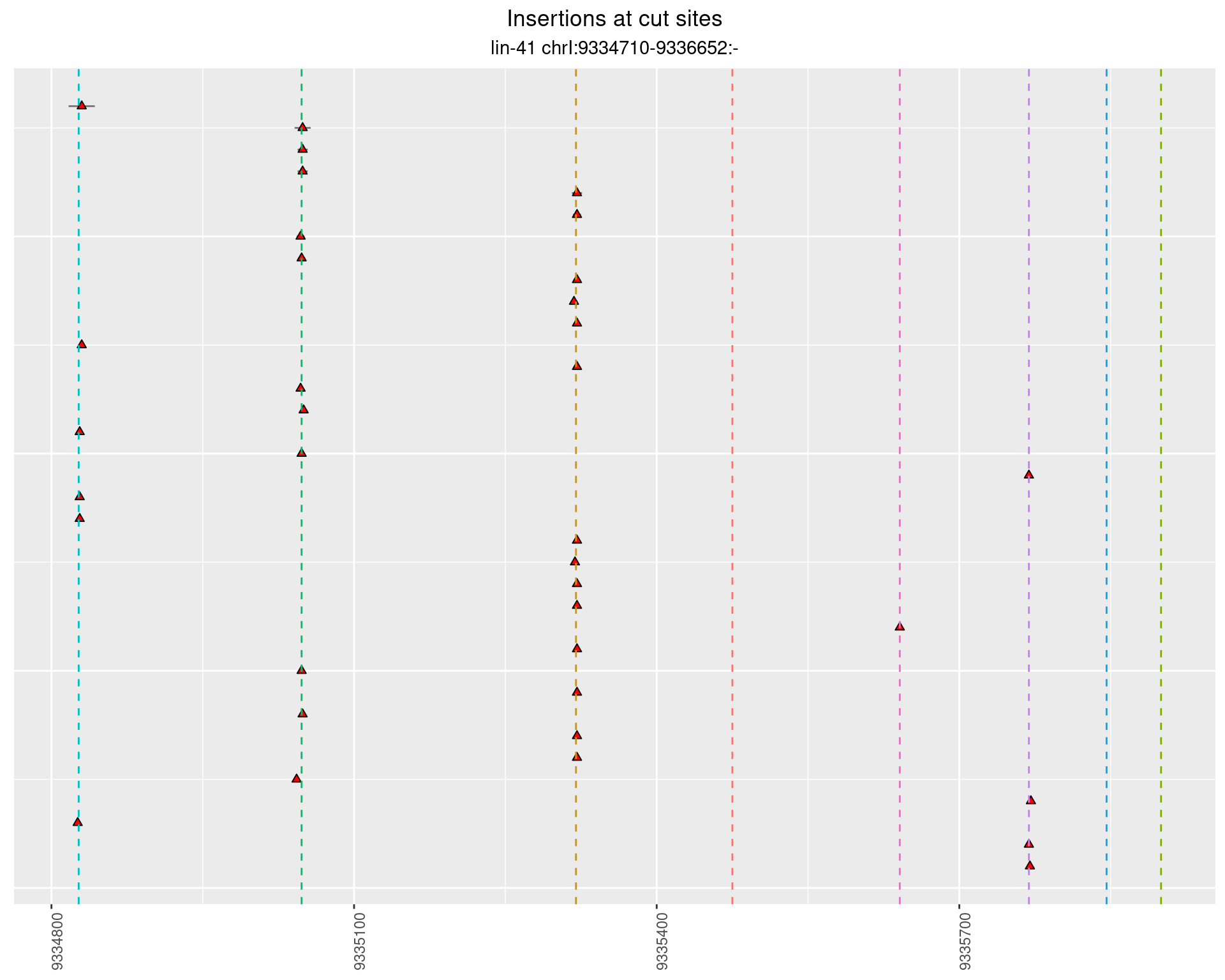
2.43 lin41_RNA_L1_N2_3end
2.43.1 Freq: 0.001 - 0.01
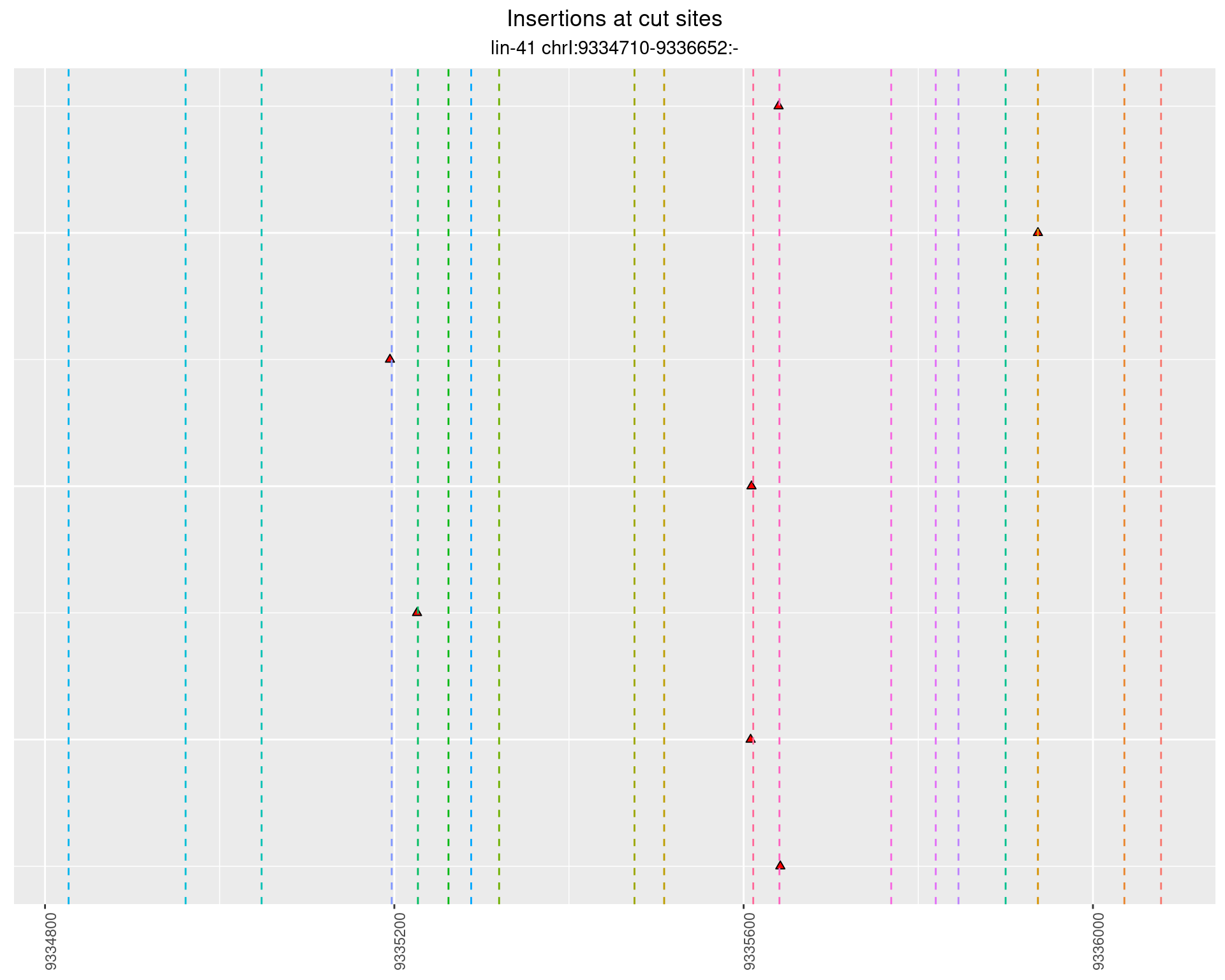
2.43.2 Freq: 1e-05 - 1e-04
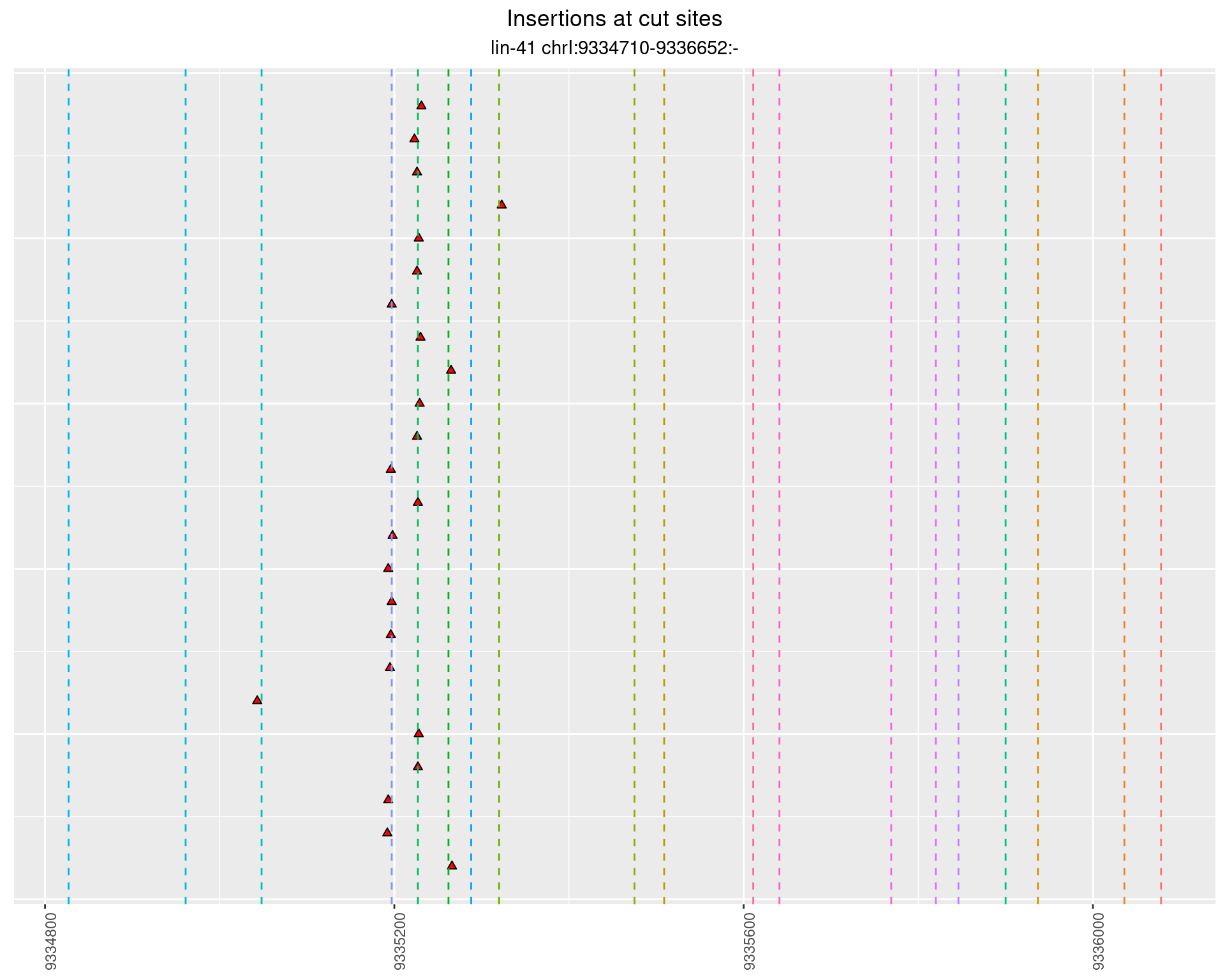
2.43.3 Freq: 1e-04 - 0.001
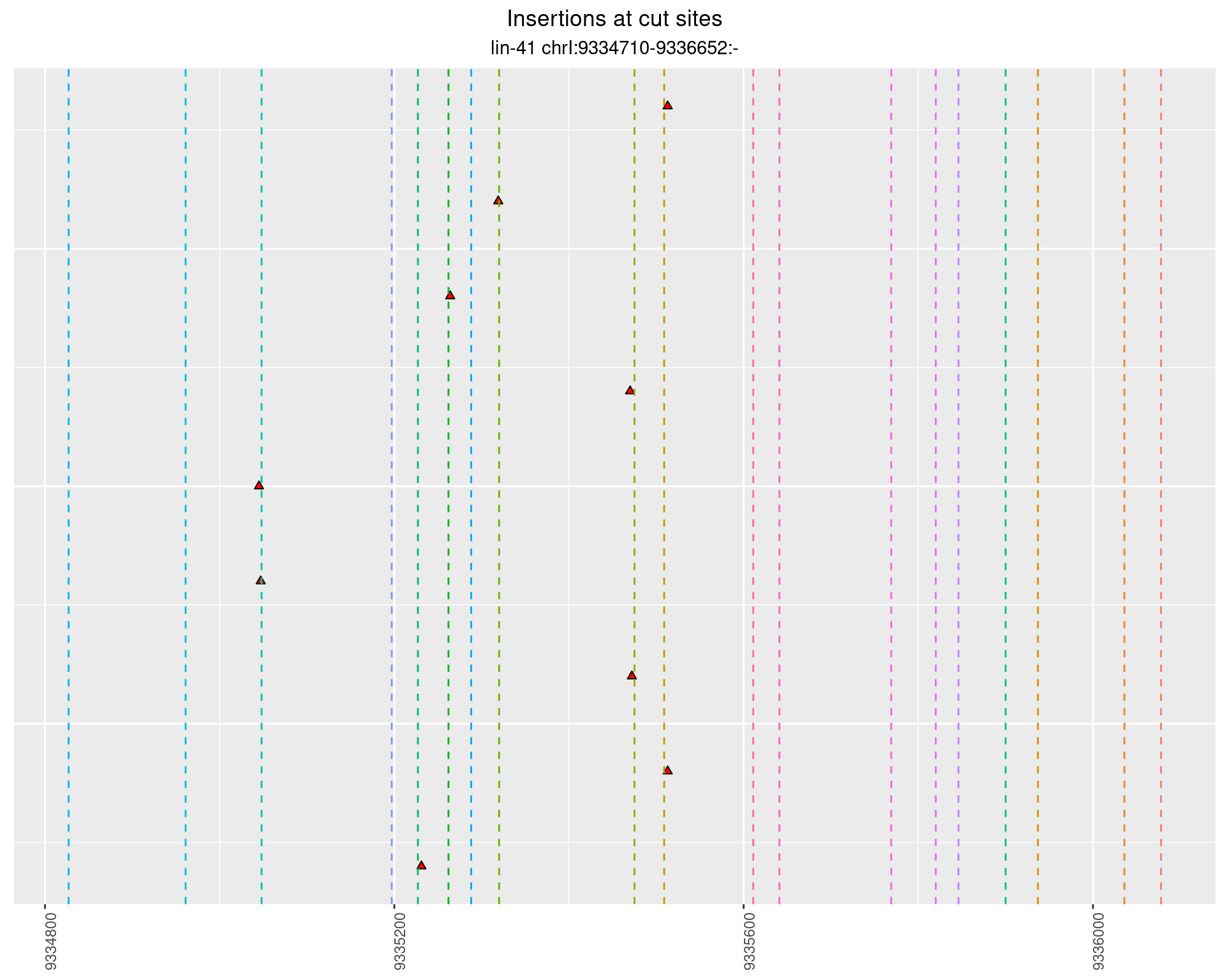
2.43.4 Freq: 0.01 - 0.1
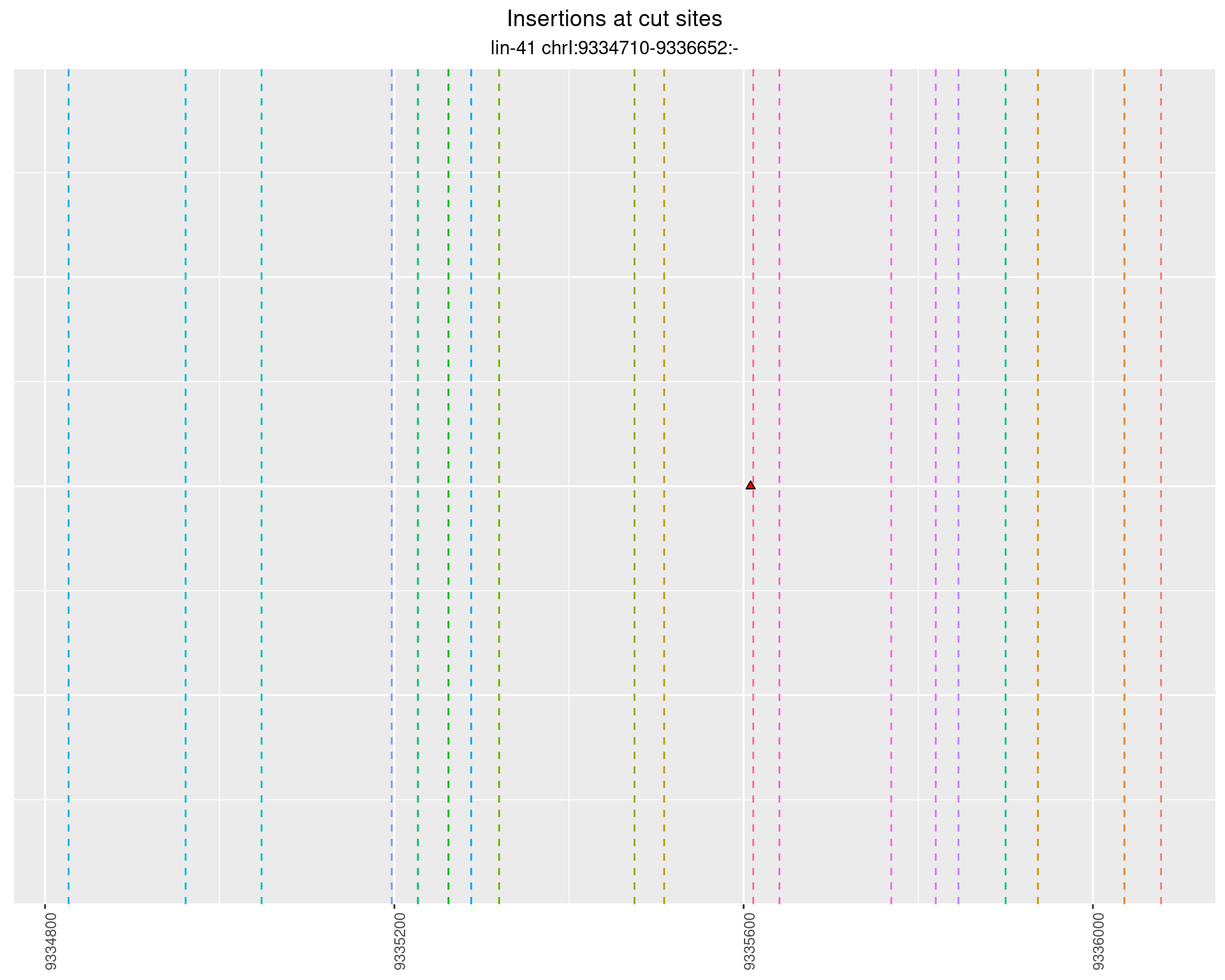
2.43.5 Freq: 1e-06 - 1e-05
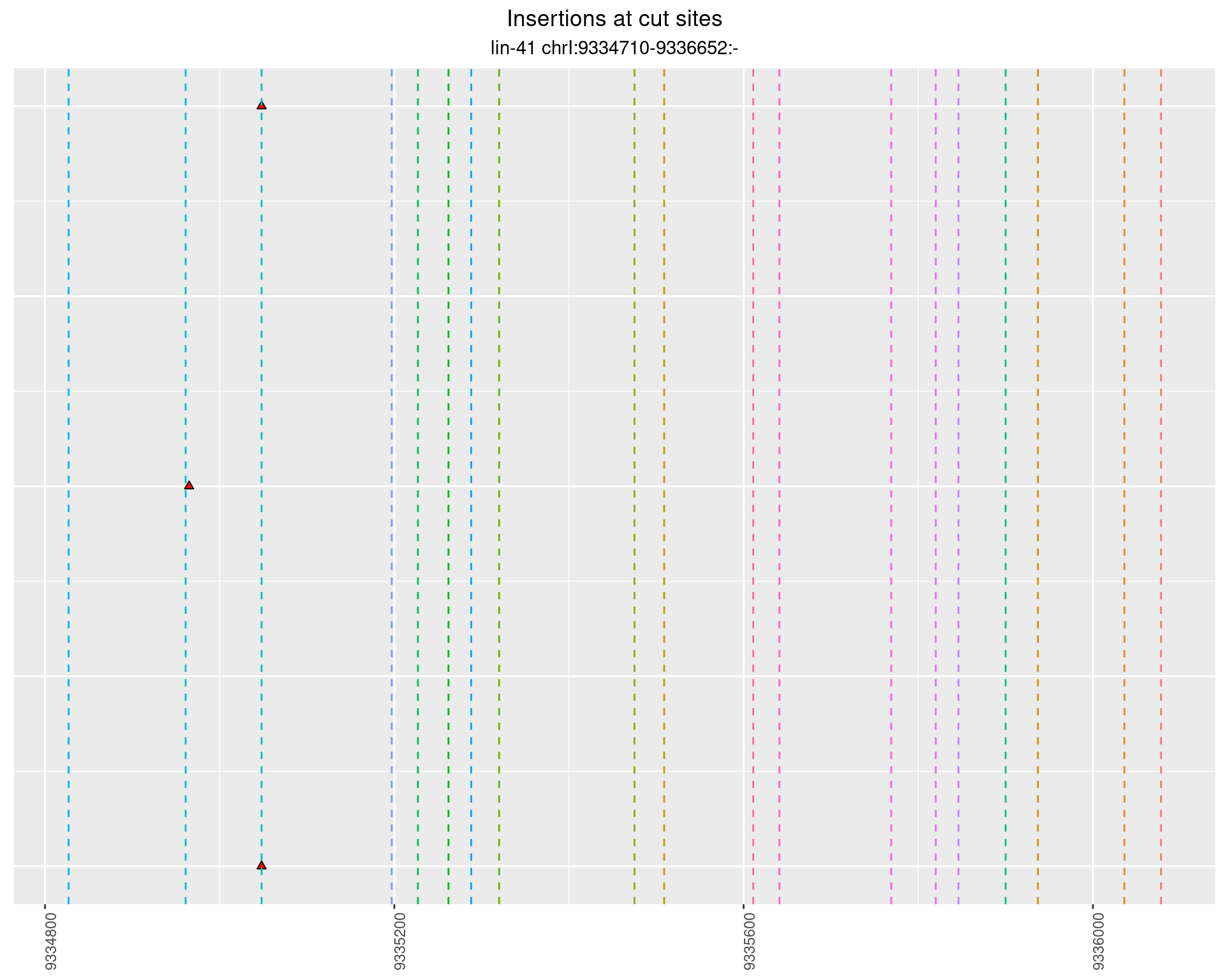
2.44 lin41_RNA_L1_1516_3end
2.44.1 Freq: 1e-04 - 0.001
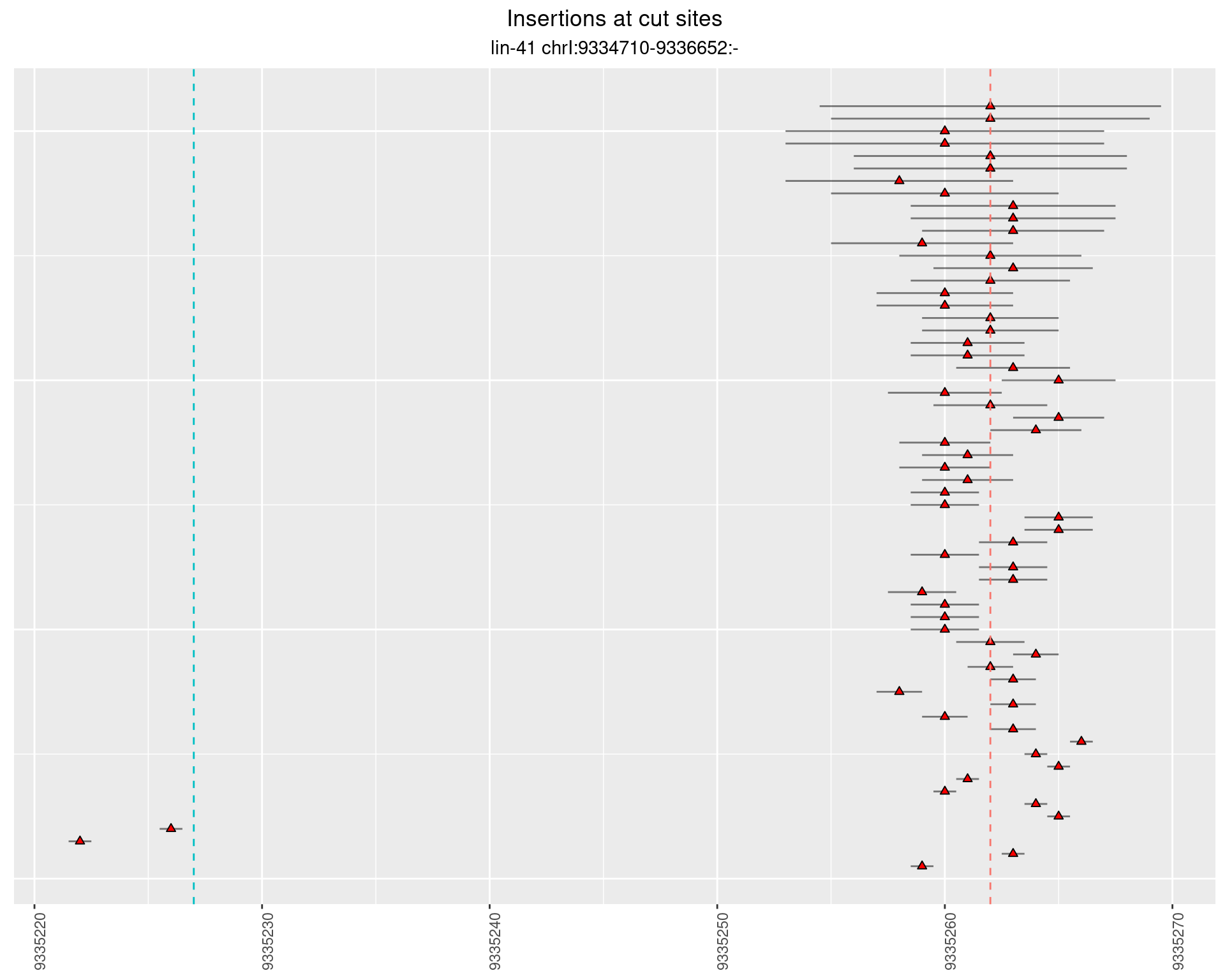
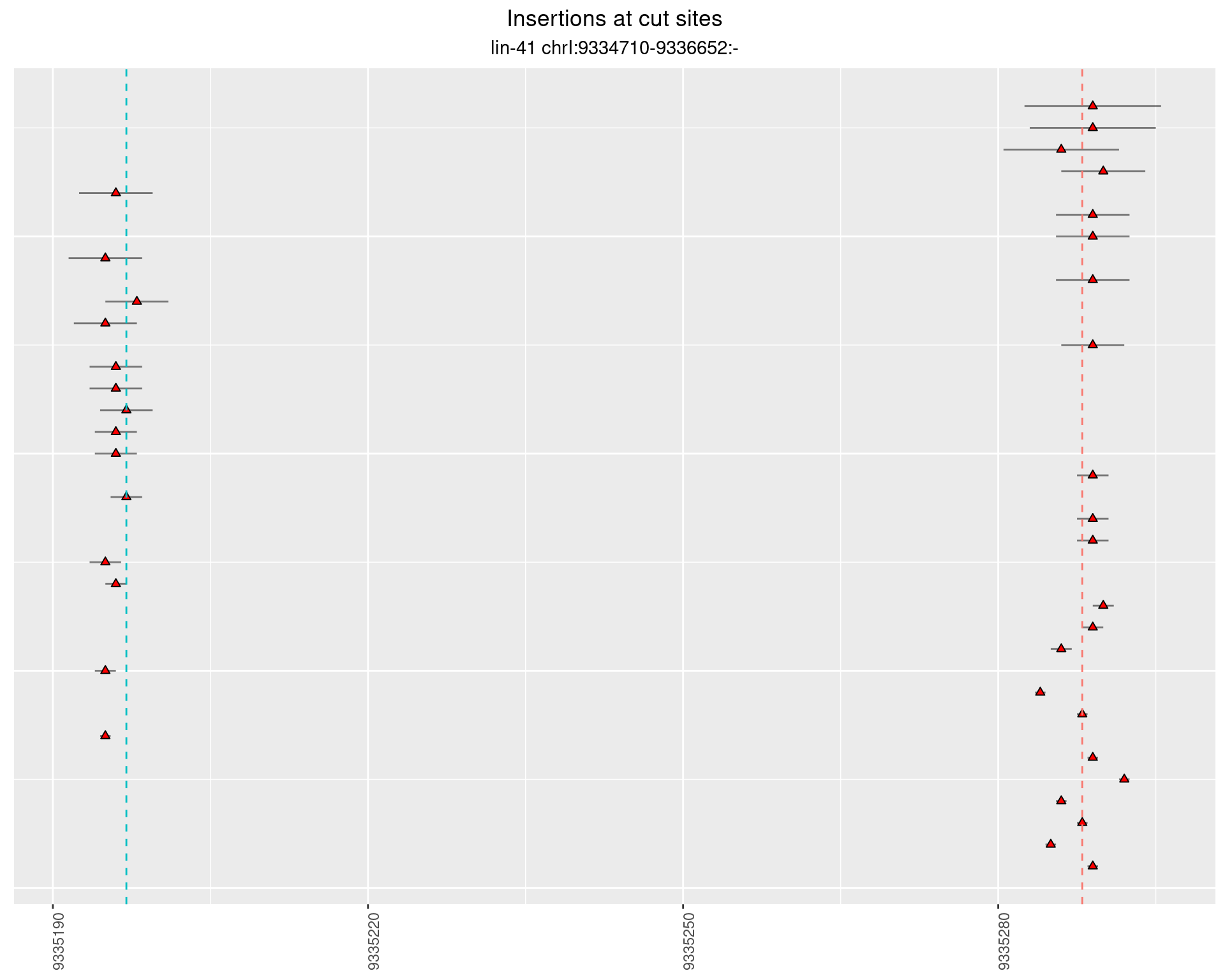
2.44.2 Freq: 1e-05 - 1e-04
2.44.3 Freq: 1e-06 - 1e-05
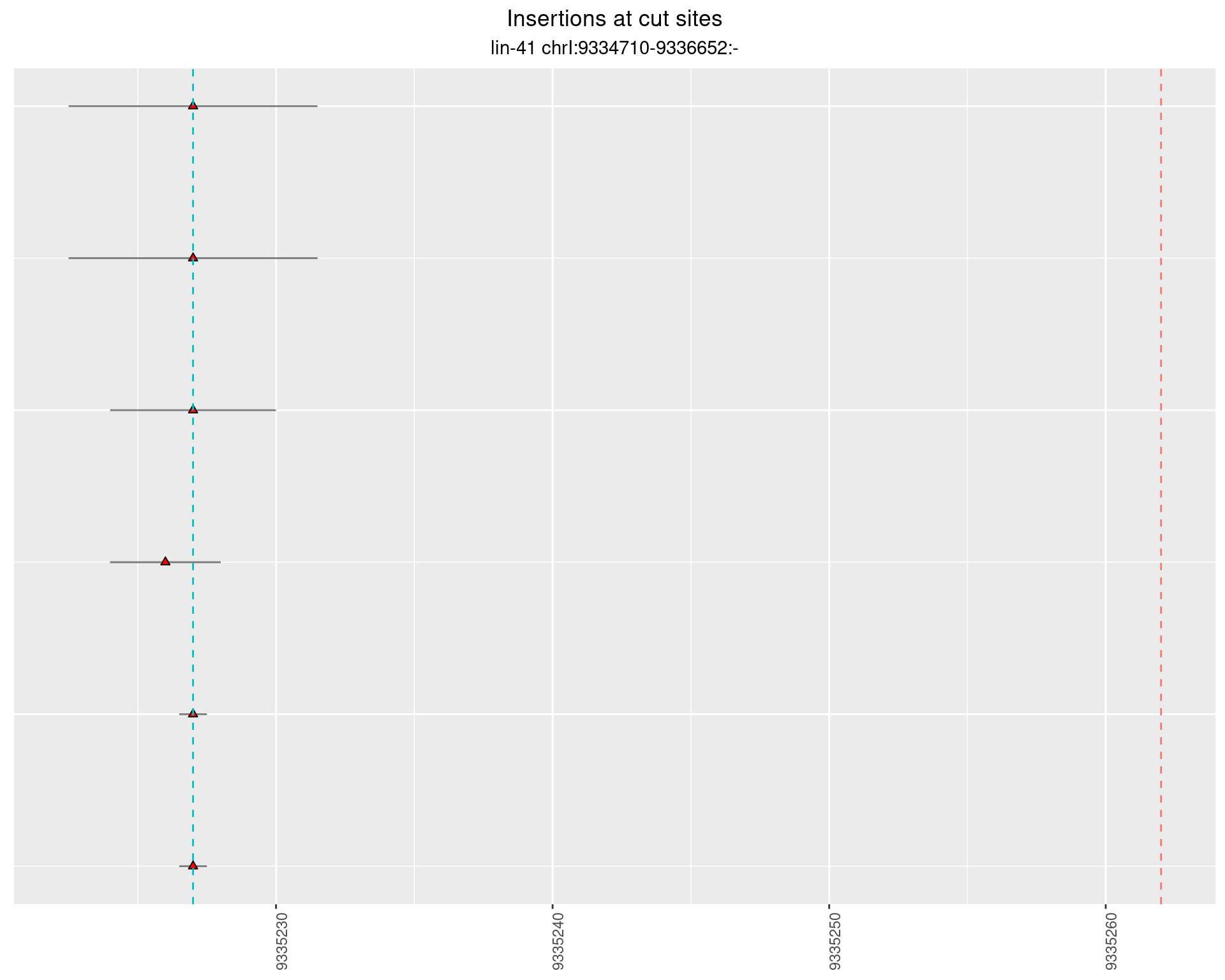
2.45 lin41_RNA_L1_2627_3end
2.45.1 Freq: 1e-04 - 0.001
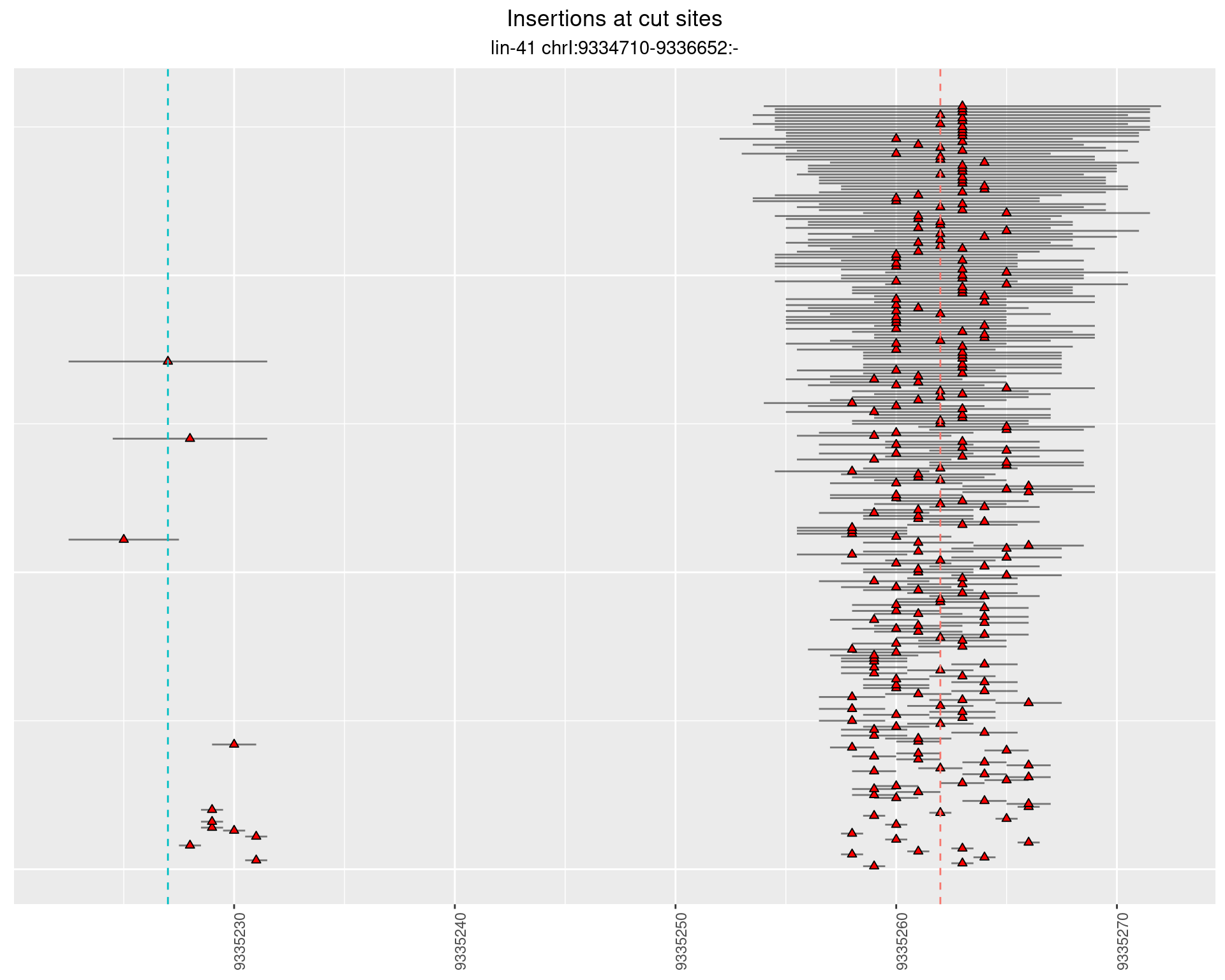
2.45.2 Freq: 1e-05 - 1e-04
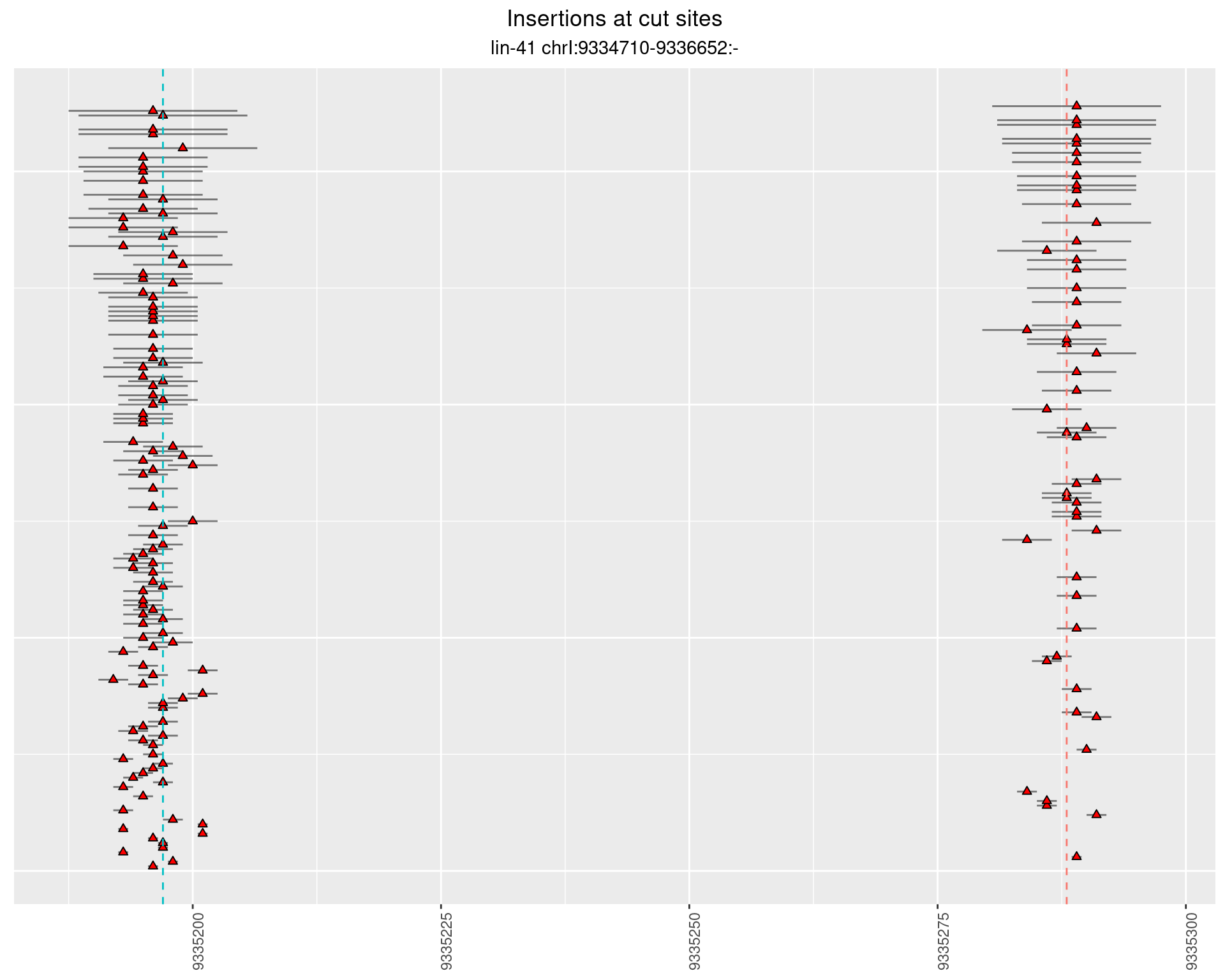
2.45.3 Freq: 0.001 - 0.01
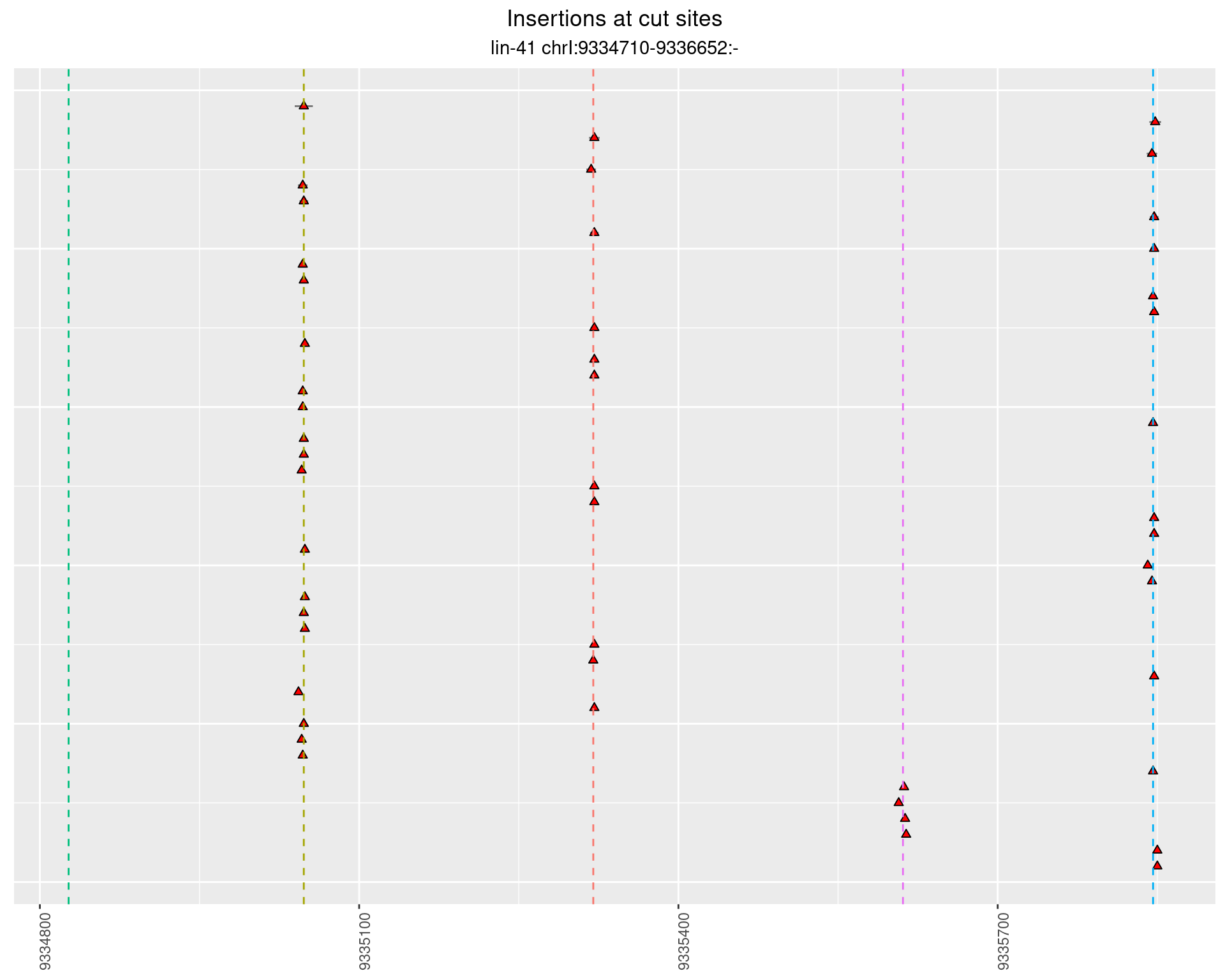
2.46 lin41_RNA_L1_pool2_3end
2.46.1 Freq: 1e-05 - 1e-04
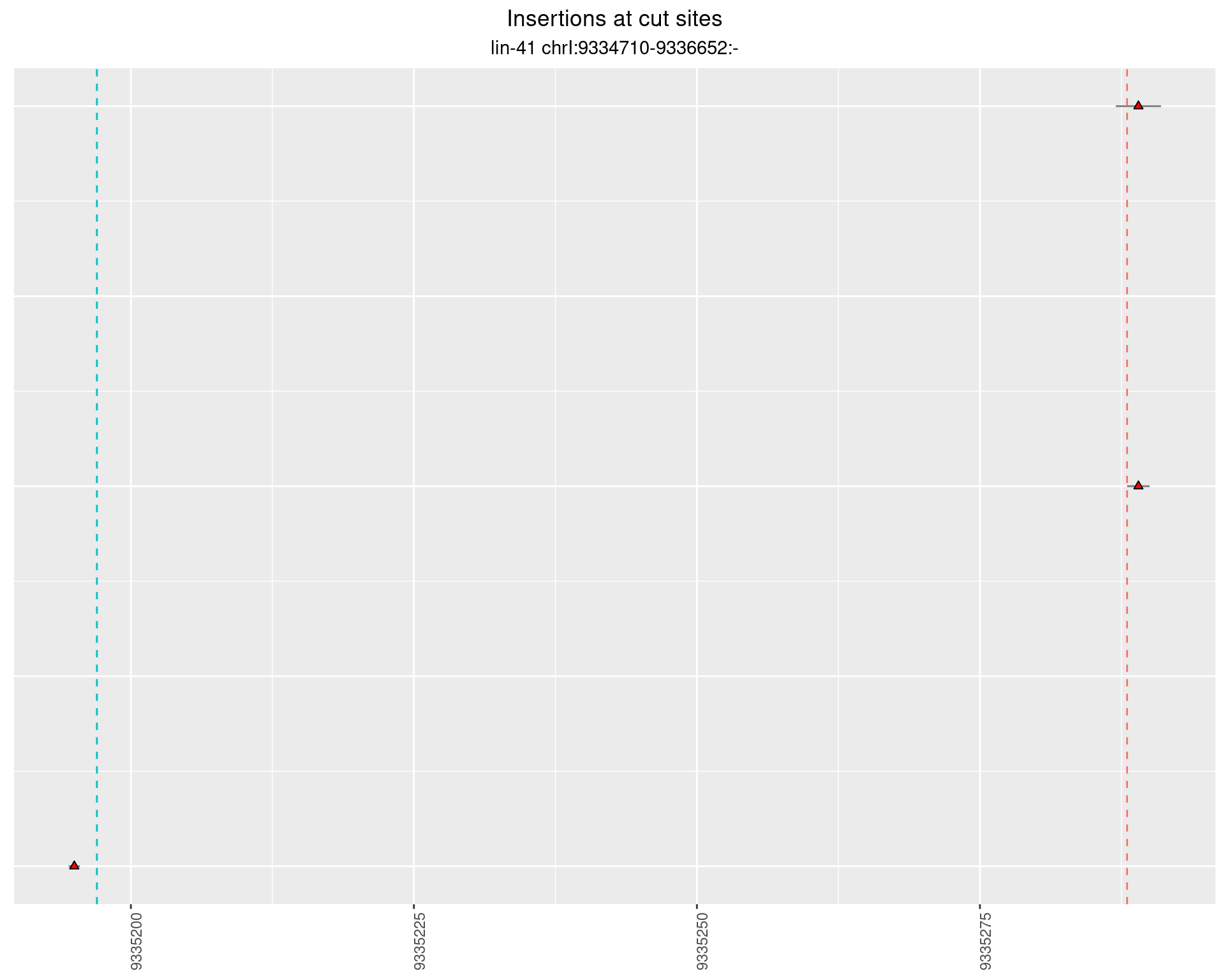
2.46.2 Freq: 1e-04 - 0.001
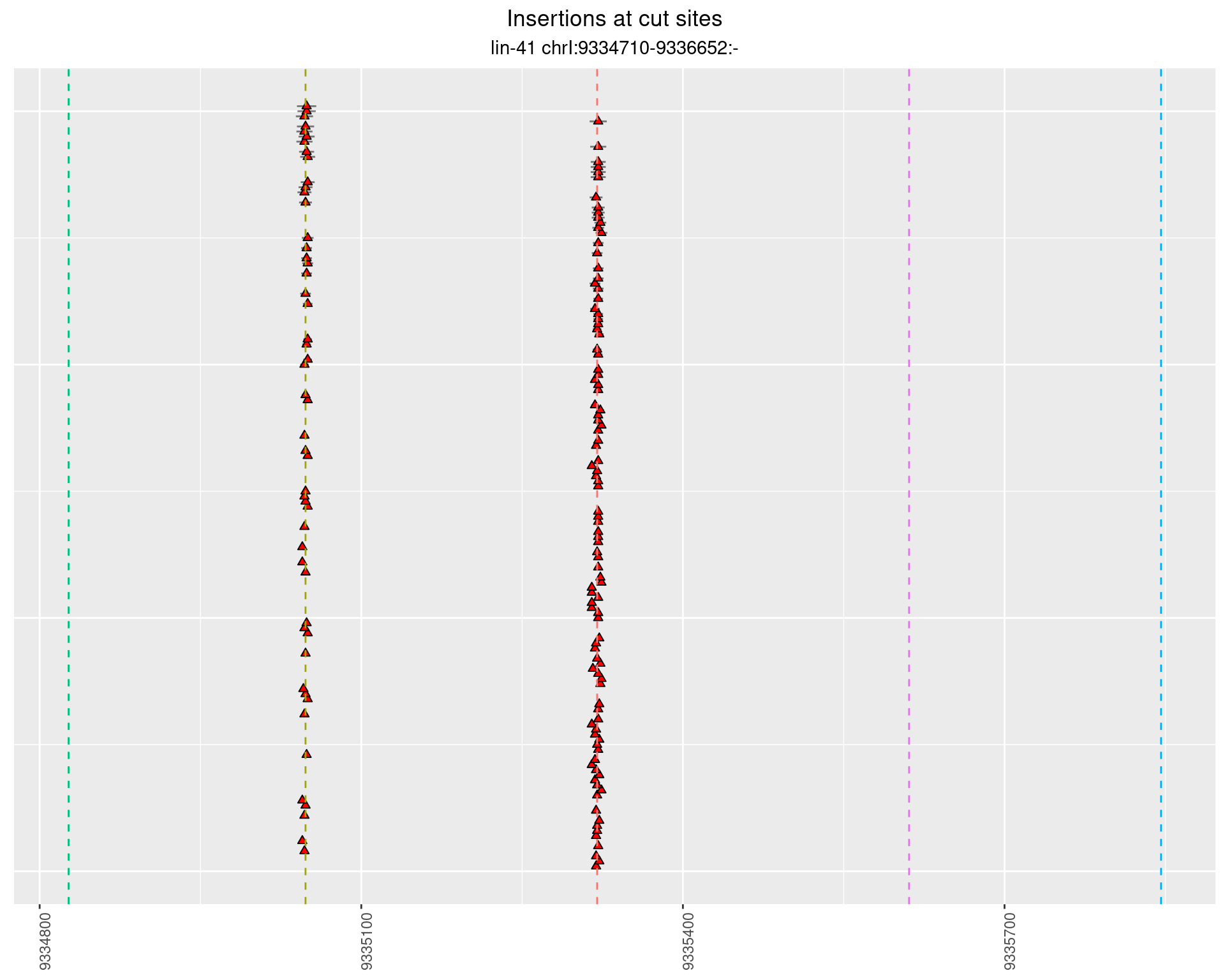
2.46.3 Freq: 0.001 - 0.01
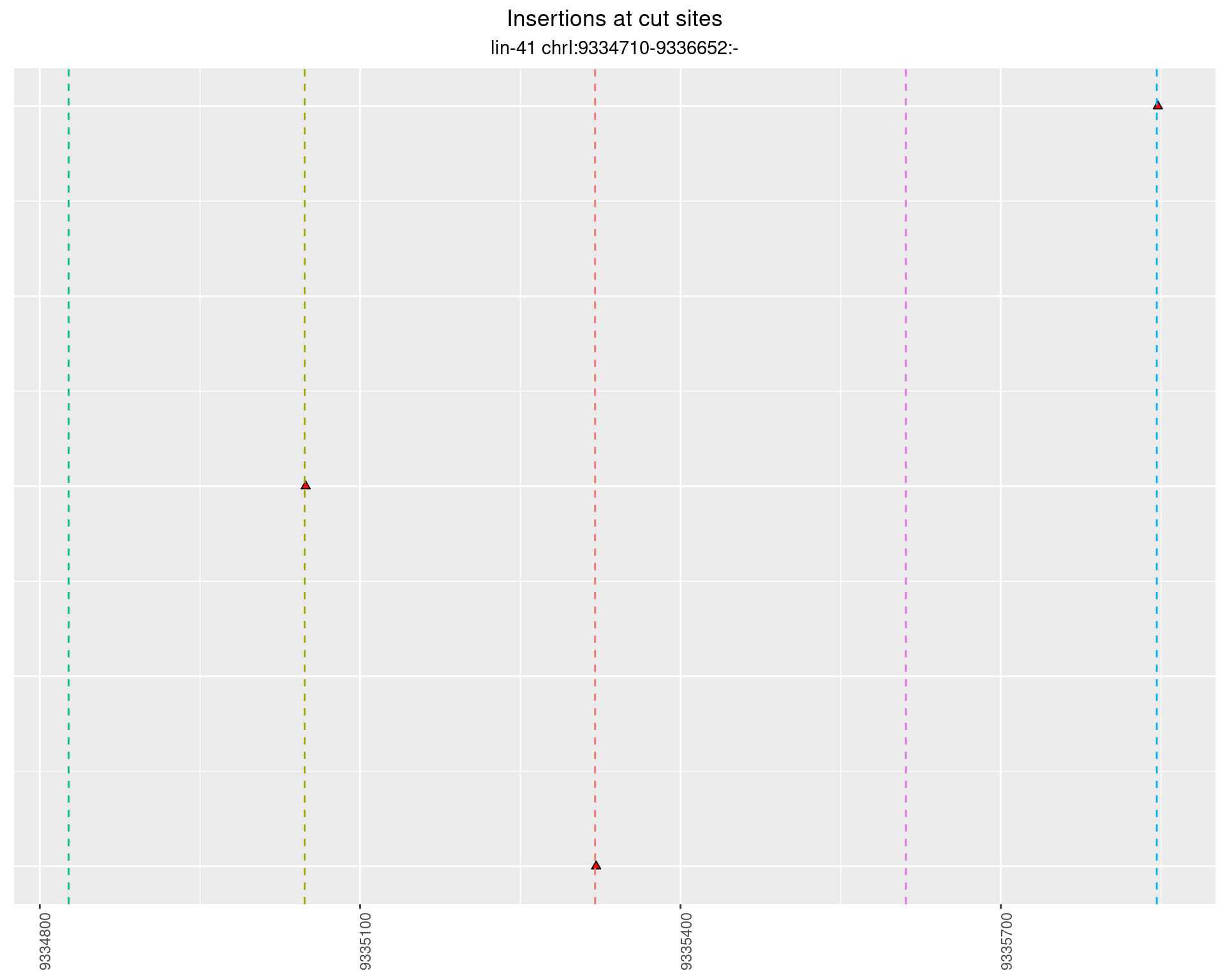
2.46.4 Freq: 1e-06 - 1e-05
2.46.5 Freq: 0.01 - 0.1
2.47 lin41_RNA_L1_pool3_3end
2.47.1 Freq: 1e-04 - 0.001
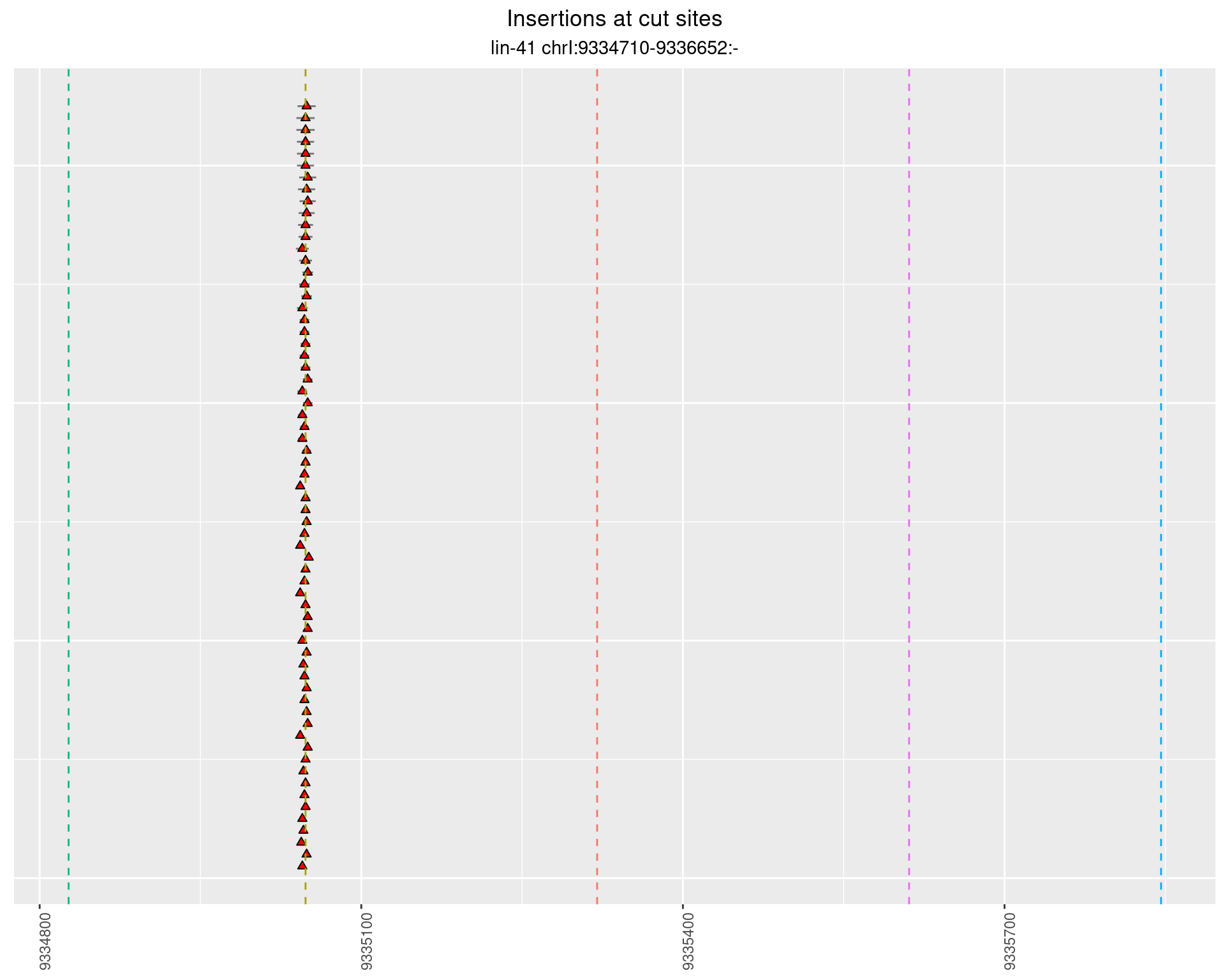
2.47.2 Freq: 1e-05 - 1e-04
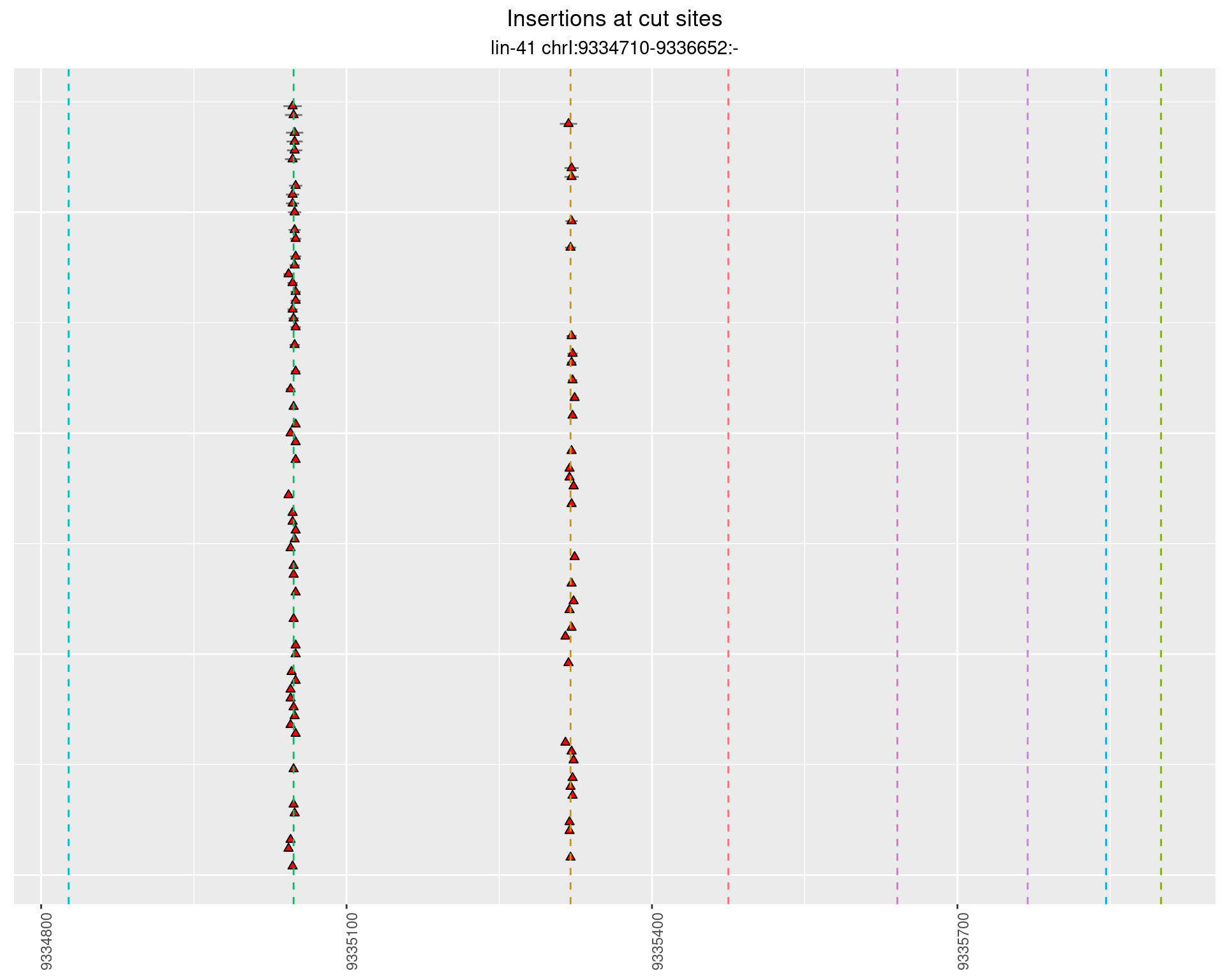
2.47.3 Freq: 0.001 - 0.01
2.47.4 Freq: 1e-06 - 1e-05
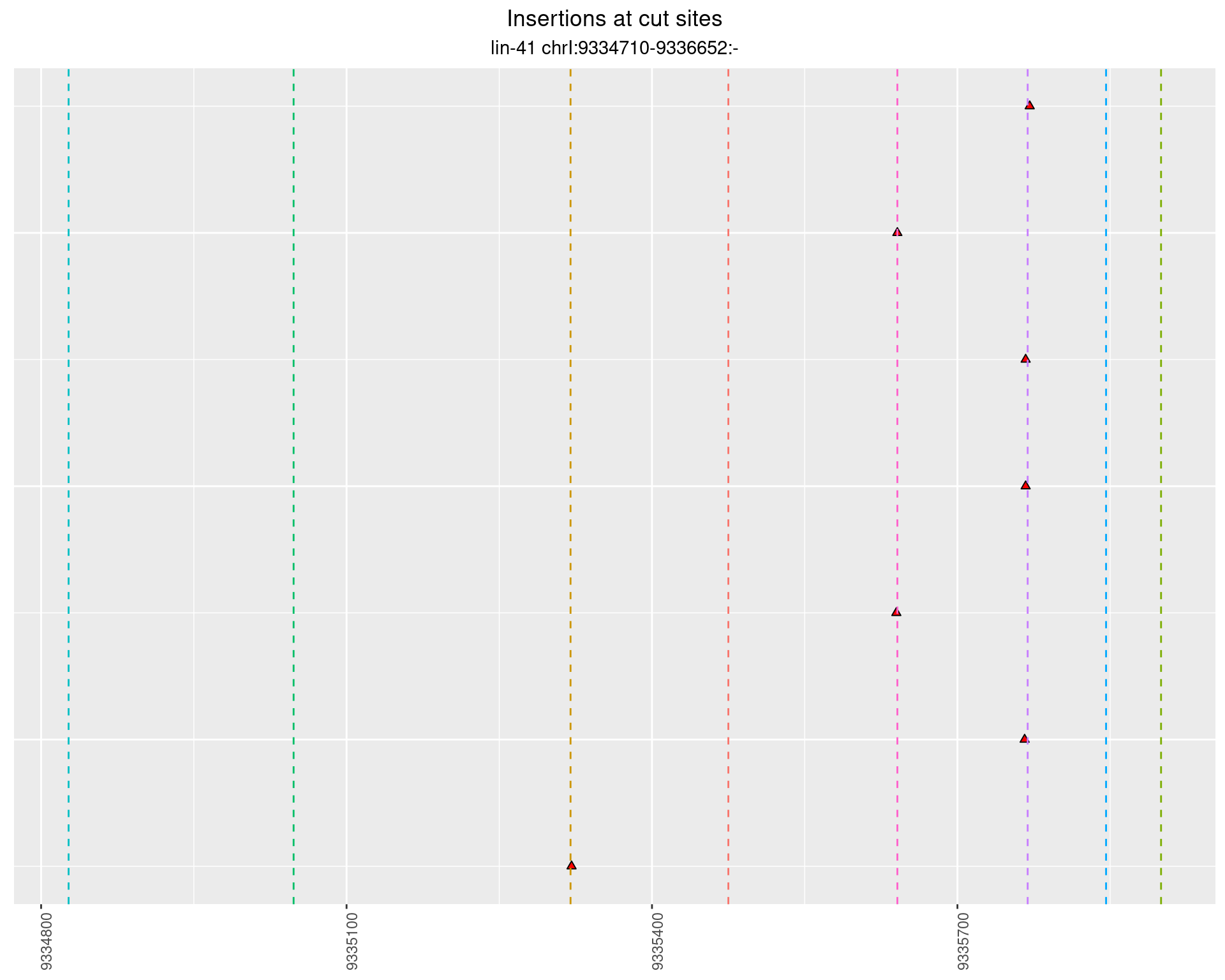
2.48 lin41_RNA_L4_N2_3end
2.48.1 Freq: 0.001 - 0.01
2.48.2 Freq: 1e-05 - 1e-04
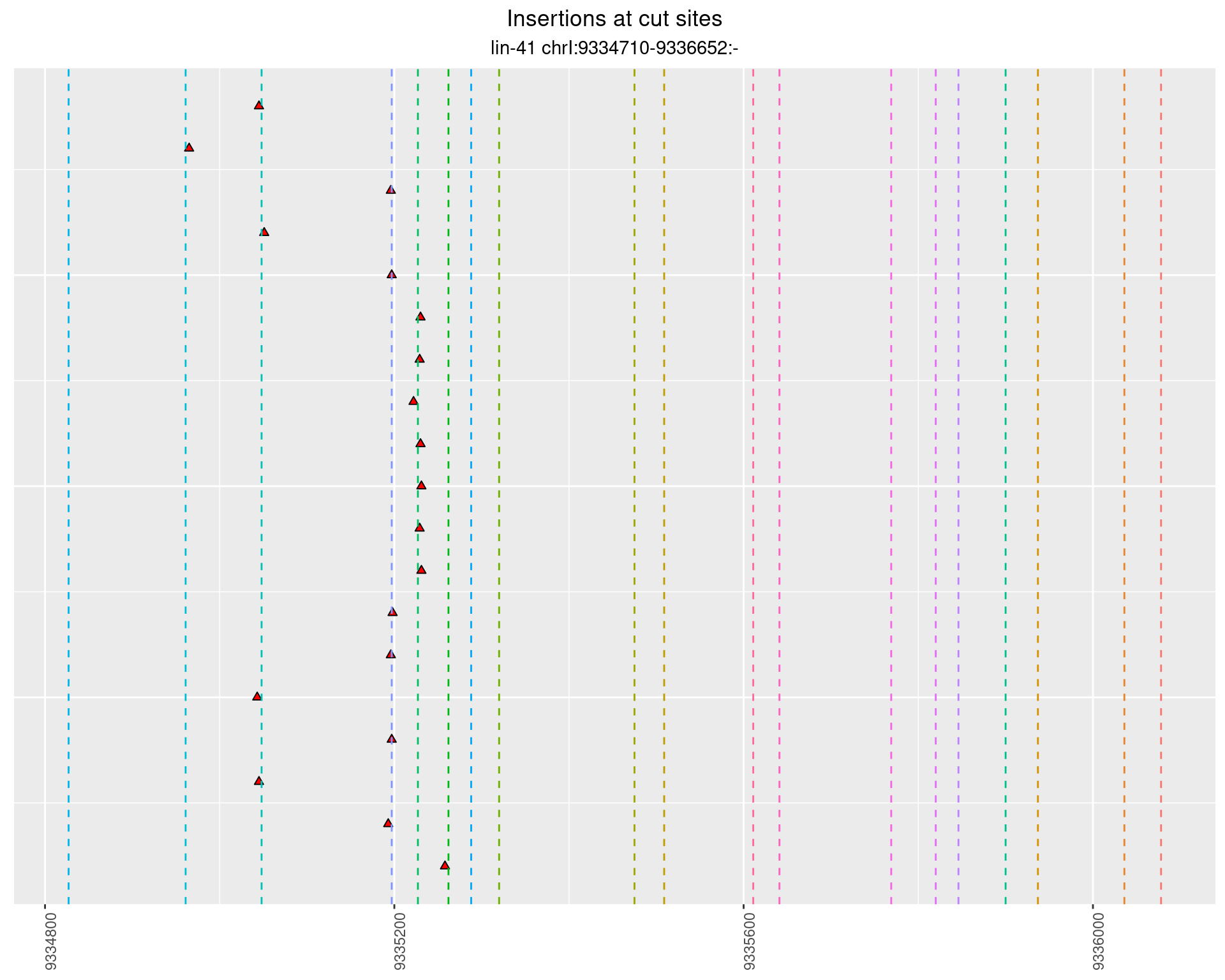
2.48.3 Freq: 0.01 - 0.1
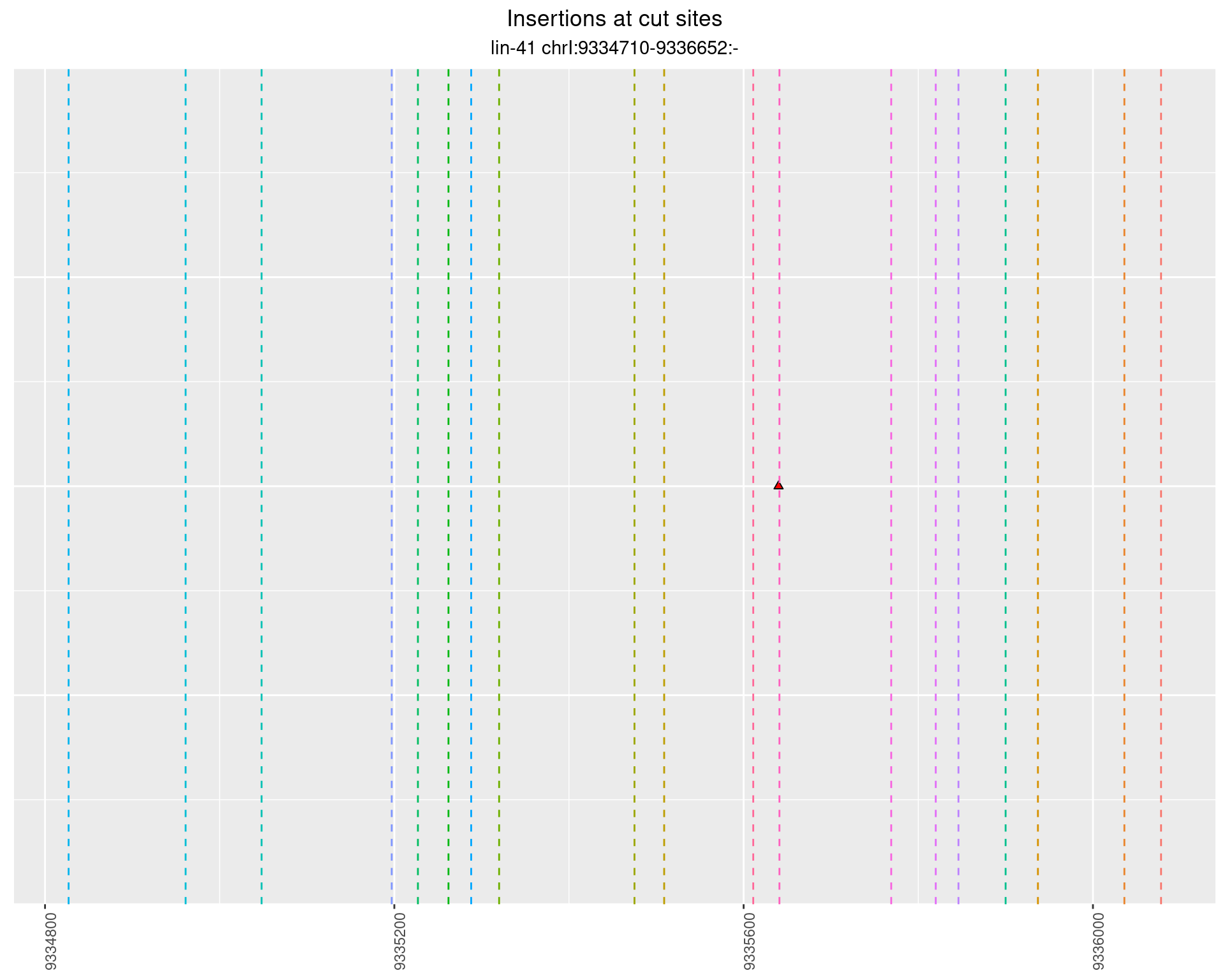
2.48.4 Freq: 1e-06 - 1e-05
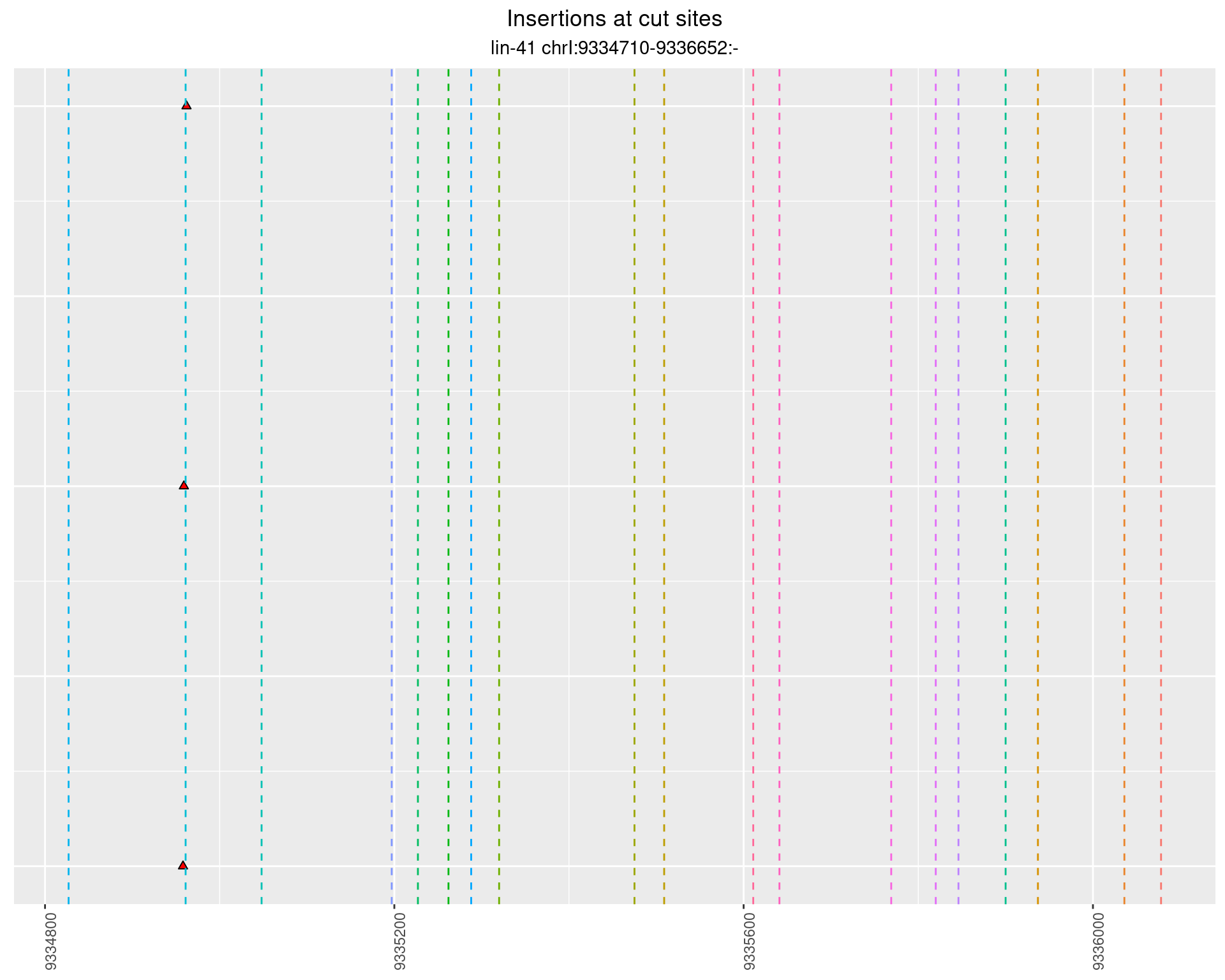
2.48.5 Freq: 1e-04 - 0.001
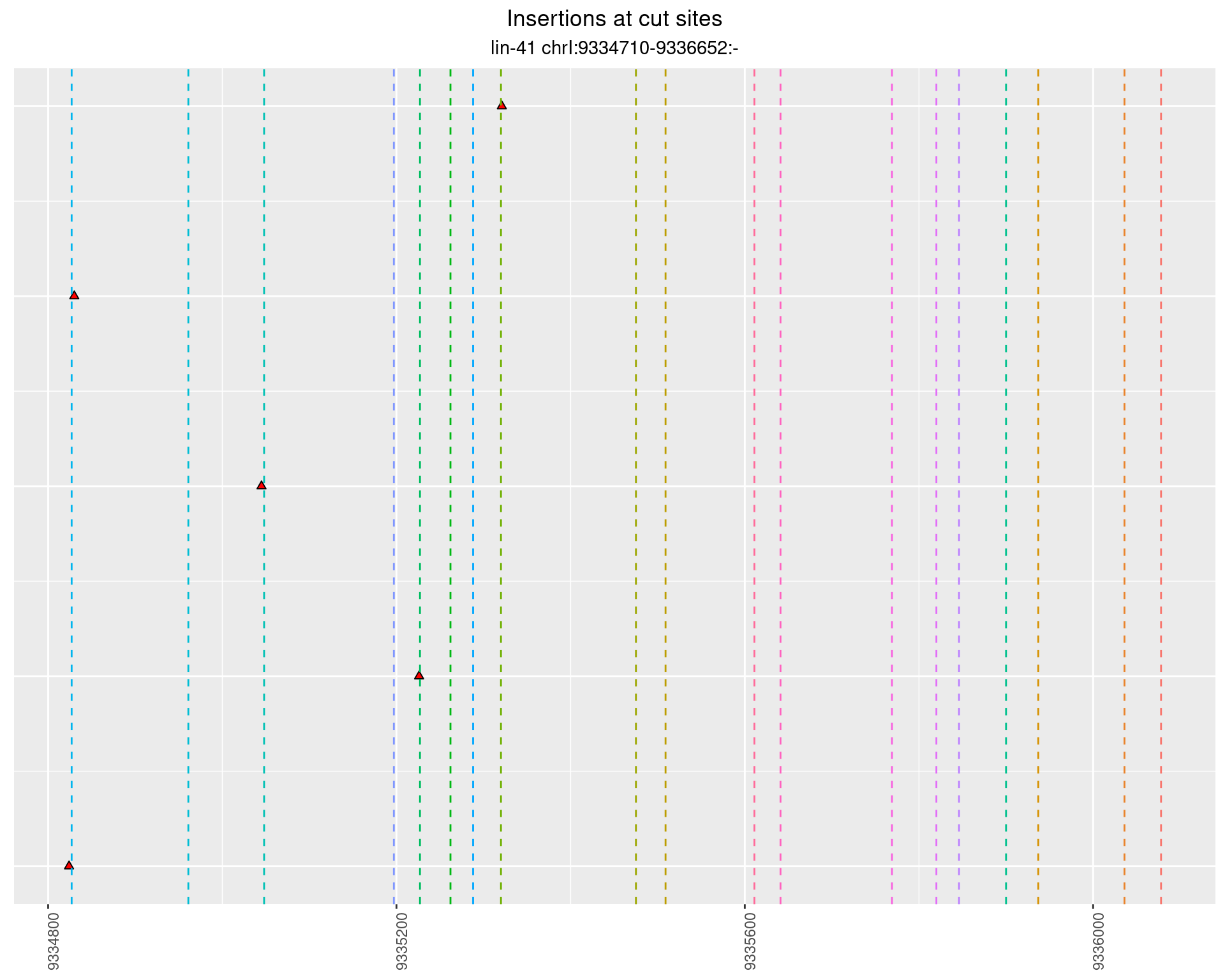
2.49 lin41_RNA_L4_1516_3end
2.49.1 Freq: 1e-05 - 1e-04
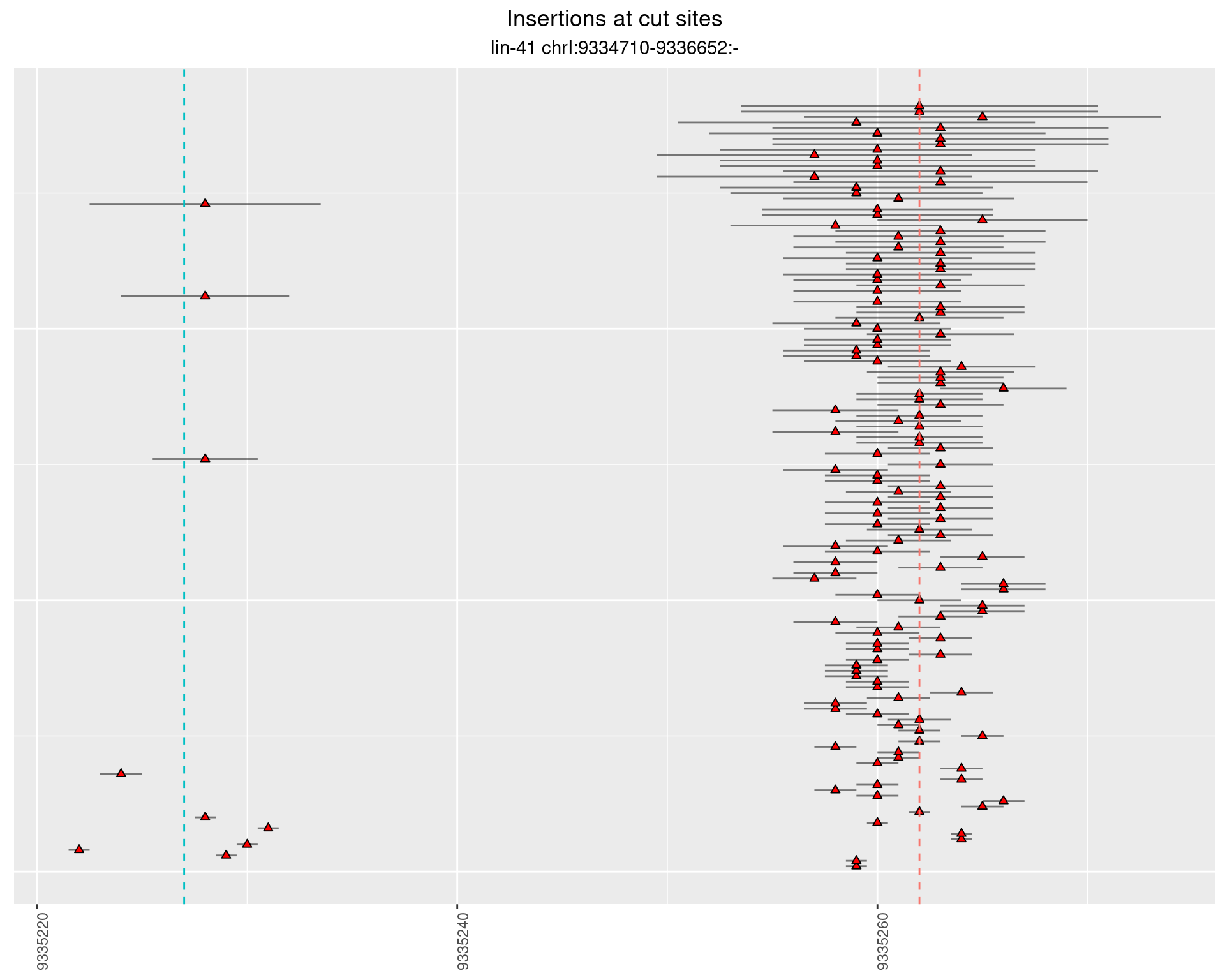
2.49.2 Freq: 1e-04 - 0.001
2.49.3 Freq: 0.001 - 0.01
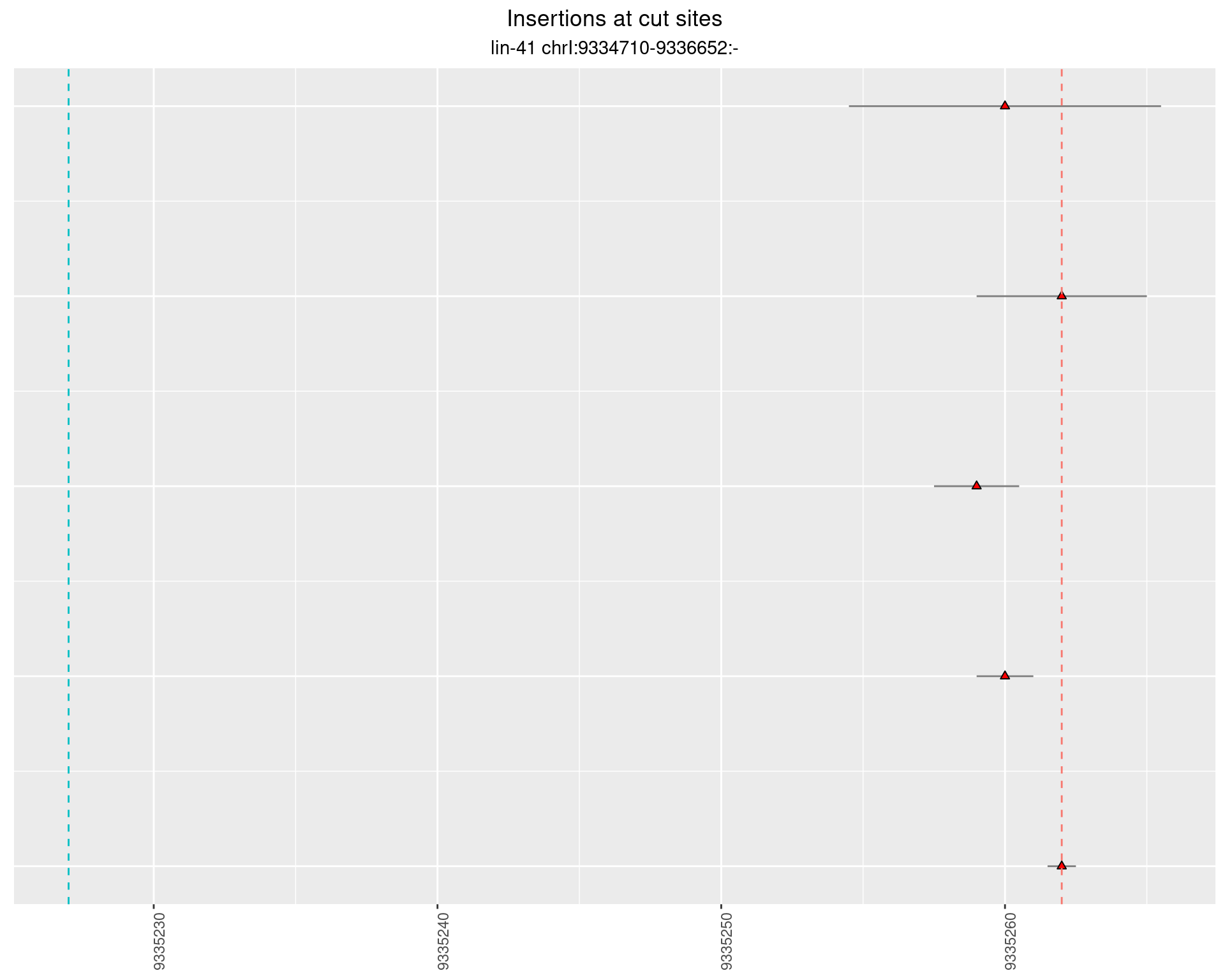
2.50 lin41_RNA_L4_2627_3end
2.50.1 Freq: 1e-05 - 1e-04
2.50.2 Freq: 0.001 - 0.01
2.50.3 Freq: 1e-04 - 0.001
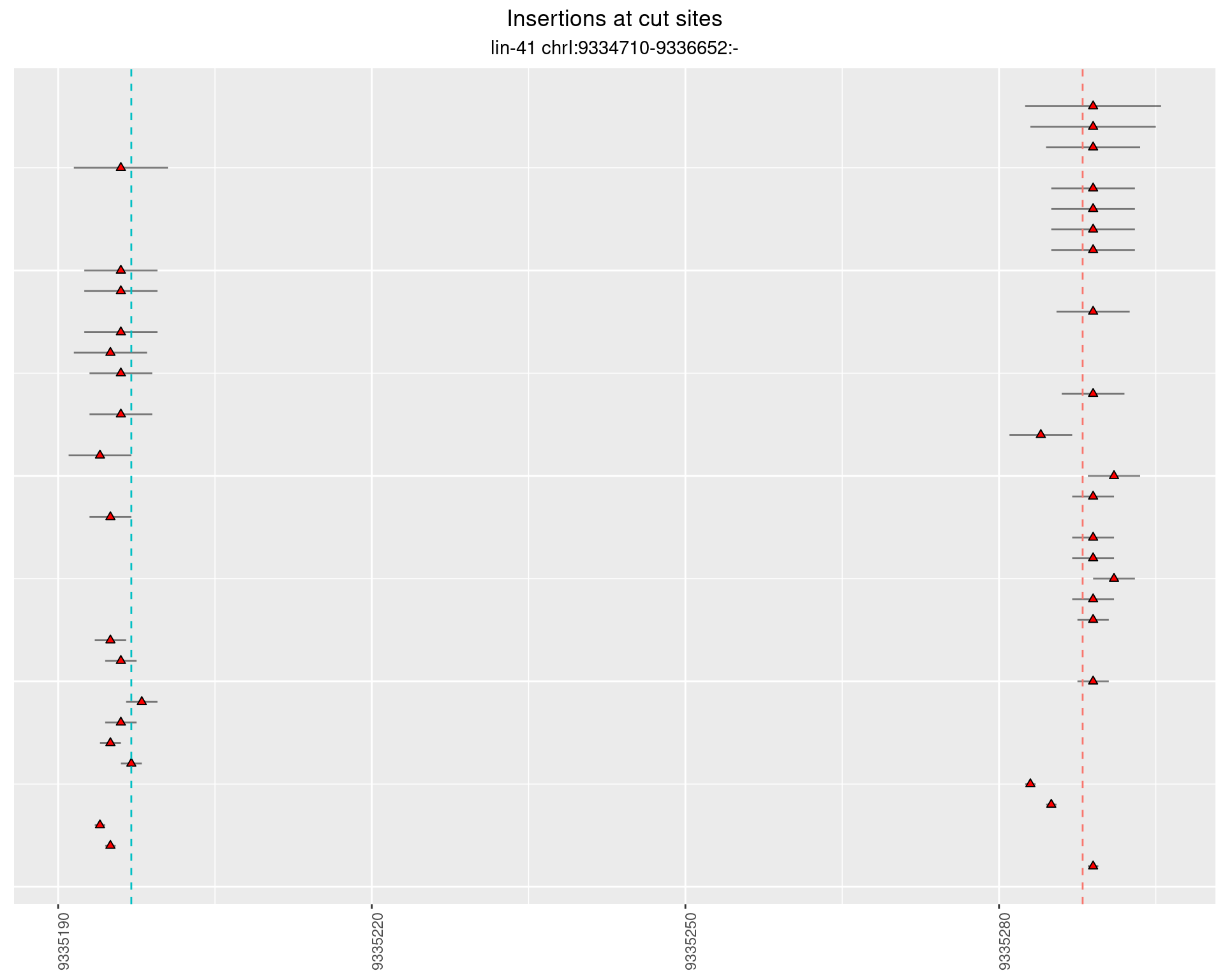
2.51 lin41_RNA_L4_pool2_3end
2.51.1 Freq: 1e-04 - 0.001
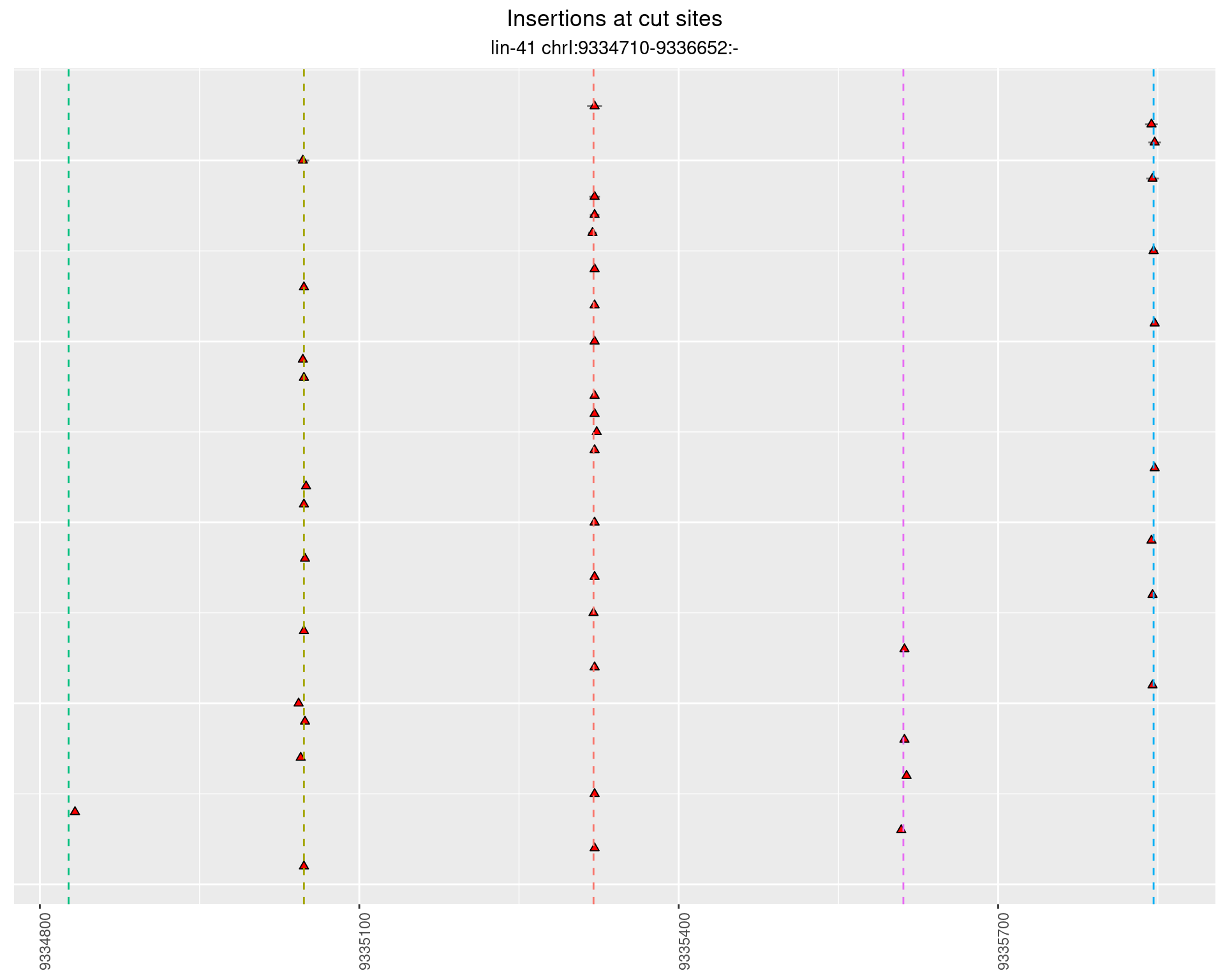
2.51.2 Freq: 0.001 - 0.01
2.51.3 Freq: 1e-05 - 1e-04
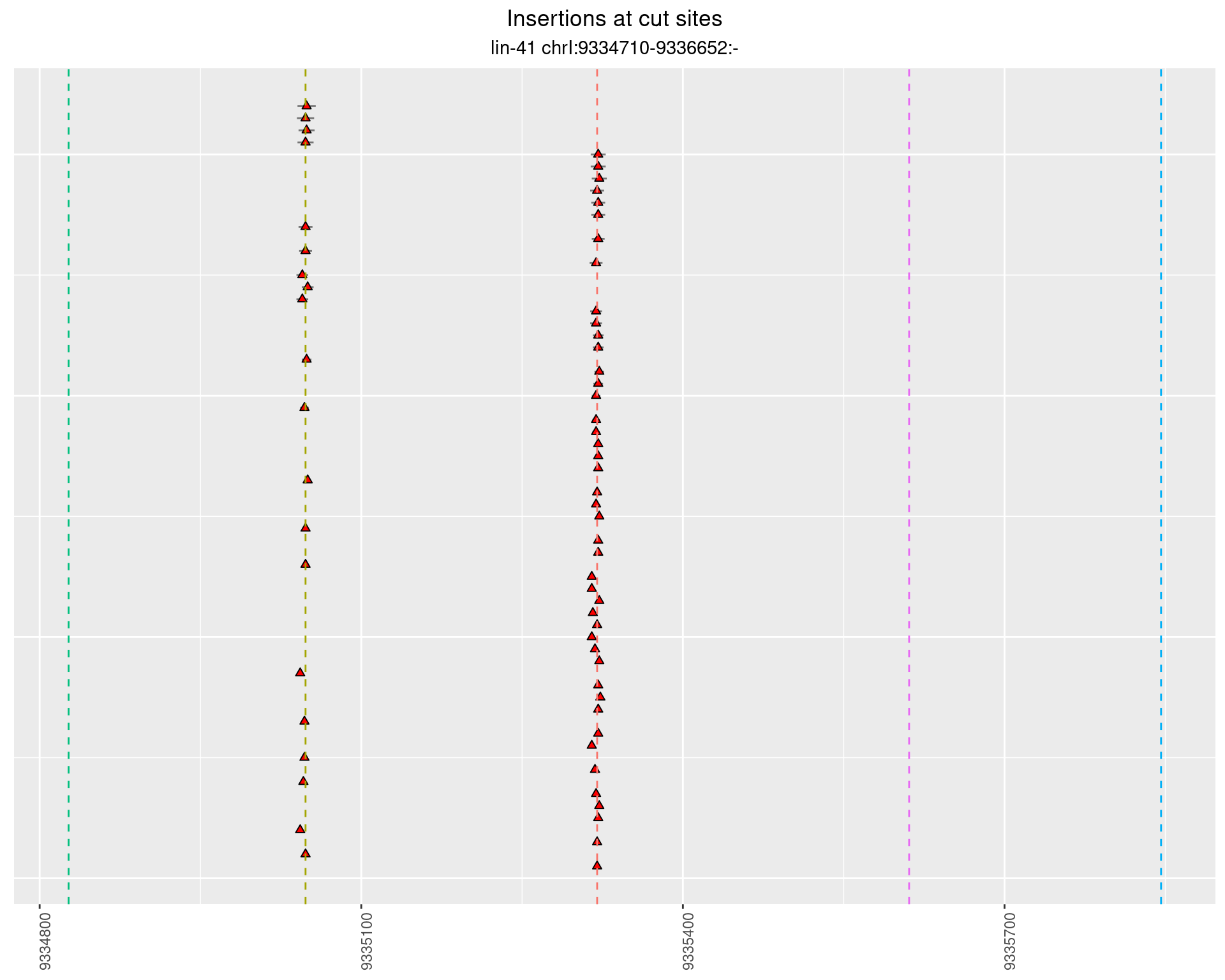
2.51.4 Freq: 0.01 - 0.1
2.51.5 Freq: 1e-06 - 1e-05
2.52 lin41_RNA_L1_N2_middle
2.52.1 Freq: 1e-04 - 0.001
2.52.2 Freq: 0.01 - 0.1
2.52.3 Freq: 1e-05 - 1e-04
2.52.4 Freq: 0.001 - 0.01
2.52.5 Freq: 1e-06 - 1e-05
2.53 lin41_RNA_L1_1516_middle
2.54 lin41_RNA_L1_2627_middle
2.55 lin41_RNA_L1_pool2_middle
2.55.1 Freq: 0.01 - 0.1
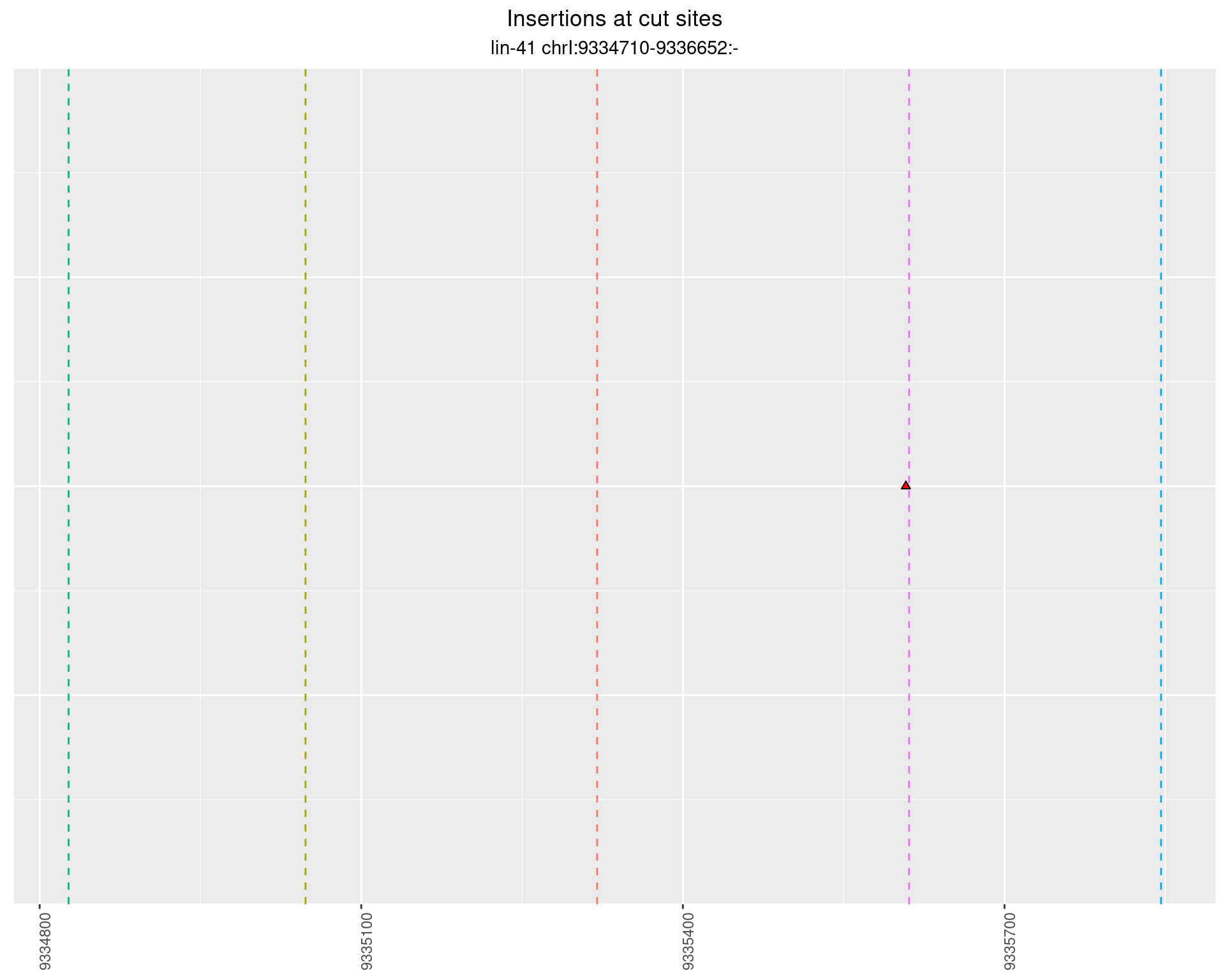
2.55.2 Freq: 1e-05 - 1e-04
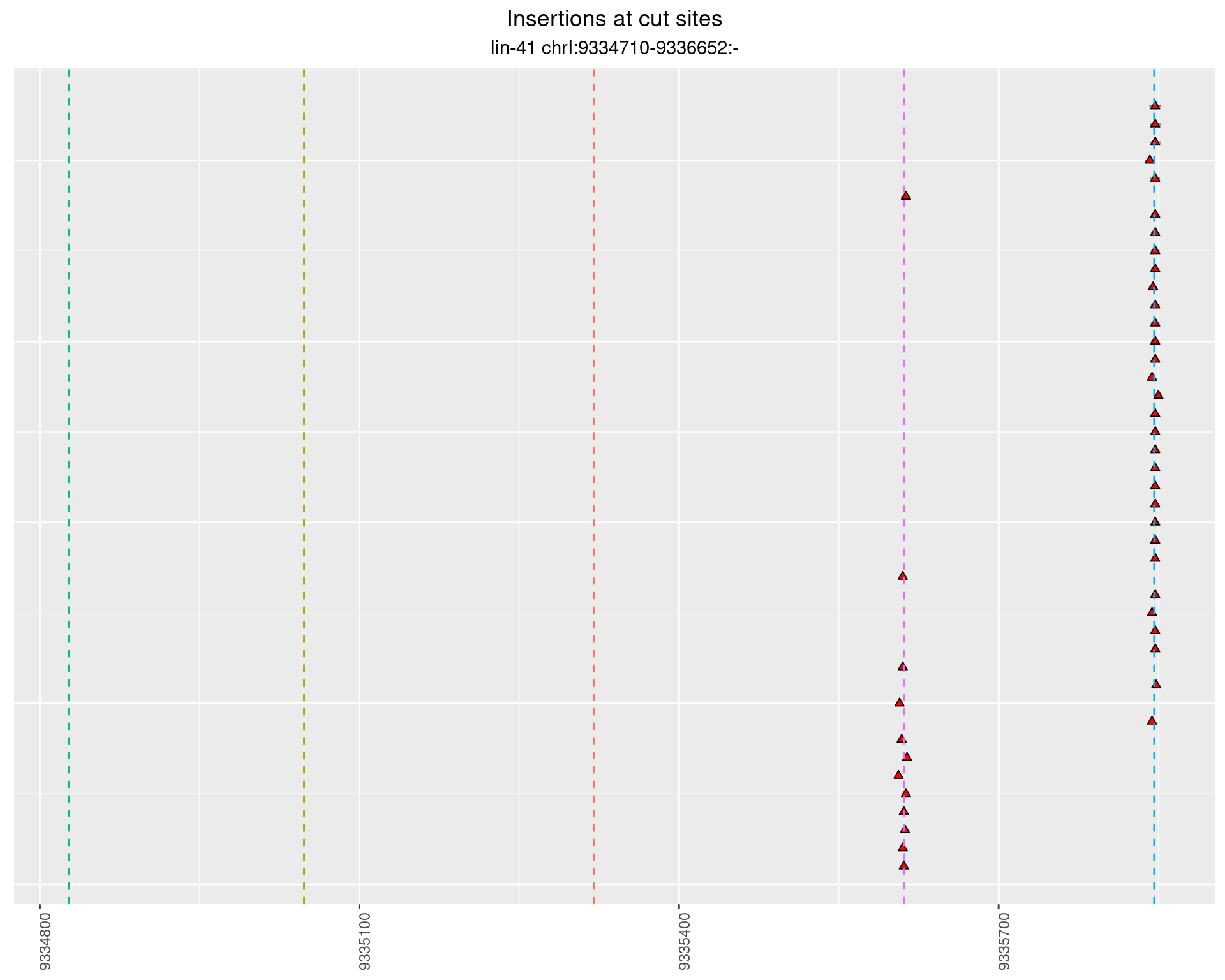
2.55.3 Freq: 1e-04 - 0.001
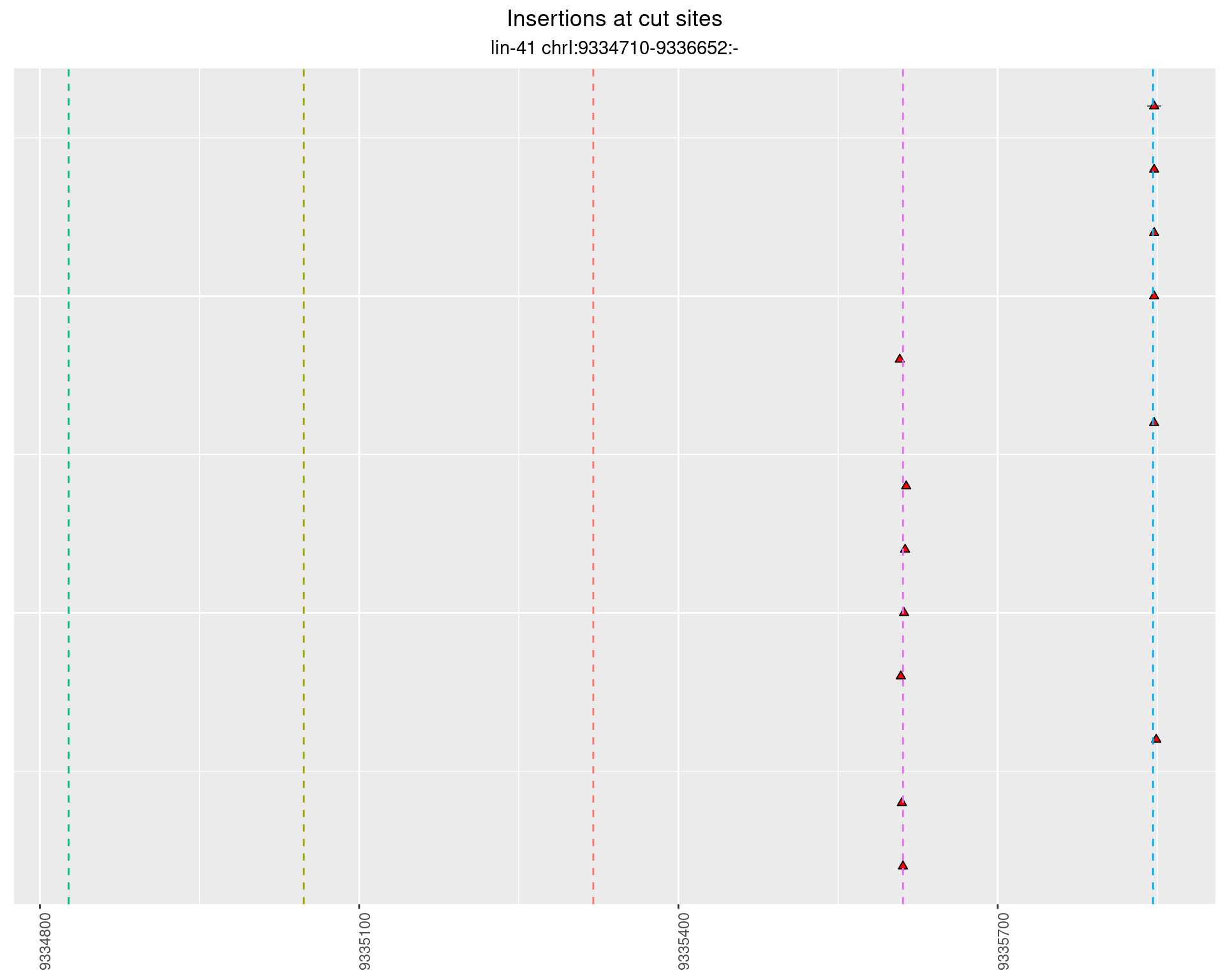
2.55.4 Freq: 1e-06 - 1e-05
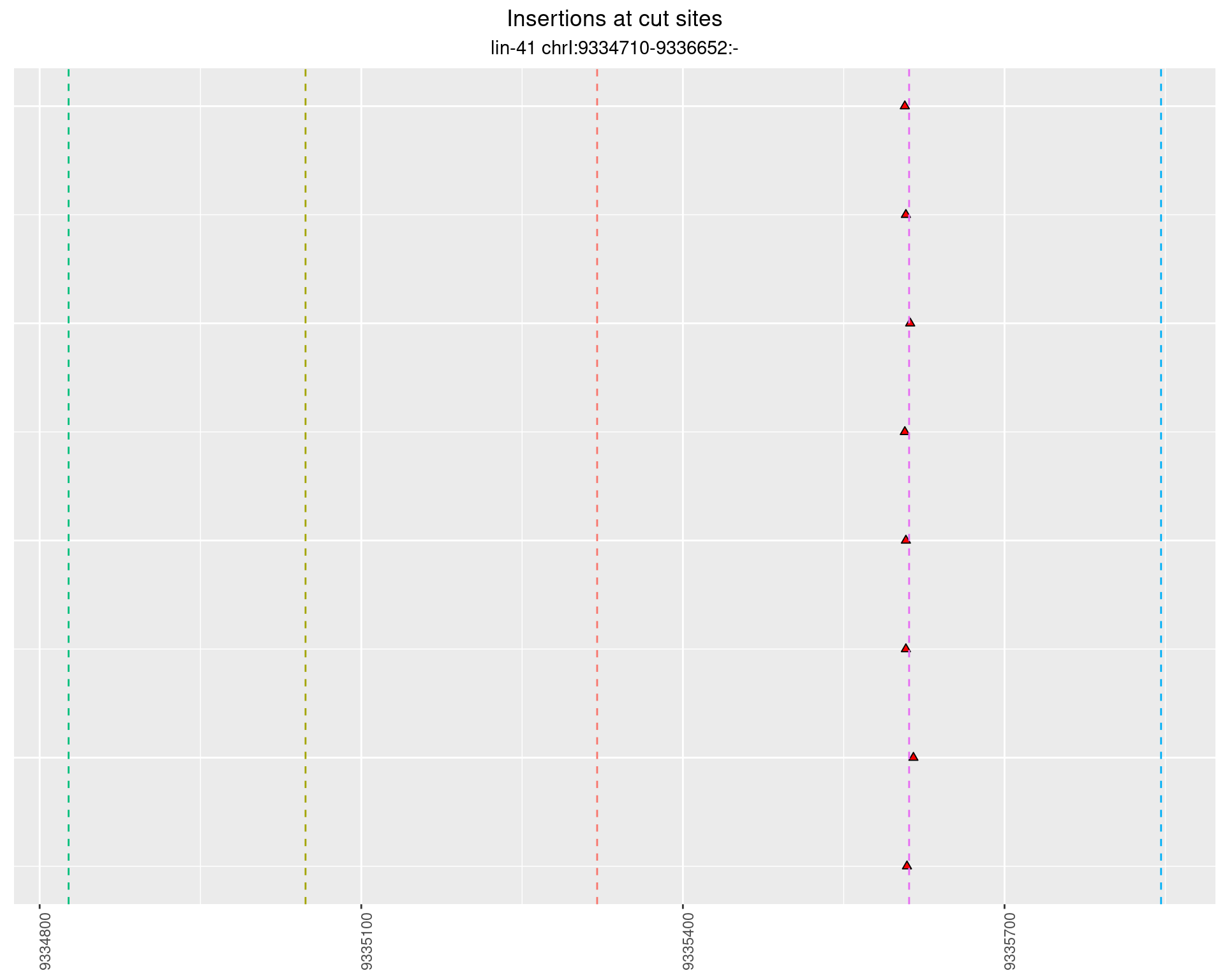
2.56 lin41_RNA_L1_pool3_middle
2.56.1 Freq: 1e-04 - 0.001
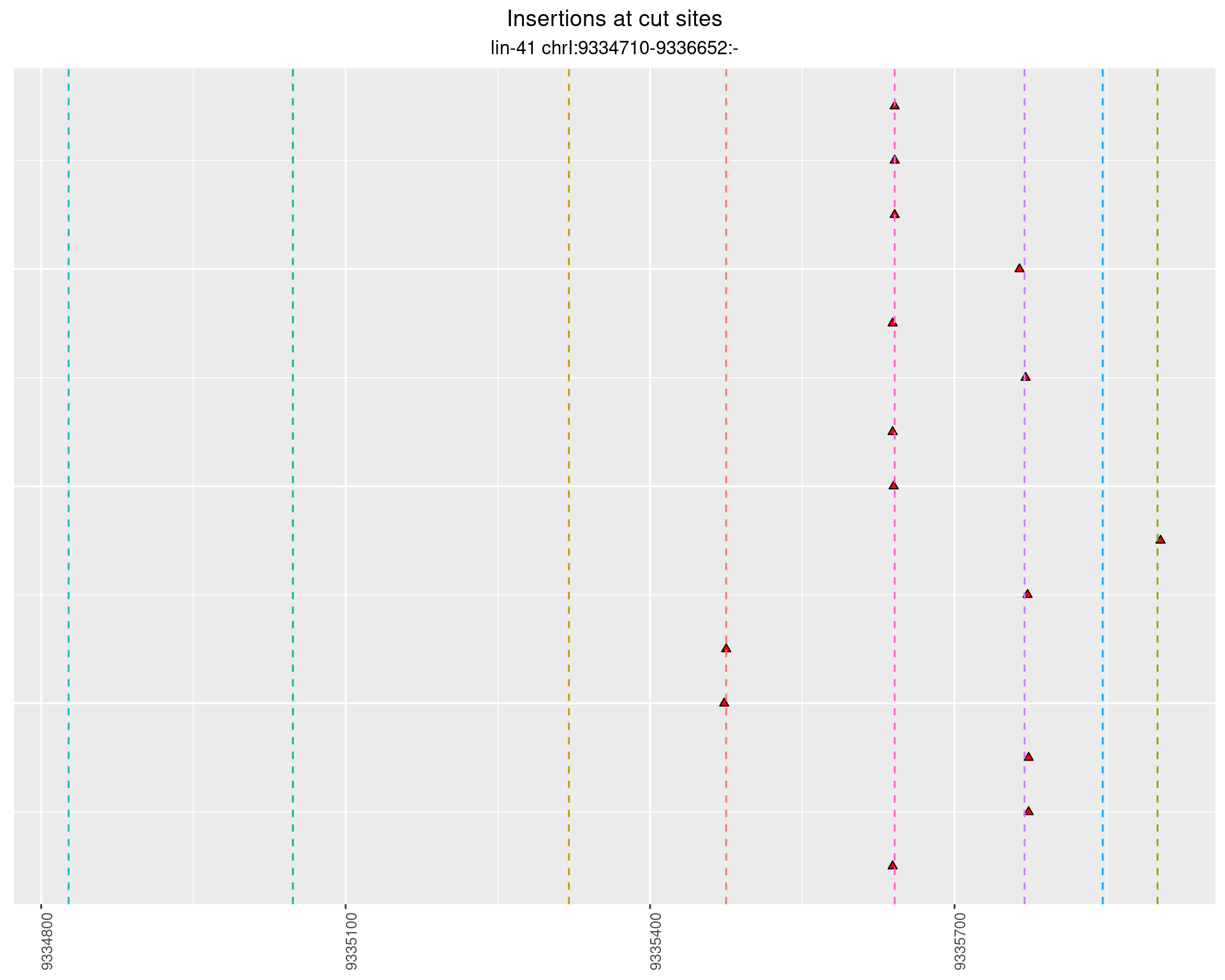
2.56.2 Freq: 1e-05 - 1e-04
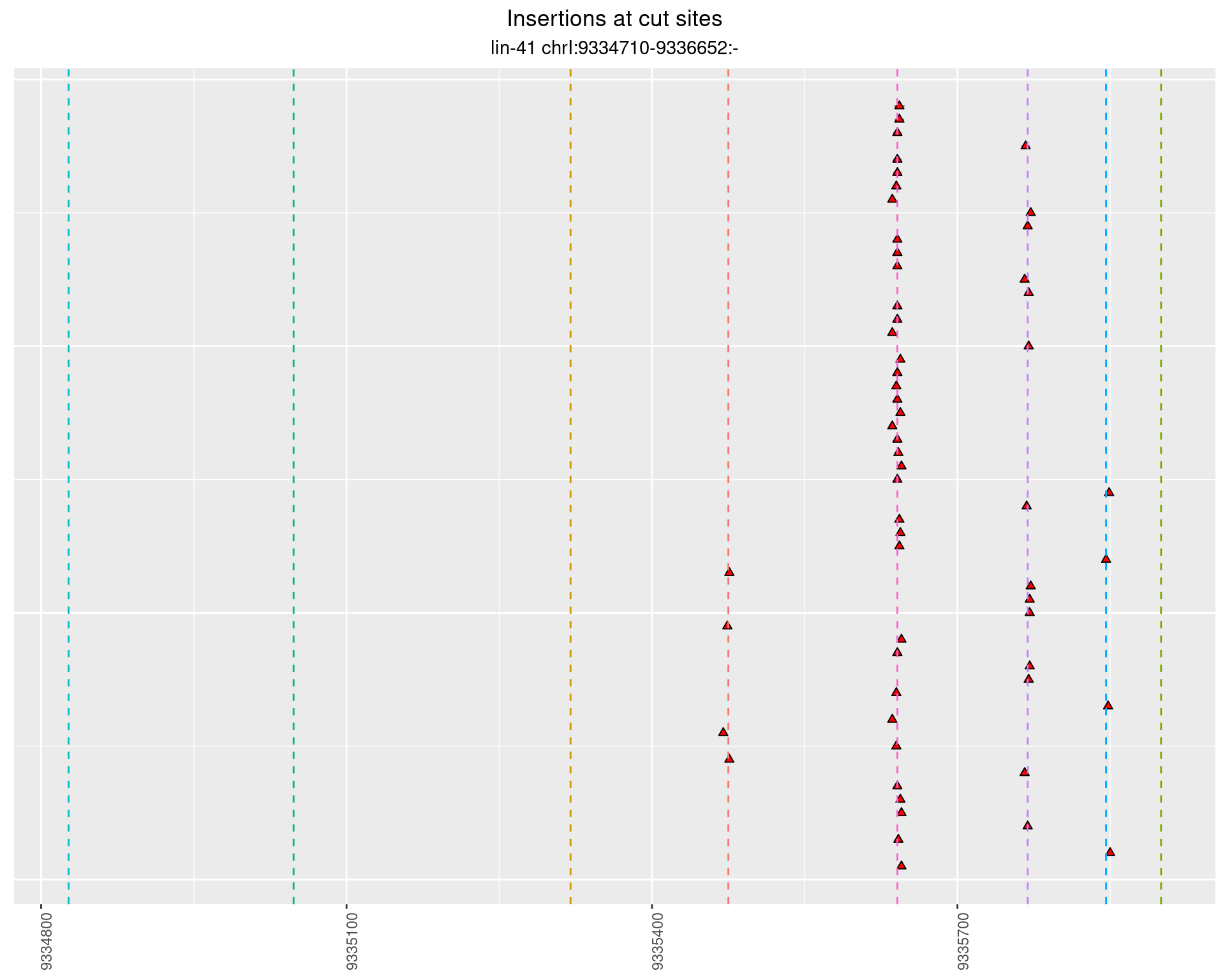
2.56.3 Freq: 0.001 - 0.01
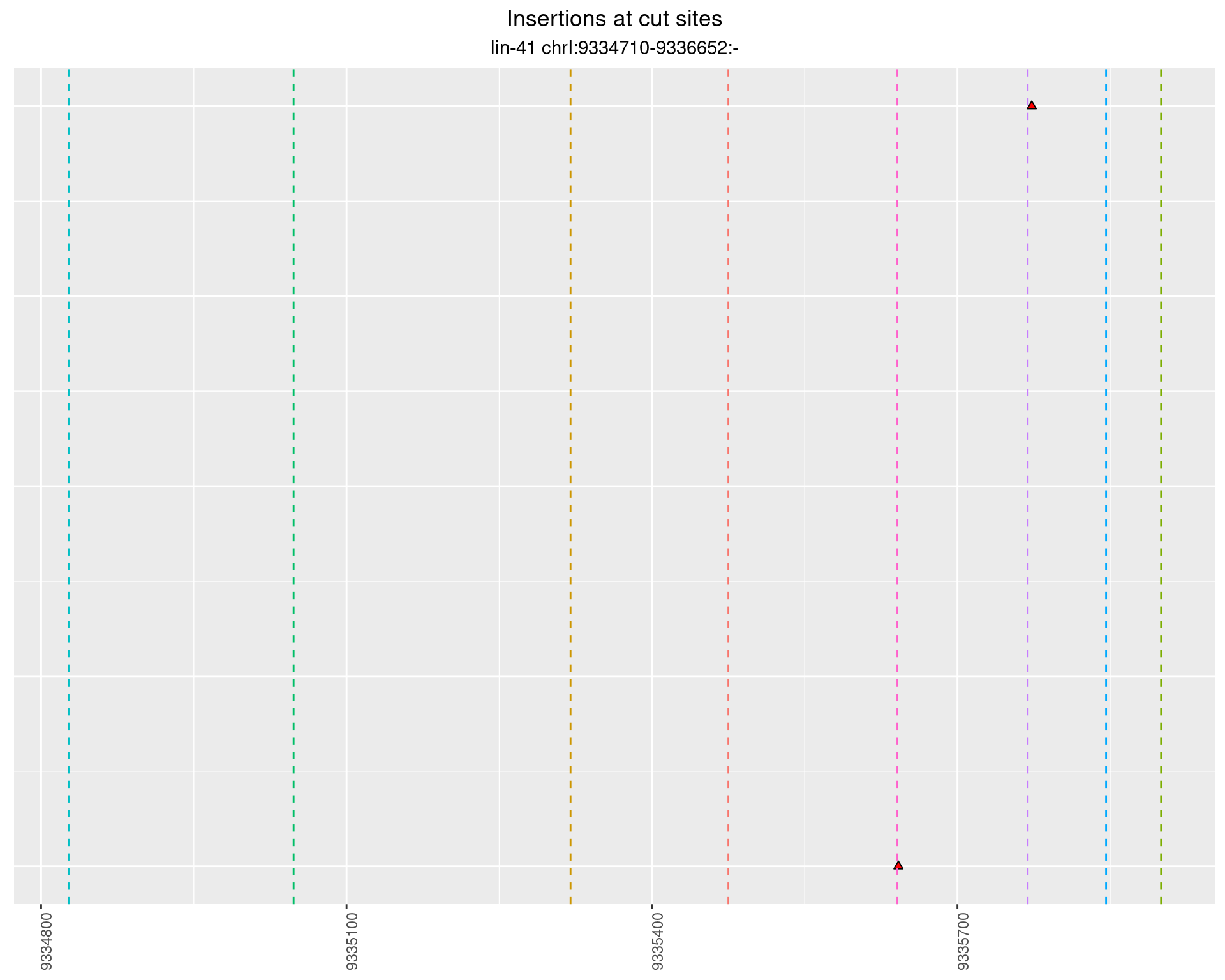
2.56.4 Freq: 1e-06 - 1e-05
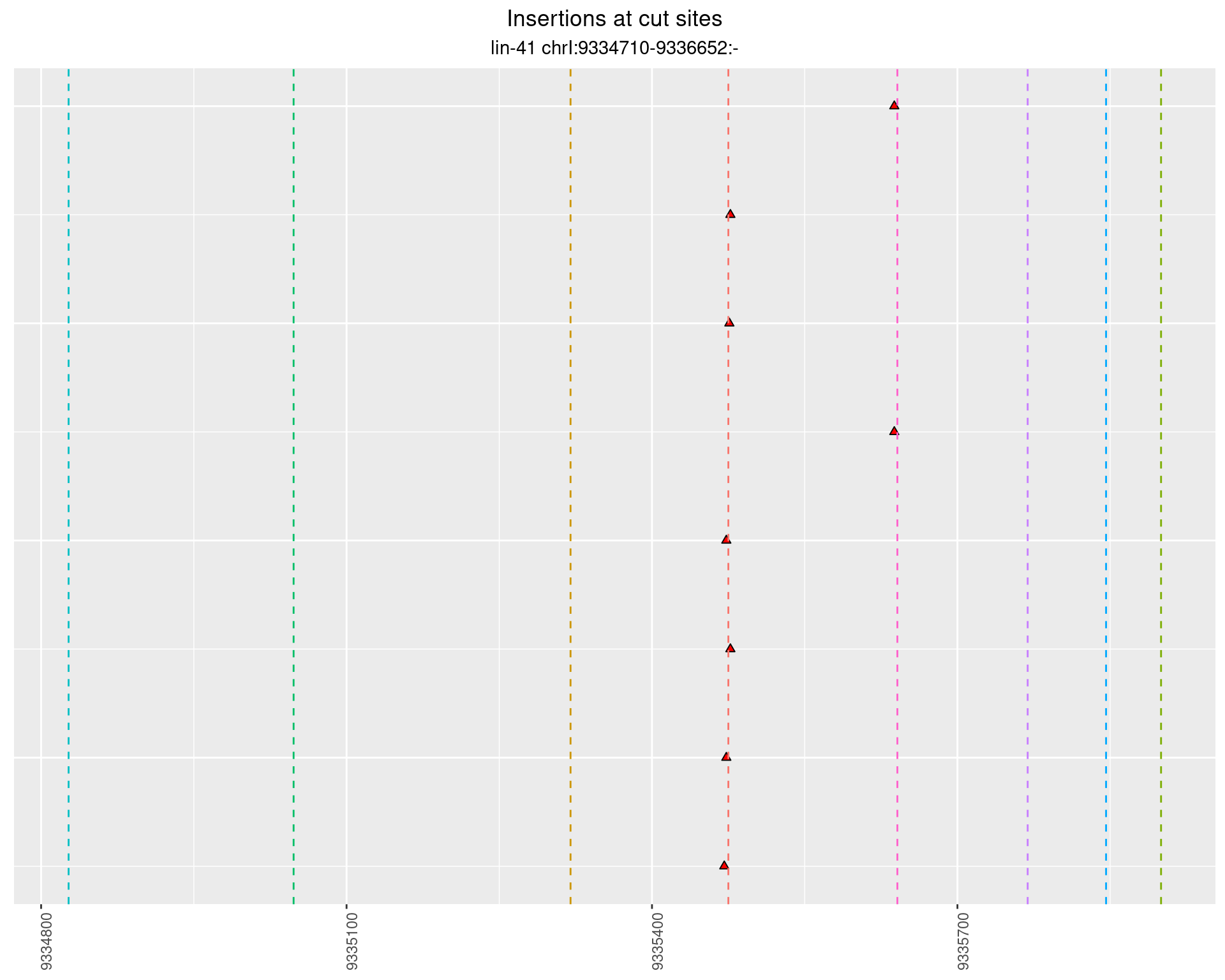
2.57 lin41_RNA_L4_N2_middle
2.57.1 Freq: 0.01 - 0.1
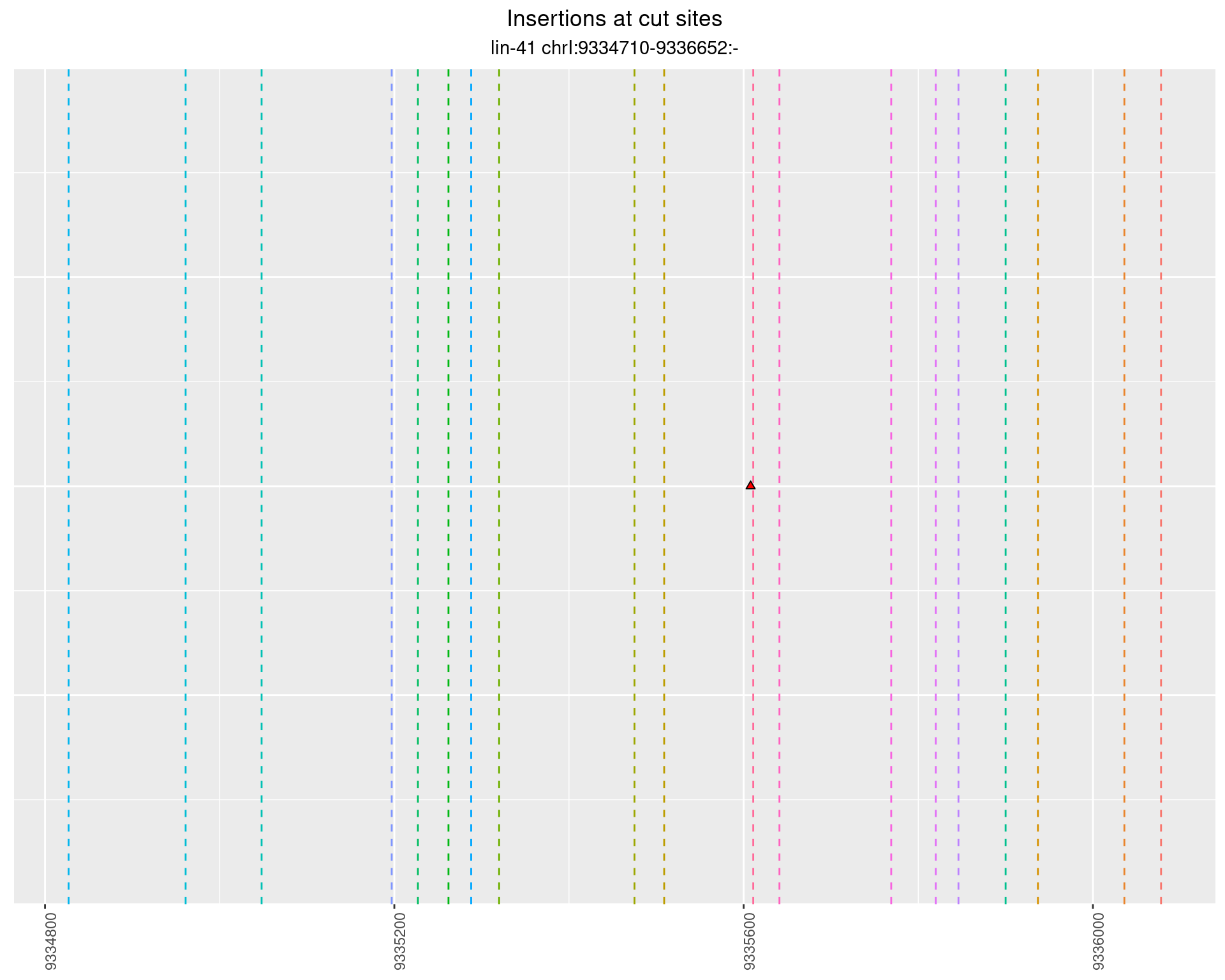
2.57.2 Freq: 1e-04 - 0.001
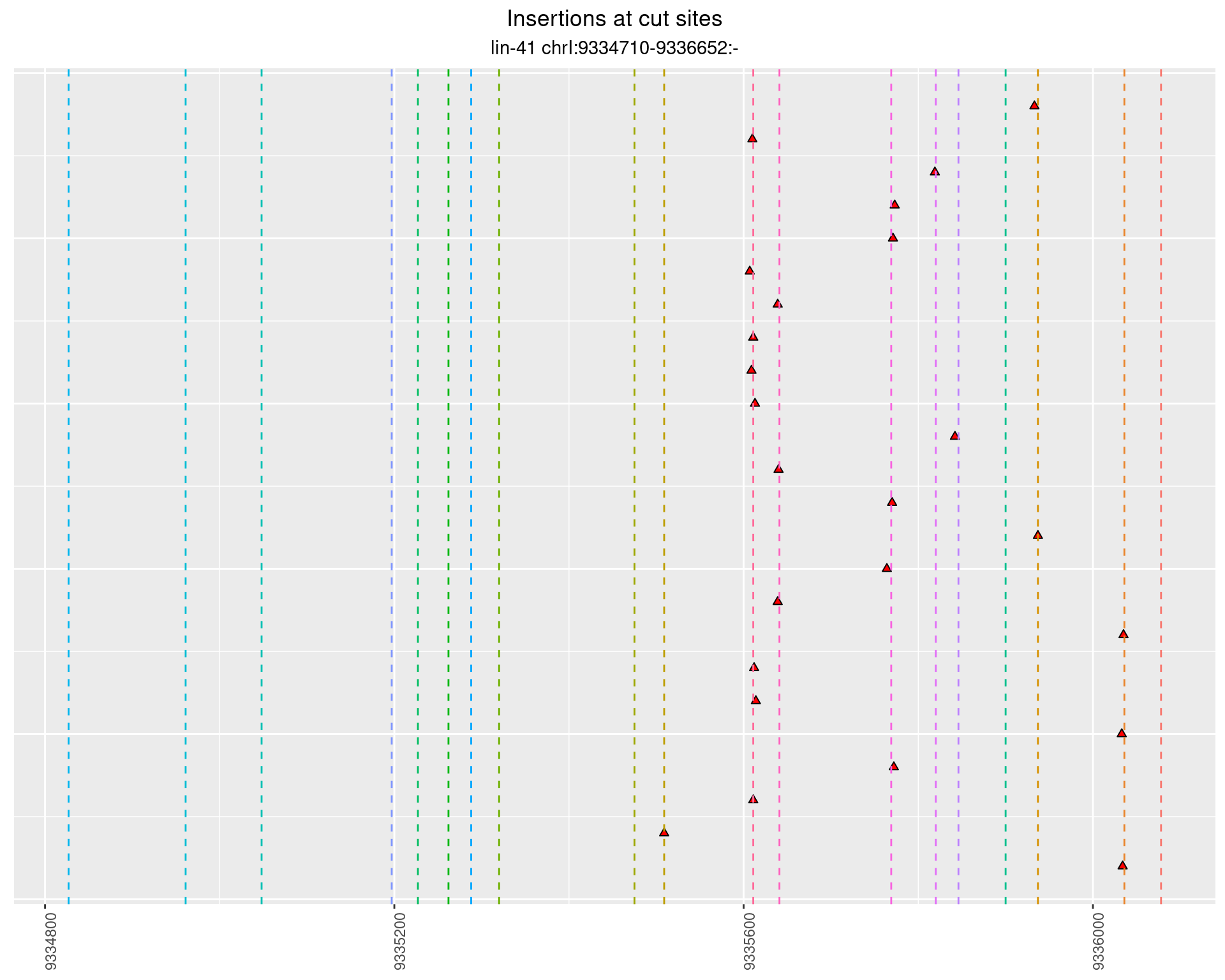
2.57.3 Freq: 0.001 - 0.01
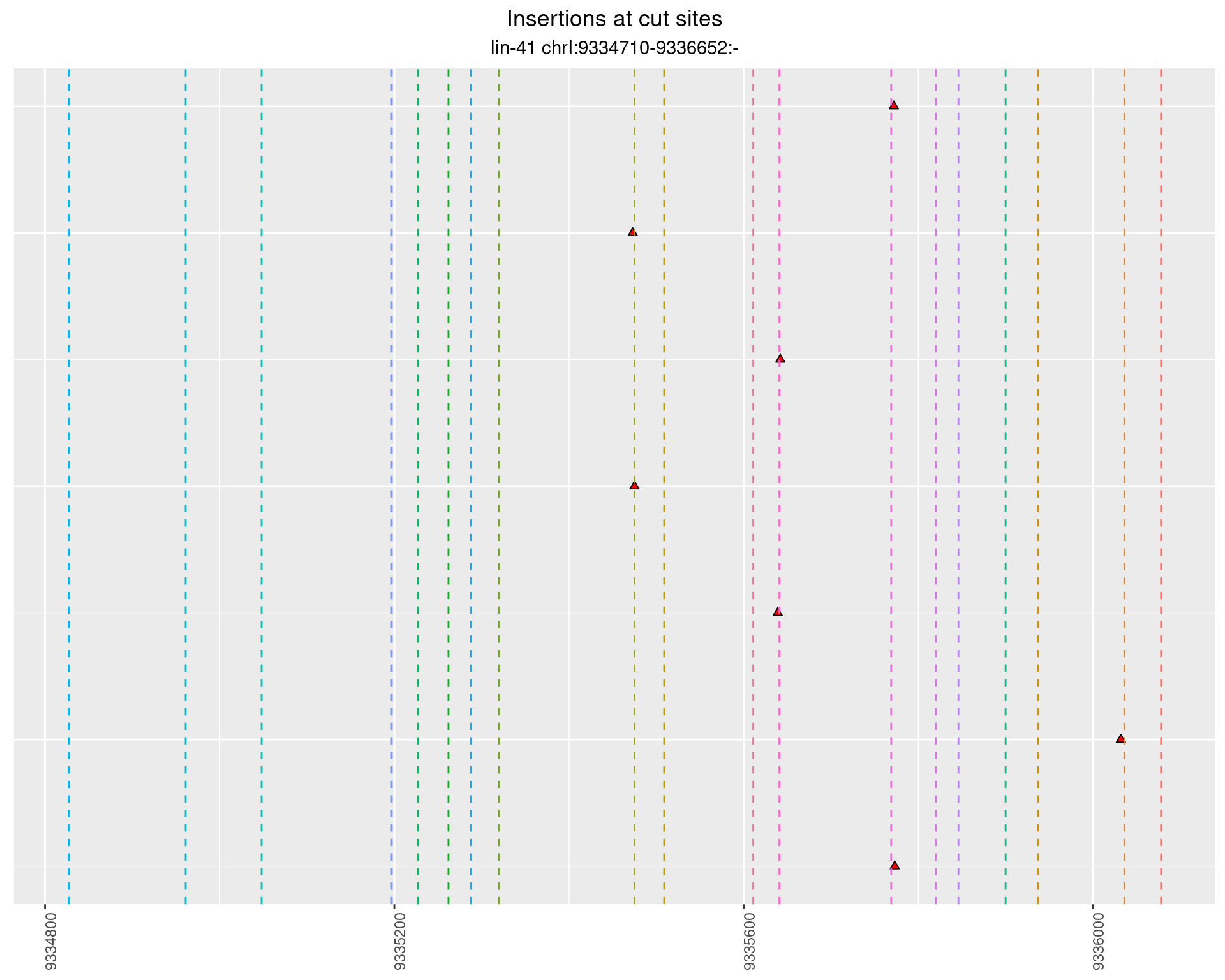
2.57.4 Freq: 1e-05 - 1e-04
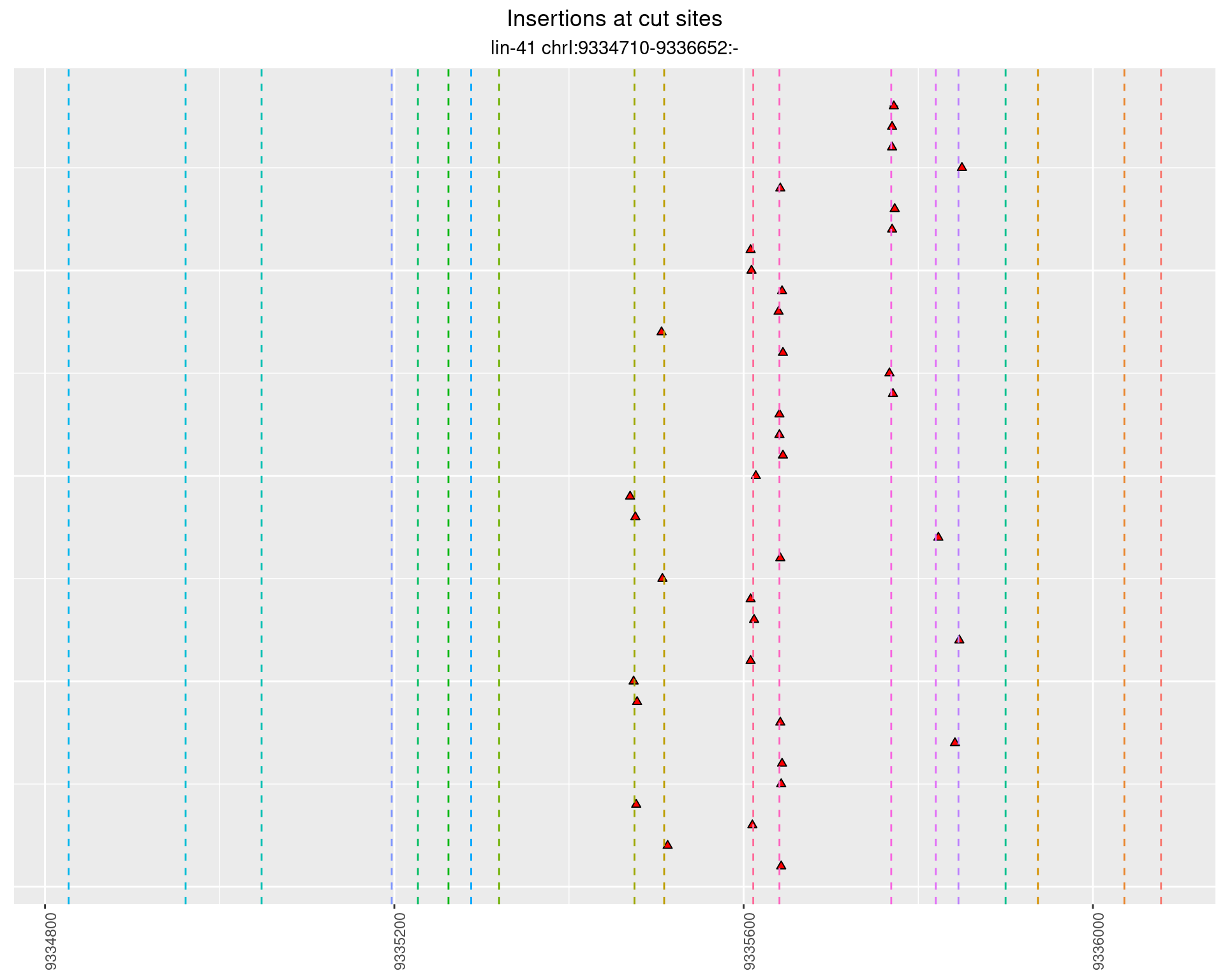
2.57.5 Freq: 1e-06 - 1e-05
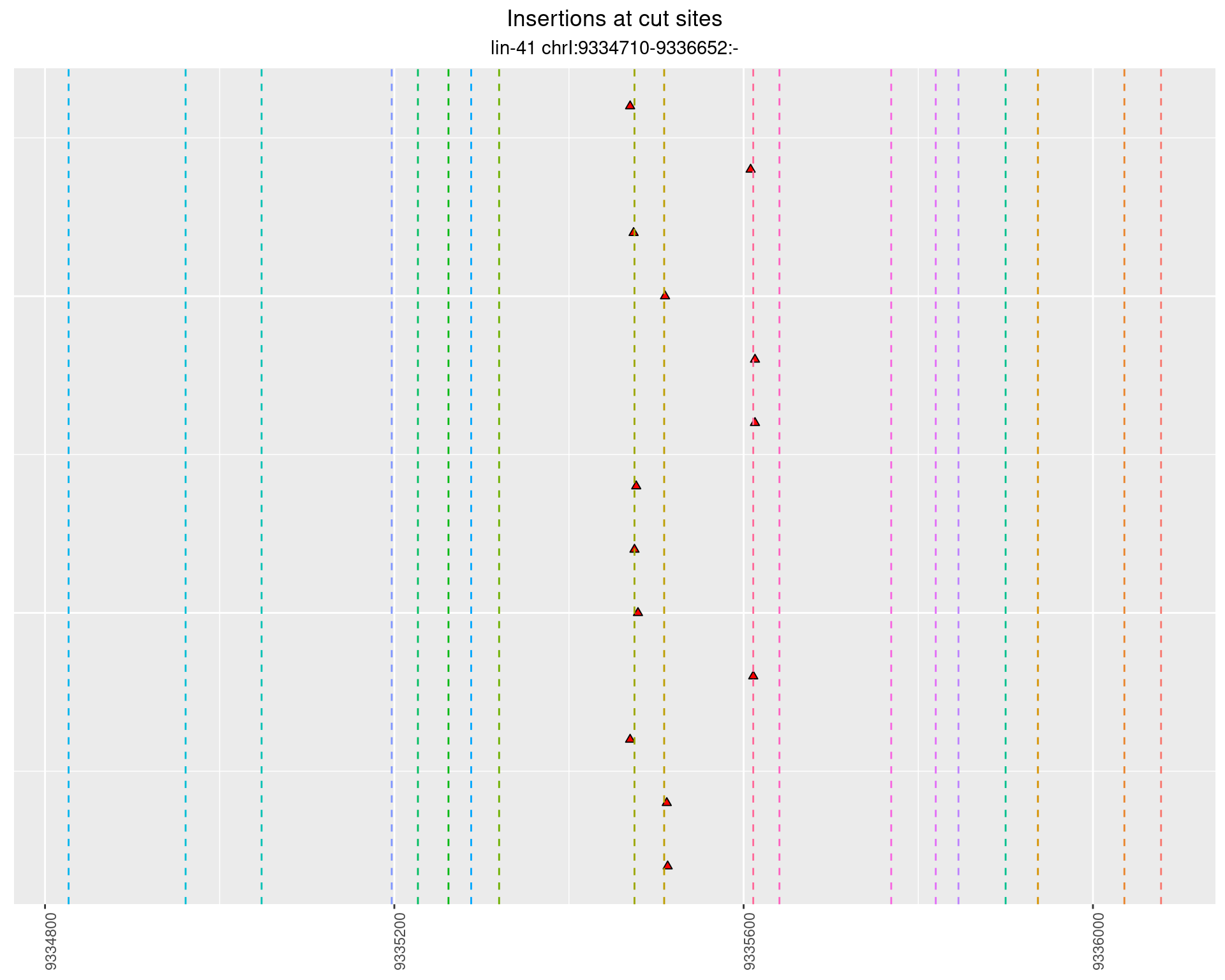
2.58 lin41_RNA_L4_1516_middle
2.58.1 Freq: 0.1 - 1
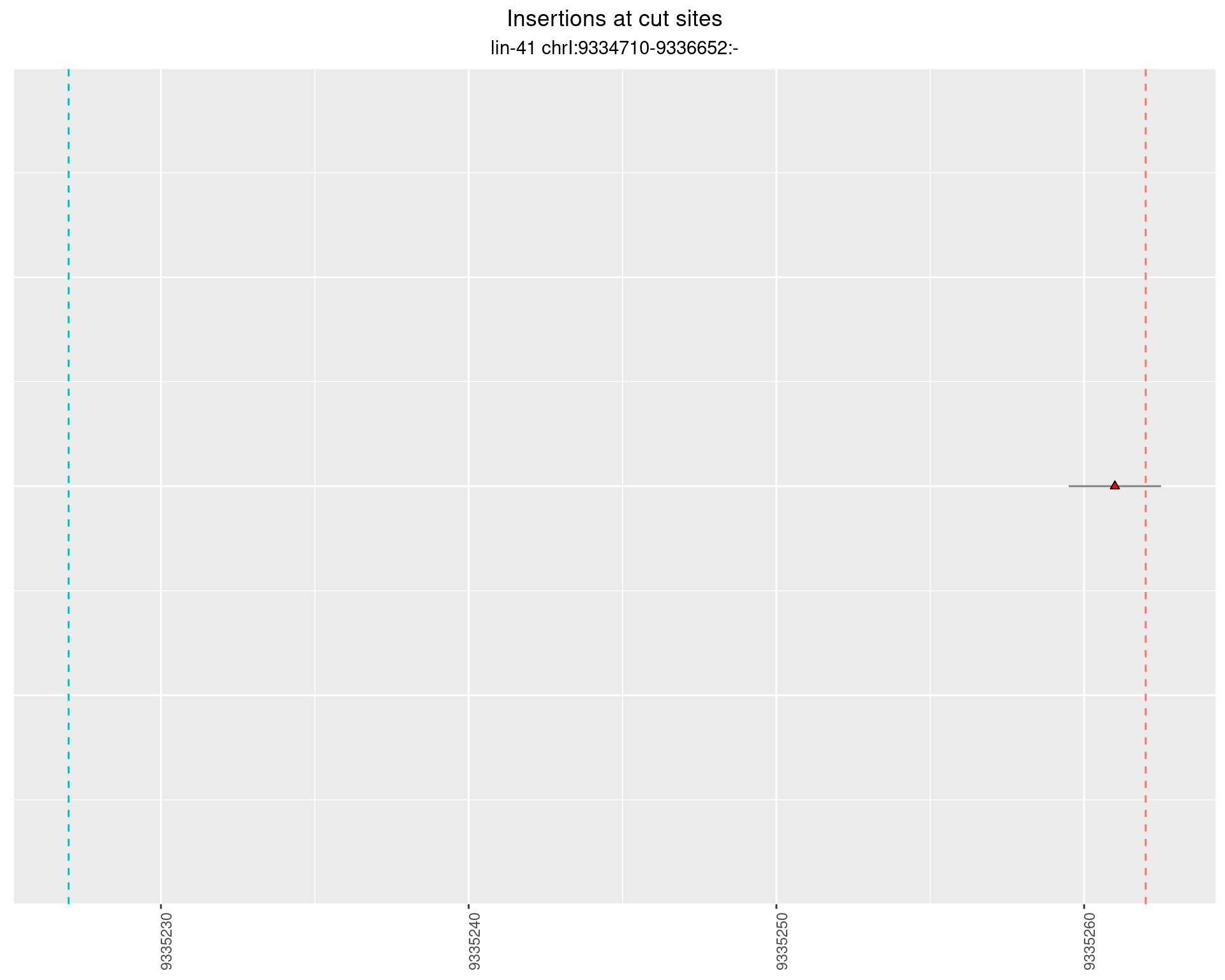
2.59 lin41_RNA_L4_2627_middle
2.60 lin41_RNA_L4_pool2_middle
2.60.1 Freq: 0.01 - 0.1
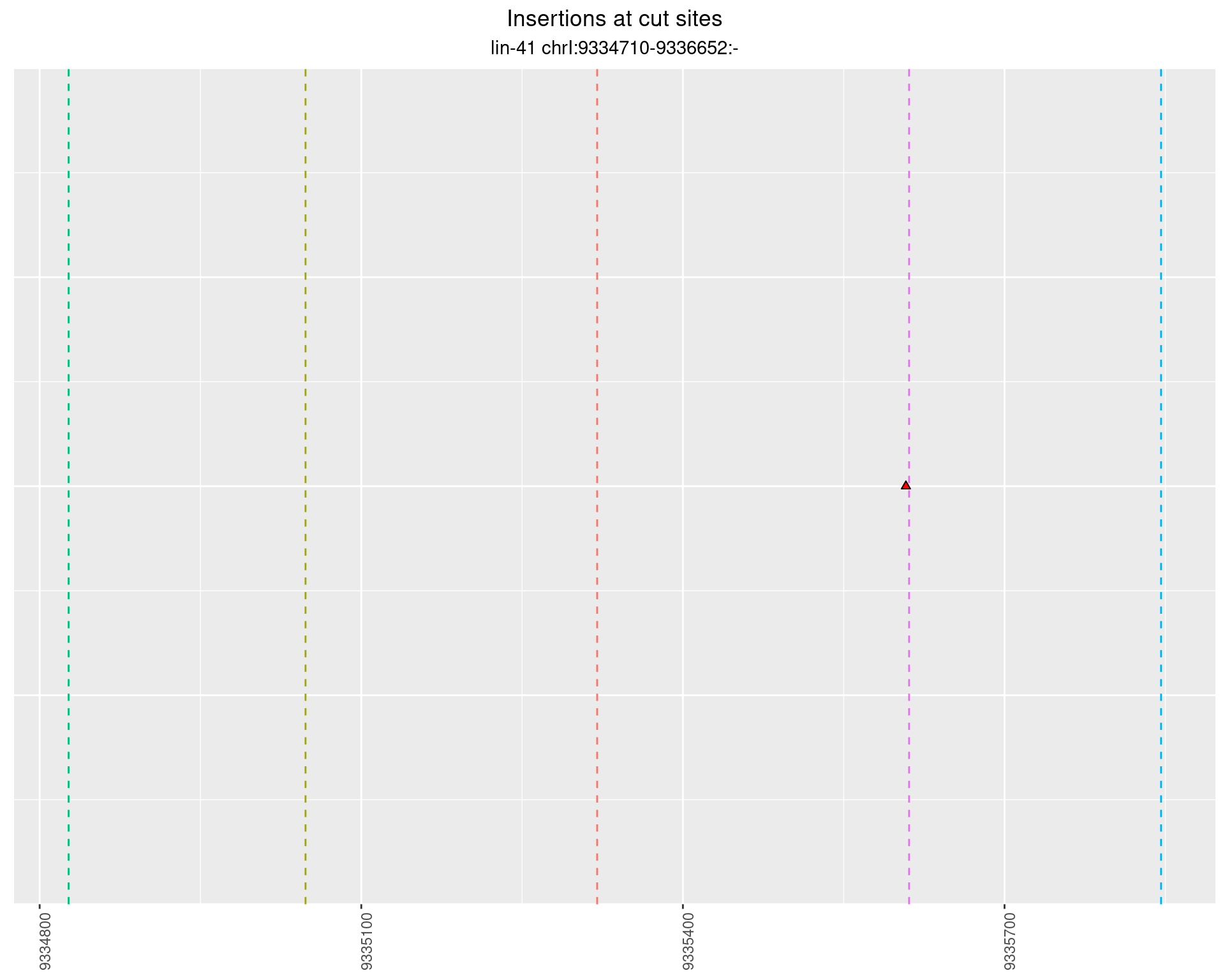
2.60.2 Freq: 1e-06 - 1e-05
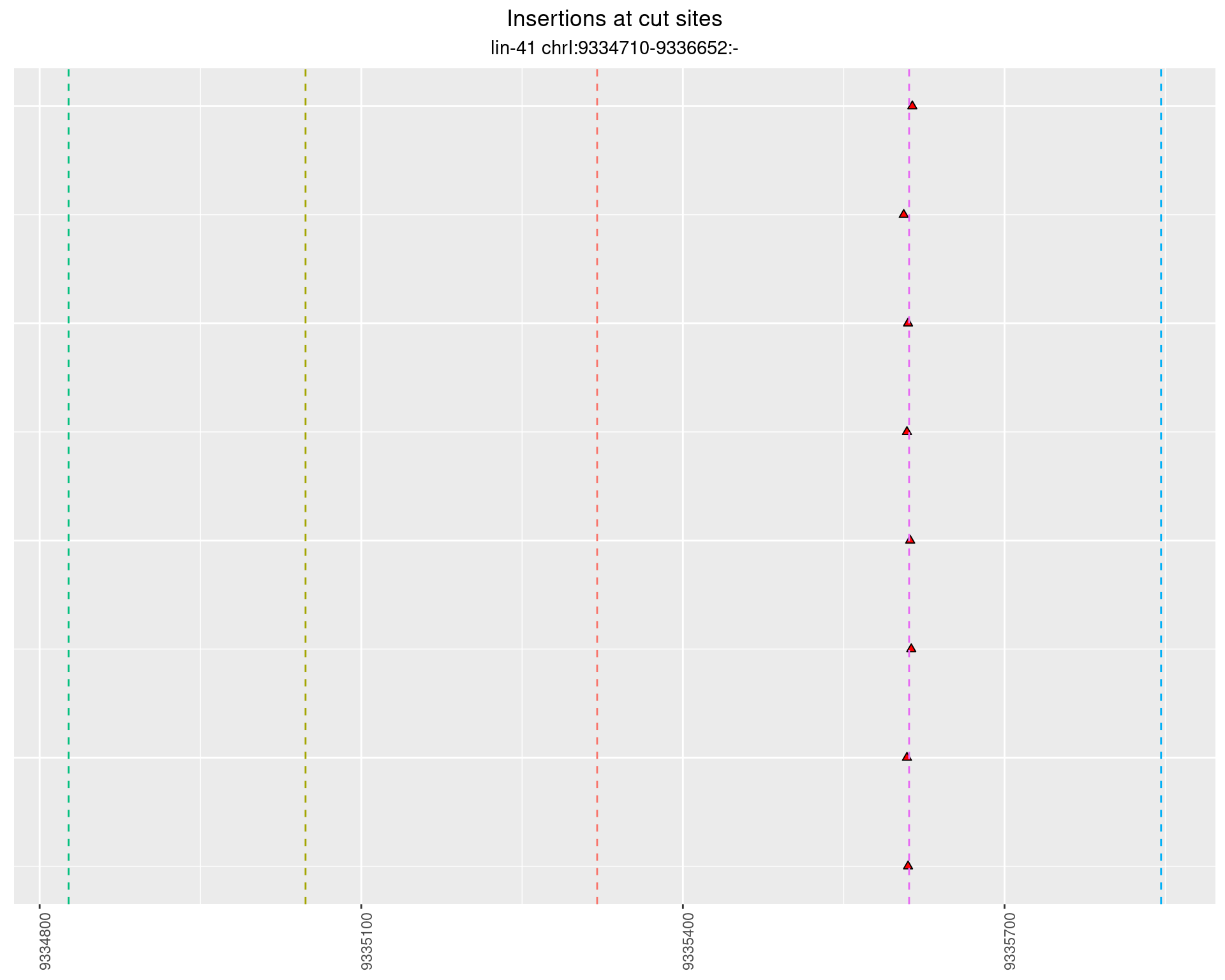
2.60.3 Freq: 1e-04 - 0.001
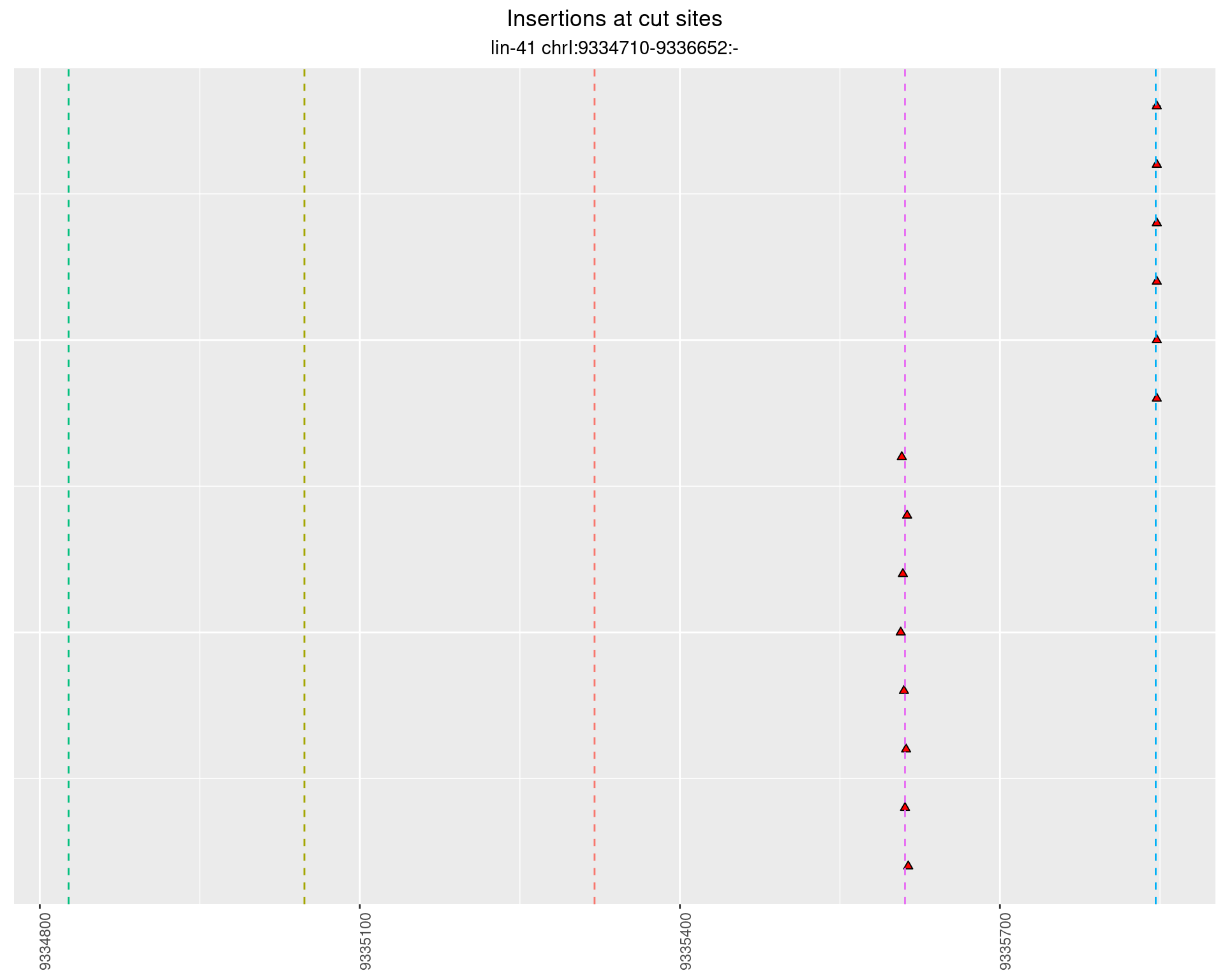
2.60.4 Freq: 1e-05 - 1e-04
2.60.5 Freq: 0.001 - 0.01
2.61 lin41_RNA_L4_pool3_middle
2.61.1 Freq: 1e-04 - 0.001
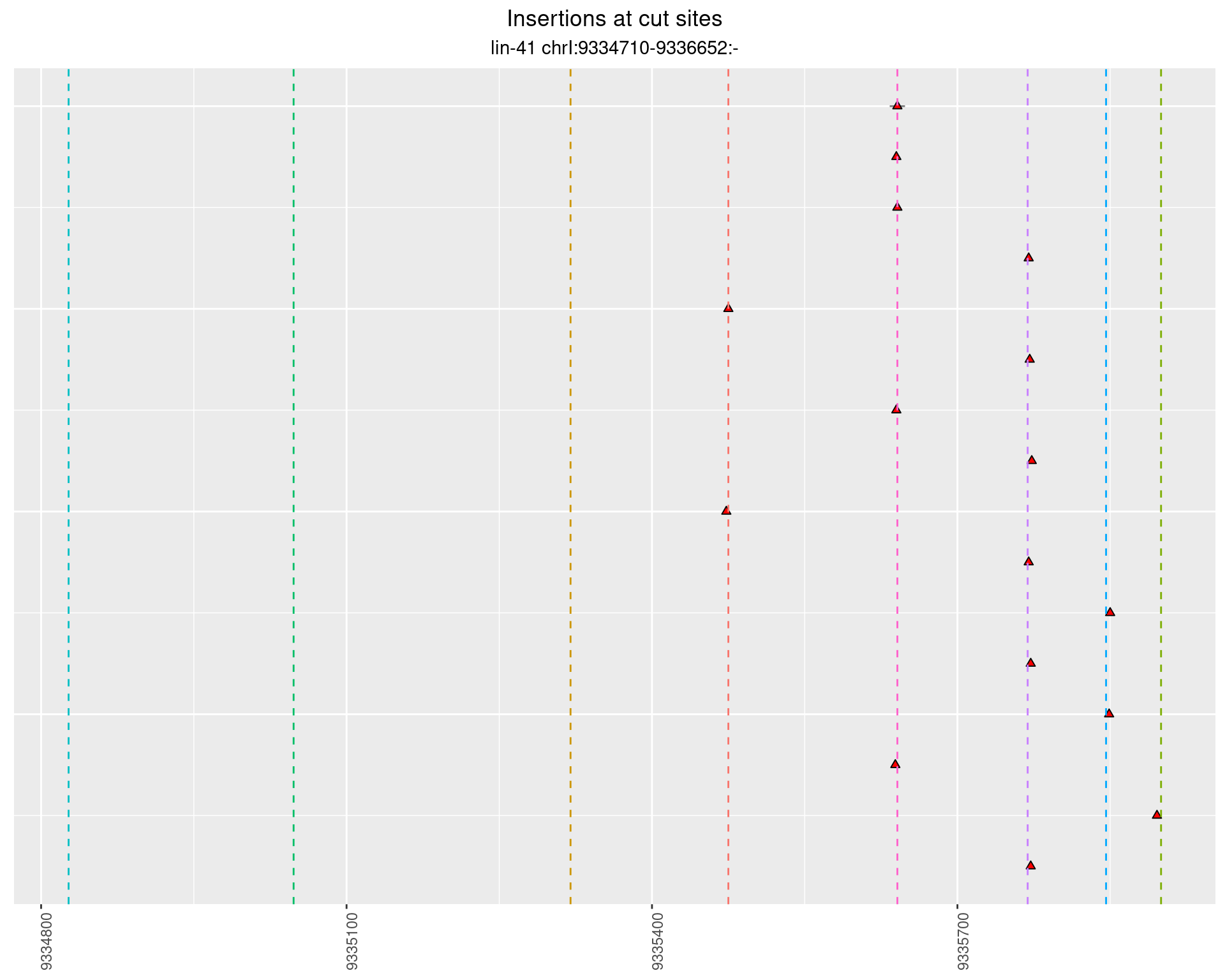
2.61.2 Freq: 1e-05 - 1e-04
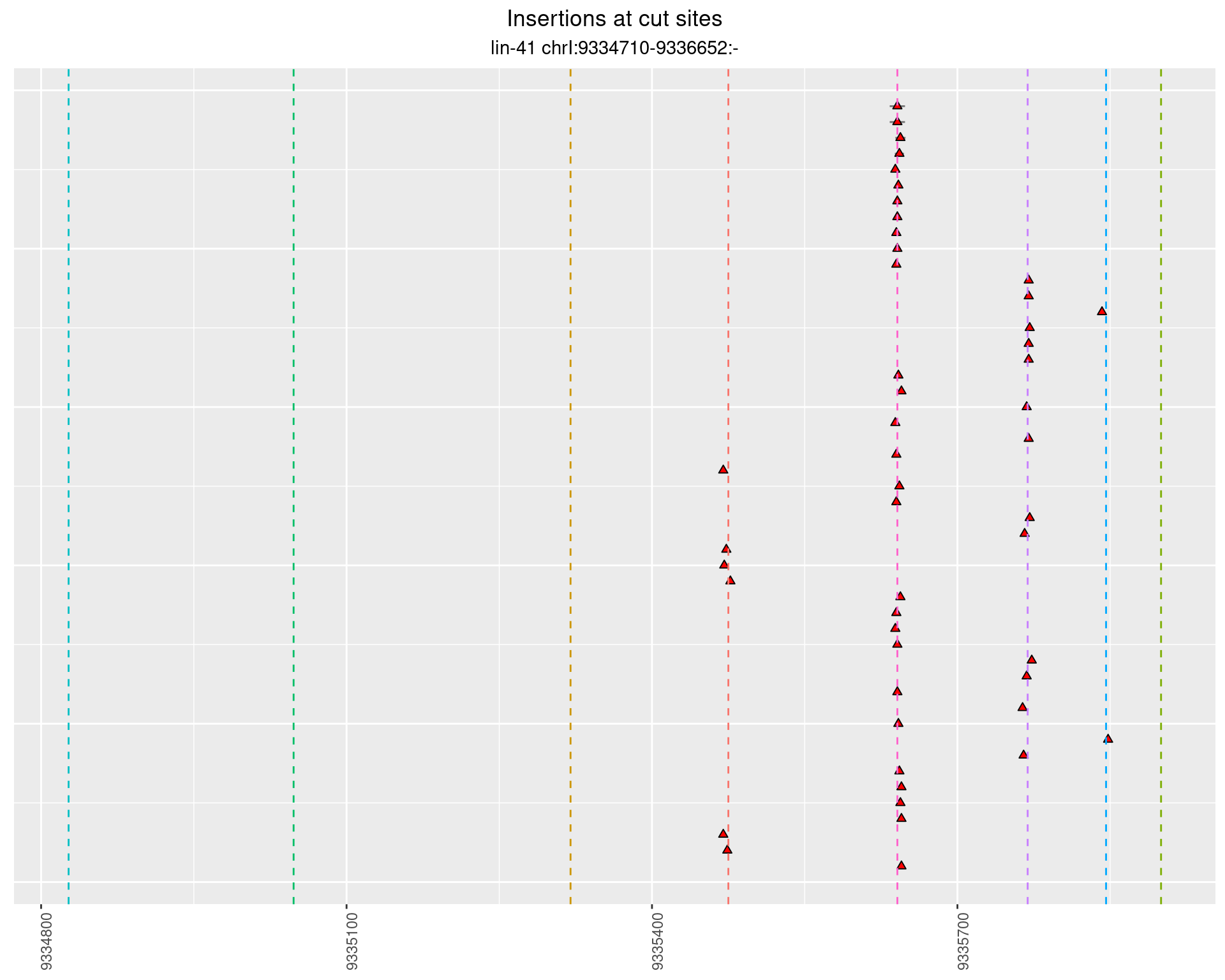
2.61.3 Freq: 0.001 - 0.01
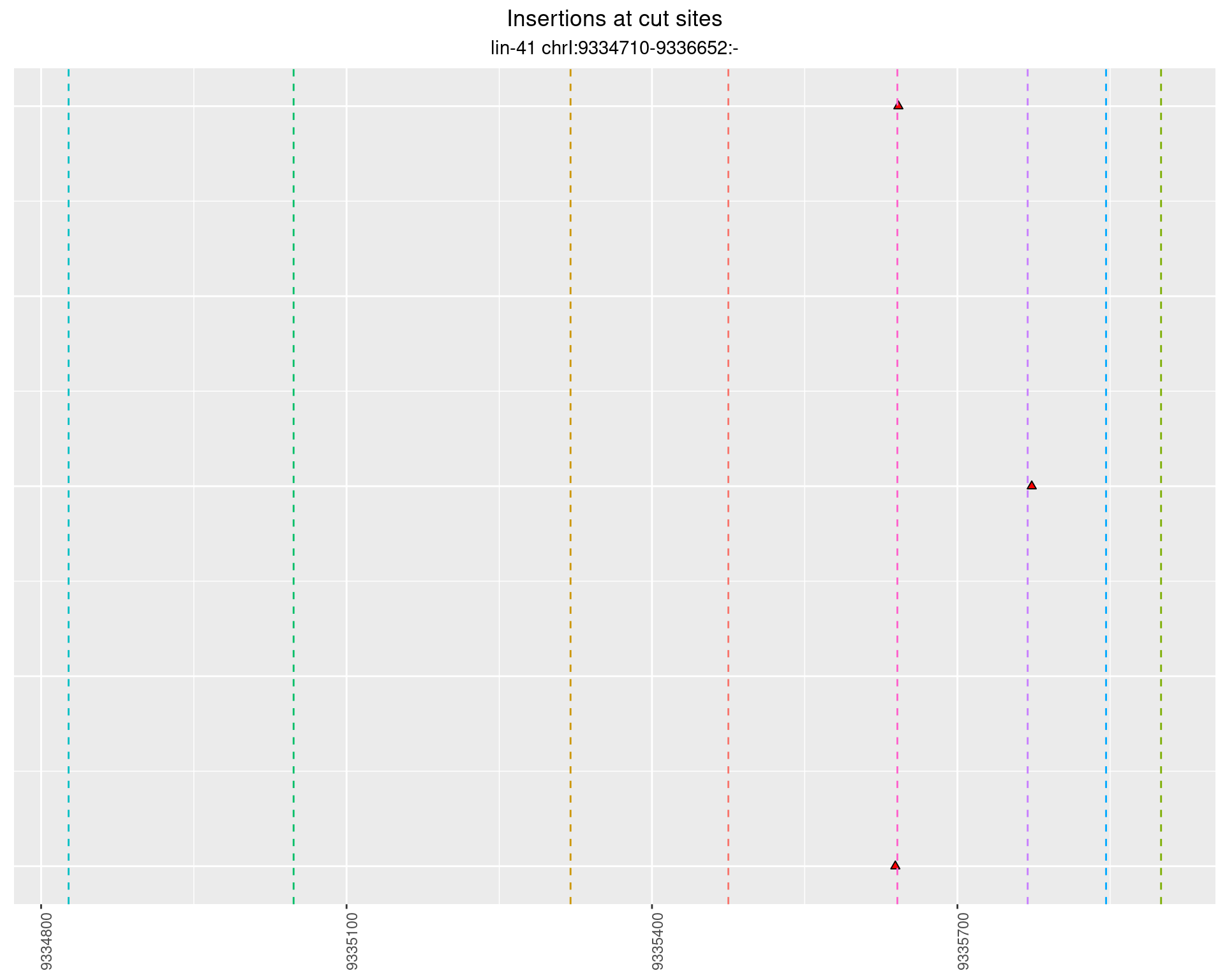
2.61.4 Freq: 1e-06 - 1e-05
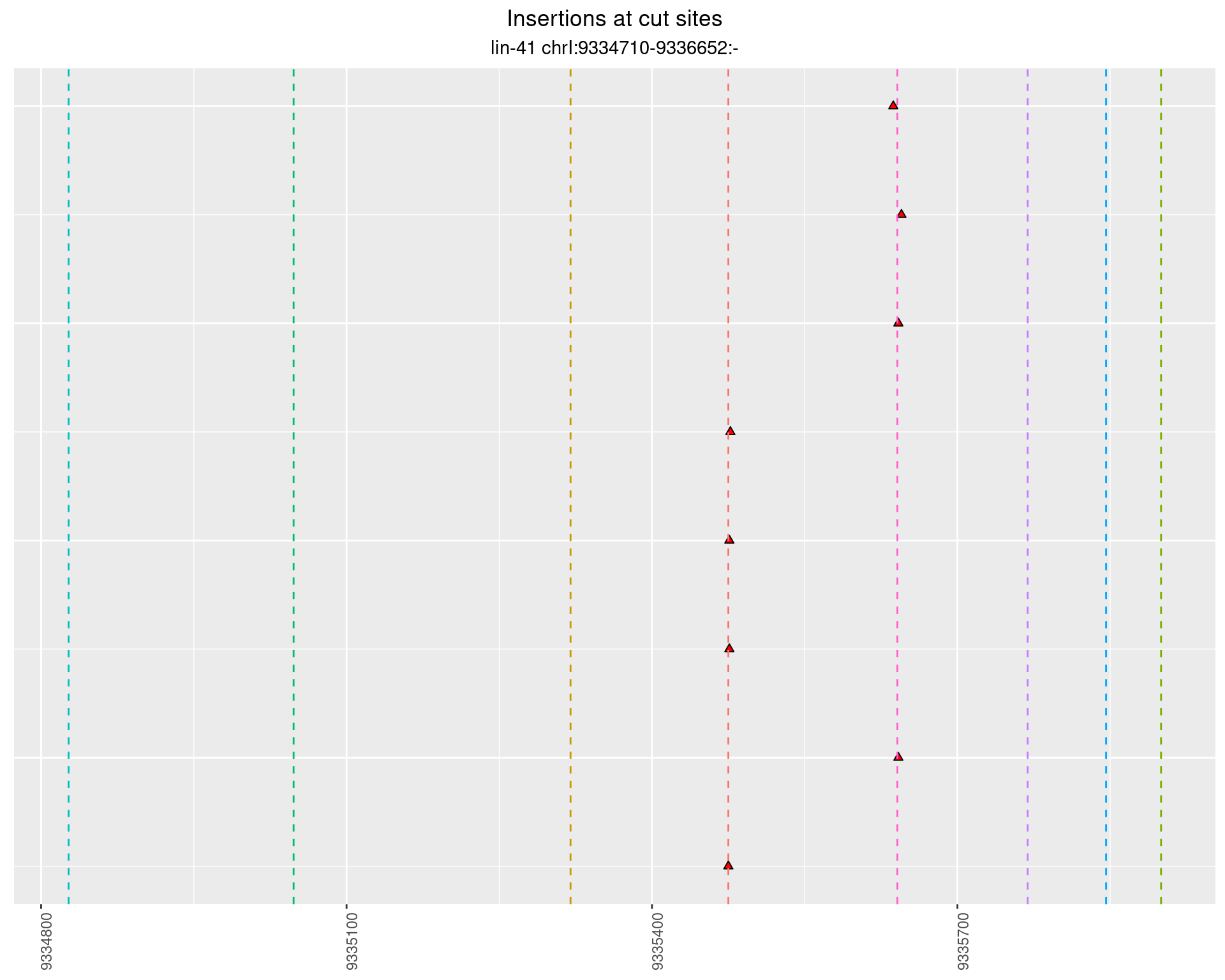
2.62 lin41_RNA_L1_N2_3end_UMI
2.62.1 Freq: 0.001 - 0.01
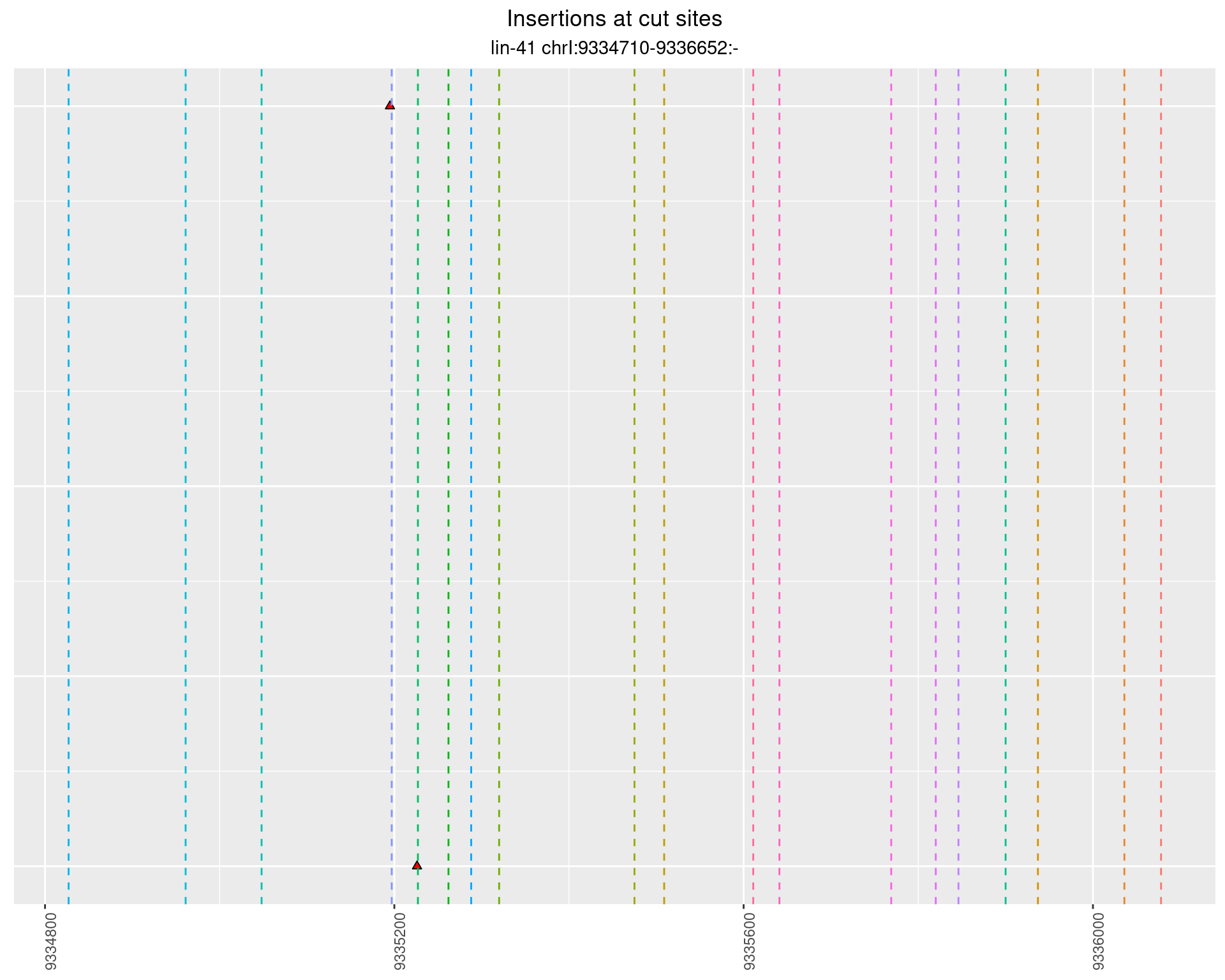
2.62.2 Freq: 1e-04 - 0.001
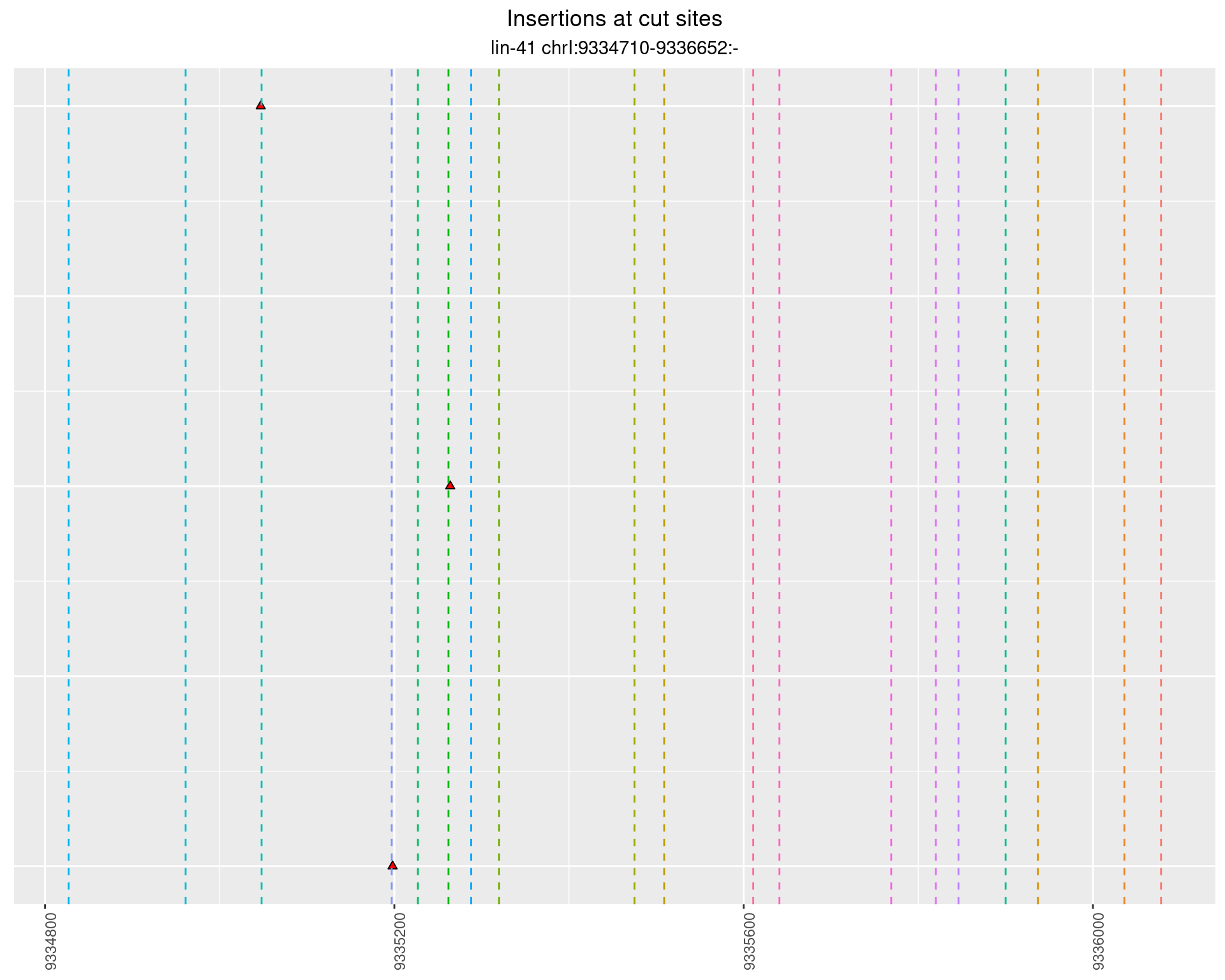
2.62.3 Freq: 0.01 - 0.1
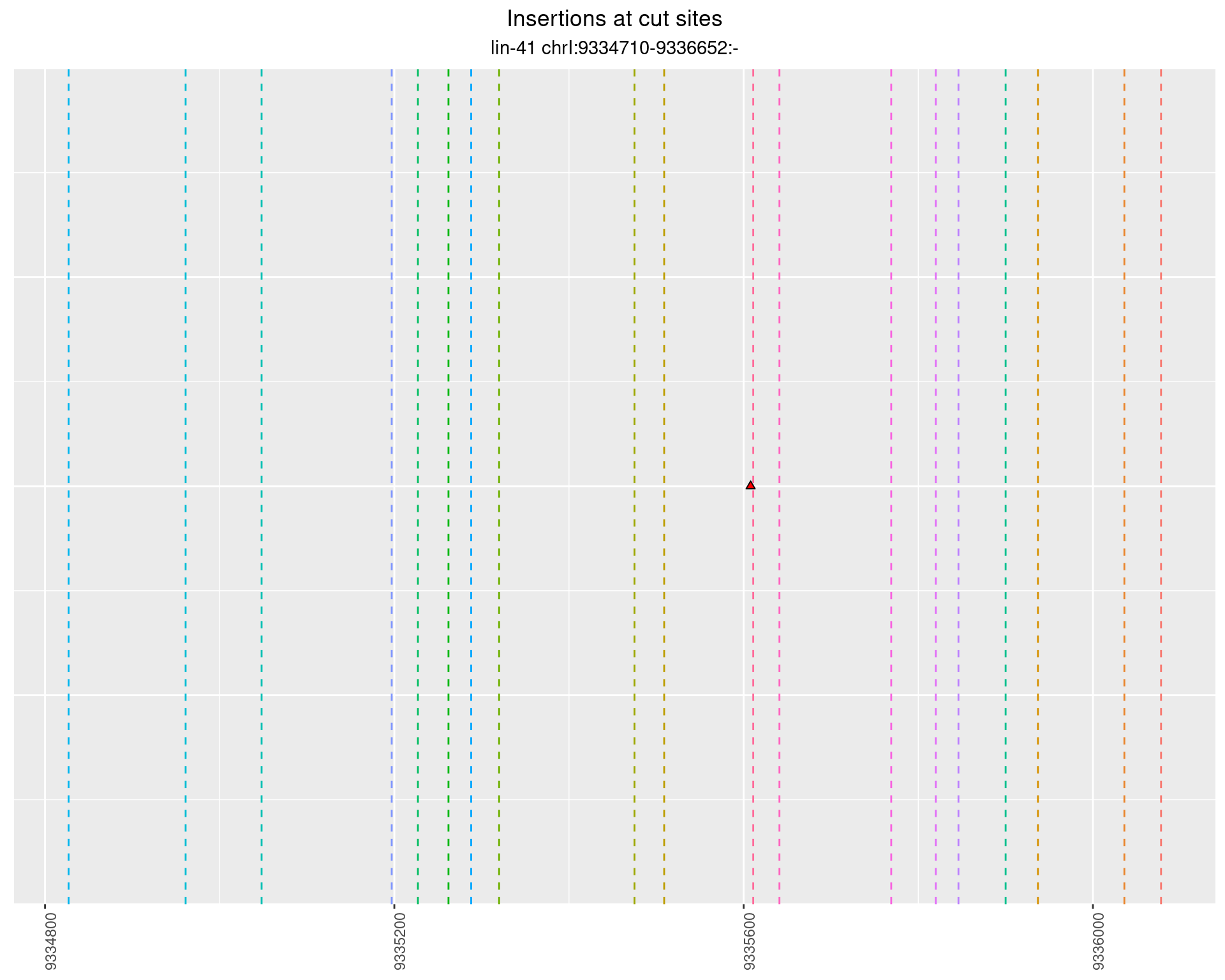
2.63 lin41_RNA_L1_1516_3end_UMI
2.63.1 Freq: 1e-04 - 0.001
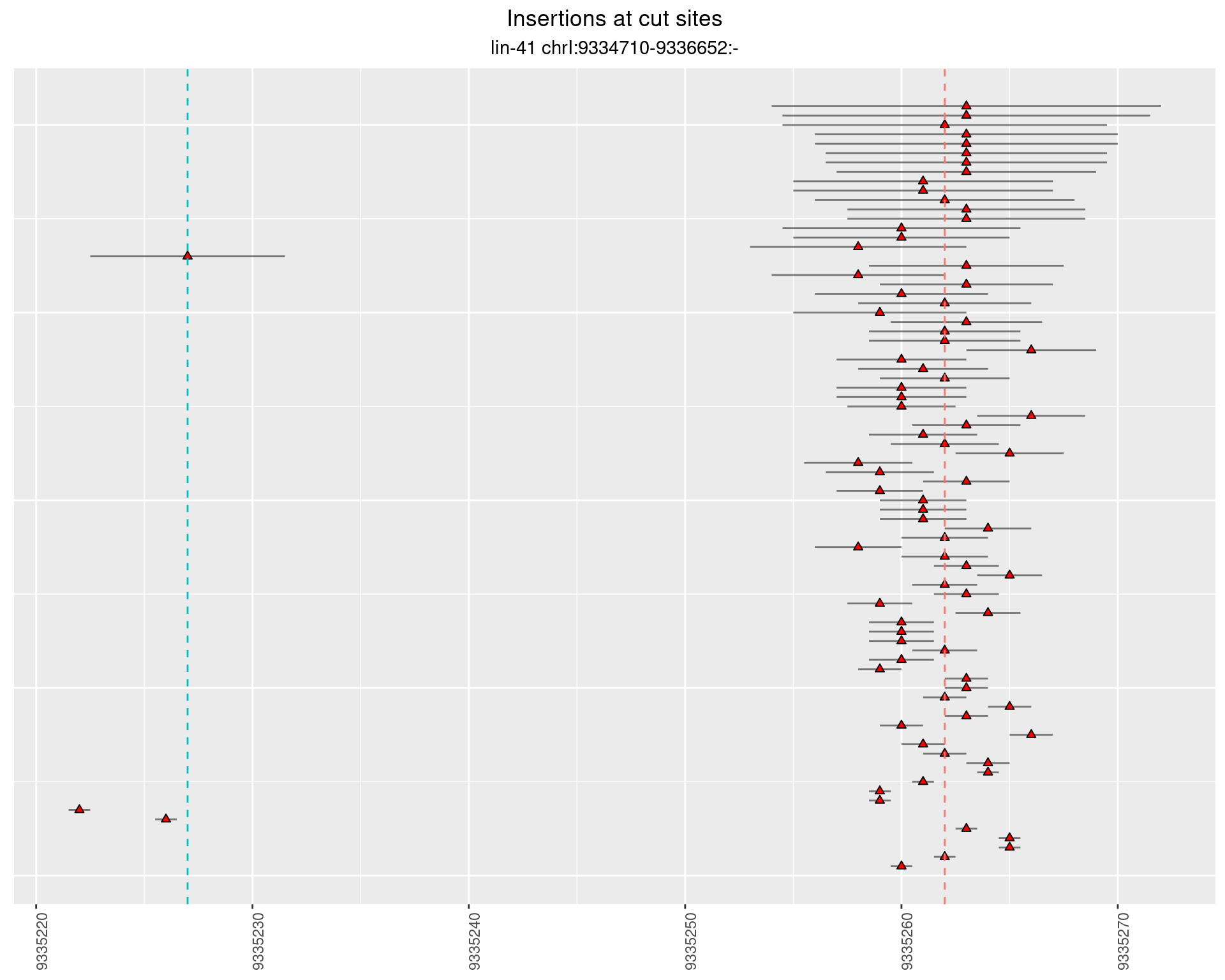
2.63.2 Freq: 0.001 - 0.01
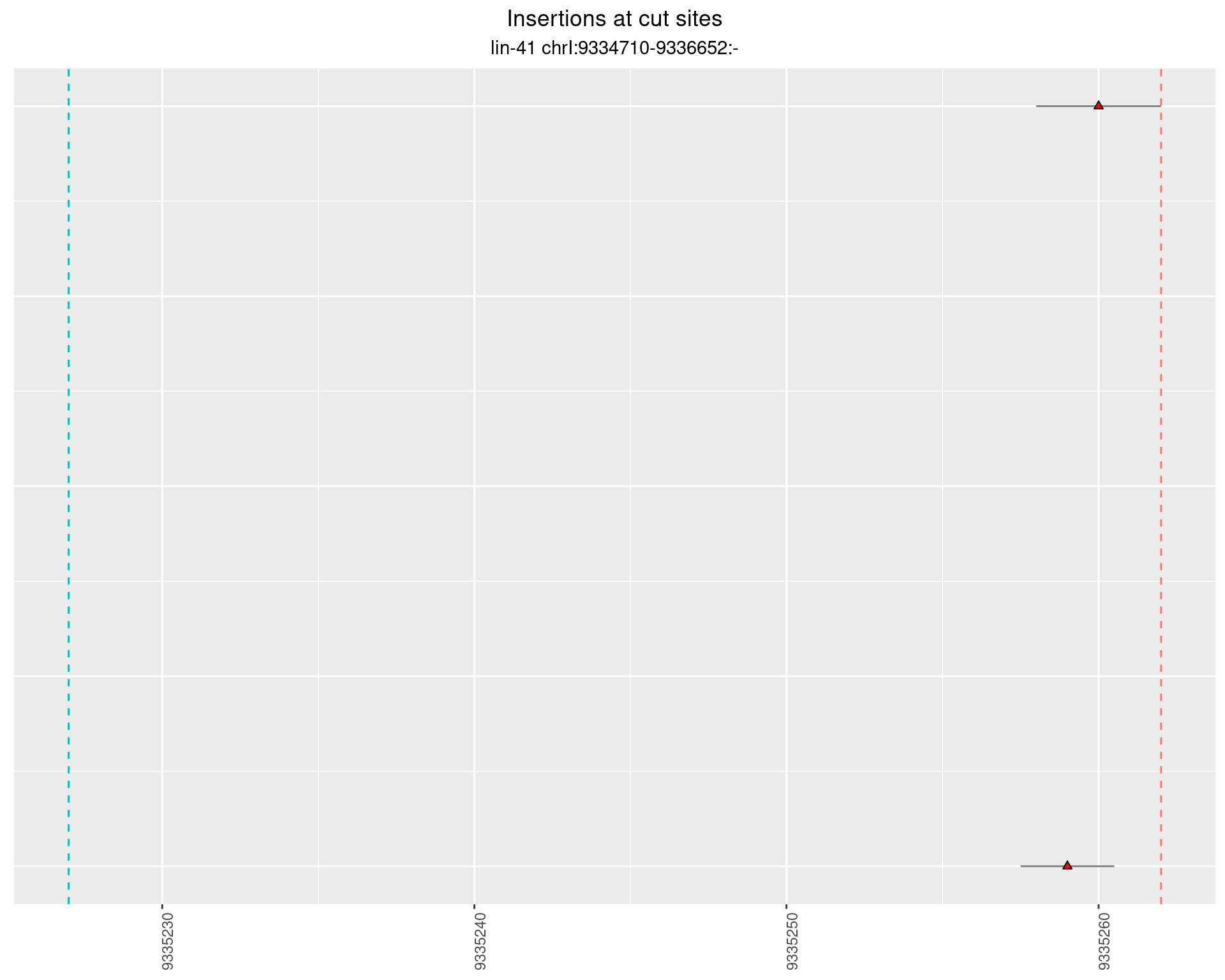
2.64 lin41_RNA_L1_2627_3end_UMI
2.64.1 Freq: 1e-04 - 0.001
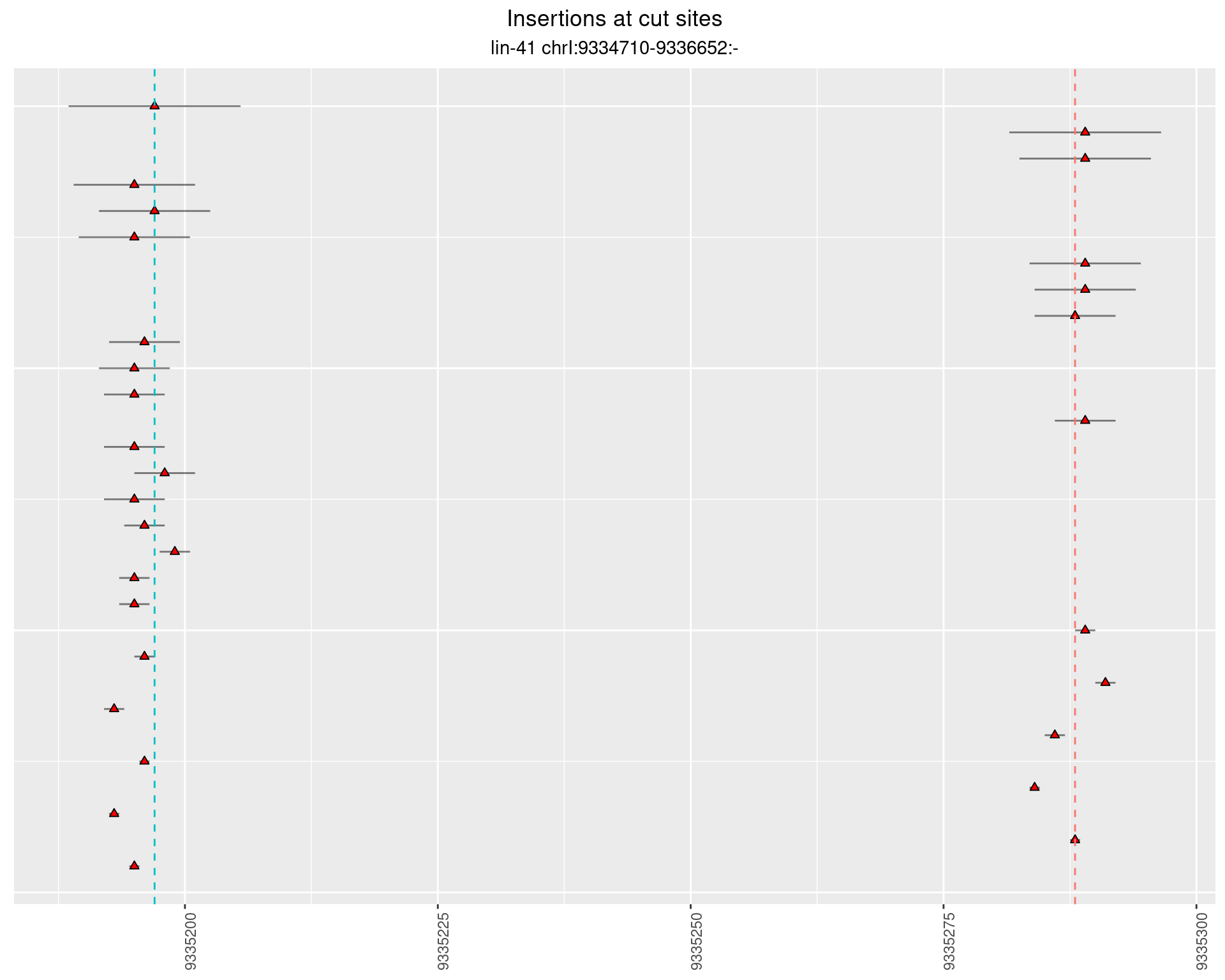
2.64.2 Freq: 0.001 - 0.01
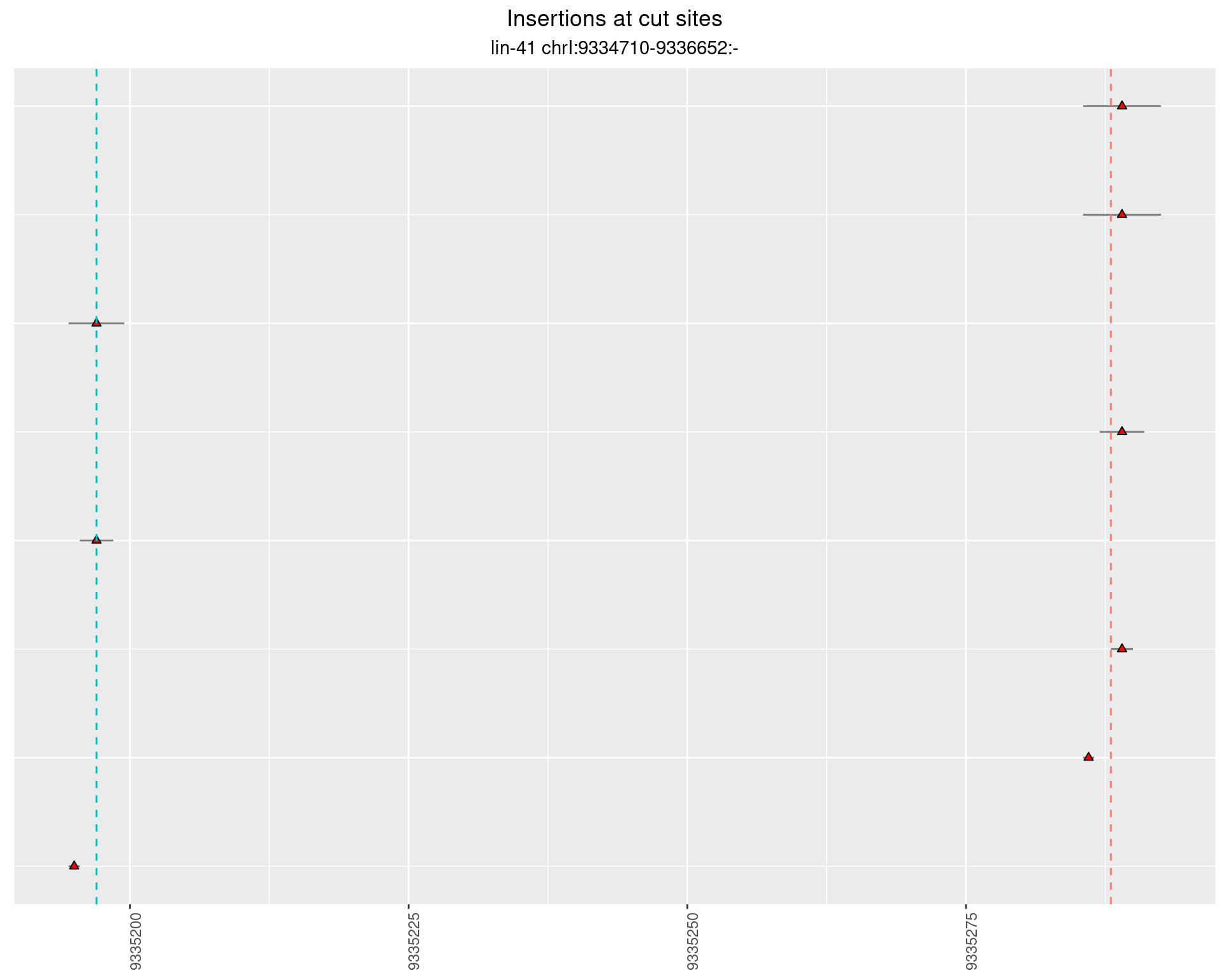
2.65 lin41_RNA_L1_pool2_3end_UMI
2.65.1 Freq: 1e-05 - 1e-04
2.65.2 Freq: 1e-04 - 0.001
2.65.3 Freq: 0.001 - 0.01
2.66 lin41_RNA_L1_pool3_3end_UMI
2.66.1 Freq: 1e-04 - 0.001
2.66.2 Freq: 0.001 - 0.01
2.67 lin41_RNA_L4_N2_3end_UMI
2.67.1 Freq: 0.001 - 0.01
2.67.2 Freq: 1e-04 - 0.001
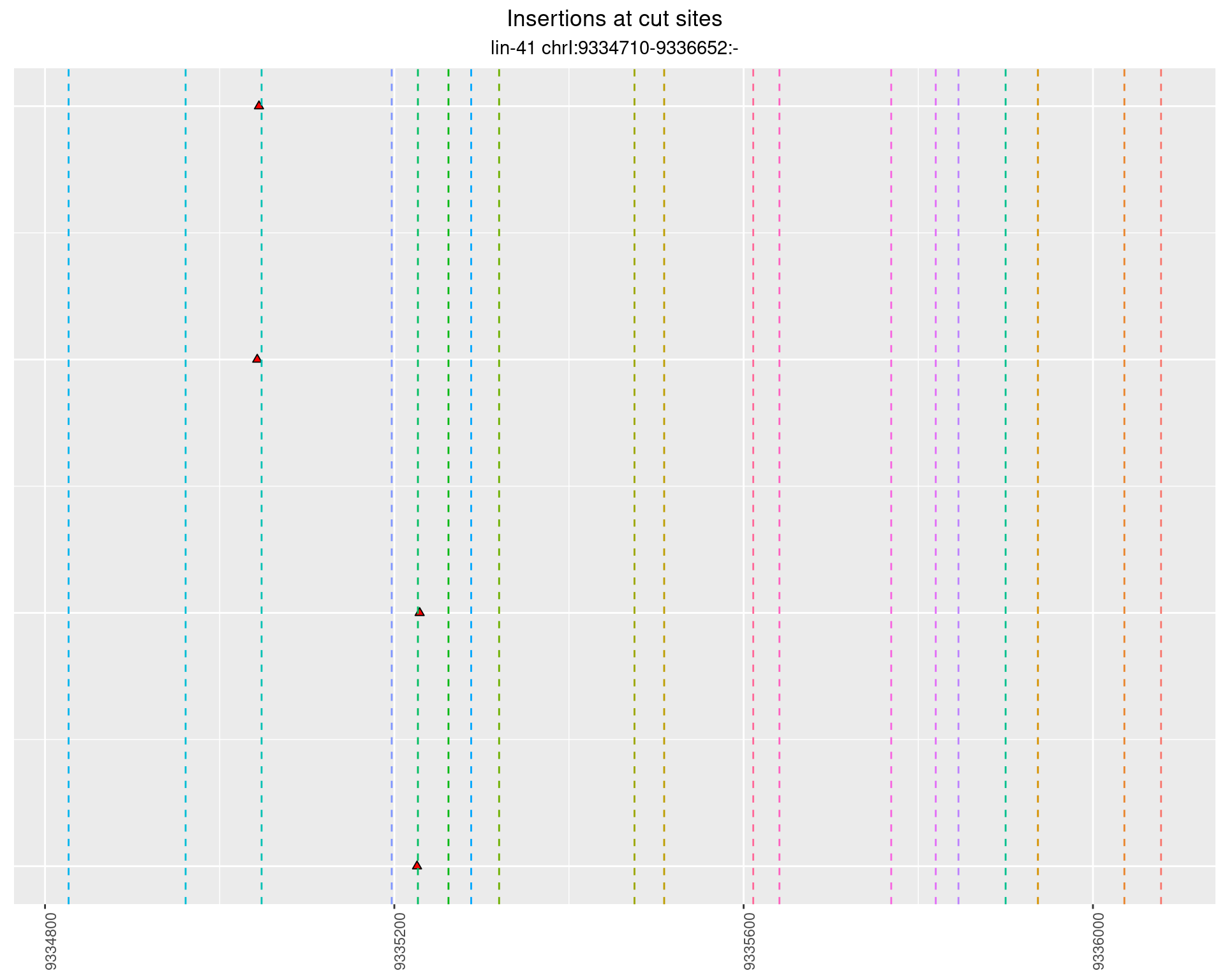
2.68 lin41_RNA_L4_1516_3end_UMI
2.68.1 Freq: 1e-04 - 0.001
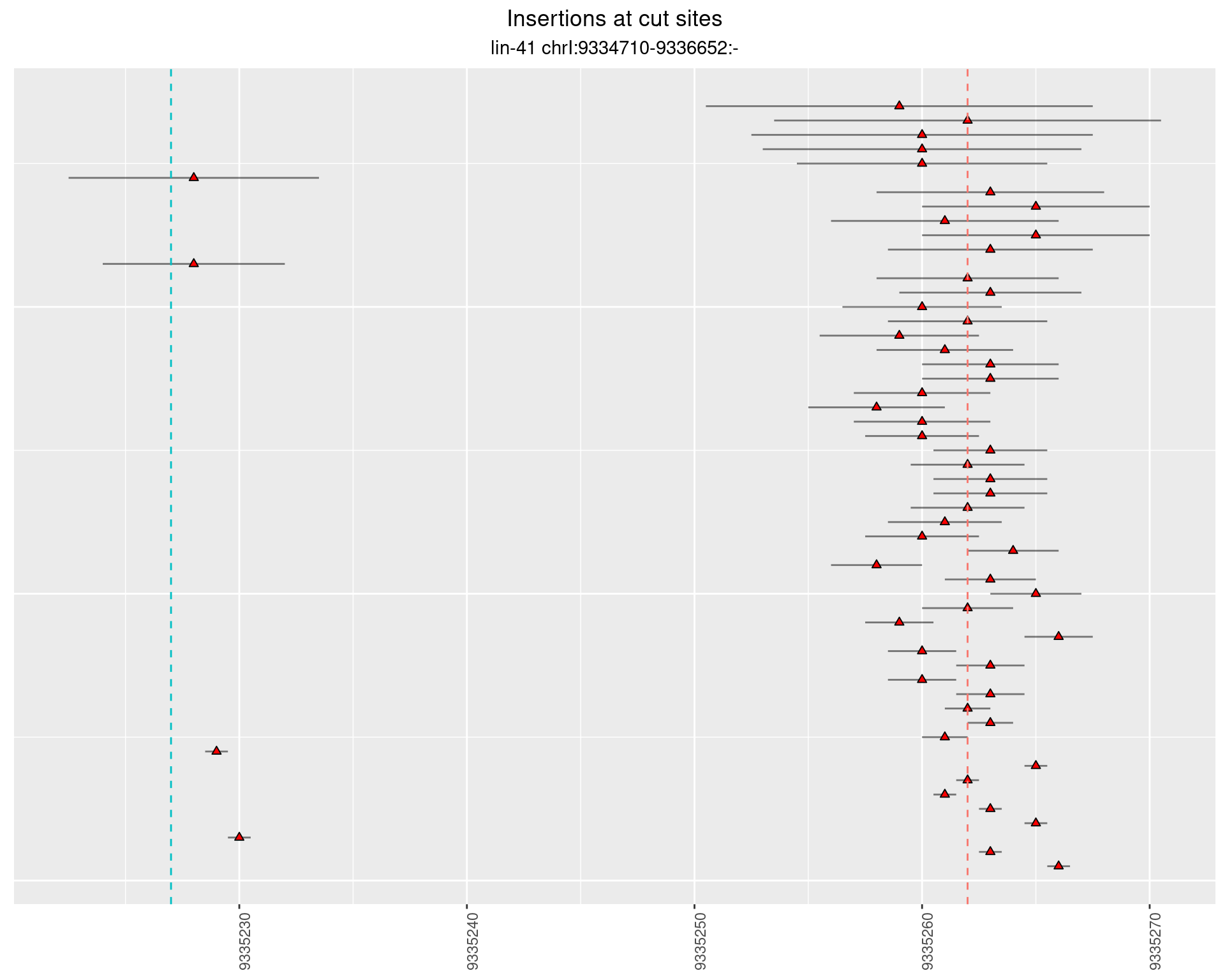
2.68.2 Freq: 0.001 - 0.01
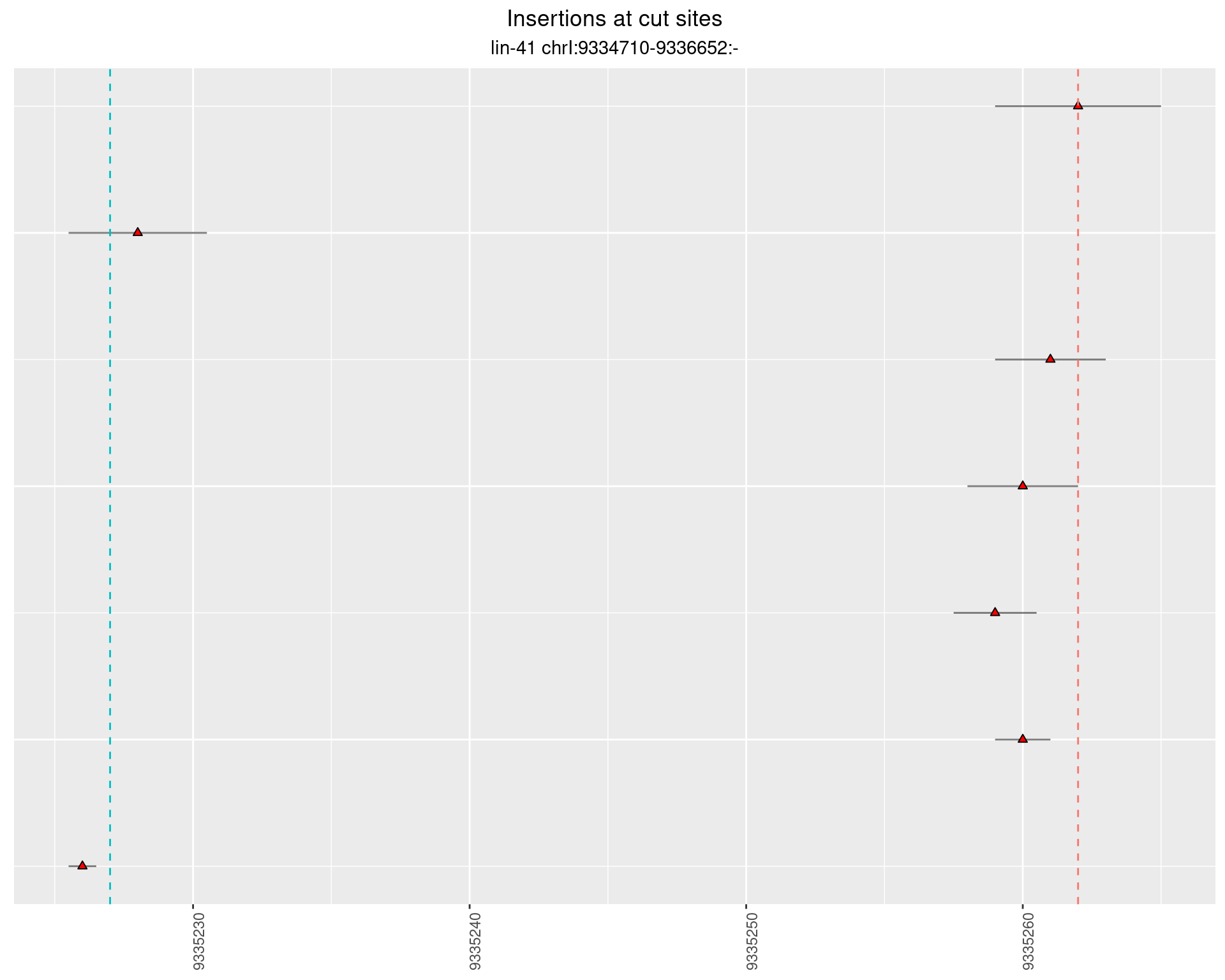
2.69 lin41_RNA_L4_2627_3end_UMI
2.69.1 Freq: 1e-04 - 0.001
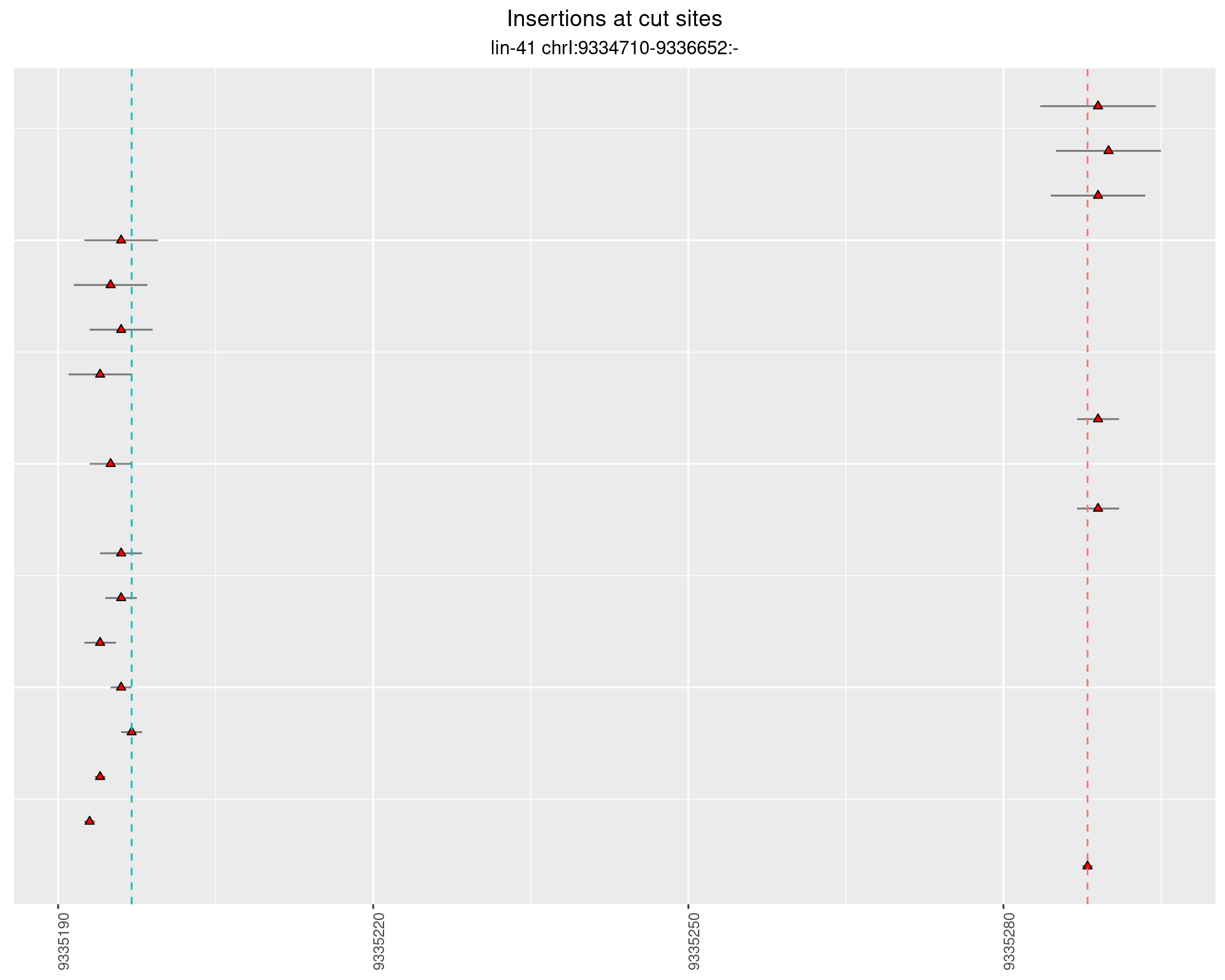
2.69.2 Freq: 0.001 - 0.01
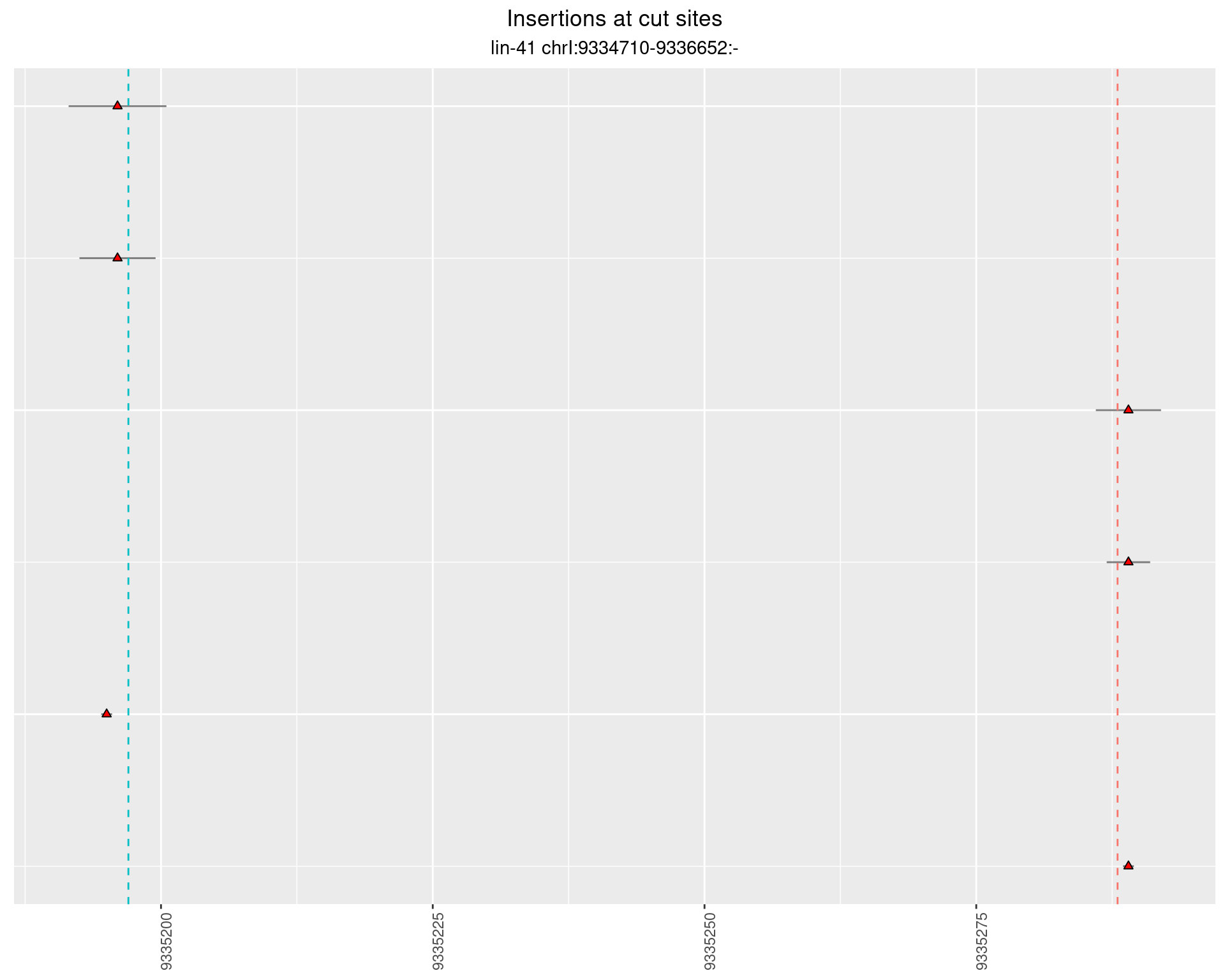
2.70 lin41_RNA_L4_pool2_3end_UMI
2.70.1 Freq: 1e-04 - 0.001
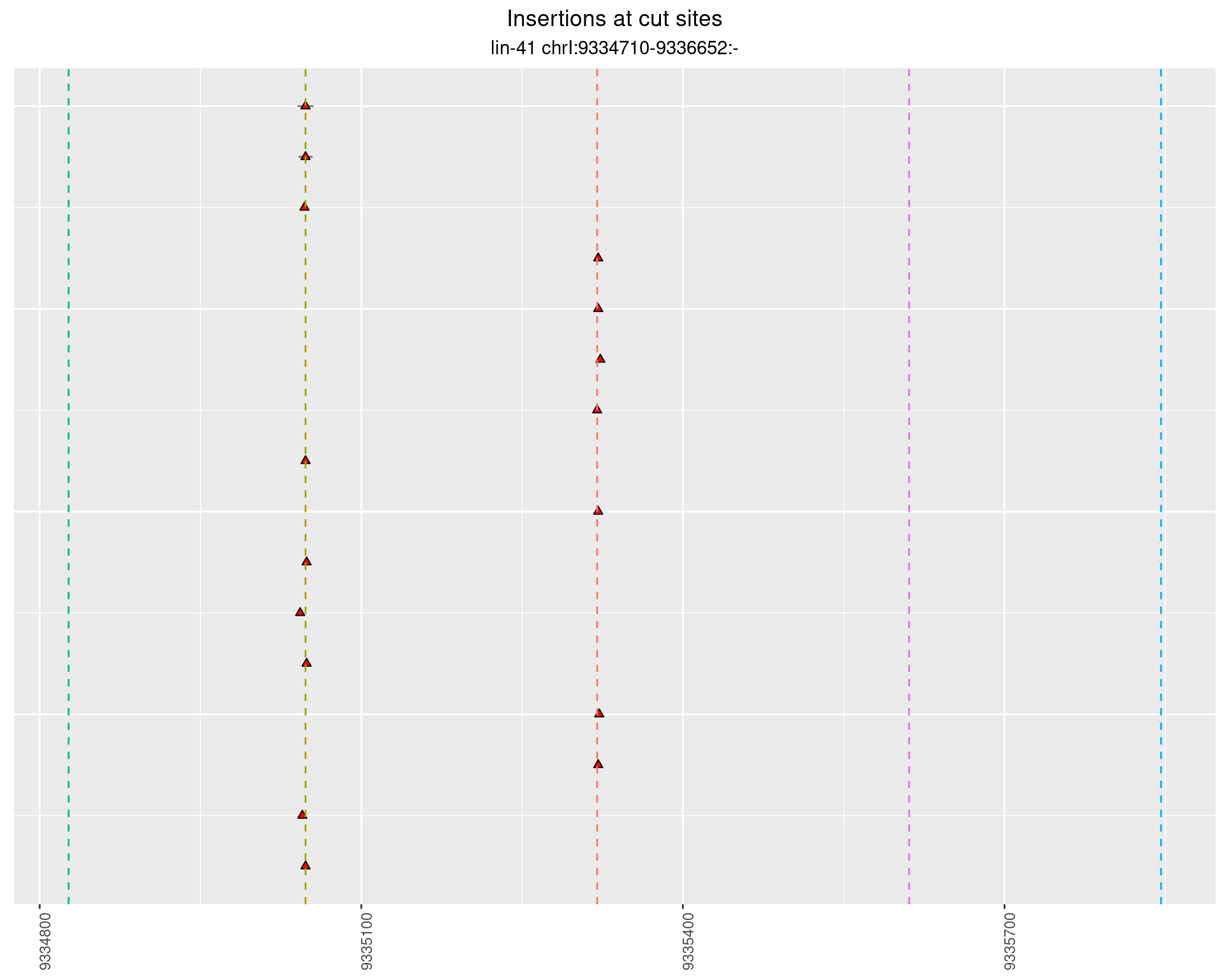
2.70.2 Freq: 0.001 - 0.01
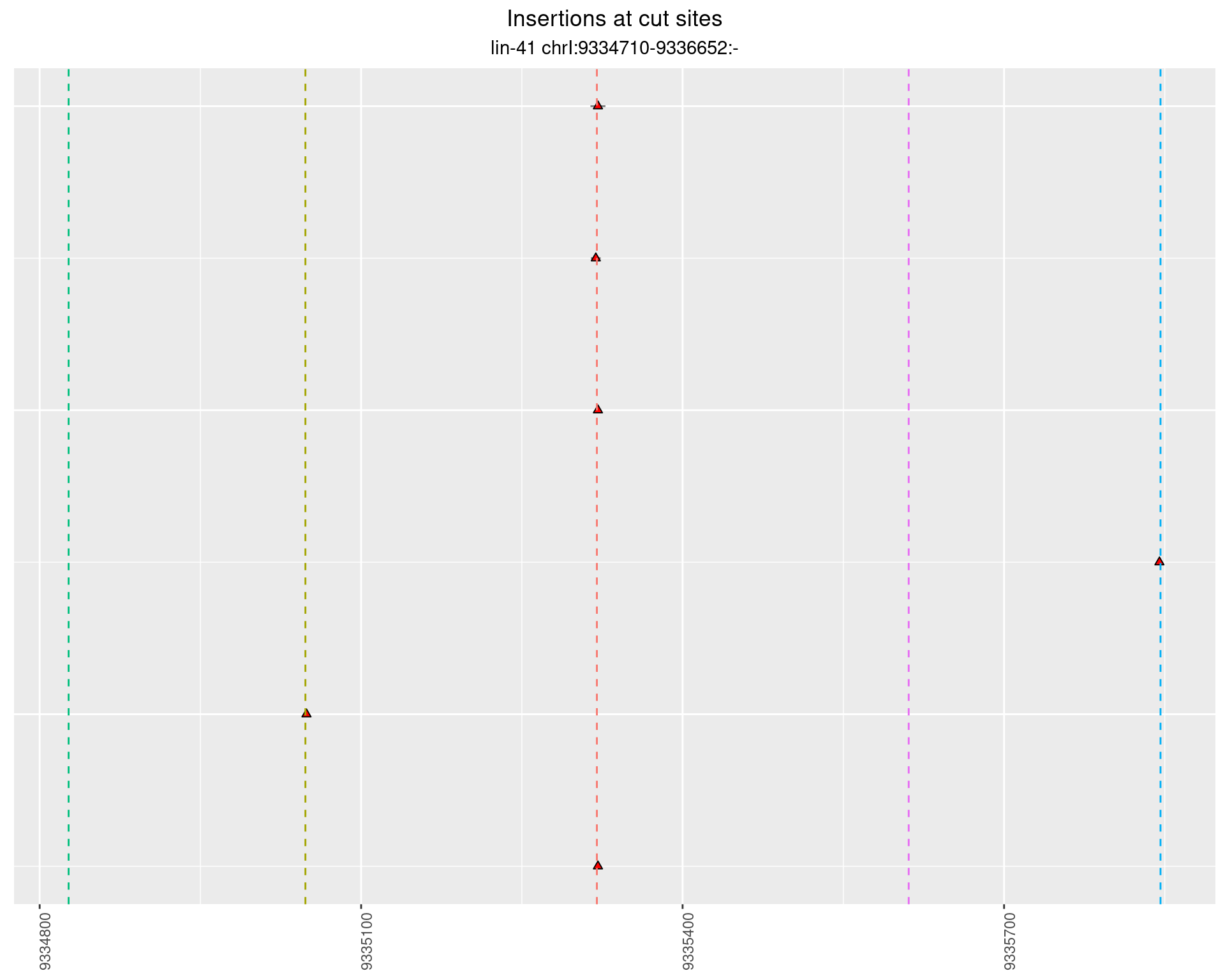
2.70.3 Freq: 1e-05 - 1e-04
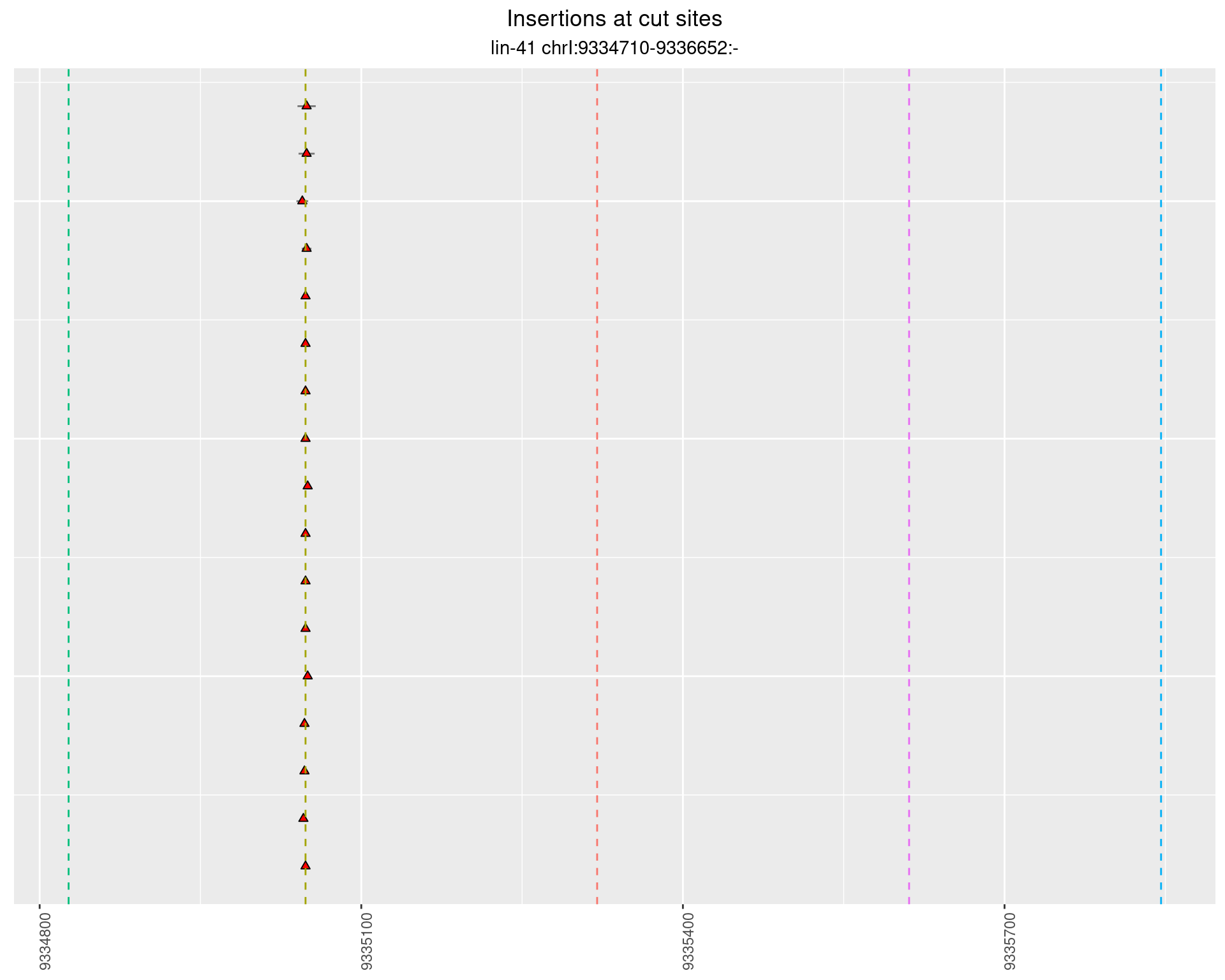
2.70.4 Freq: 0.01 - 0.1
2.71 lin41_RNA_L4_pool3_3end_UMI
2.71.1 Freq: 1e-04 - 0.001
2.71.2 Freq: 0.001 - 0.01
2.71.3 Freq: 0.01 - 0.1
2.72 lin41_RNA_L1_N2_middle_UMI
2.72.1 Freq: 1e-04 - 0.001
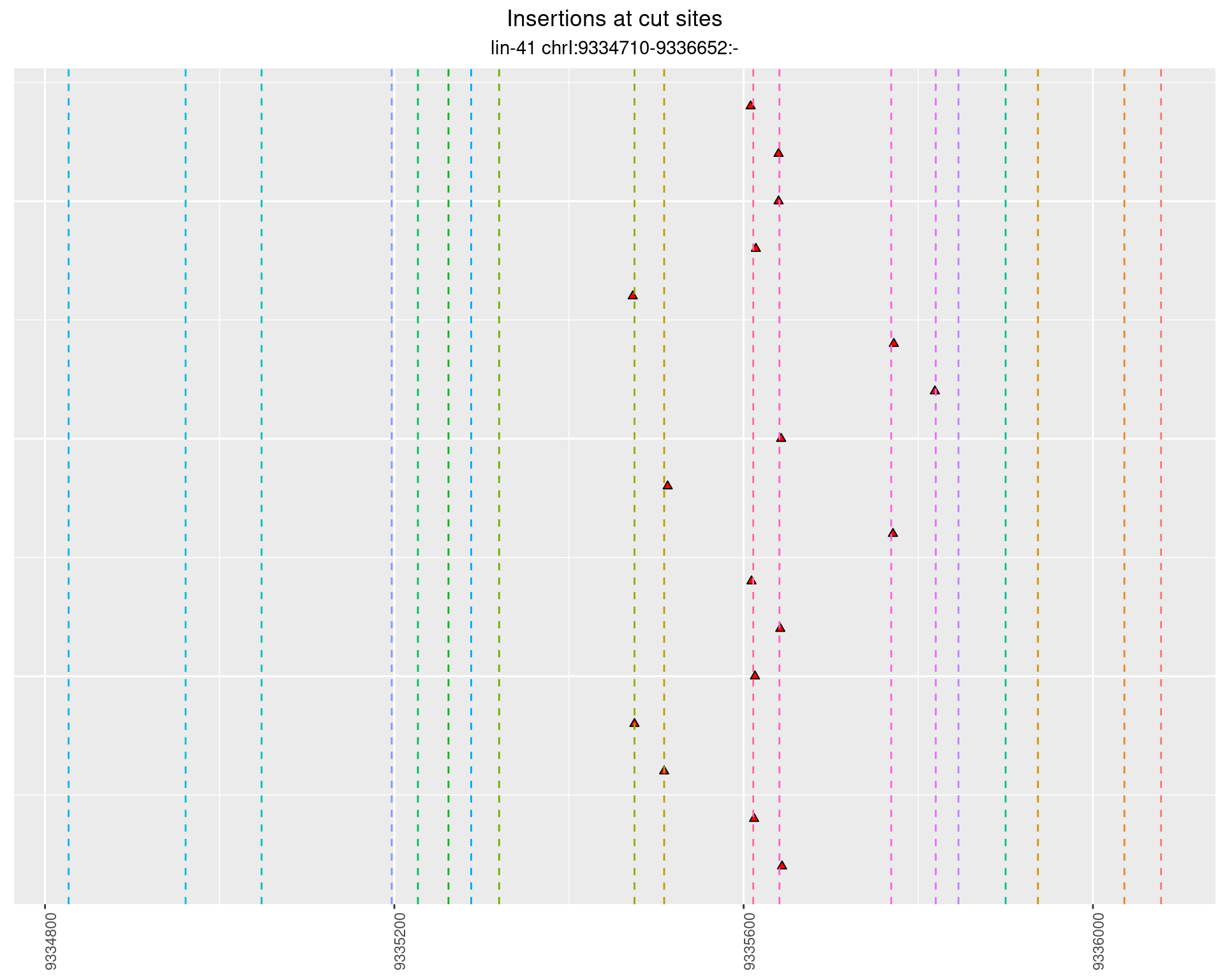
2.72.2 Freq: 0.01 - 0.1
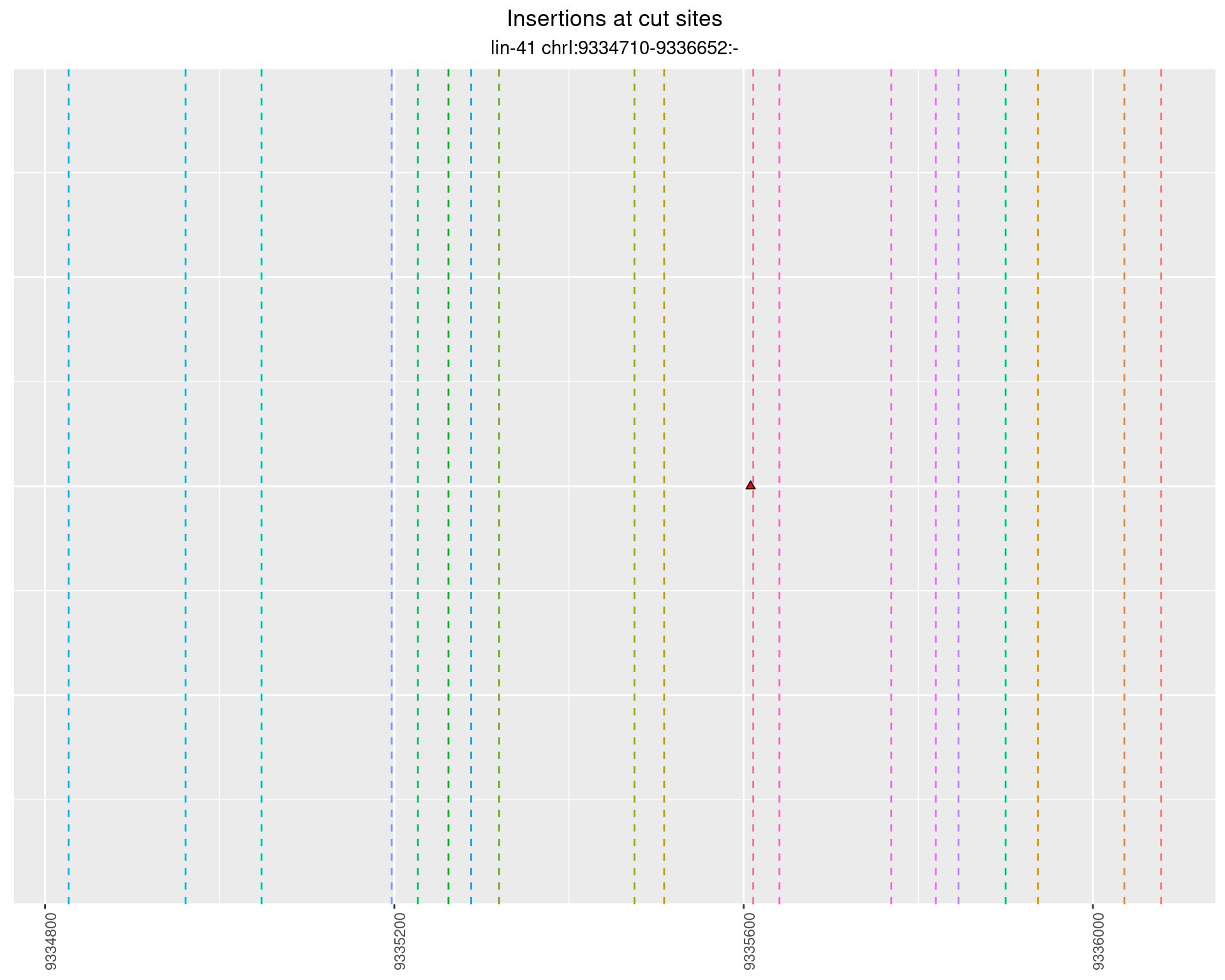
2.72.3 Freq: 1e-05 - 1e-04
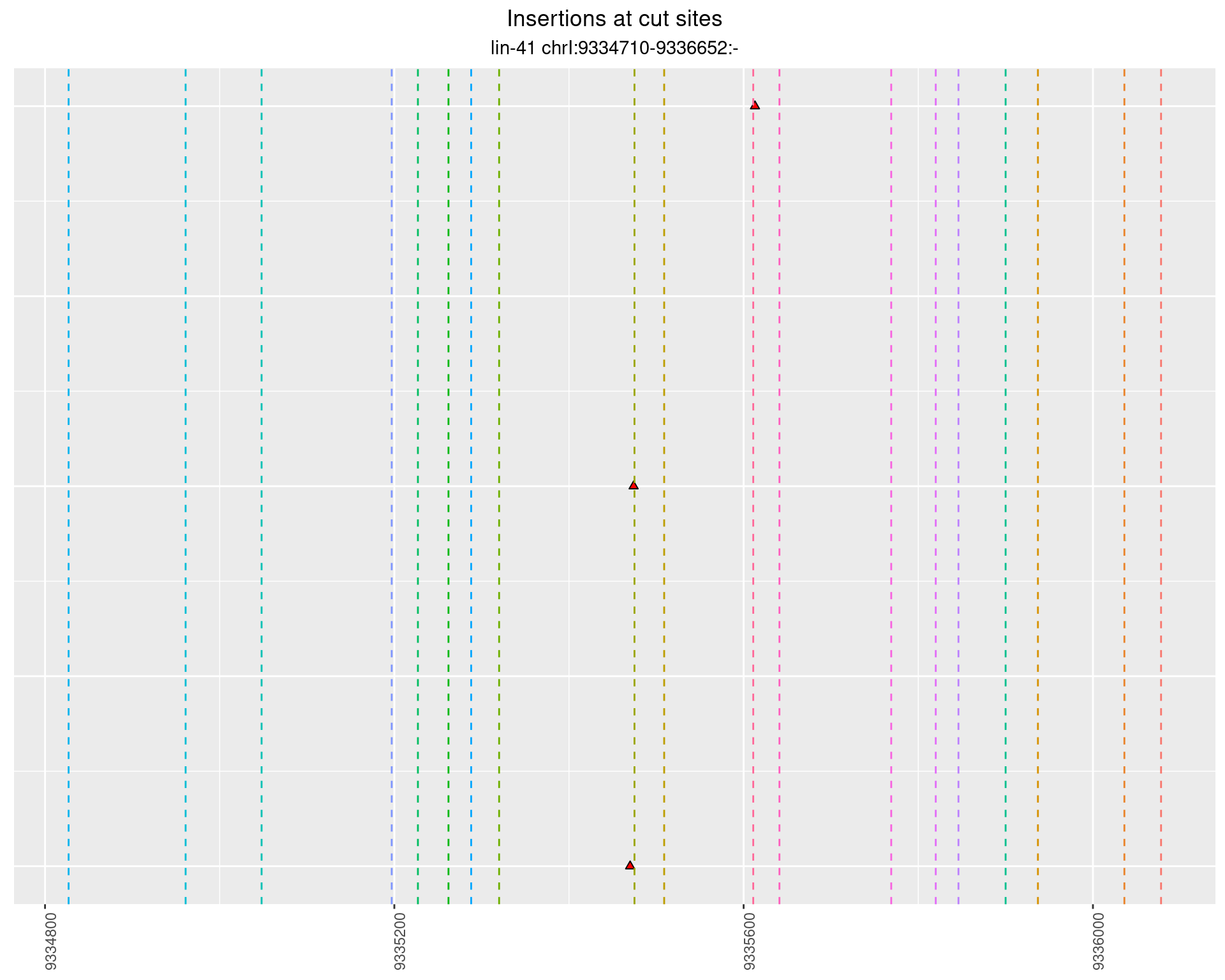
2.72.4 Freq: 0.001 - 0.01
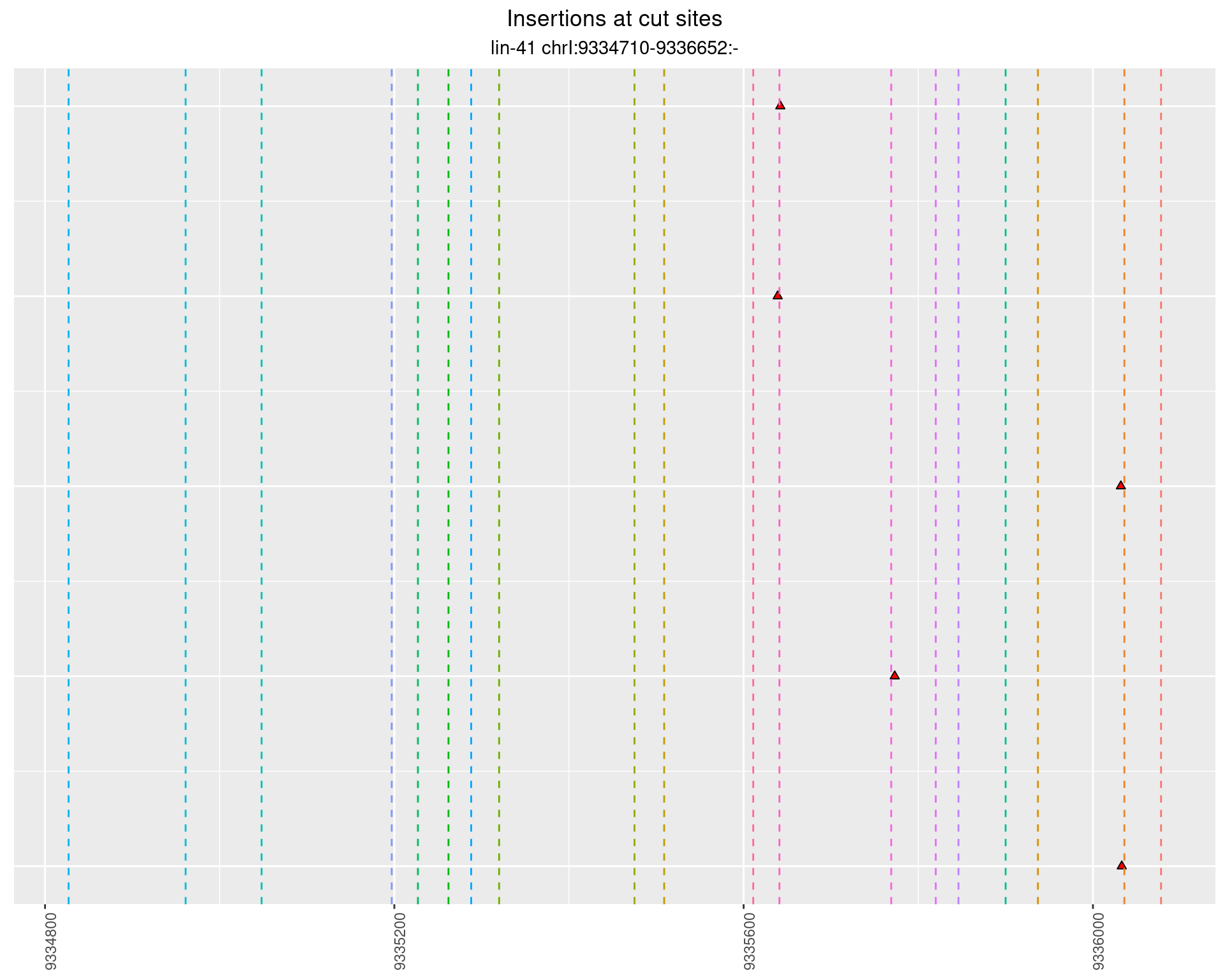
2.73 lin41_RNA_L1_1516_middle_UMI
2.74 lin41_RNA_L1_2627_middle_UMI
2.75 lin41_RNA_L1_pool2_middle_UMI
2.75.1 Freq: 0.01 - 0.1
2.75.2 Freq: 1e-04 - 0.001
2.75.3 Freq: 1e-05 - 1e-04
2.75.4 Freq: 0.001 - 0.01
2.76 lin41_RNA_L1_pool3_middle_UMI
2.76.1 Freq: 1e-04 - 0.001
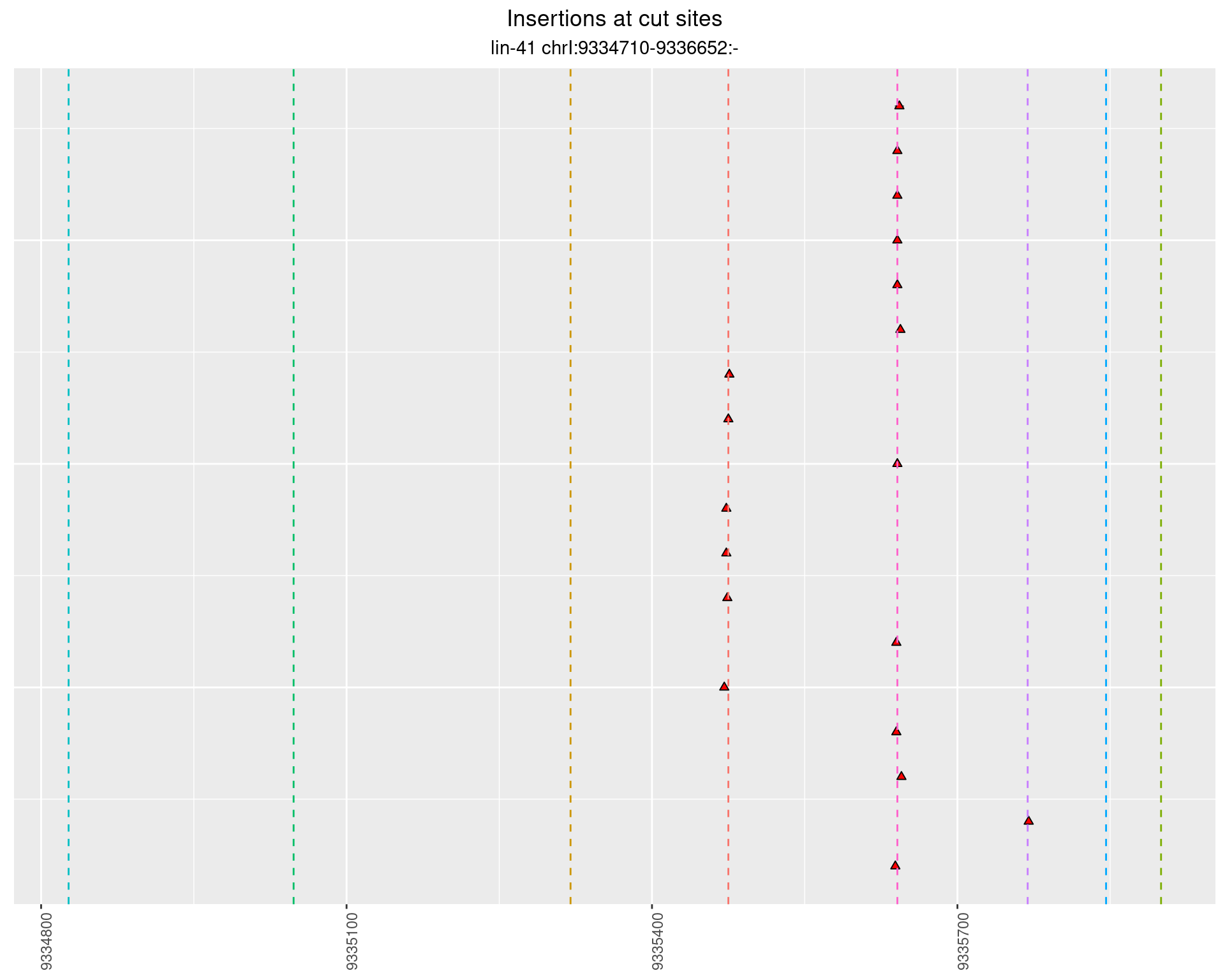
2.76.2 Freq: 0.001 - 0.01
2.77 lin41_RNA_L4_N2_middle_UMI
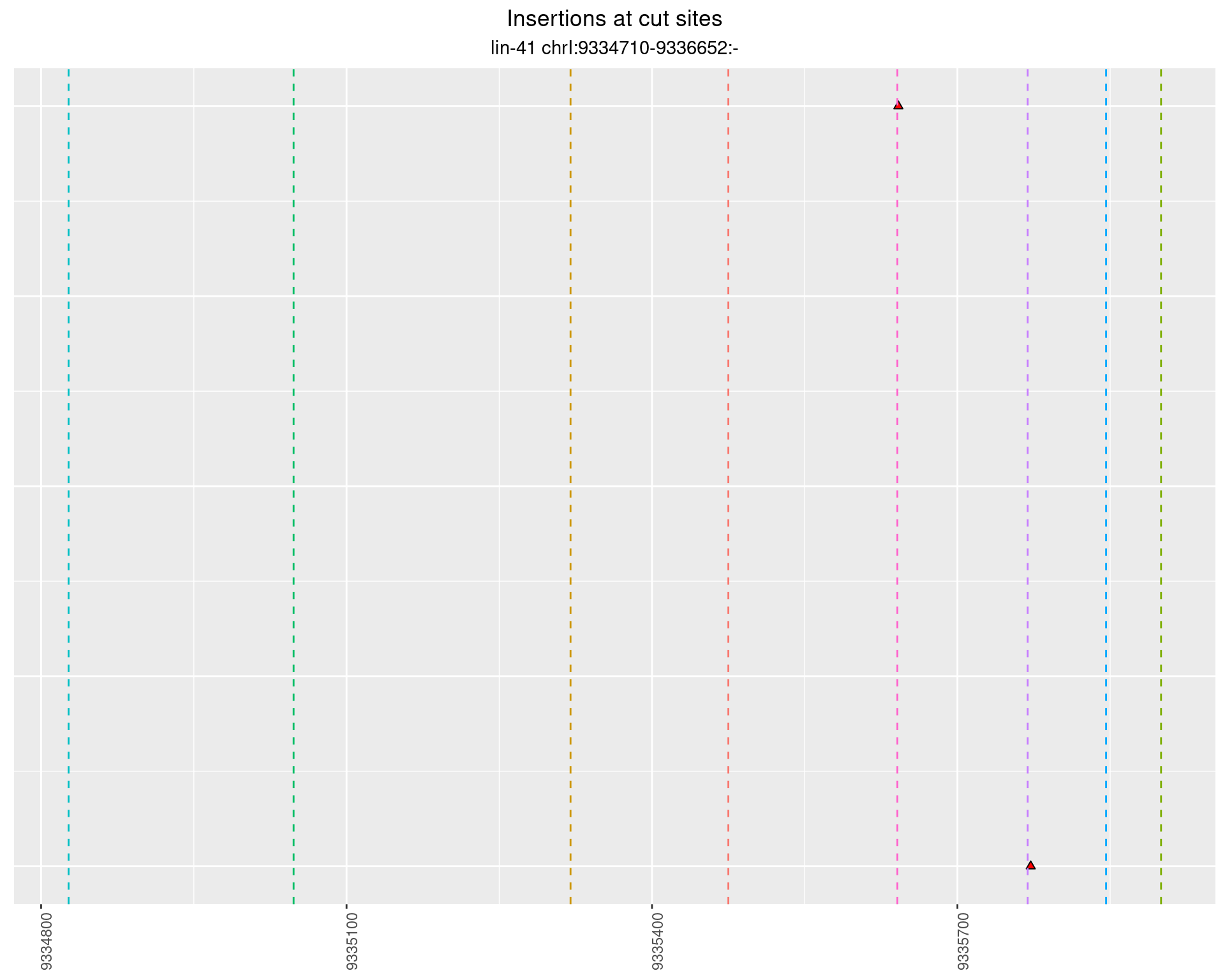
2.77.1 Freq: 0.01 - 0.1
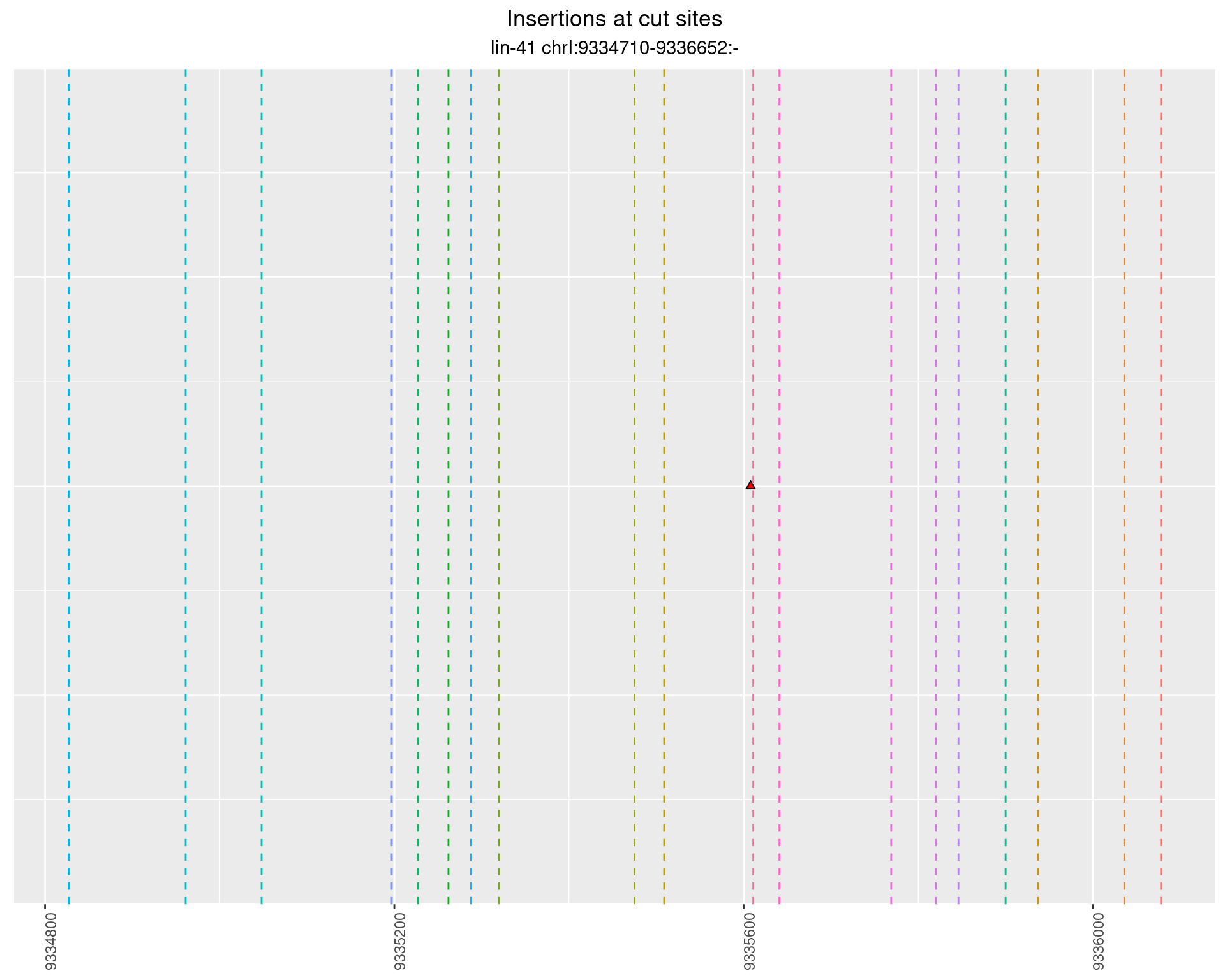
2.77.2 Freq: 1e-04 - 0.001
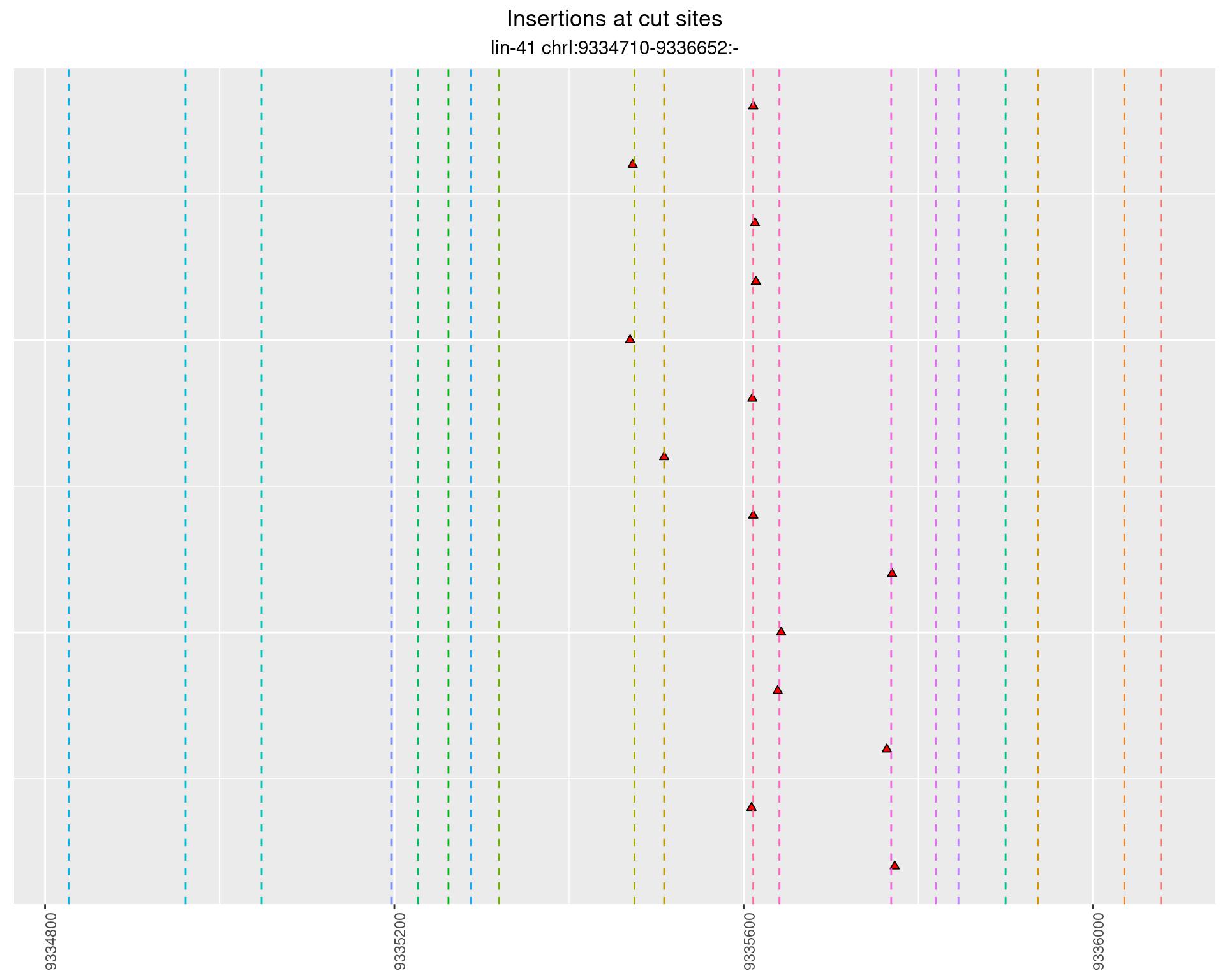
2.77.3 Freq: 0.001 - 0.01
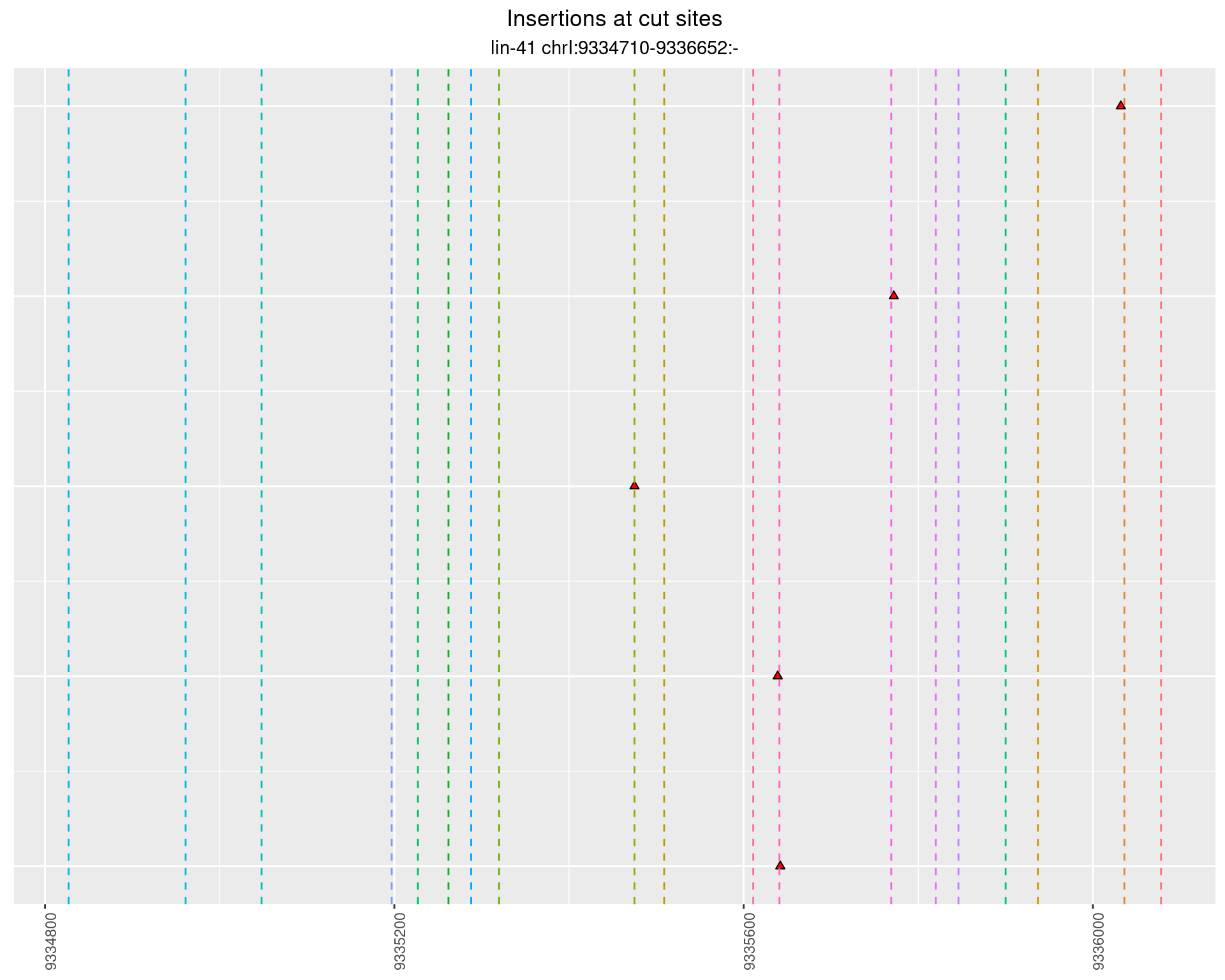
2.77.4 Freq: 1e-05 - 1e-04
2.78 lin41_RNA_L4_1516_middle_UMI
2.79 lin41_RNA_L4_2627_middle_UMI
2.80 lin41_RNA_L4_pool2_middle_UMI
2.80.1 Freq: 0.01 - 0.1
2.80.2 Freq: 1e-04 - 0.001
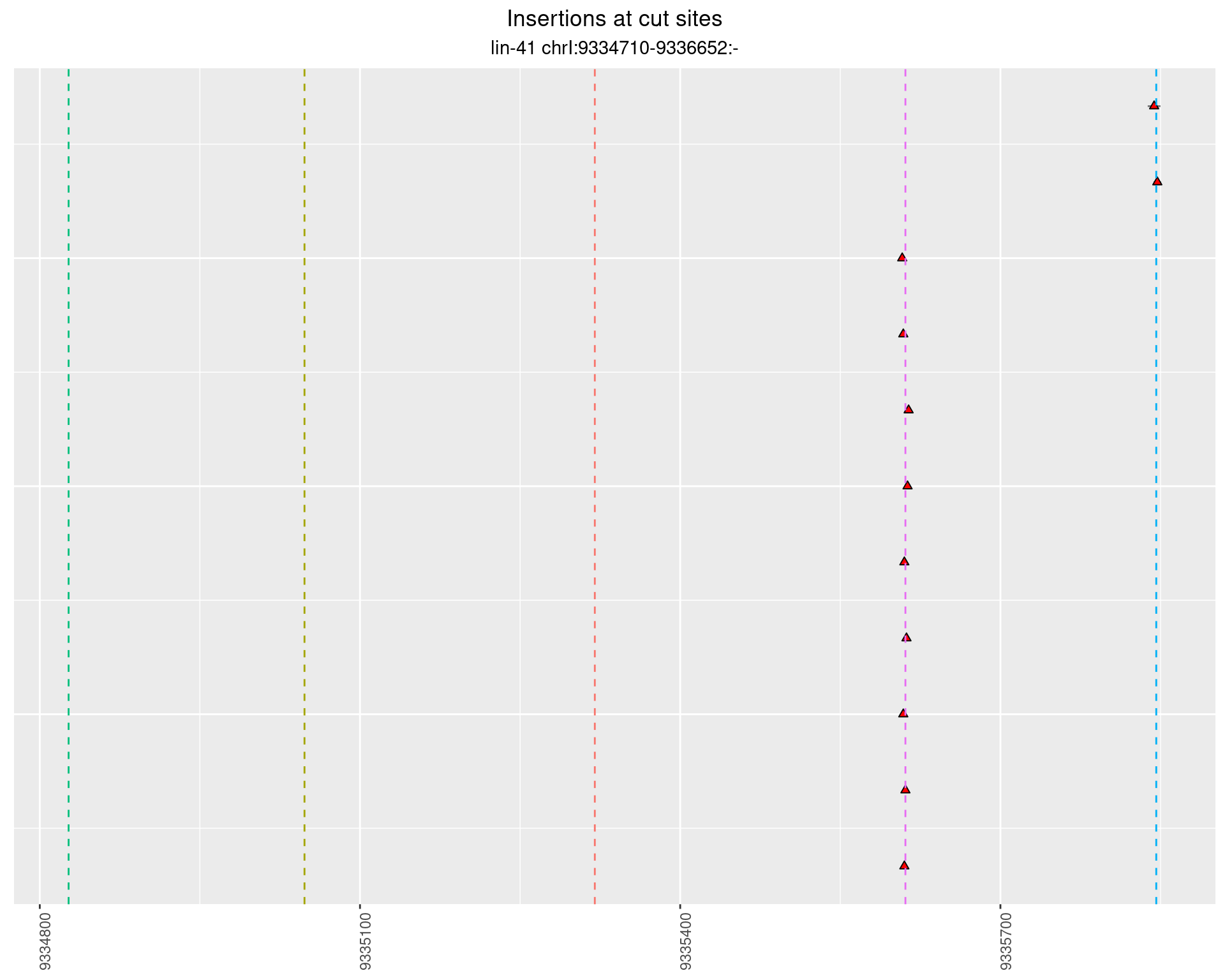
2.81 lin41_RNA_L4_pool3_middle_UMI
2.81.1 Freq: 1e-04 - 0.001
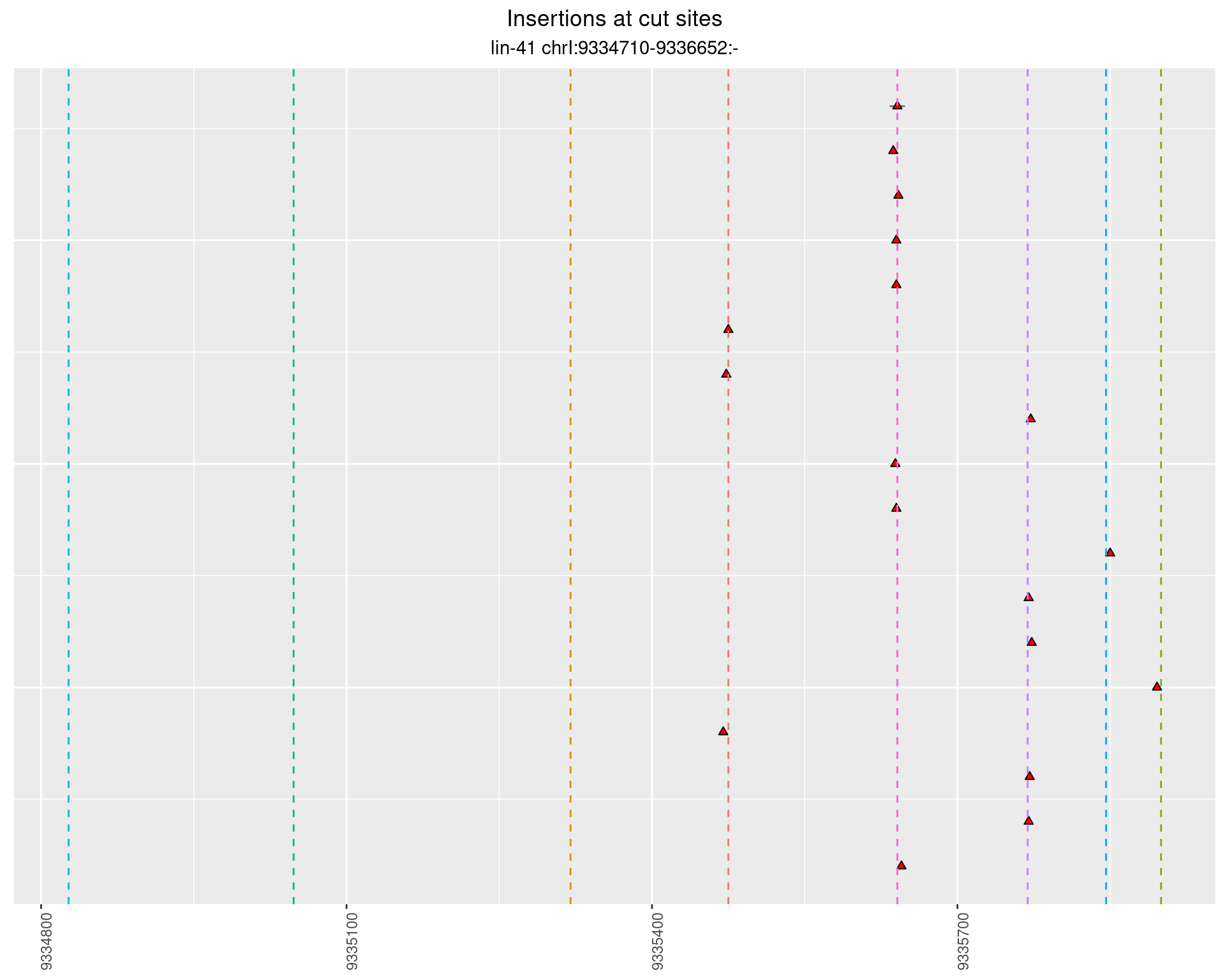
2.81.2 Freq: 0.001 - 0.01
2.82 lin41_RNA_L1_N2_3end_UMI_R2
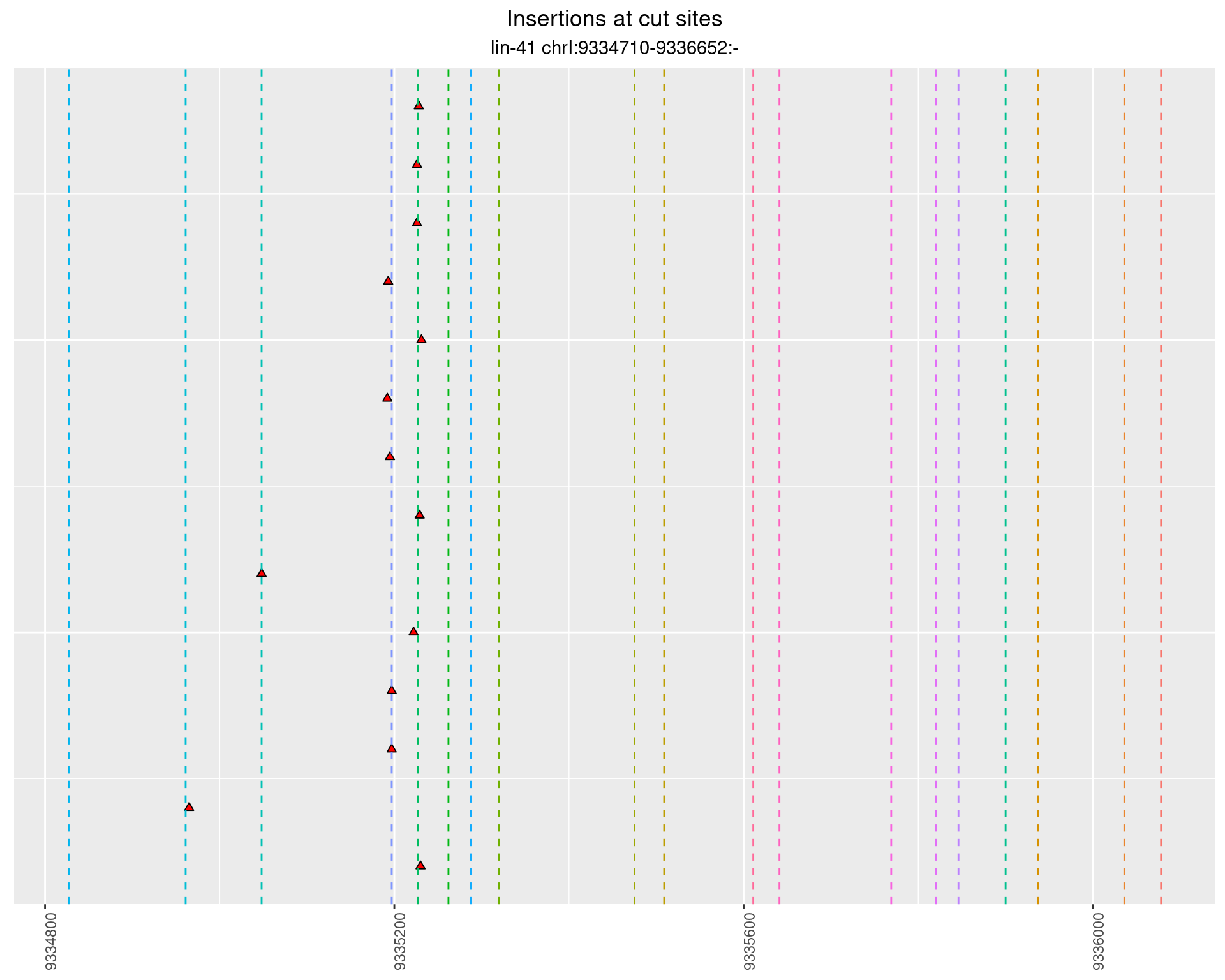
2.82.1 Freq: 0.001 - 0.01
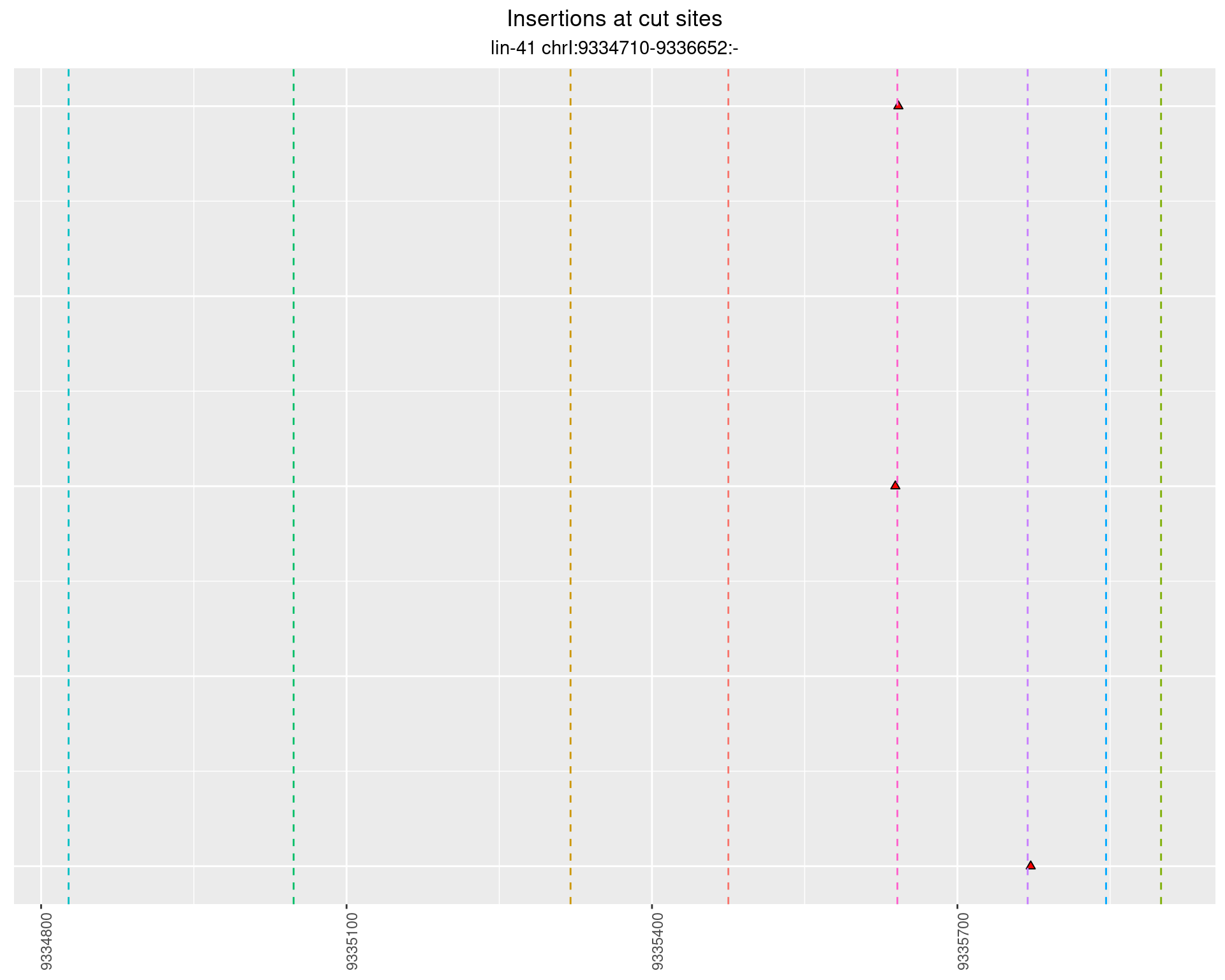
2.82.2 Freq: 1e-05 - 1e-04
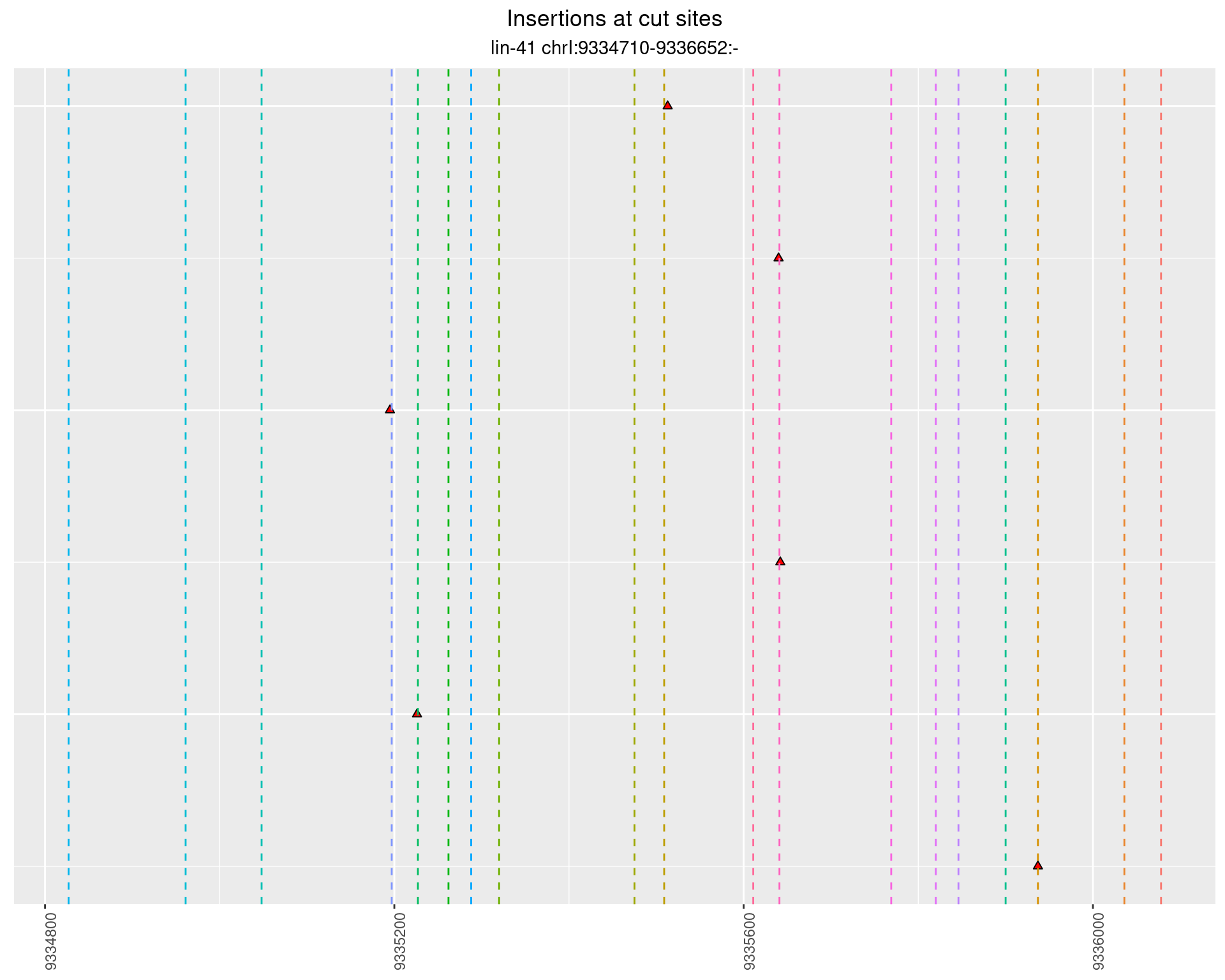
2.82.3 Freq: 1e-04 - 0.001
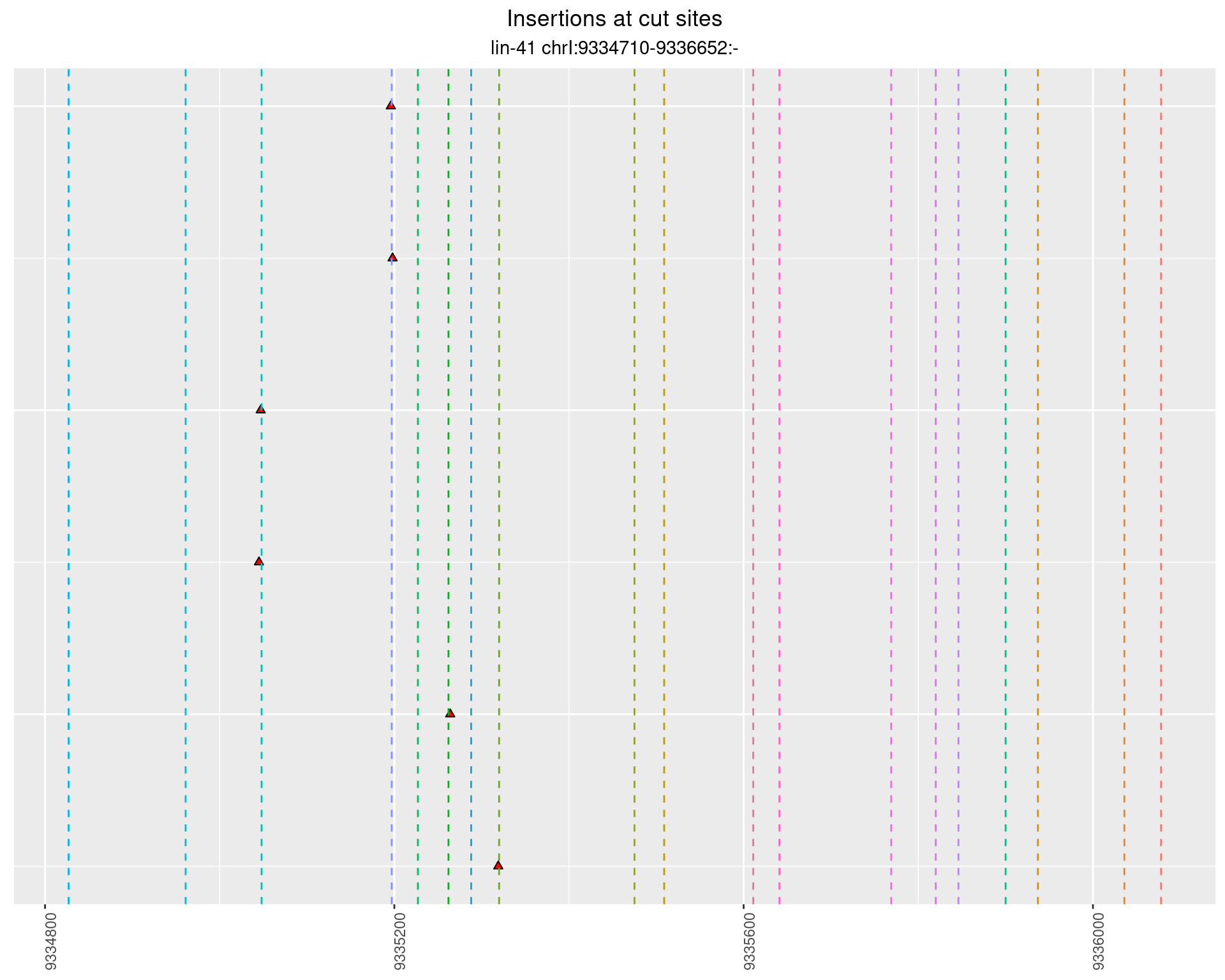
2.82.4 Freq: 0.01 - 0.1
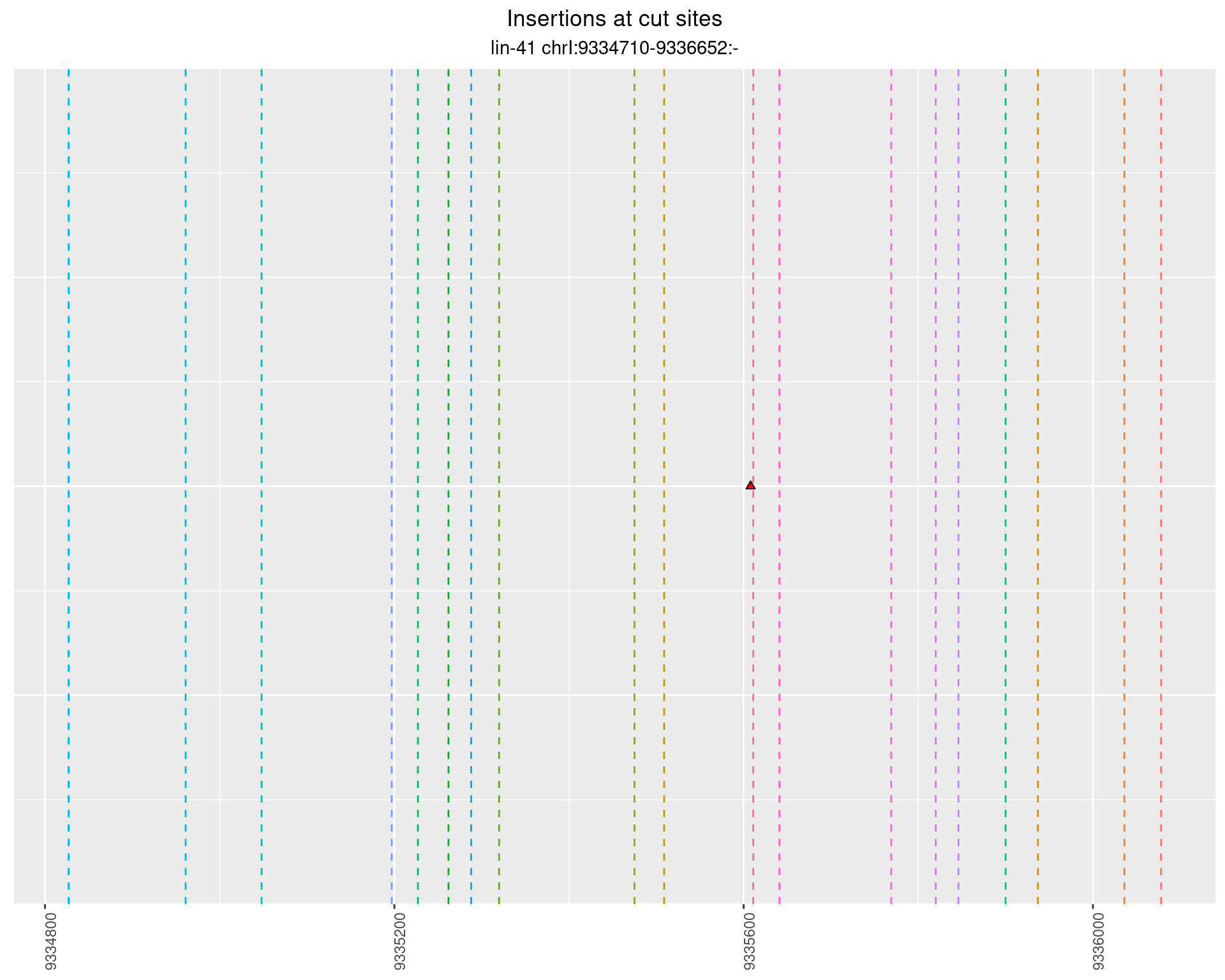
2.83 lin41_RNA_L1_1516_3end_UMI_R2
2.83.1 Freq: 1e-04 - 0.001
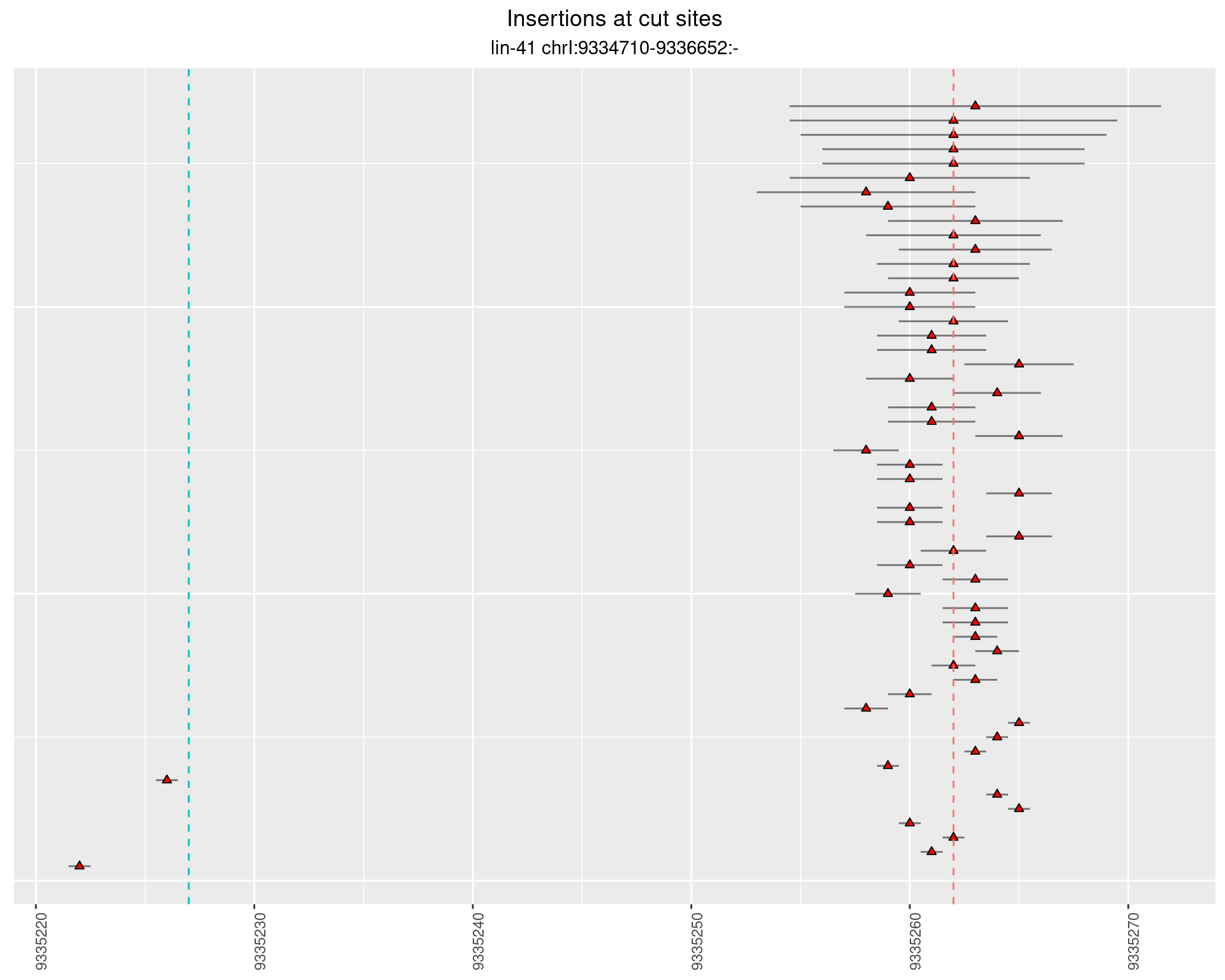
2.83.2 Freq: 1e-05 - 1e-04
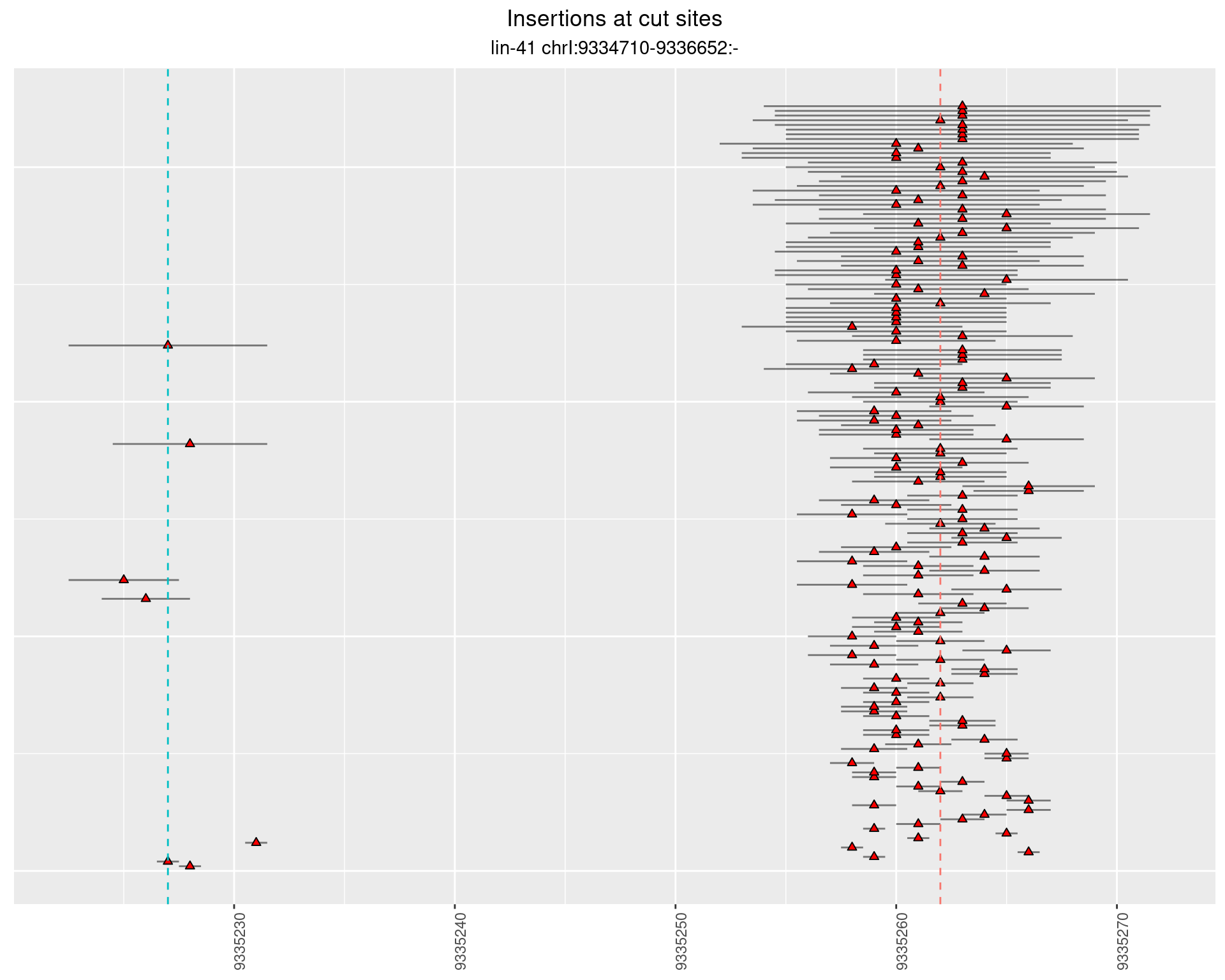
2.84 lin41_RNA_L1_2627_3end_UMI_R2
2.84.1 Freq: 1e-04 - 0.001
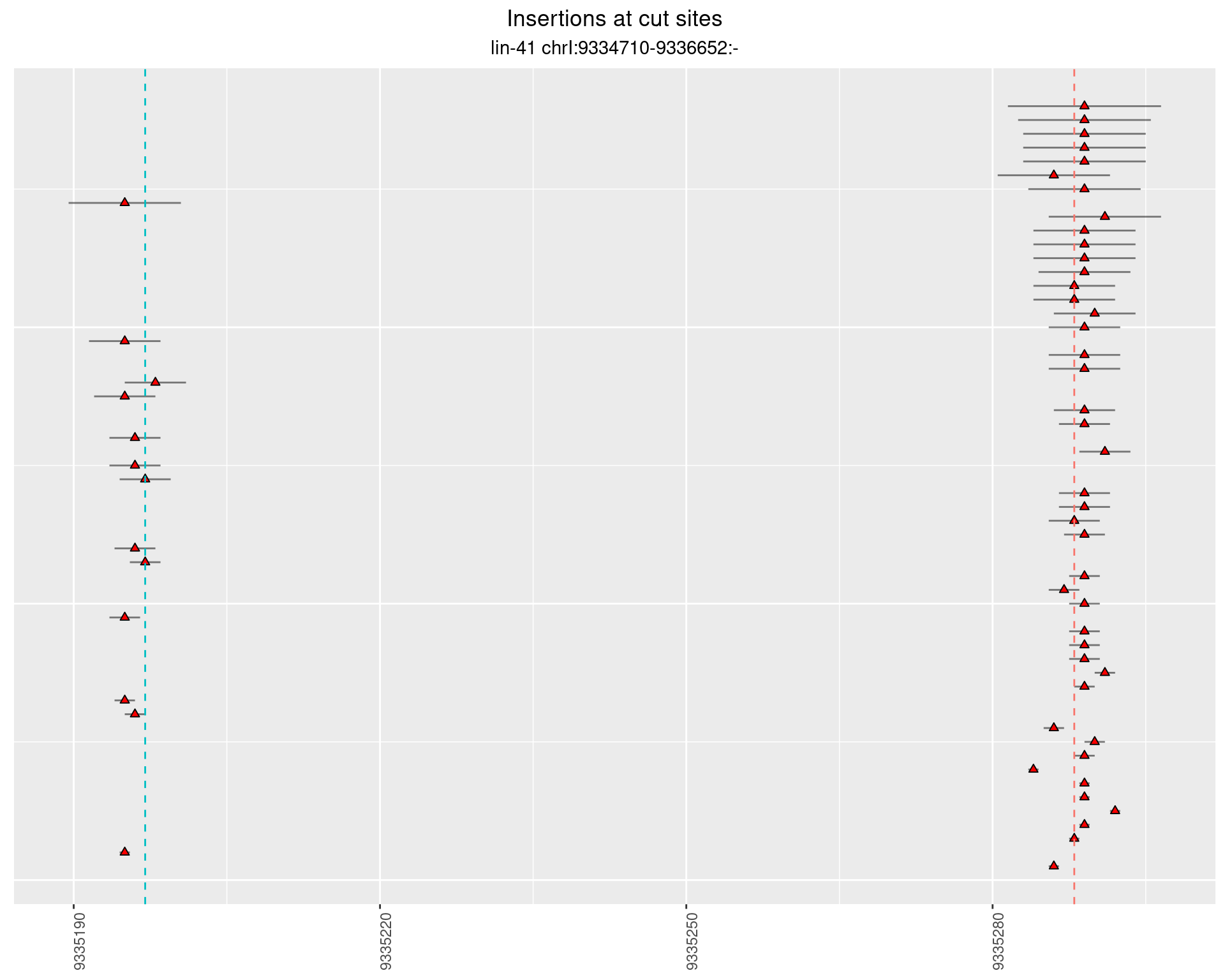
2.84.2 Freq: 1e-05 - 1e-04
2.84.3 Freq: 0.001 - 0.01
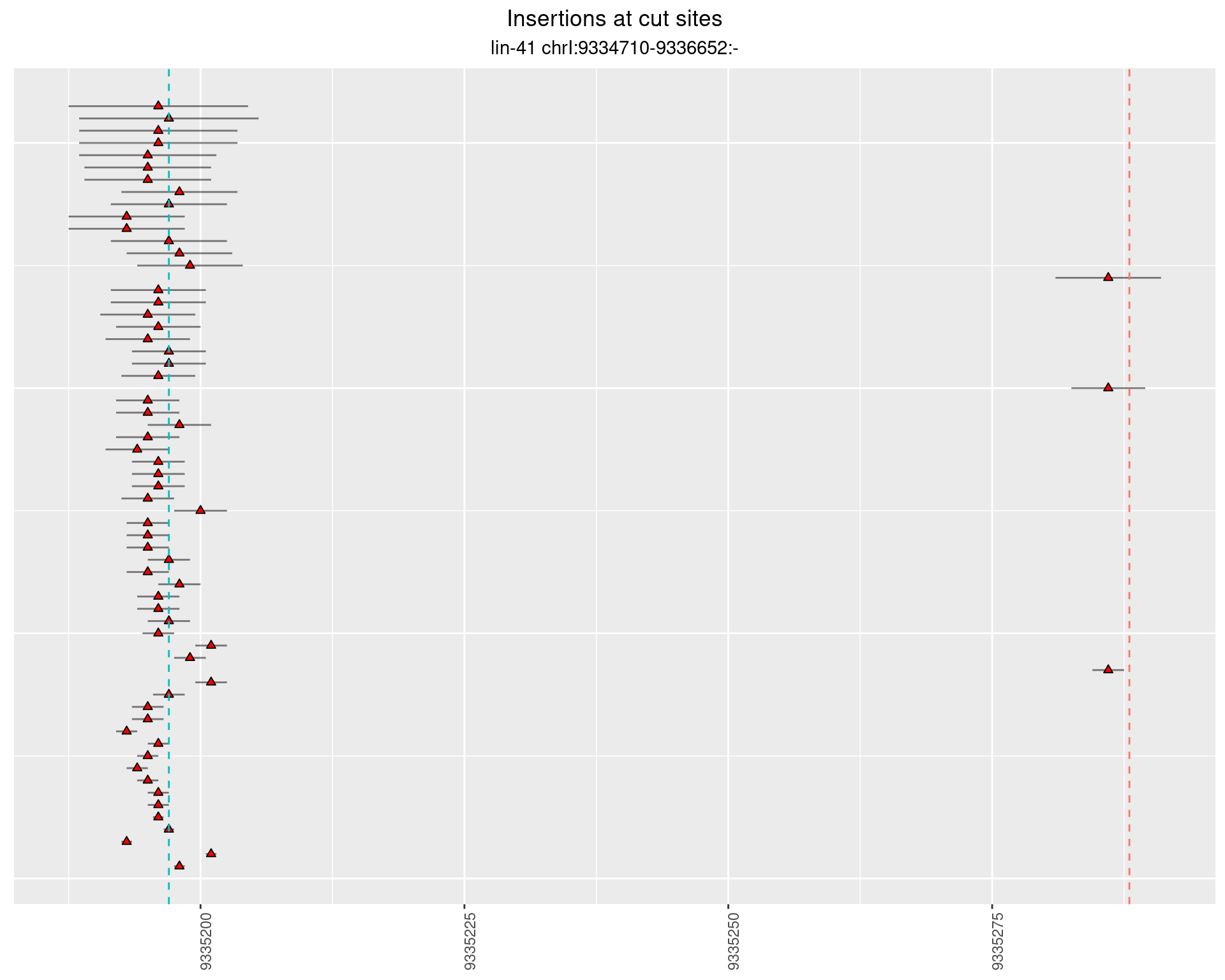
2.85 lin41_RNA_L1_pool2_3end_UMI_R2
2.85.1 Freq: 1e-05 - 1e-04
2.85.2 Freq: 1e-04 - 0.001
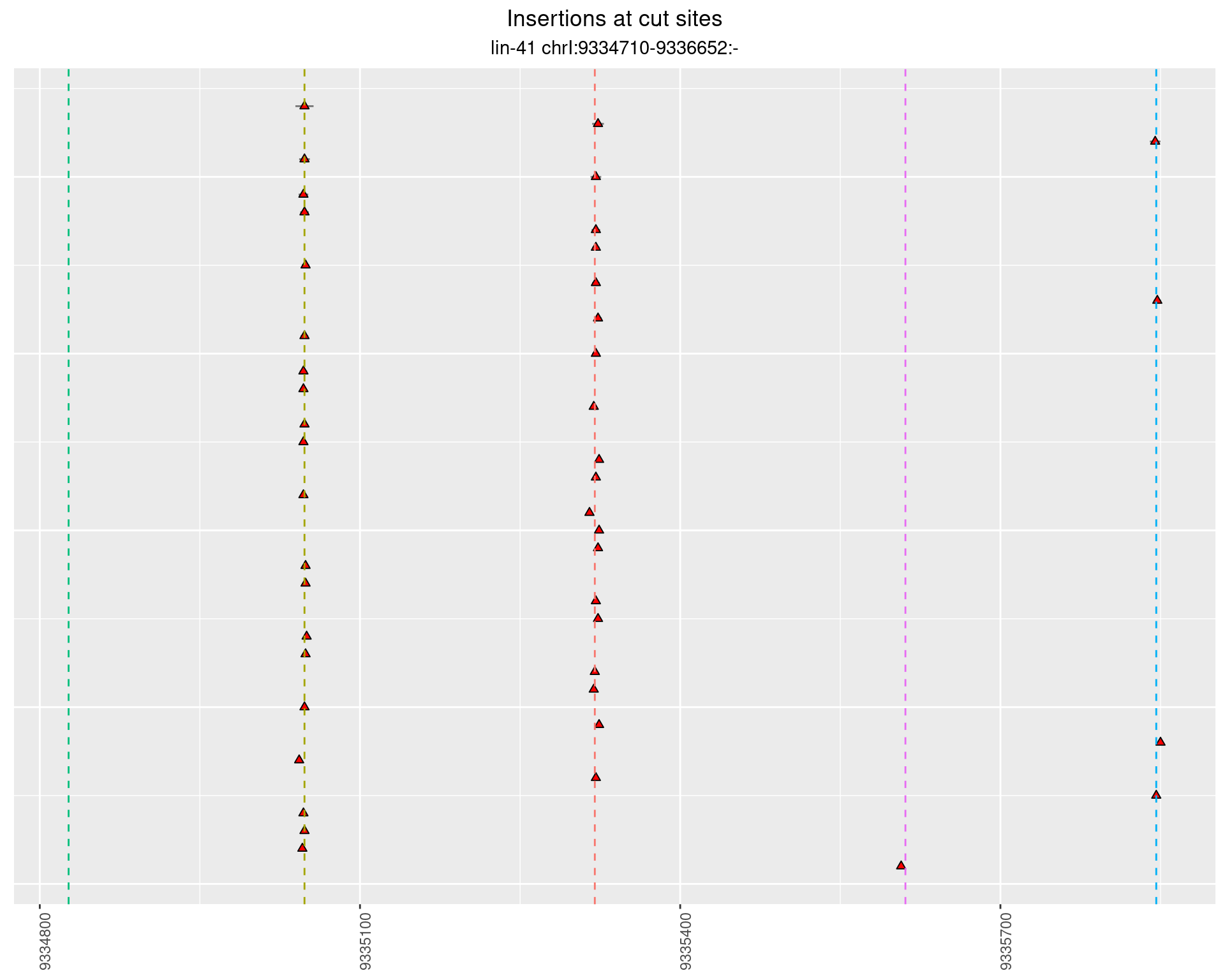
2.85.3 Freq: 0.001 - 0.01
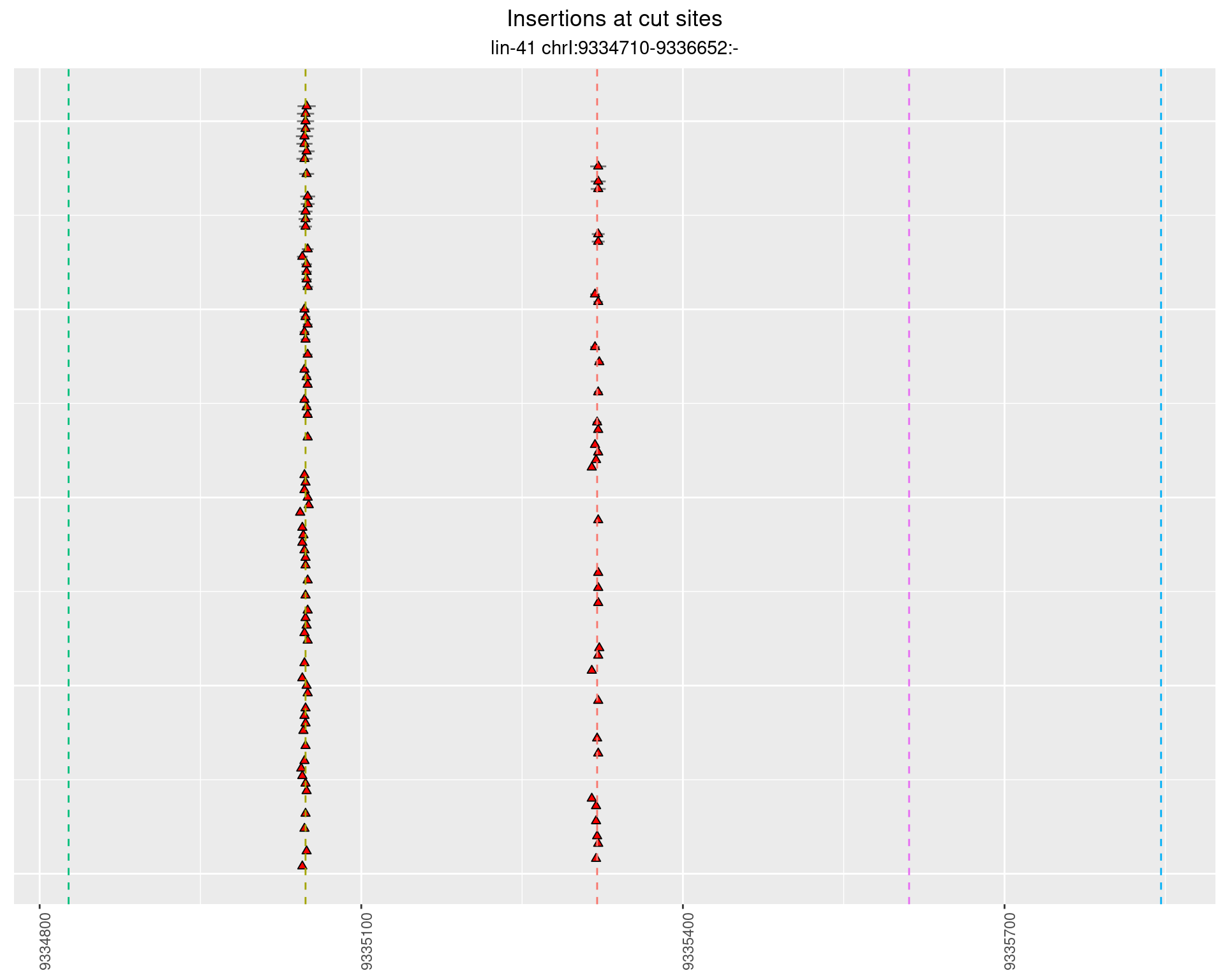
2.86 lin41_RNA_L1_pool3_3end_UMI_R2
2.86.1 Freq: 1e-04 - 0.001
2.86.2 Freq: 1e-05 - 1e-04
2.86.3 Freq: 0.001 - 0.01
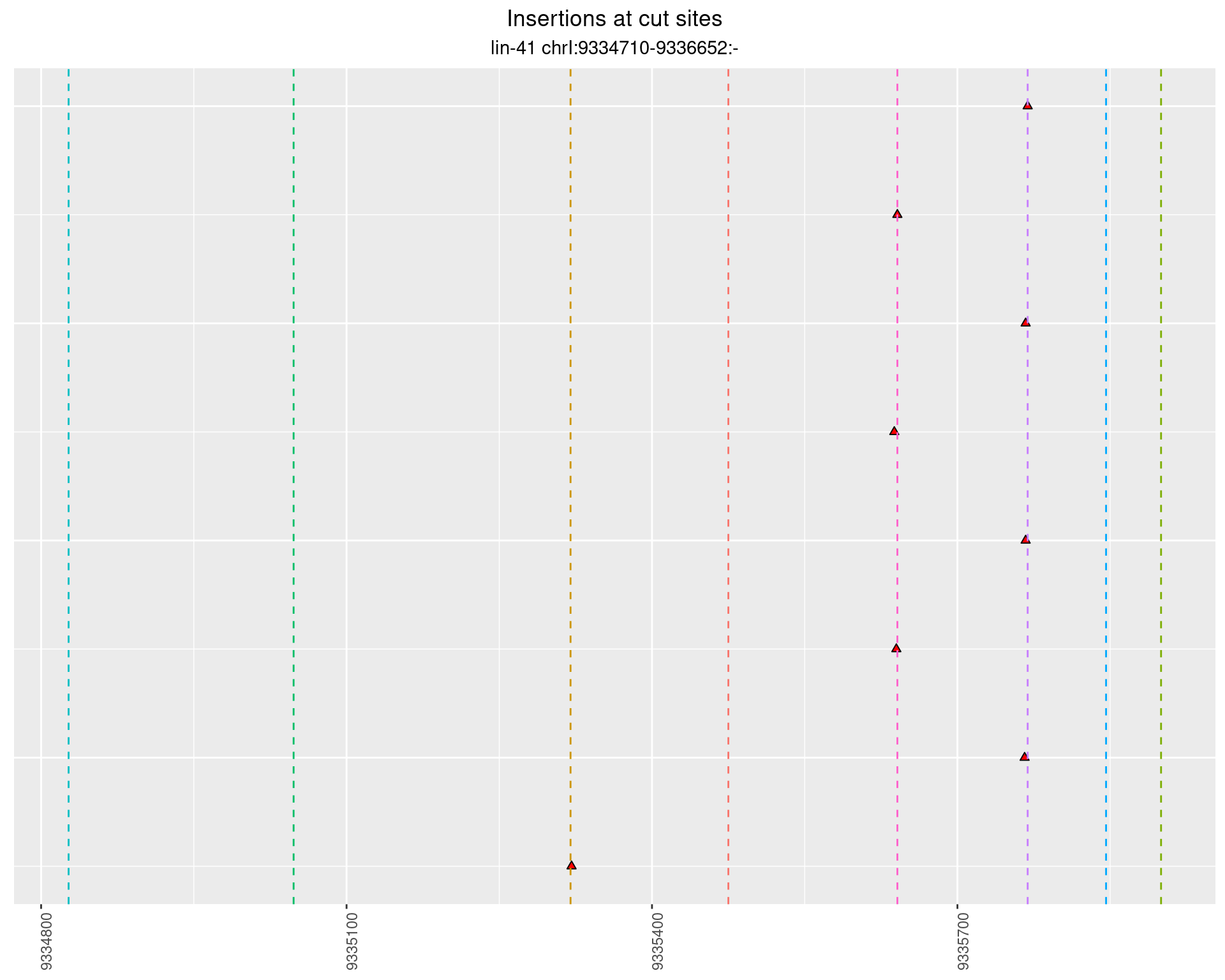
2.87 lin41_RNA_L4_N2_3end_UMI_R2
2.87.1 Freq: 0.001 - 0.01
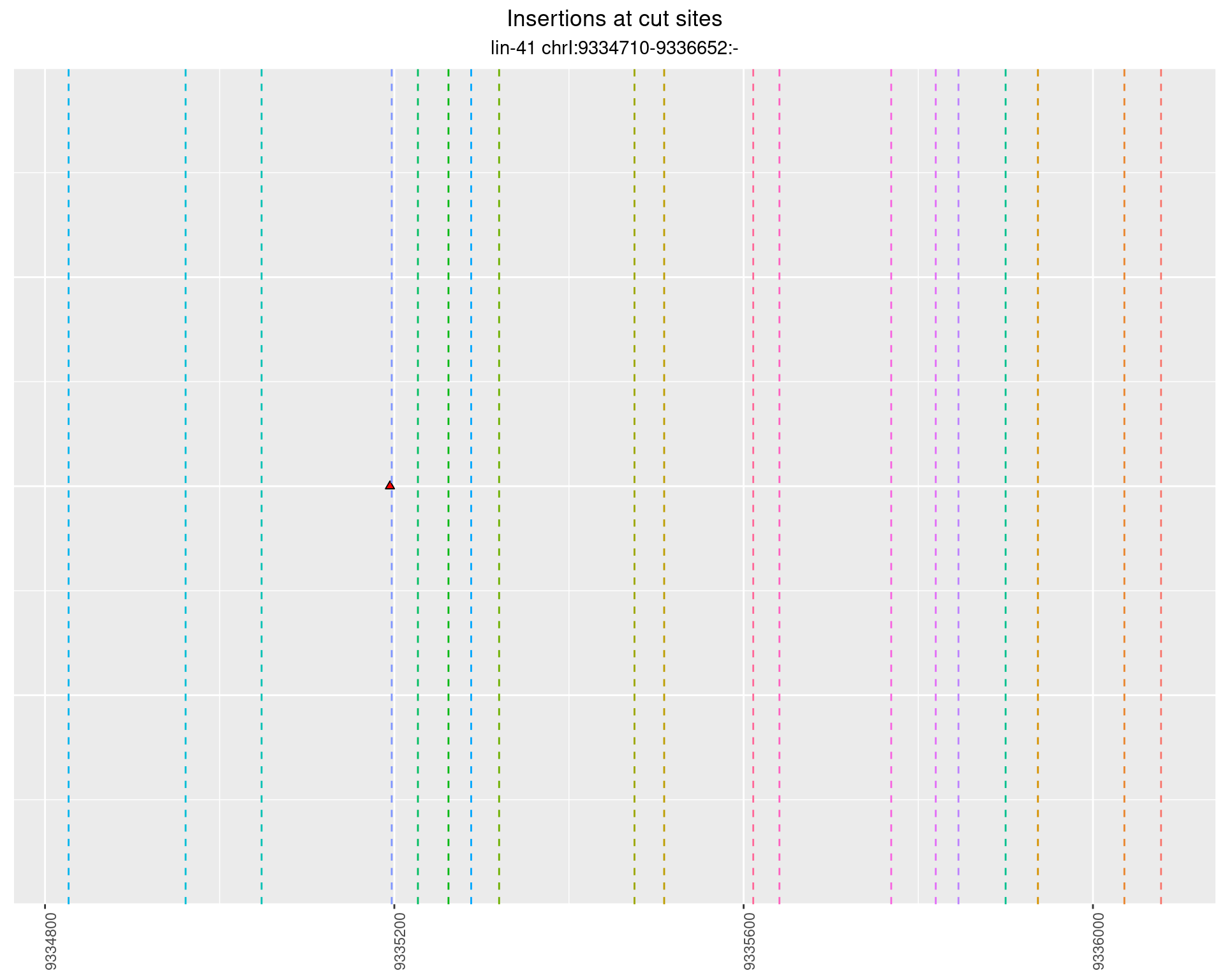
2.87.2 Freq: 1e-05 - 1e-04
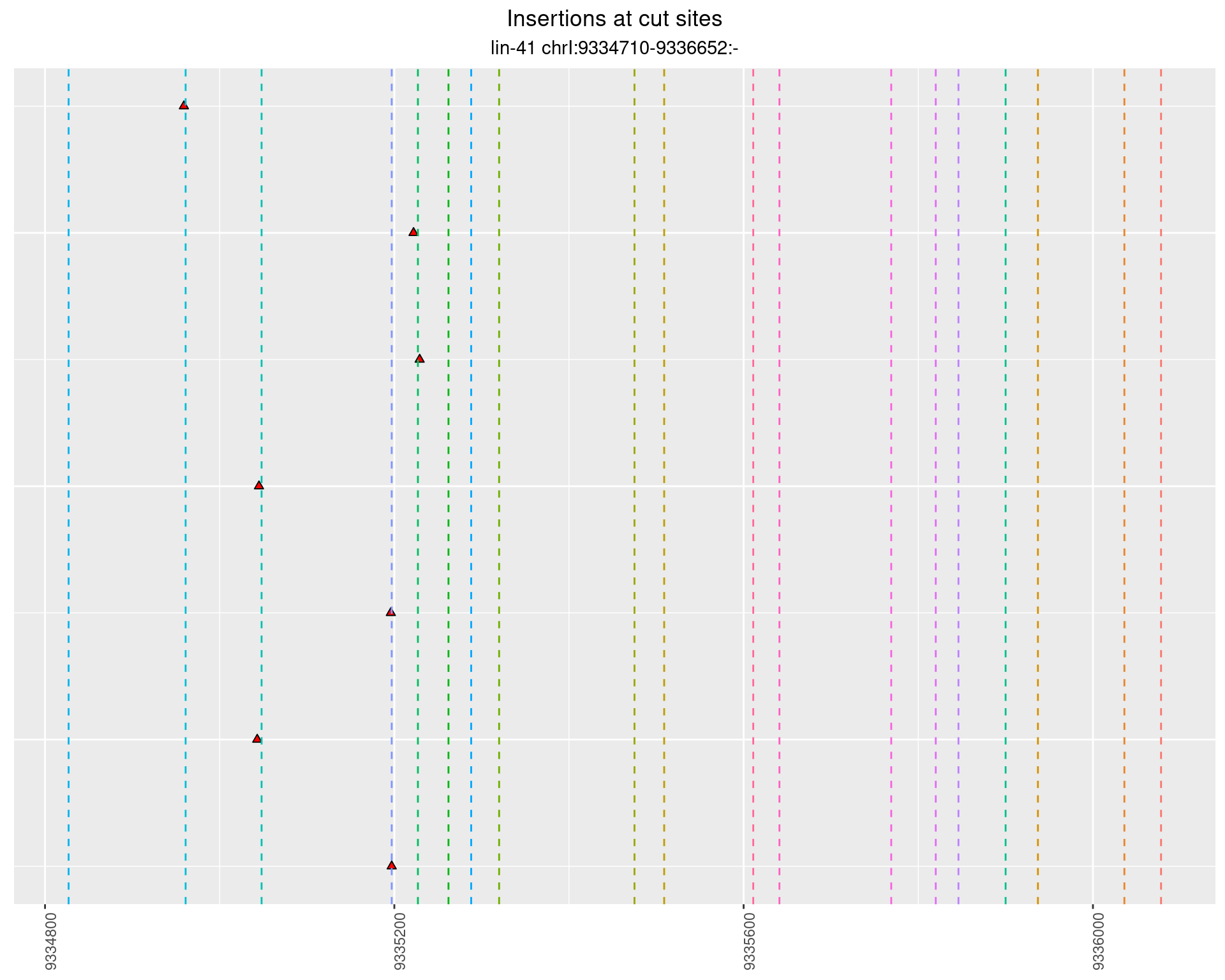
2.87.3 Freq: 1e-04 - 0.001
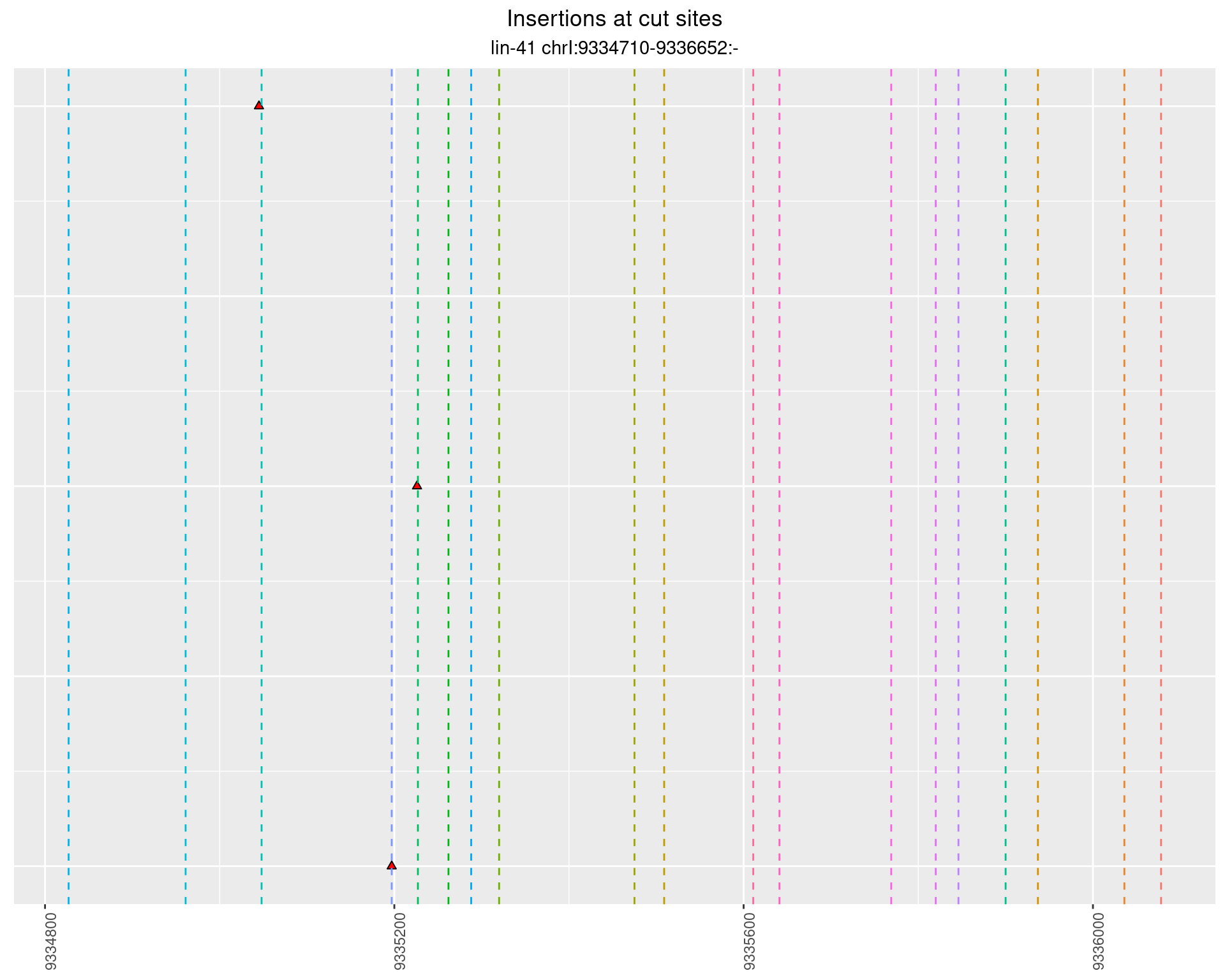
2.87.4 Freq: 0.01 - 0.1
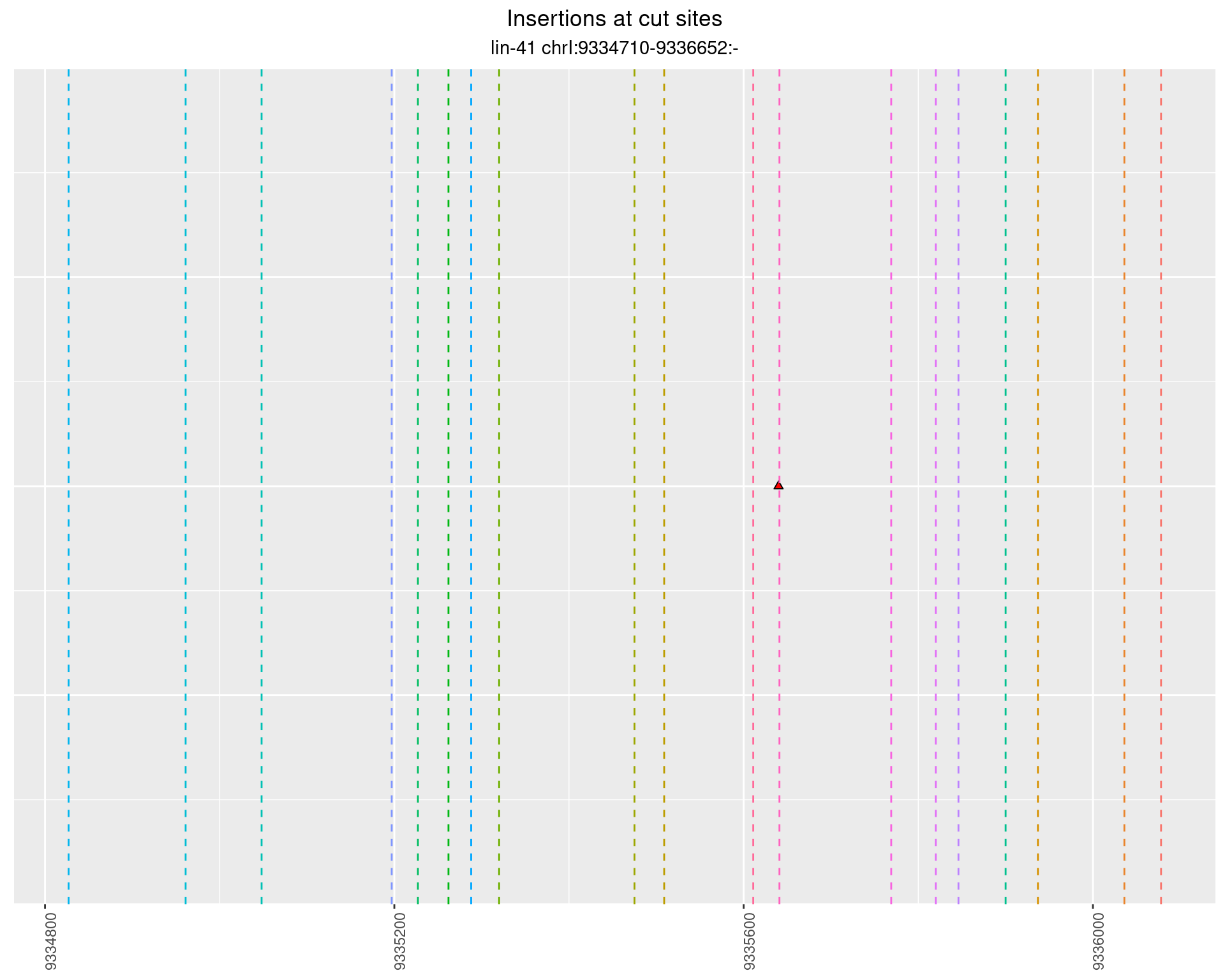
2.88 lin41_RNA_L4_1516_3end_UMI_R2
2.88.1 Freq: 1e-04 - 0.001
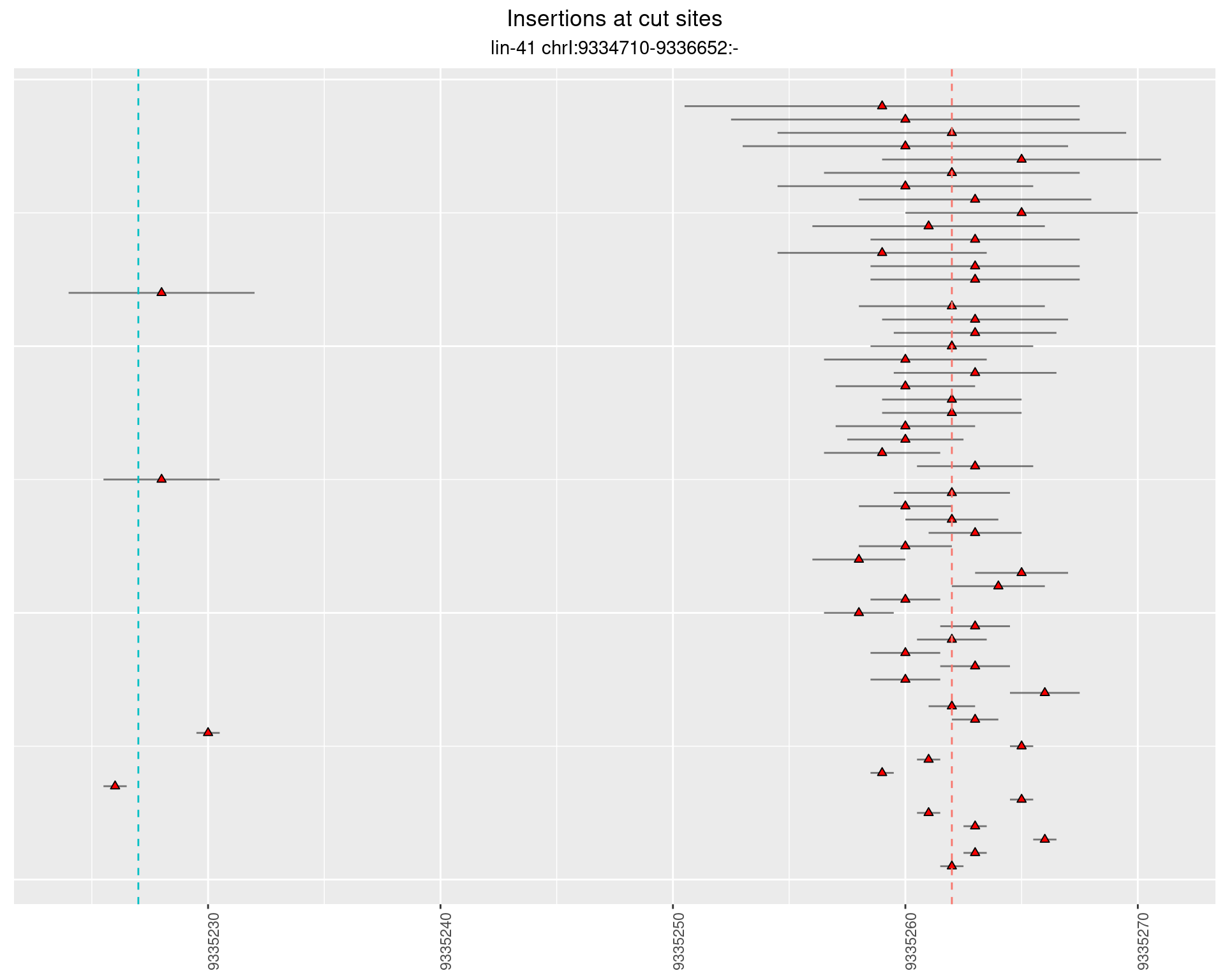
2.88.2 Freq: 0.001 - 0.01
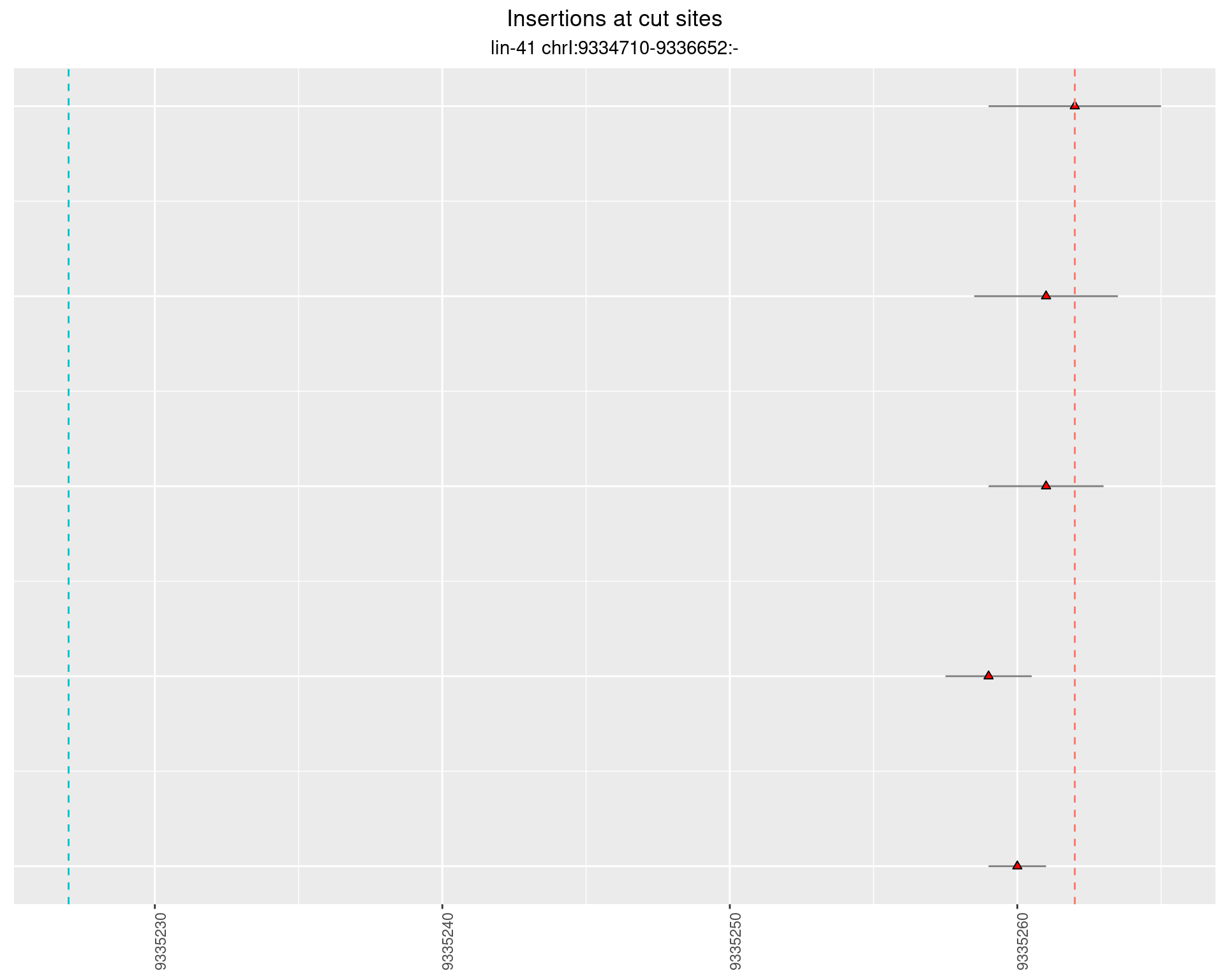
2.88.3 Freq: 1e-05 - 1e-04
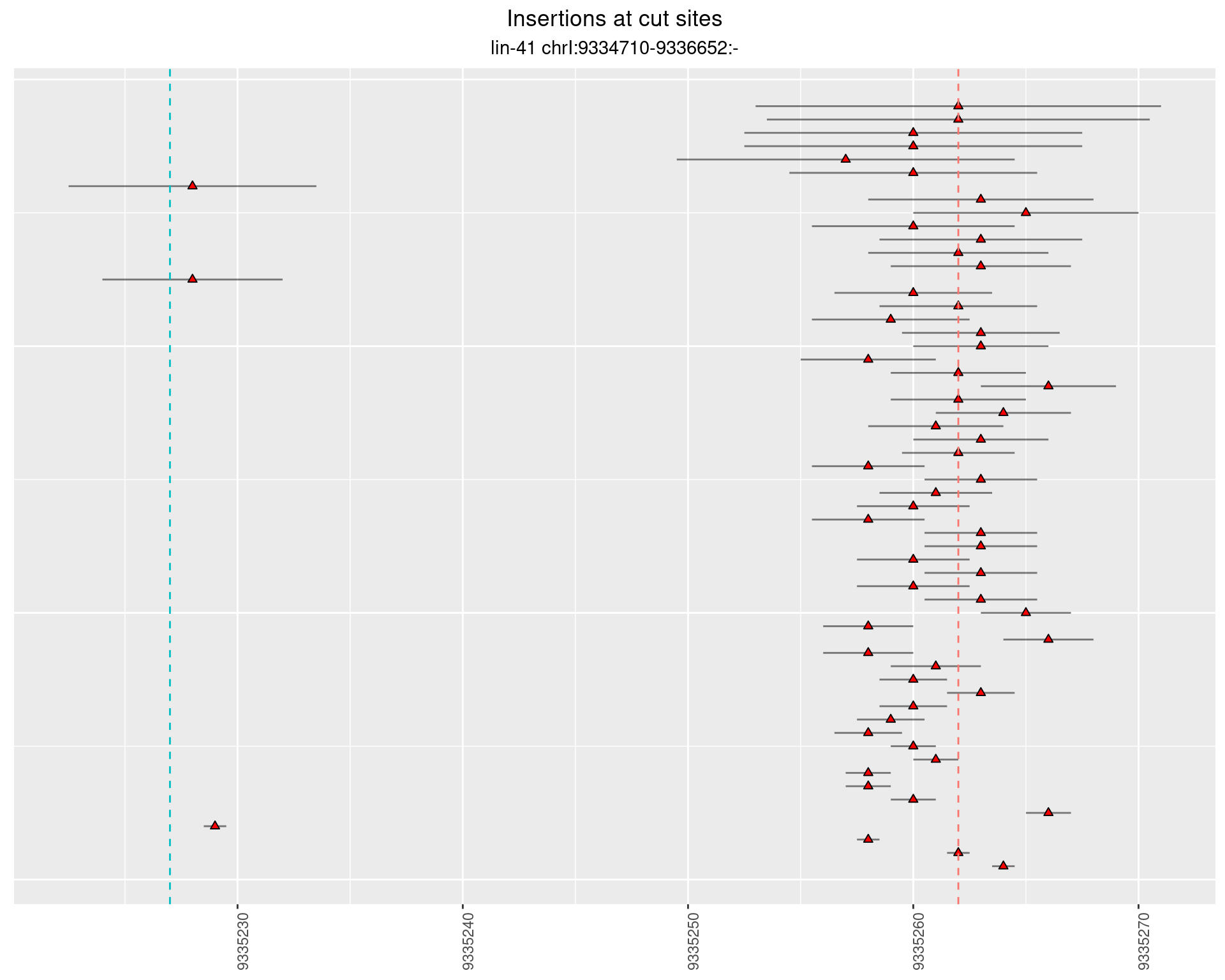
2.89 lin41_RNA_L4_2627_3end_UMI_R2
2.89.1 Freq: 1e-05 - 1e-04
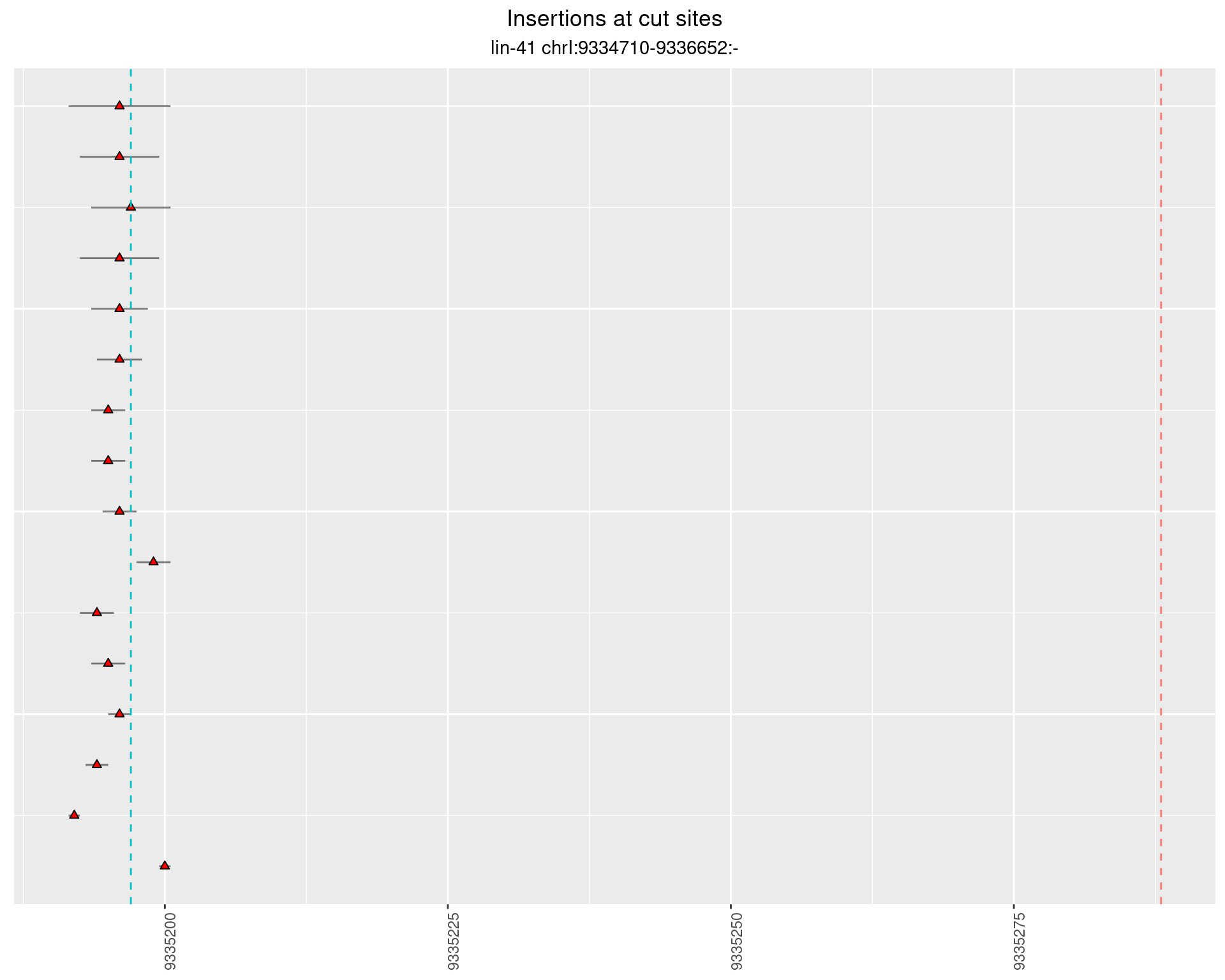
2.89.2 Freq: 0.001 - 0.01
2.89.3 Freq: 1e-04 - 0.001
2.90 lin41_RNA_L4_pool2_3end_UMI_R2
2.90.1 Freq: 1e-04 - 0.001
2.90.2 Freq: 0.001 - 0.01
2.90.3 Freq: 1e-05 - 1e-04
2.90.4 Freq: 0.01 - 0.1
2.91 lin41_RNA_L4_pool3_3end_UMI_R2
2.91.1 Freq: 1e-04 - 0.001
2.91.2 Freq: 1e-05 - 1e-04
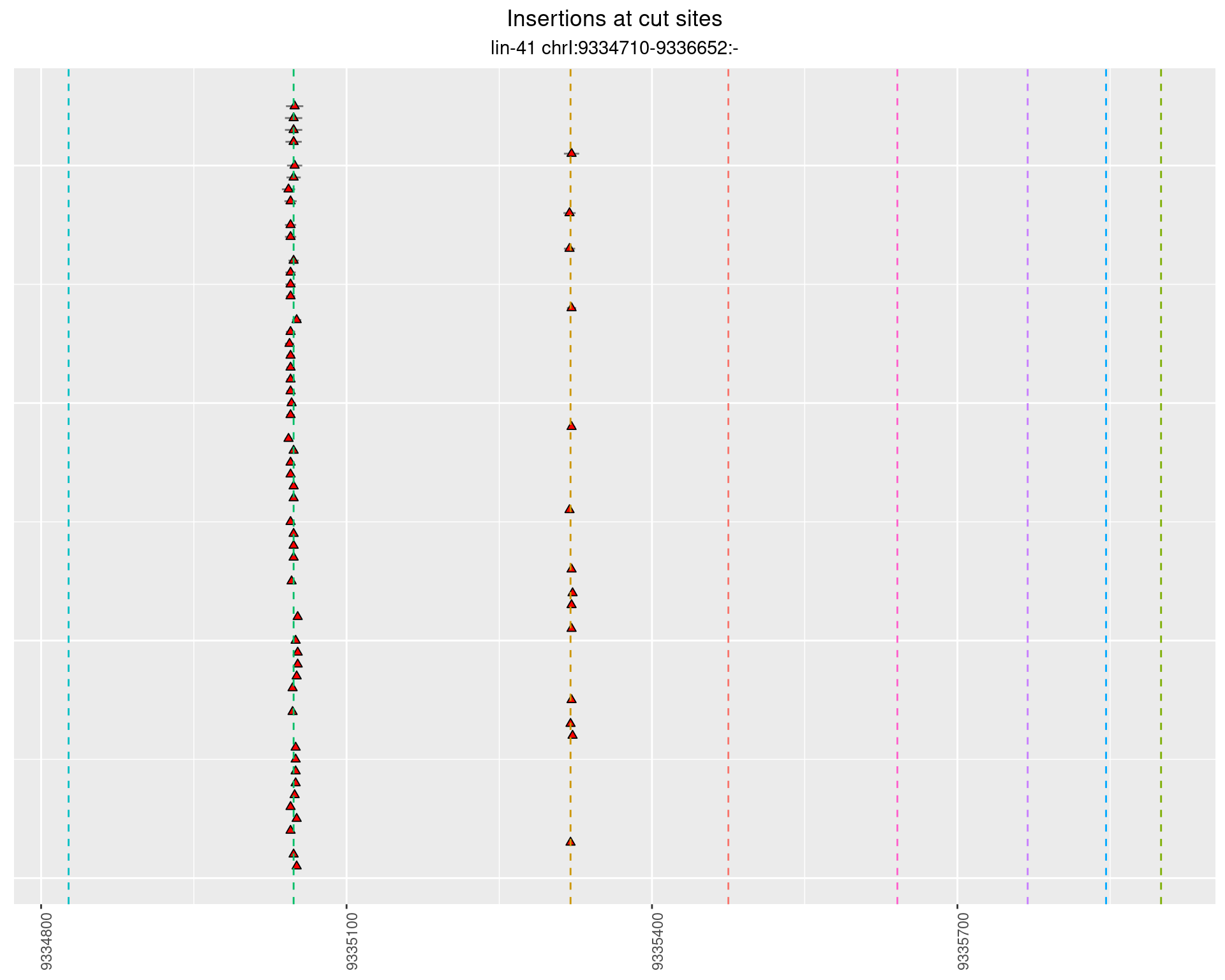
2.91.3 Freq: 0.001 - 0.01
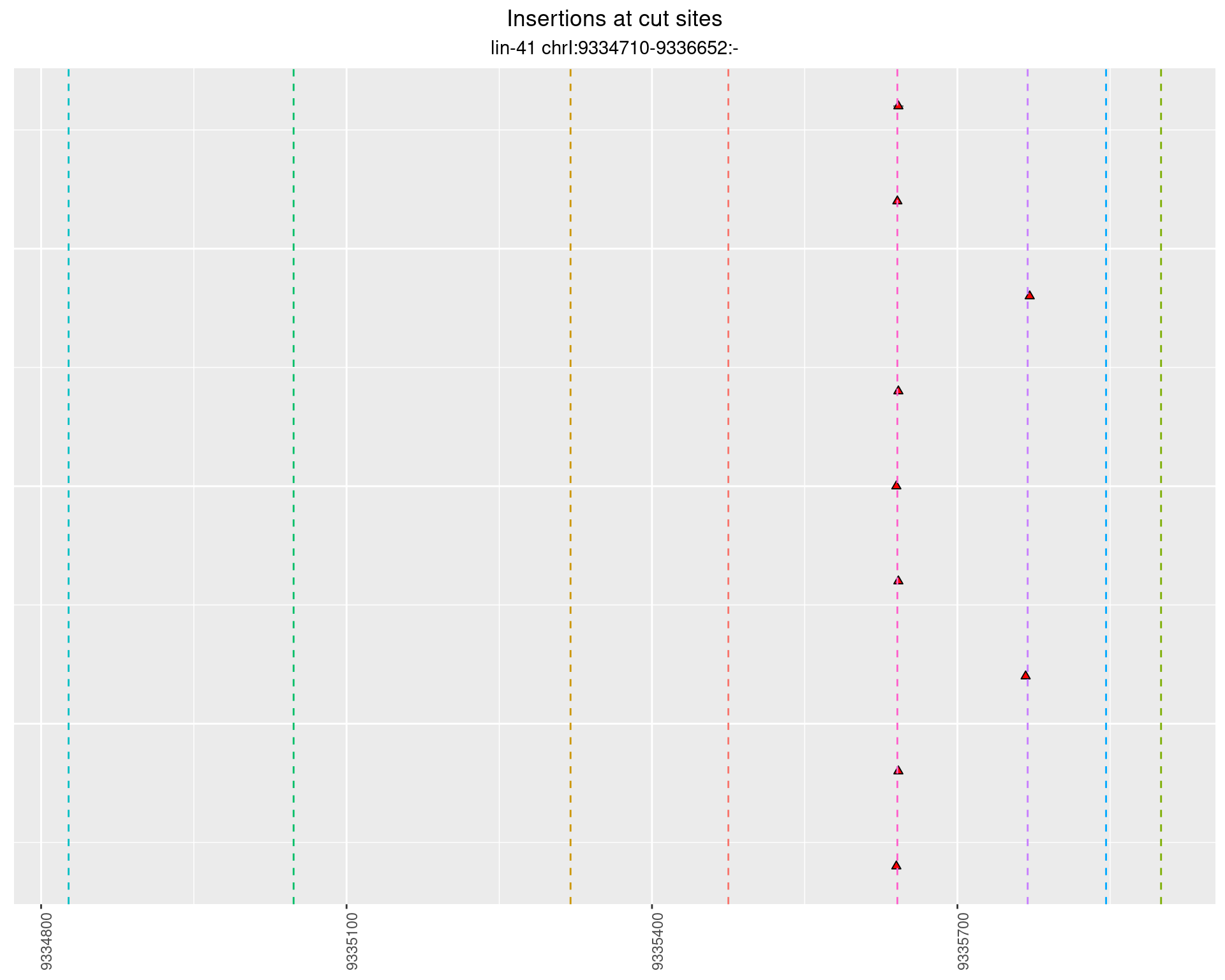
2.91.4 Freq: 0.01 - 0.1
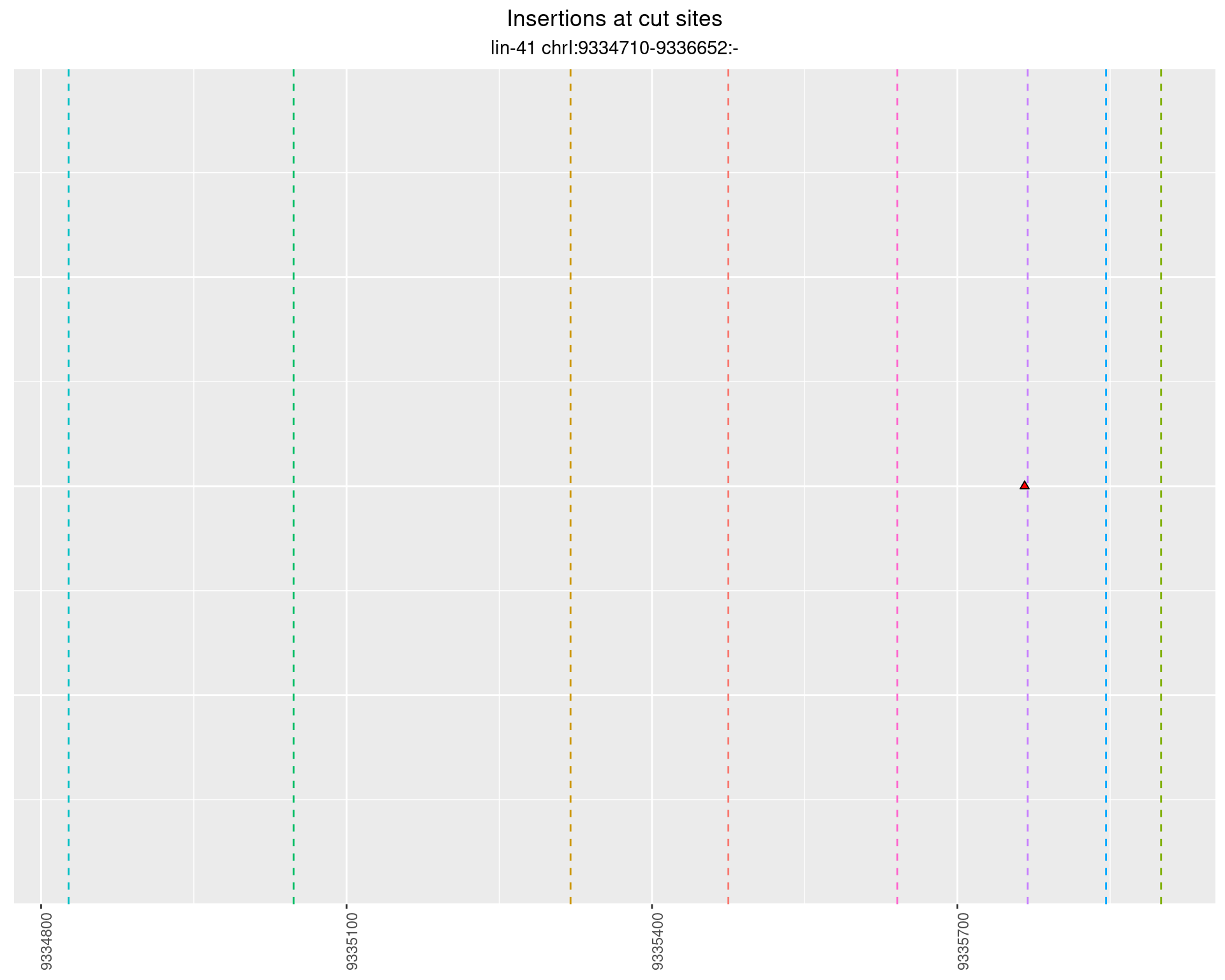
2.92 lin41_RNA_L1_N2_middle_UMI_R2
2.92.1 Freq: 1e-04 - 0.001
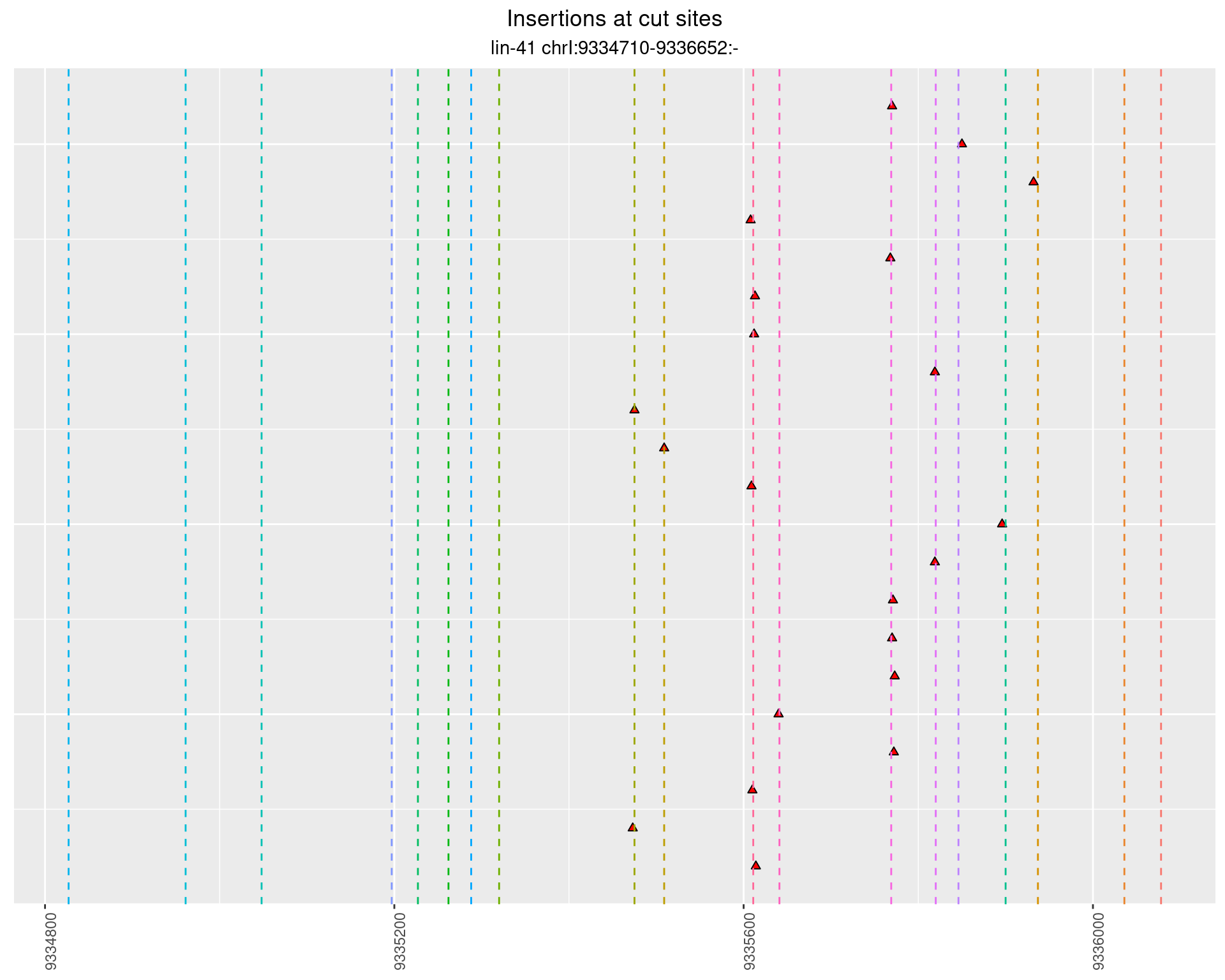
2.92.2 Freq: 0.01 - 0.1
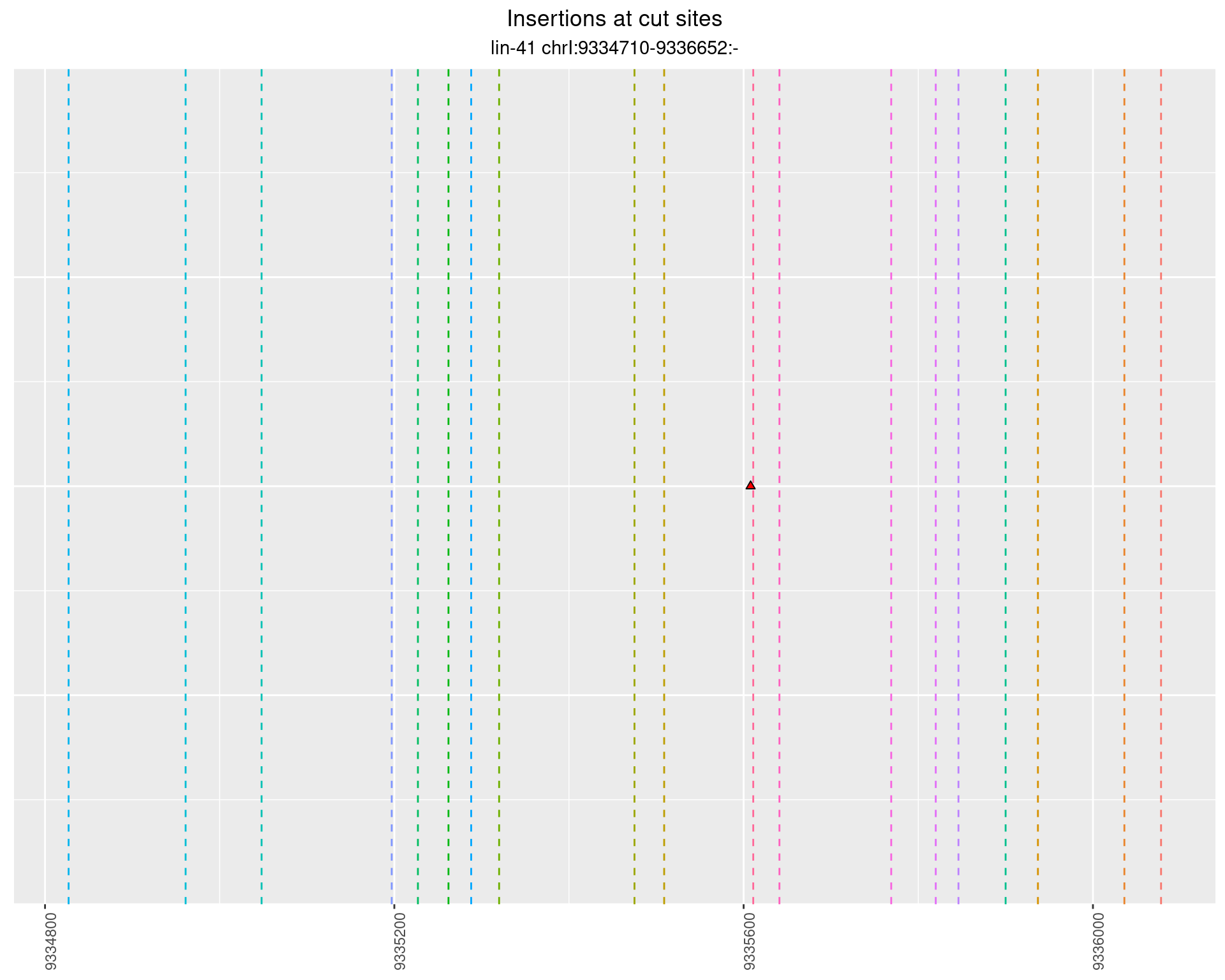
2.92.3 Freq: 1e-05 - 1e-04
2.92.4 Freq: 0.001 - 0.01
2.93 lin41_RNA_L1_1516_middle_UMI_R2
2.94 lin41_RNA_L1_2627_middle_UMI_R2
2.95 lin41_RNA_L1_pool2_middle_UMI_R2
2.95.1 Freq: 0.01 - 0.1
2.95.2 Freq: 1e-04 - 0.001
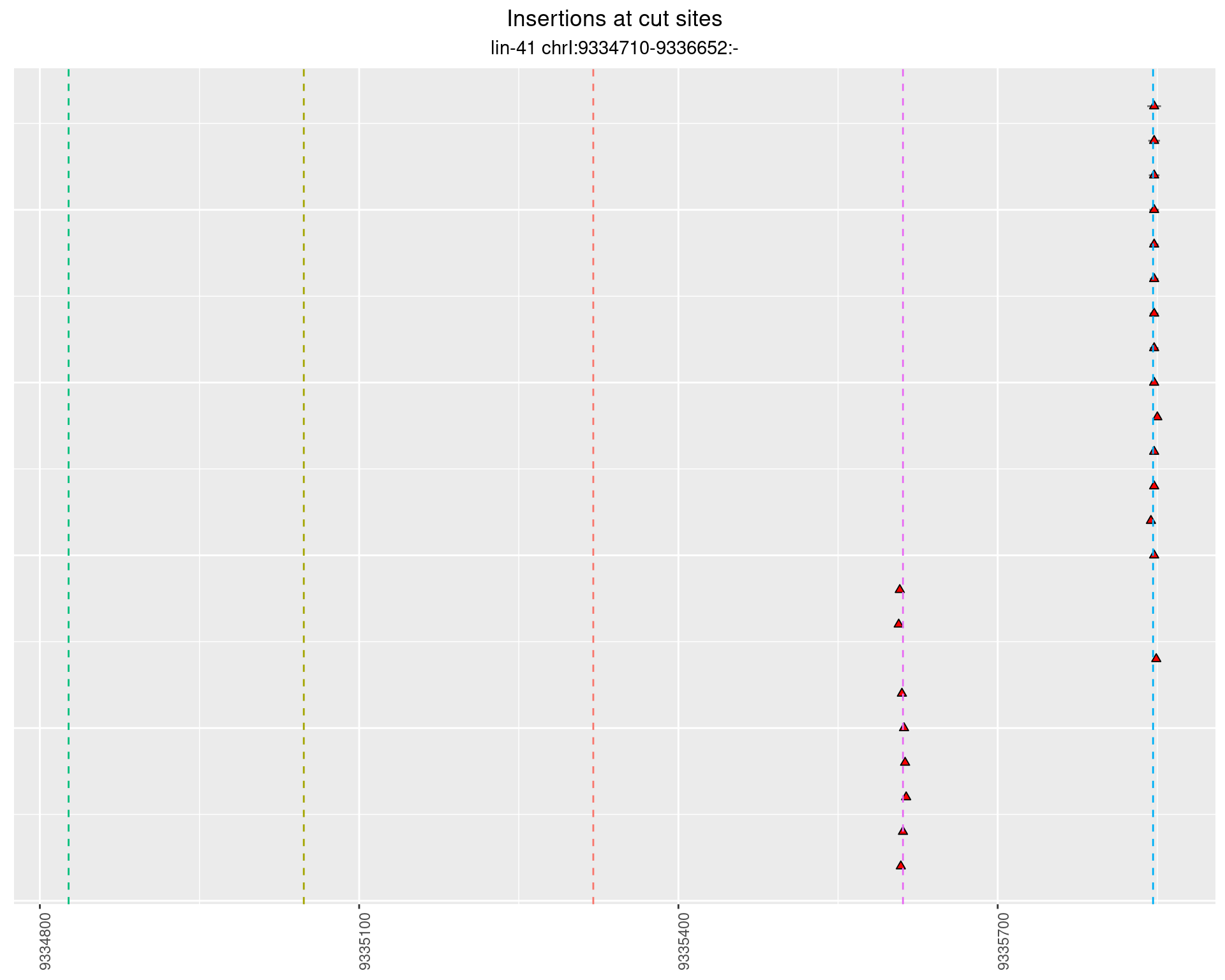
2.95.3 Freq: 1e-05 - 1e-04
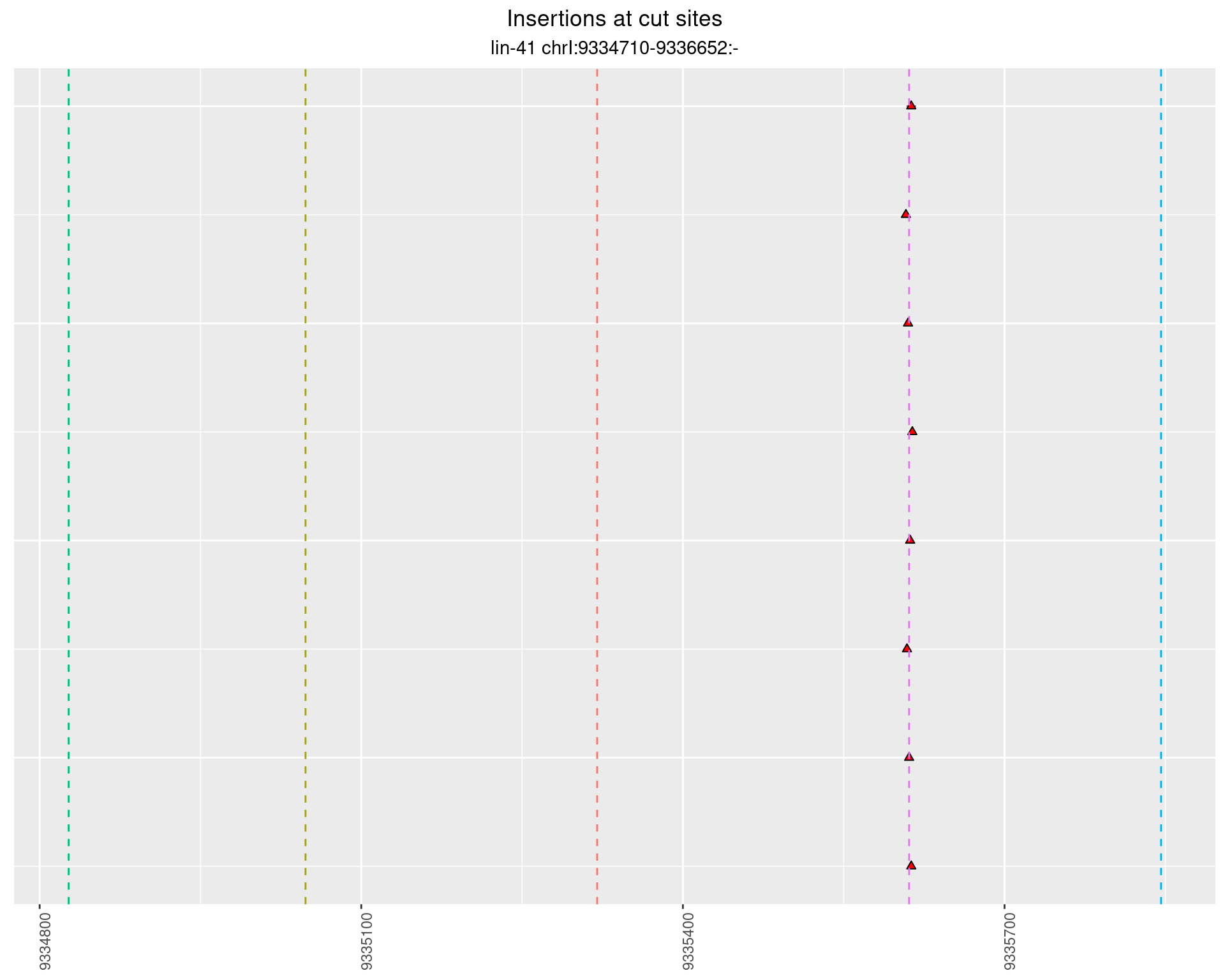
2.95.4 Freq: 0.001 - 0.01
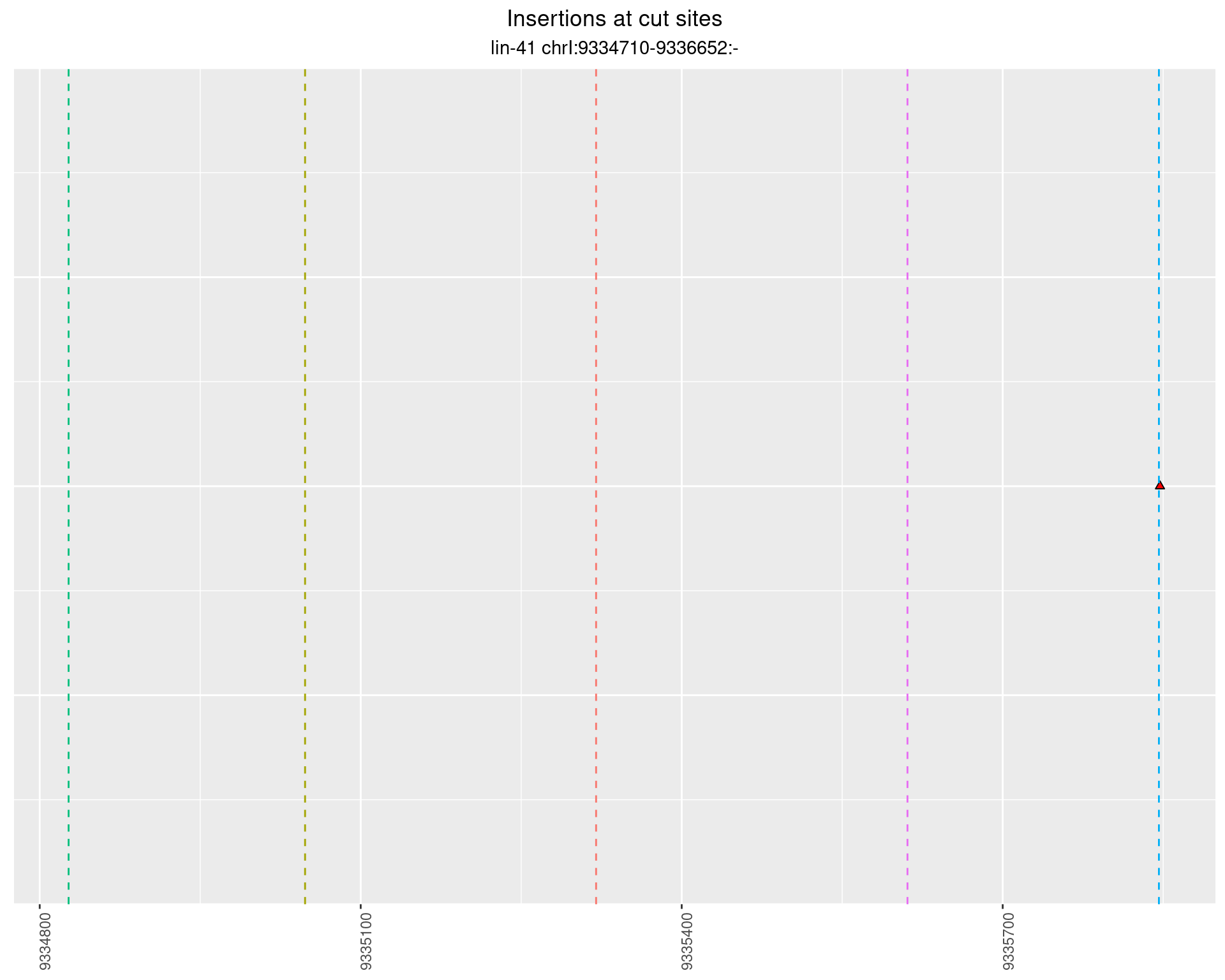
2.96 lin41_RNA_L1_pool3_middle_UMI_R2
2.96.1 Freq: 1e-04 - 0.001
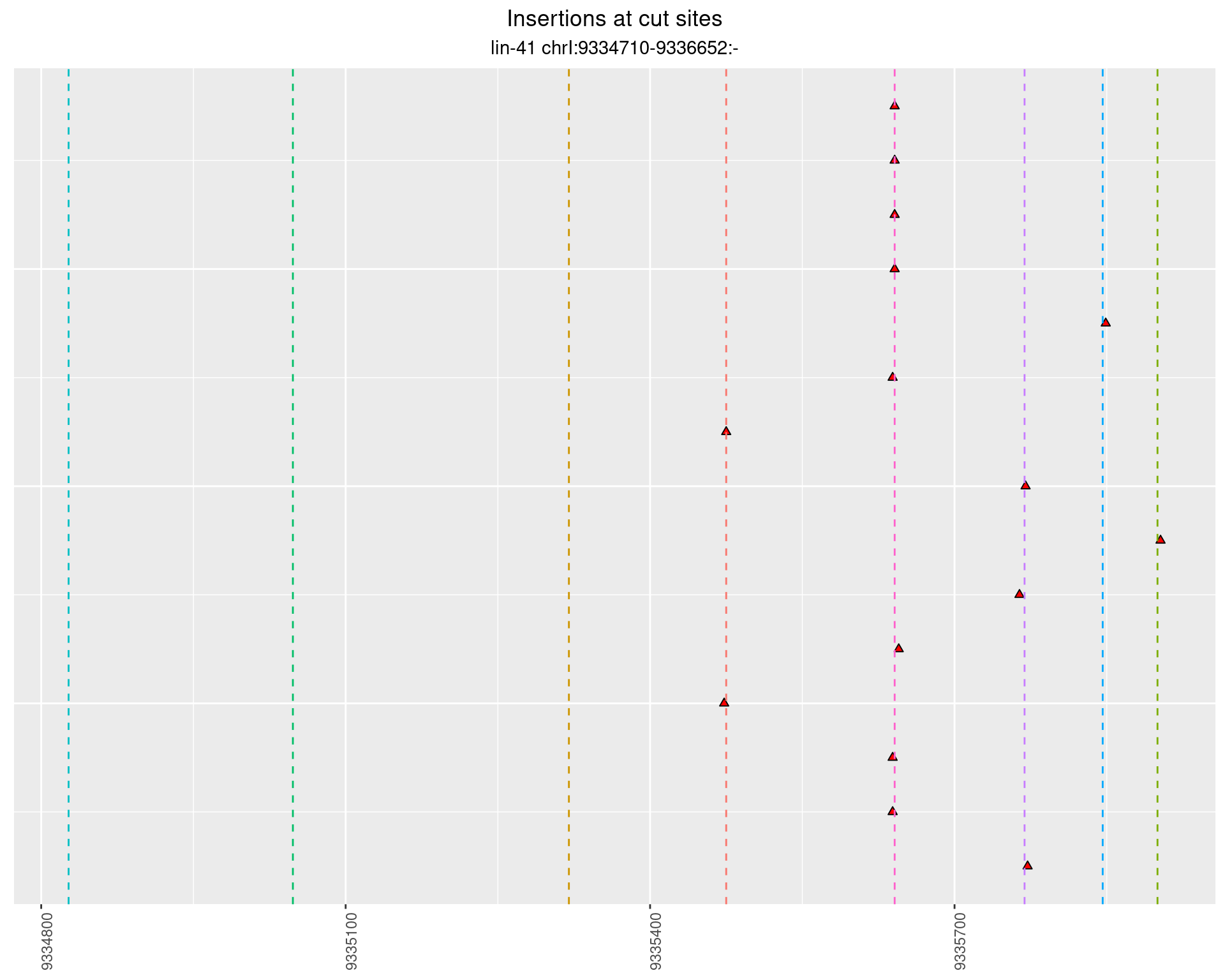
2.96.2 Freq: 1e-05 - 1e-04
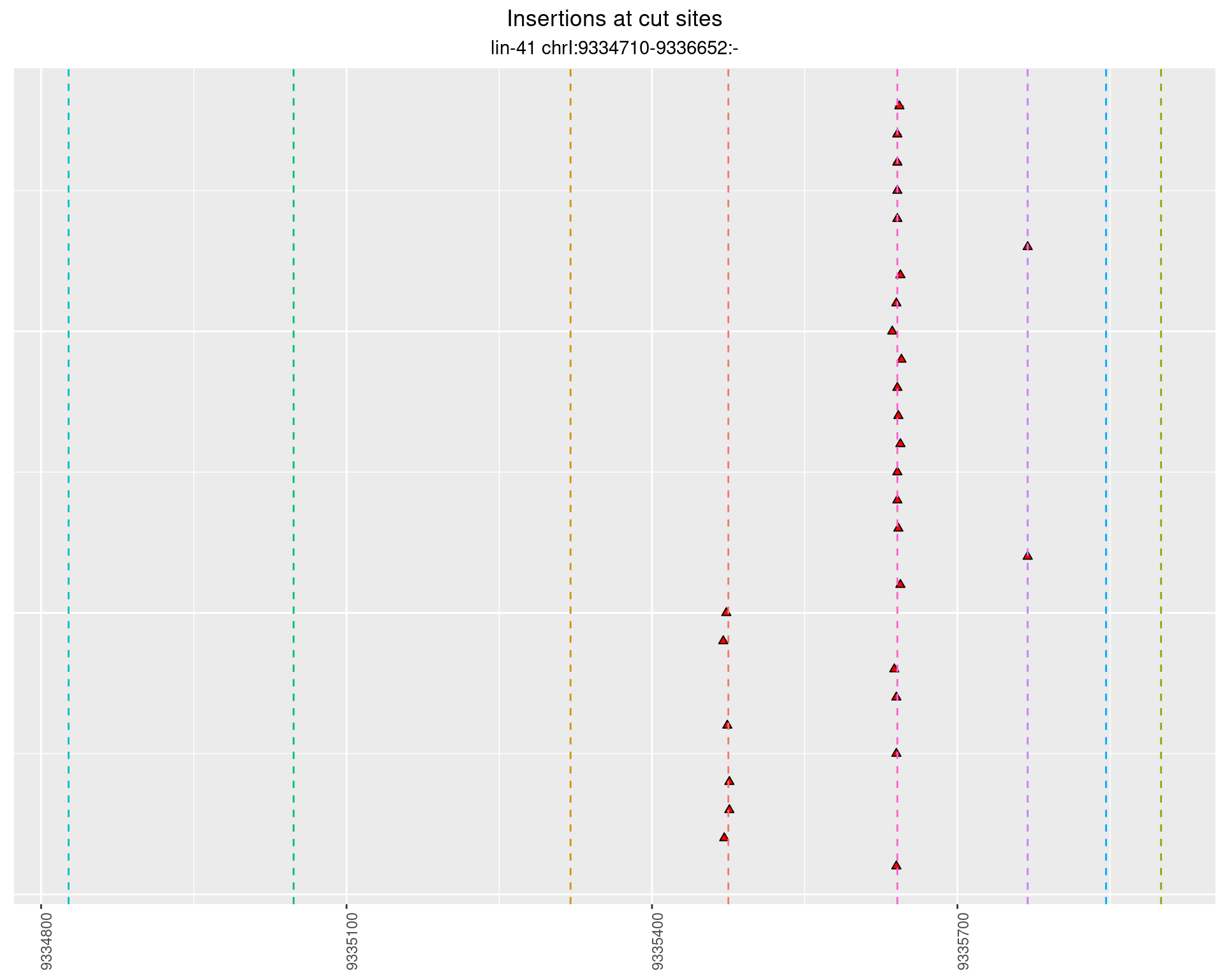
2.96.3 Freq: 0.001 - 0.01
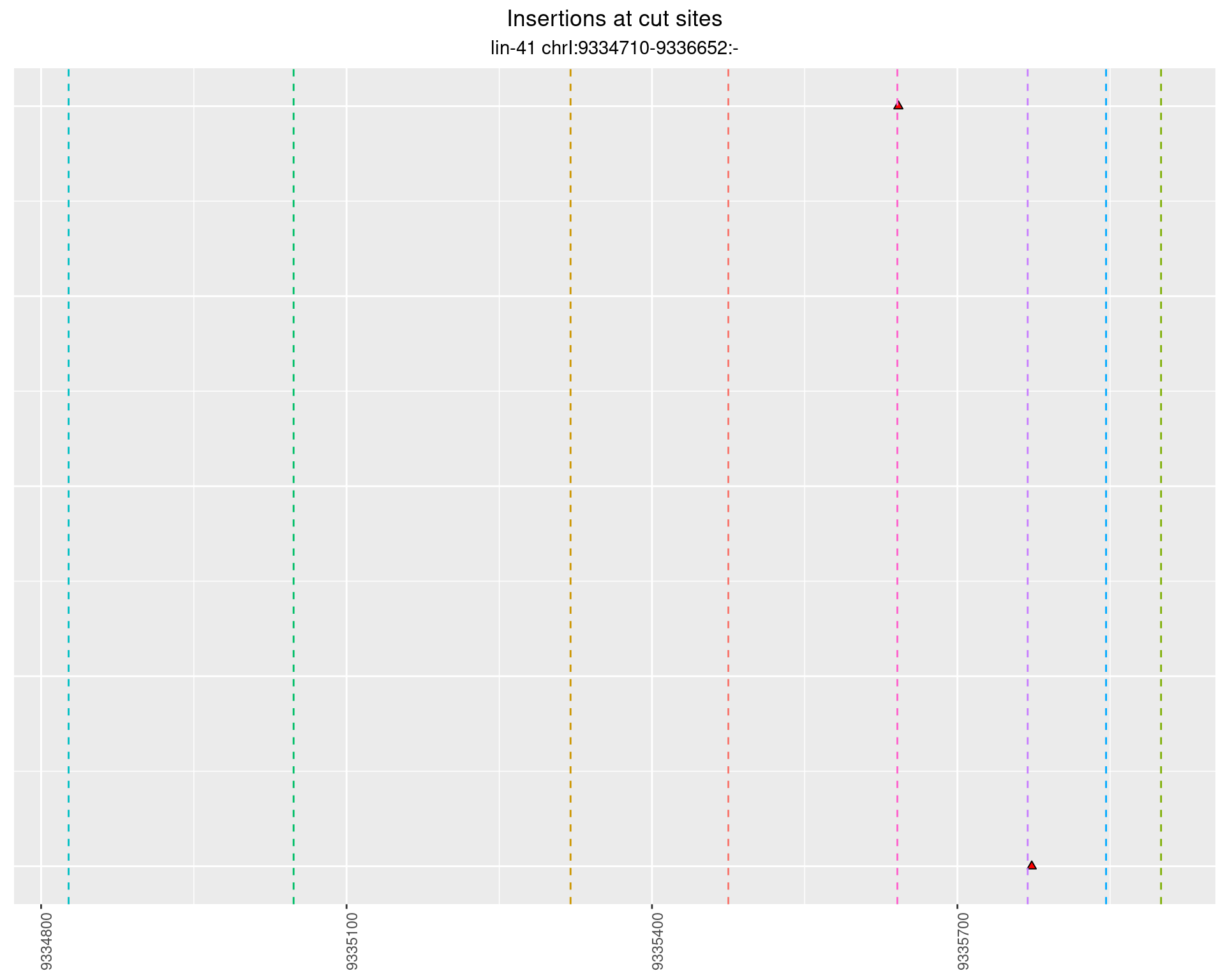
2.97 lin41_RNA_L4_N2_middle_UMI_R2
2.97.1 Freq: 0.01 - 0.1
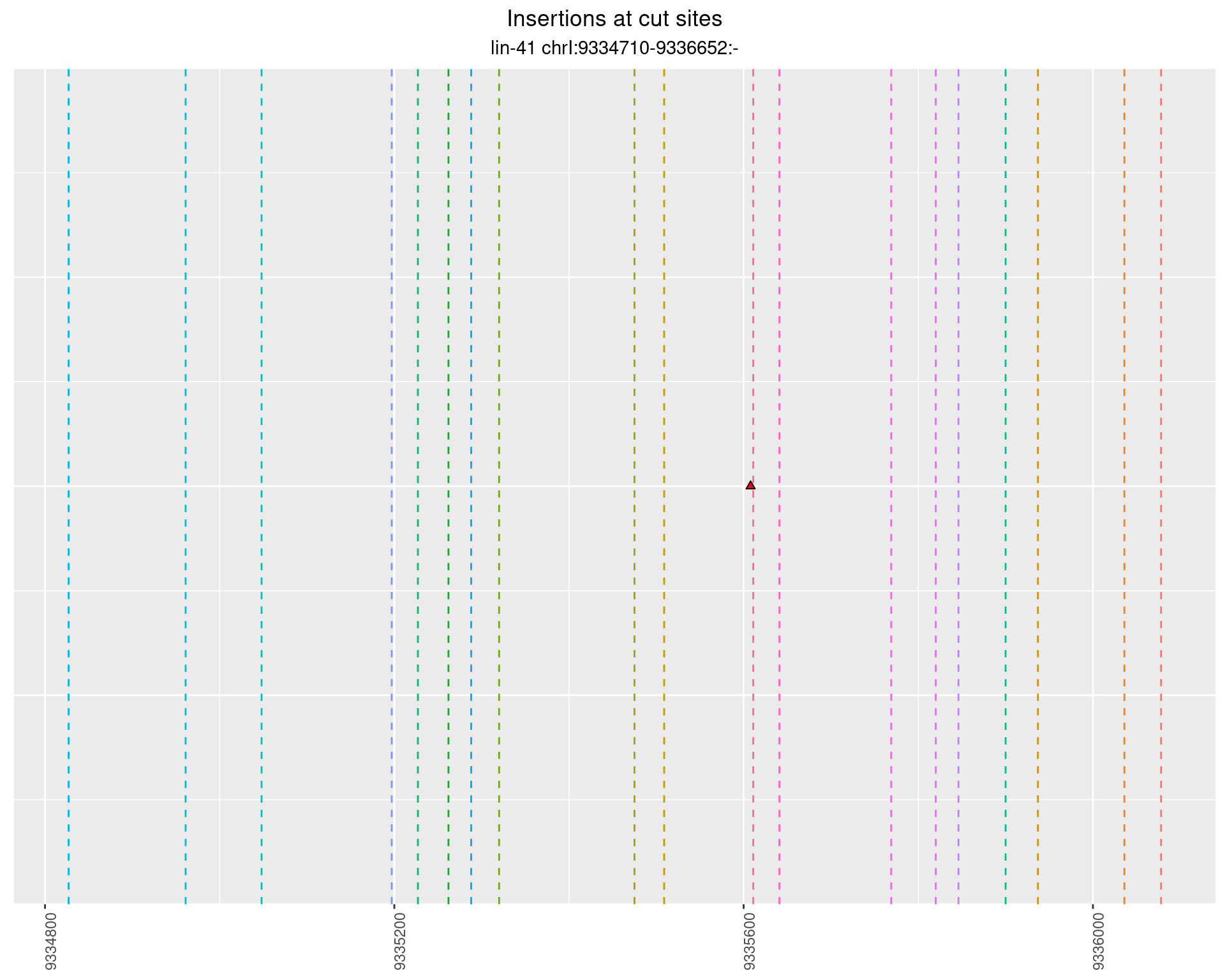
2.97.2 Freq: 1e-04 - 0.001
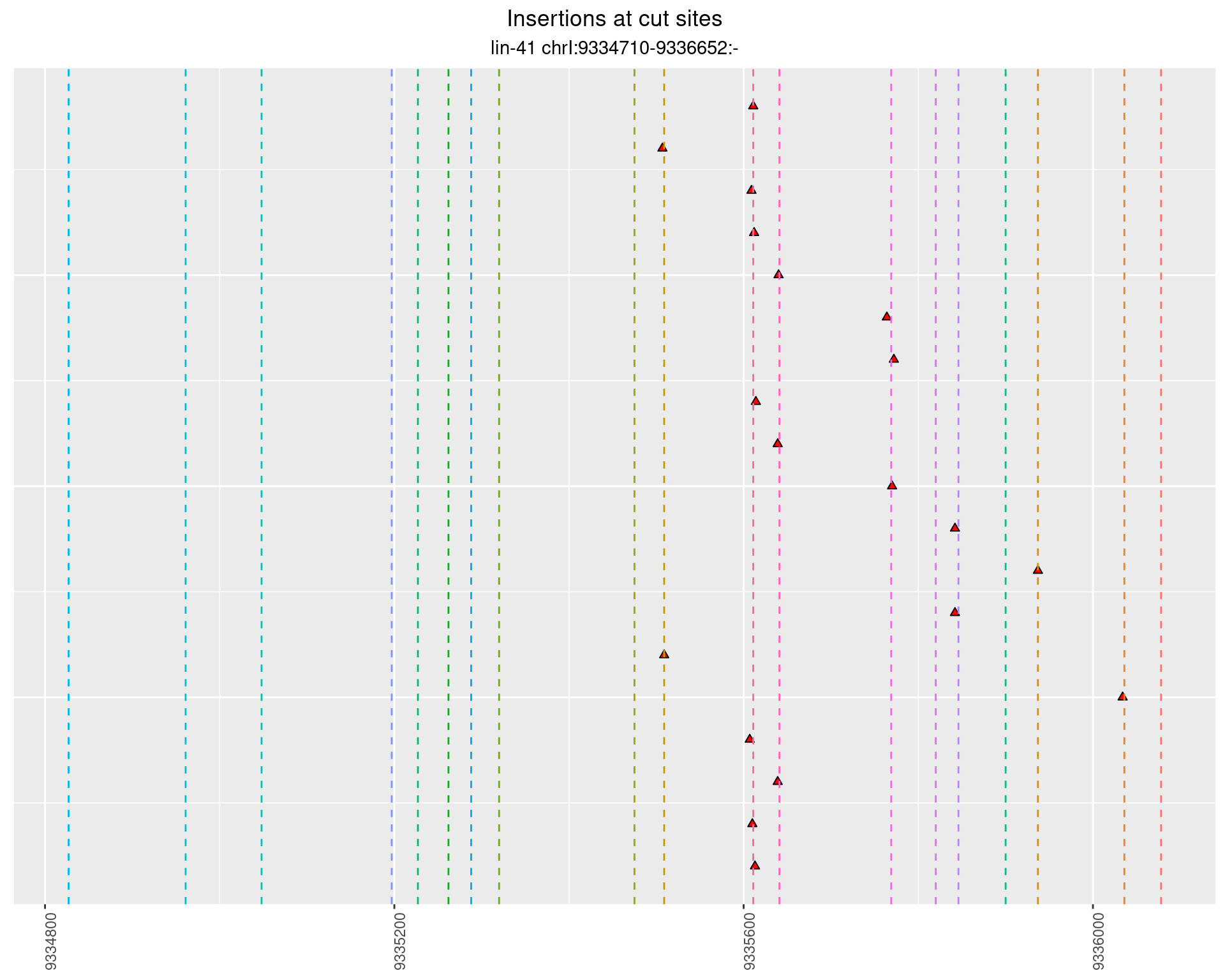
2.97.3 Freq: 0.001 - 0.01
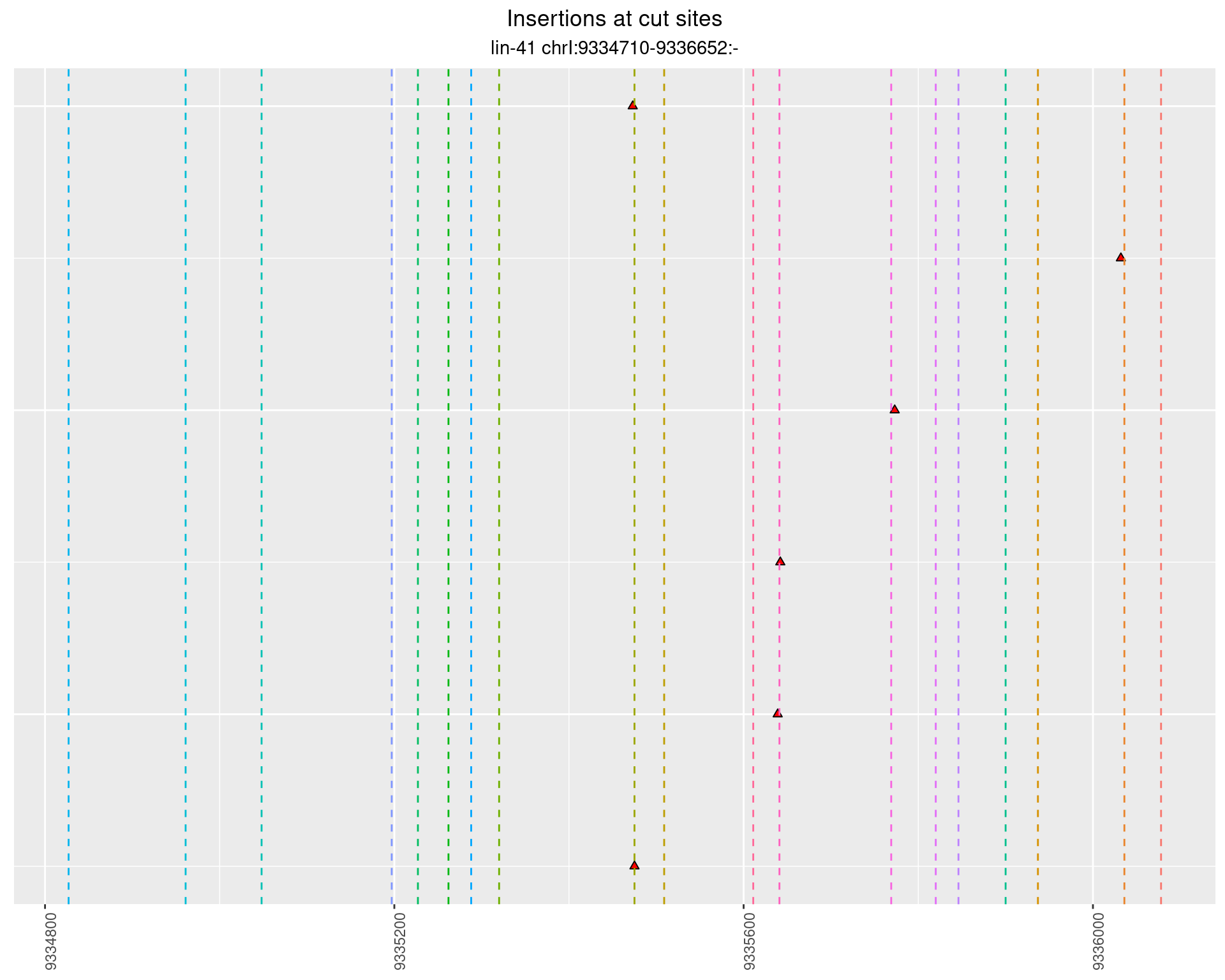
2.97.4 Freq: 1e-05 - 1e-04
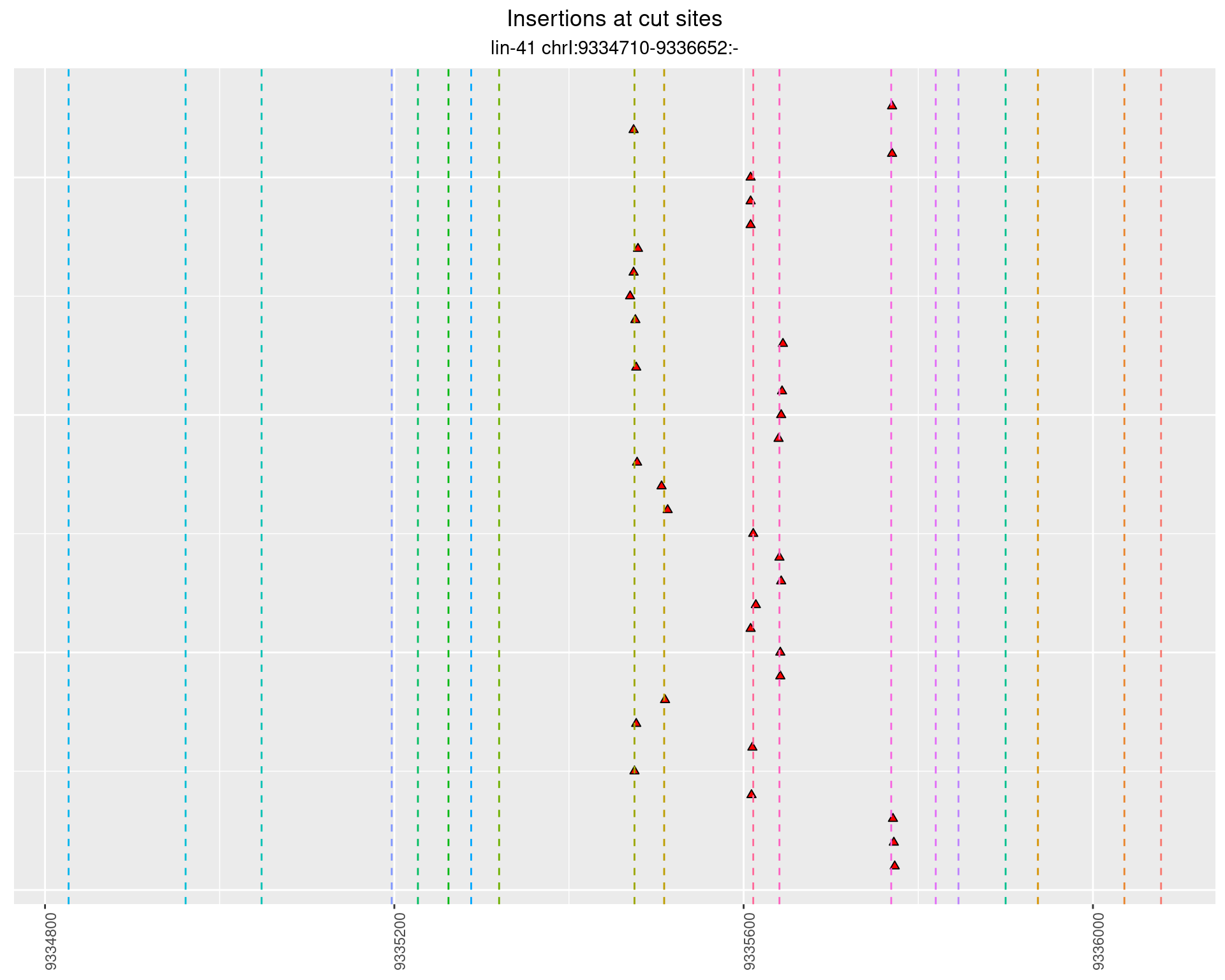
2.98 lin41_RNA_L4_1516_middle_UMI_R2
2.98.1 Freq: 0.1 - 1
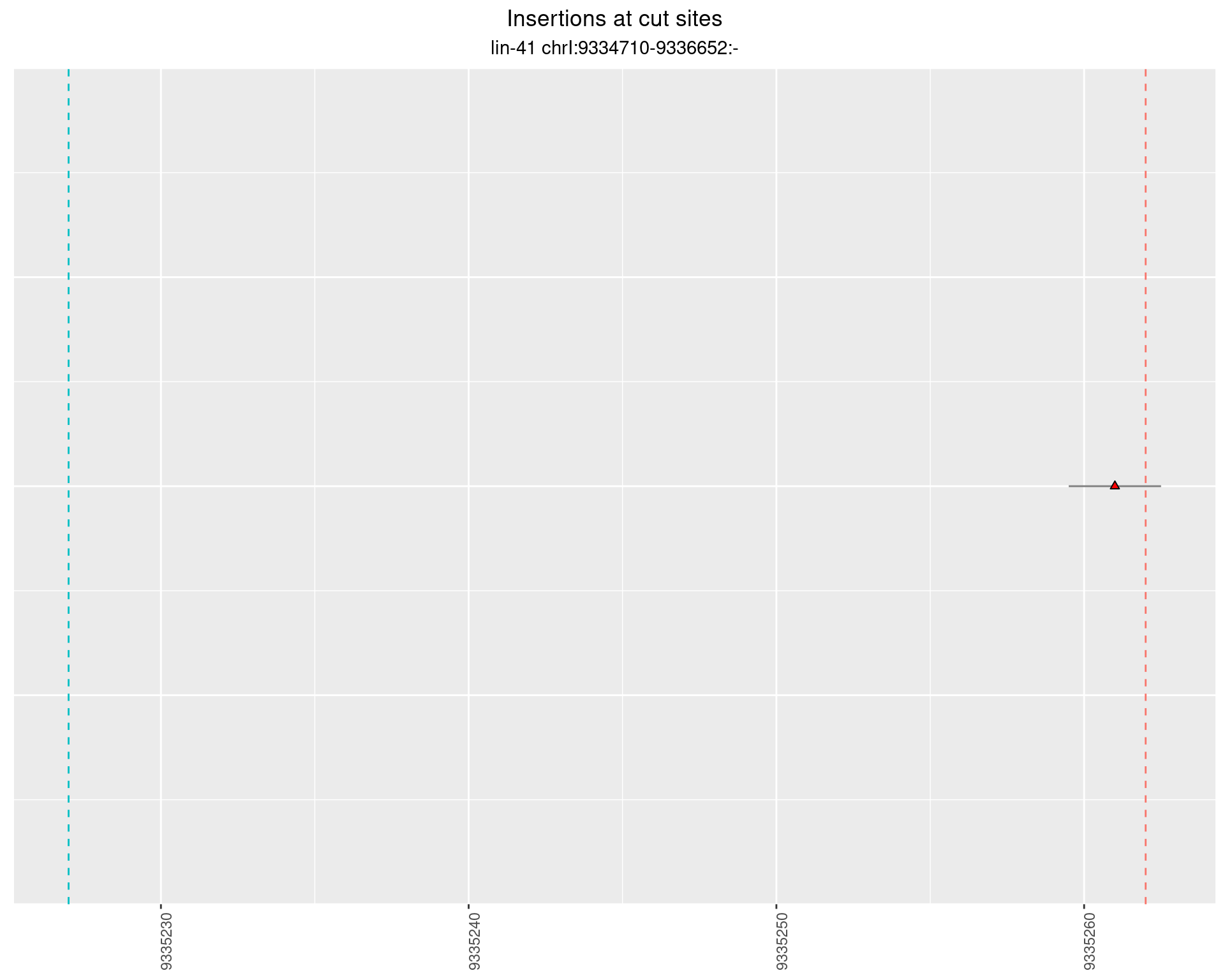
2.99 lin41_RNA_L4_2627_middle_UMI_R2
2.100 lin41_RNA_L4_pool2_middle_UMI_R2
2.100.1 Freq: 0.01 - 0.1
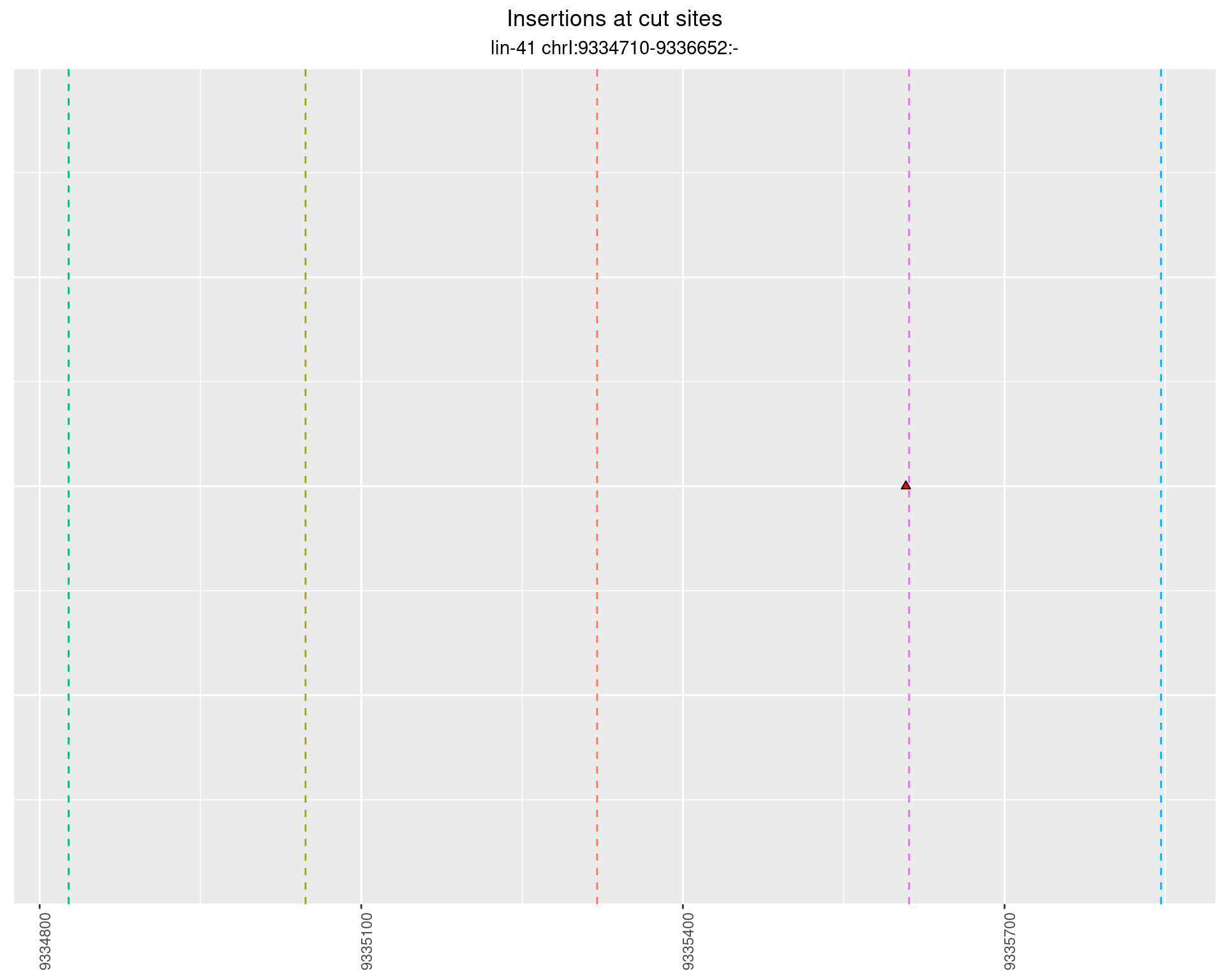
2.100.2 Freq: 1e-05 - 1e-04
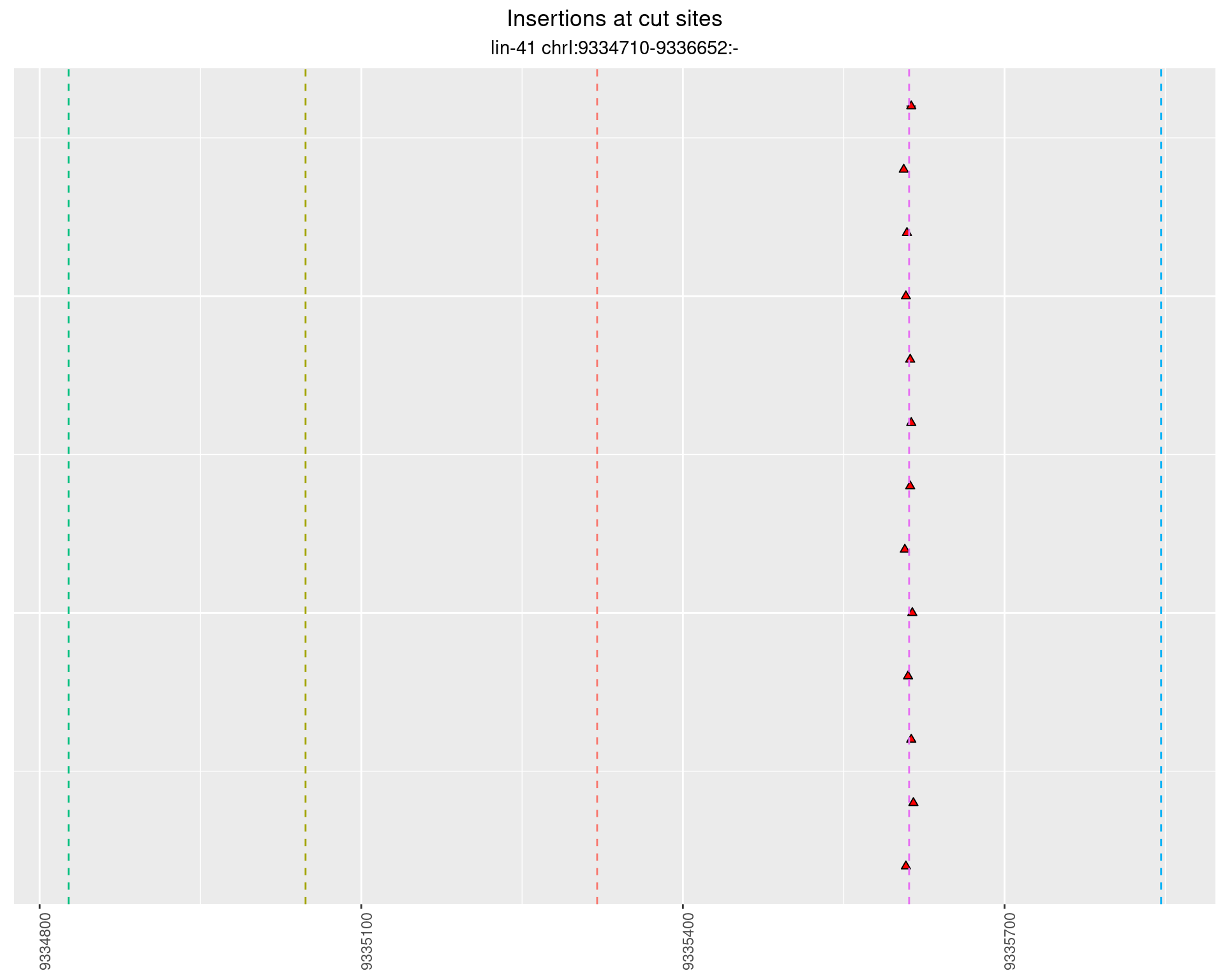
2.100.3 Freq: 1e-04 - 0.001
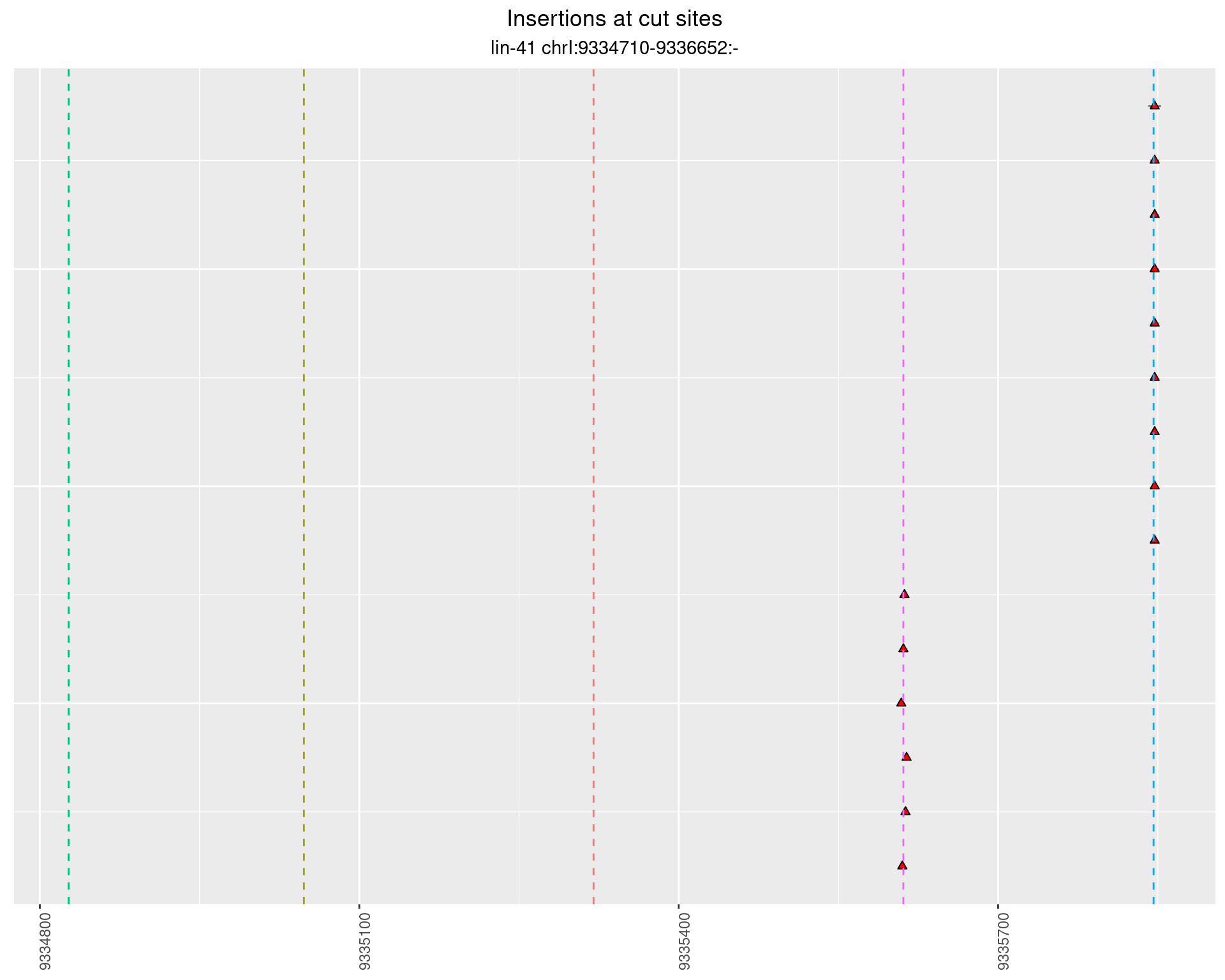
2.101 lin41_RNA_L4_pool3_middle_UMI_R2
2.101.1 Freq: 1e-04 - 0.001
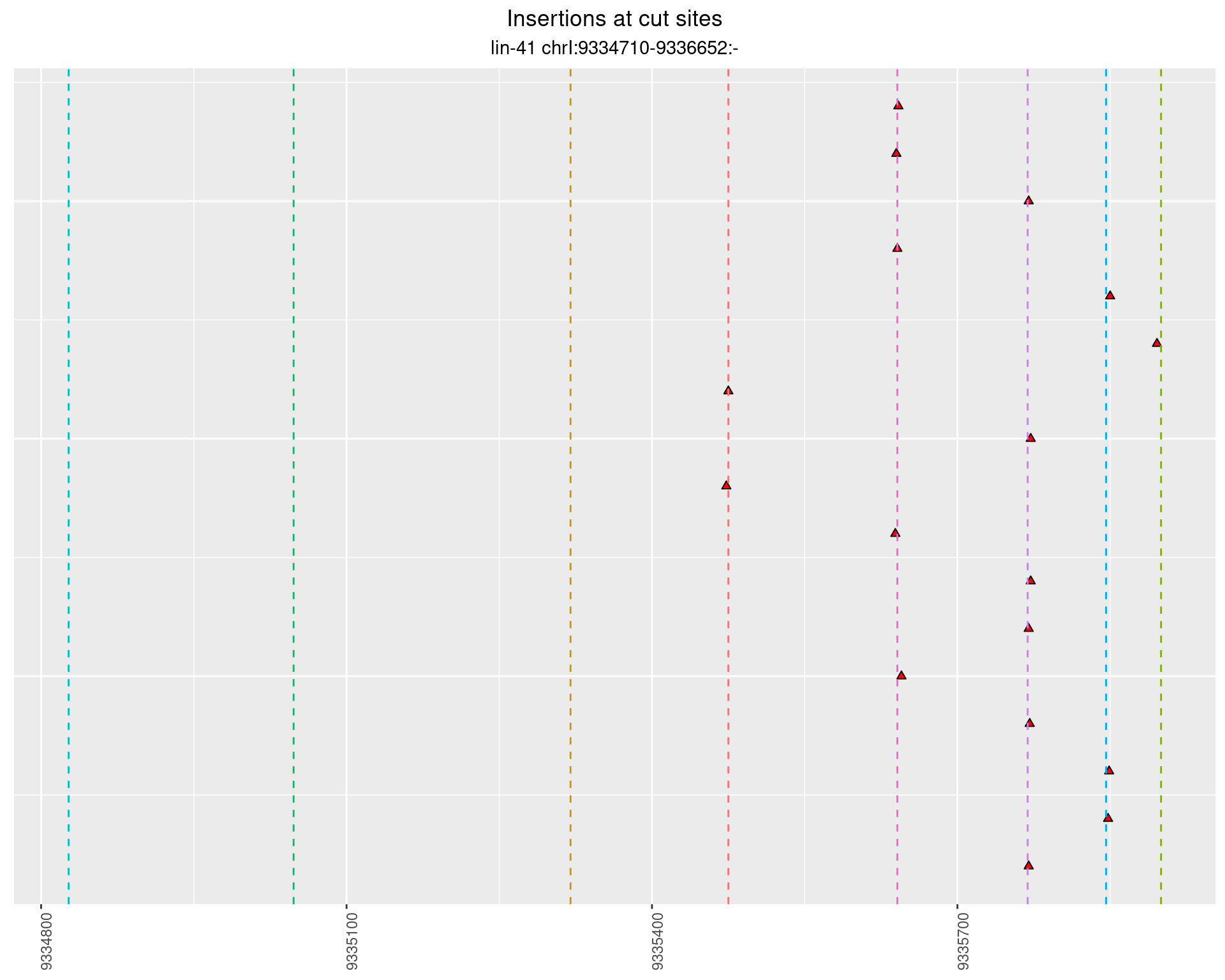
2.101.2 Freq: 0.001 - 0.01
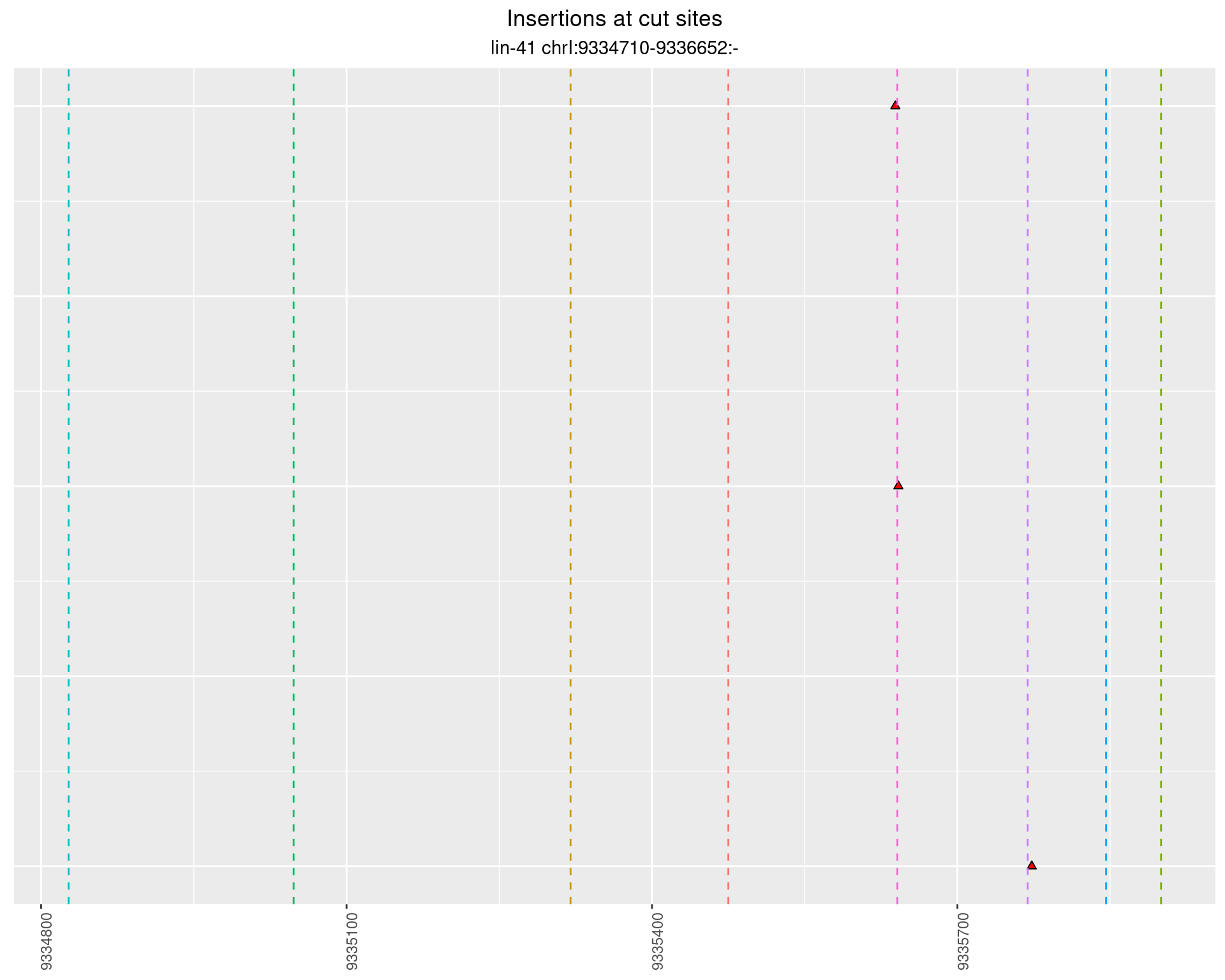
2.101.3 Freq: 1e-05 - 1e-04
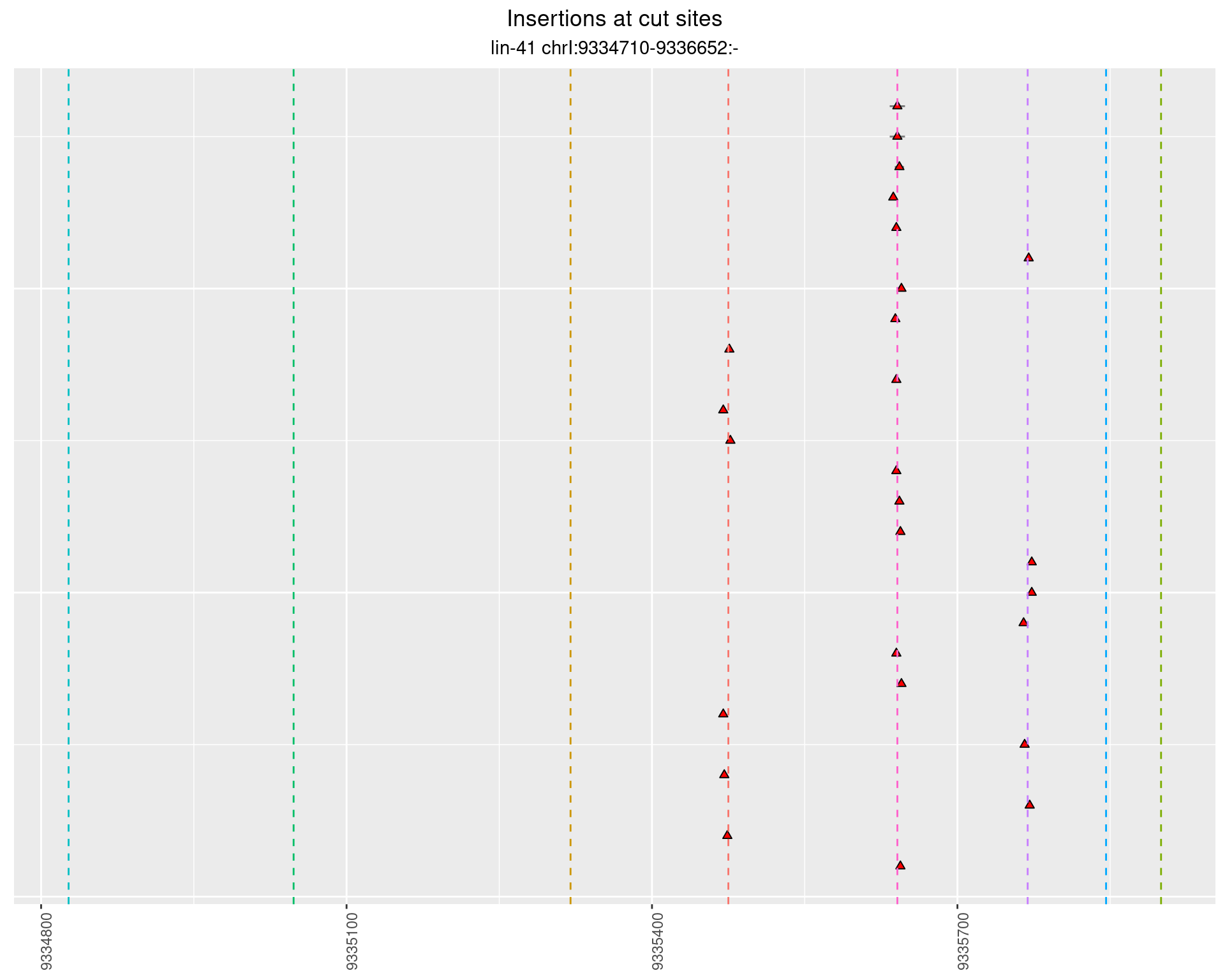
2.102 lin41_RNApacbio_L1_1516
2.102.1 Freq: 1e-04 - 0.001
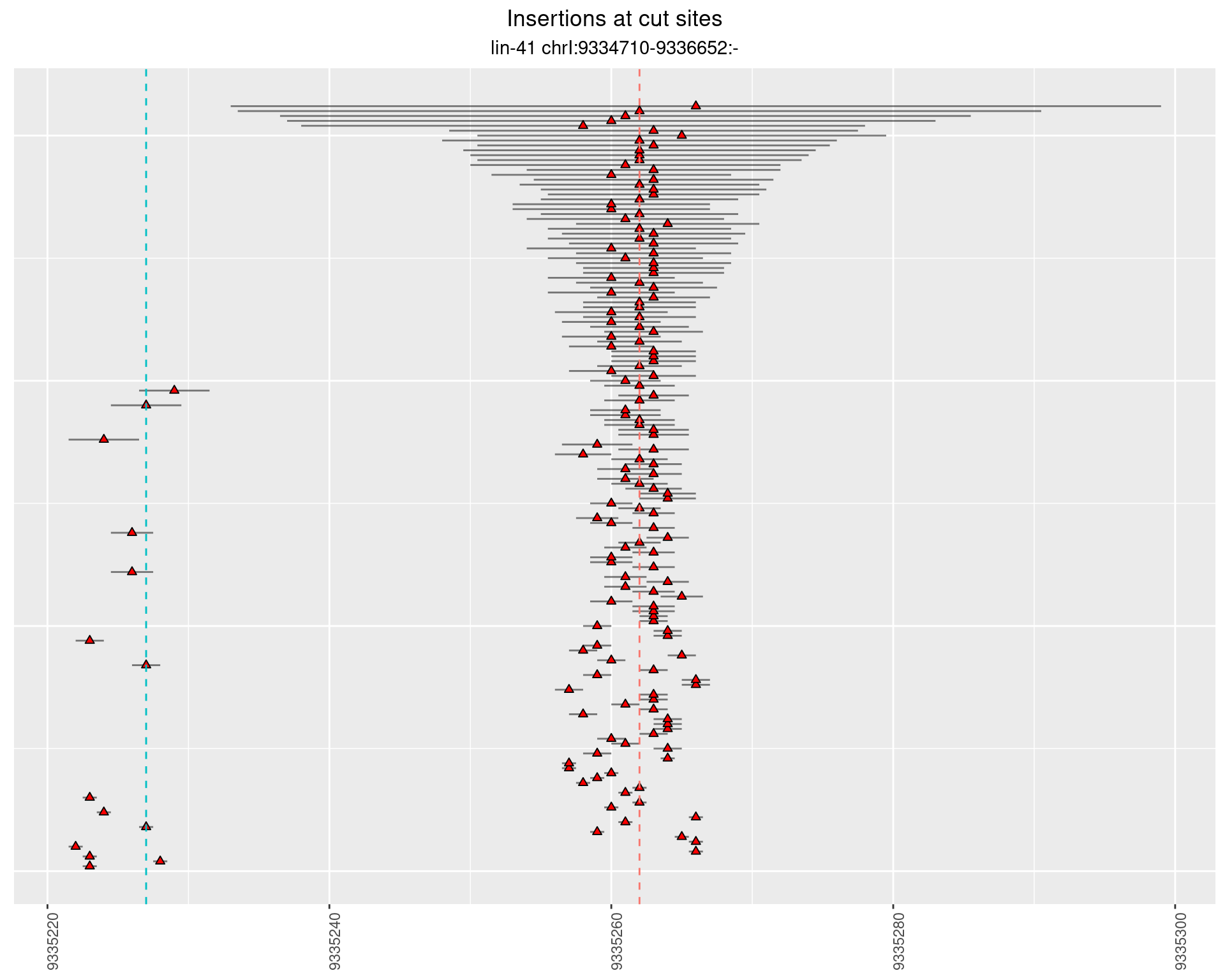
2.102.2 Freq: 0.001 - 0.01
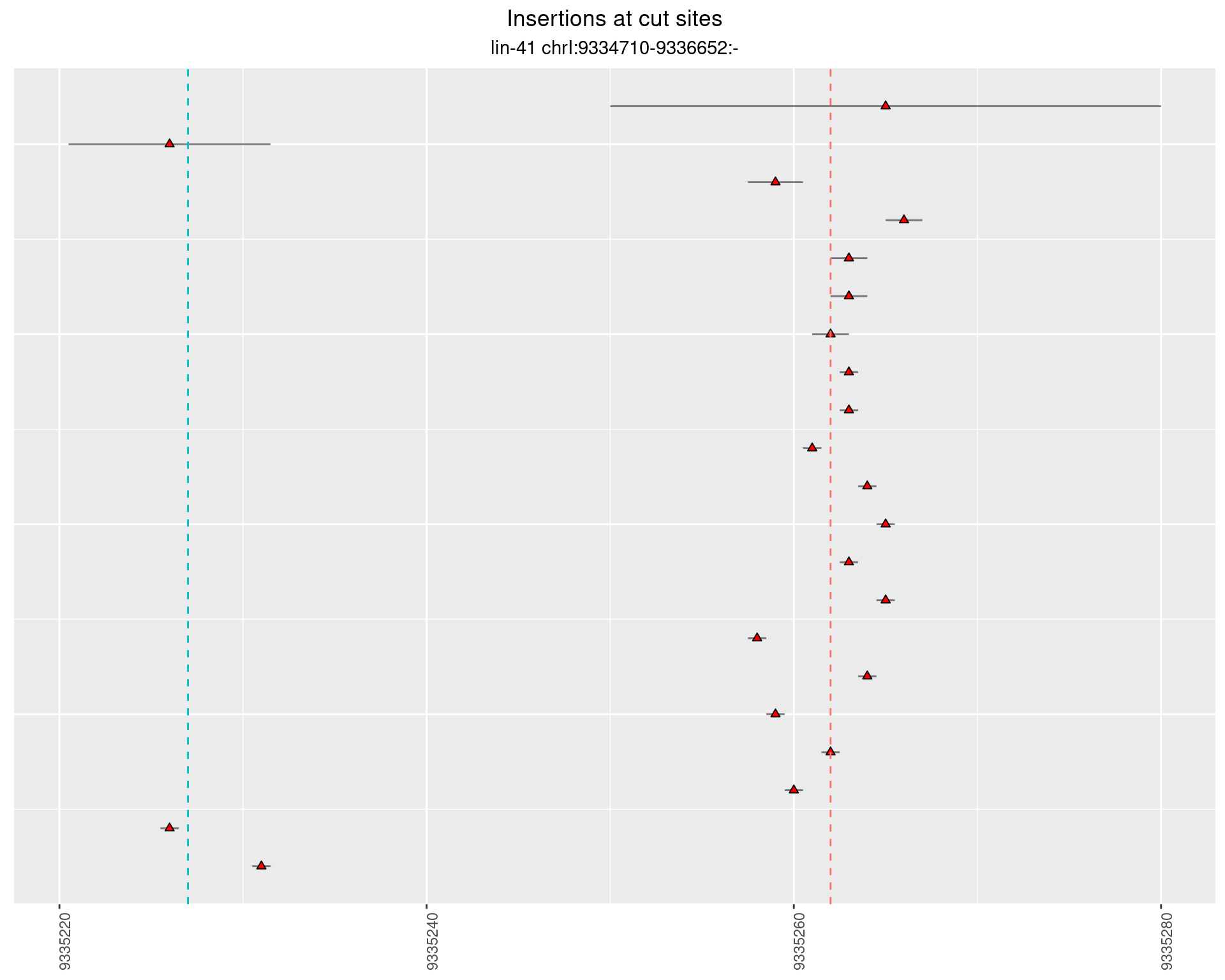
2.102.3 Freq: 1e-05 - 1e-04
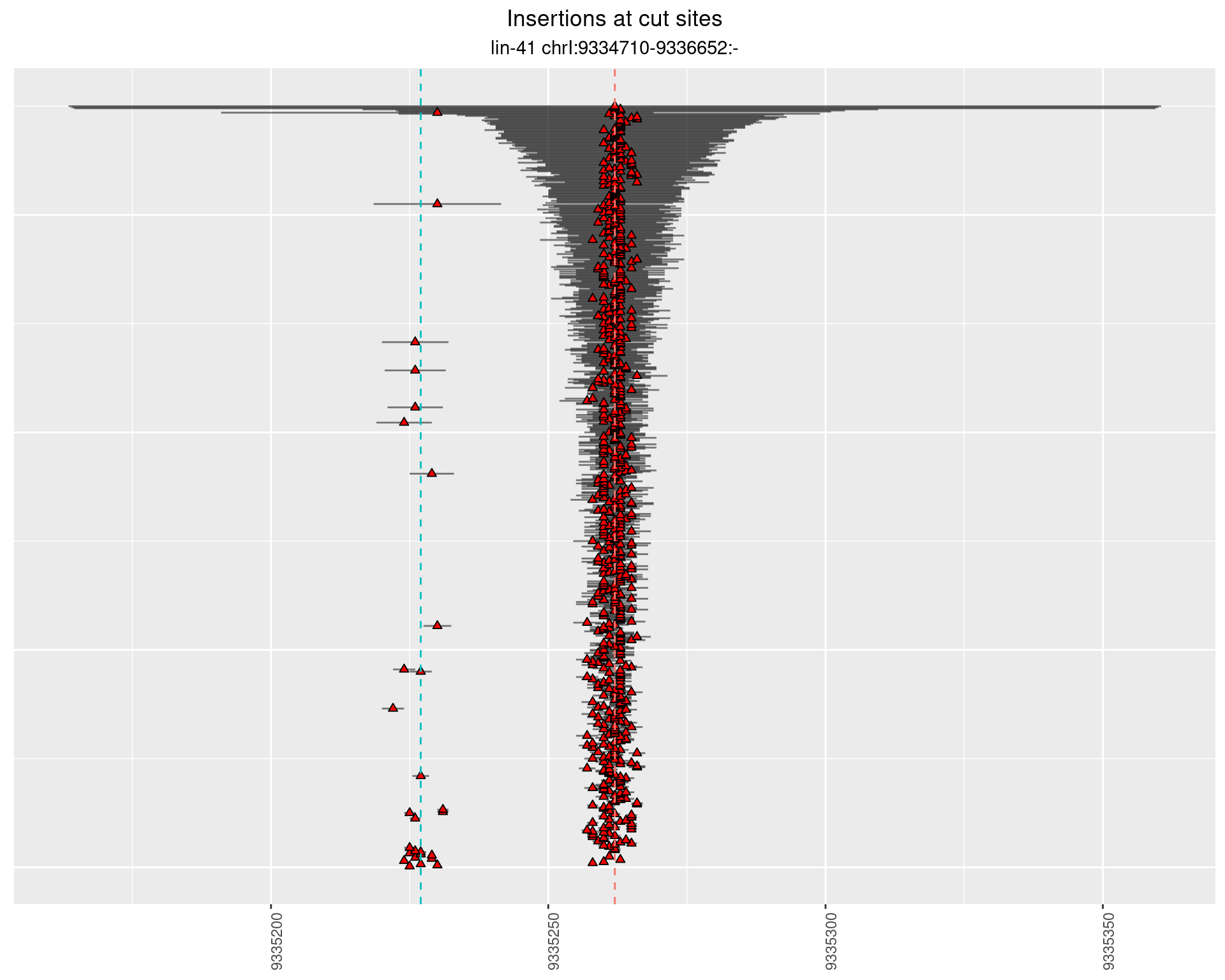
2.102.4 Freq: 0.01 - 0.1
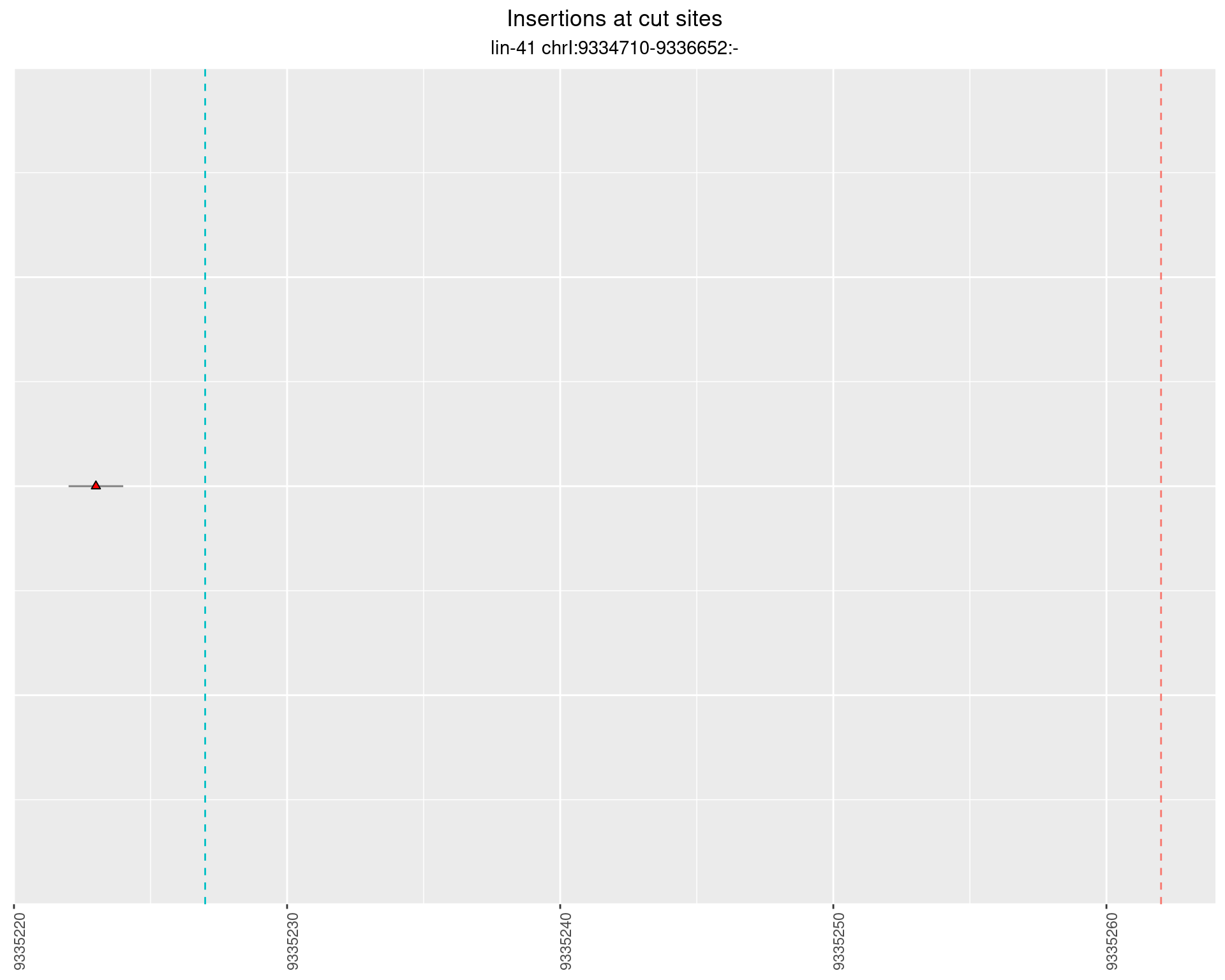
2.103 lin41_RNApacbio_L1_2627
2.103.1 Freq: 0.001 - 0.01
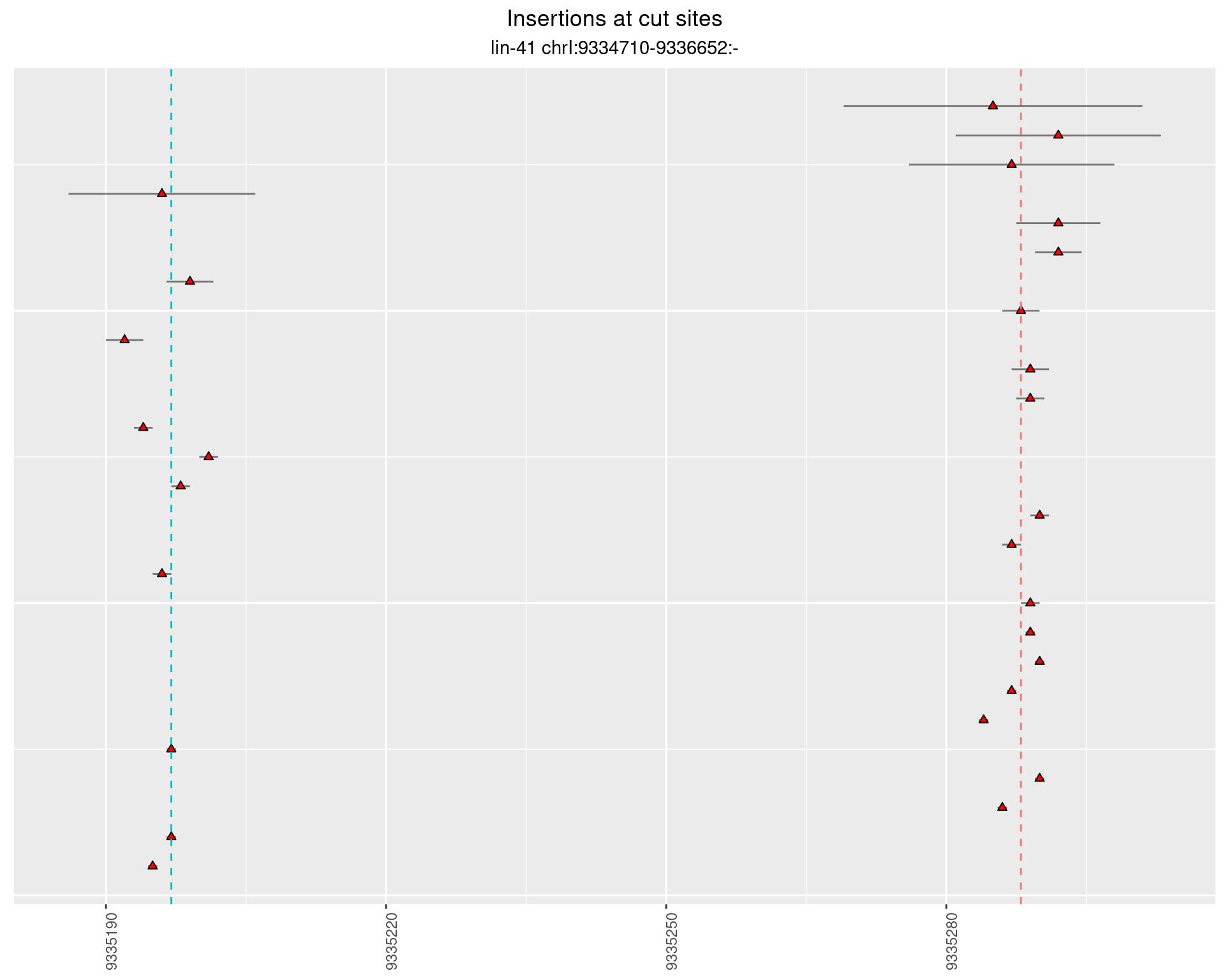
2.103.2 Freq: 0.01 - 0.1
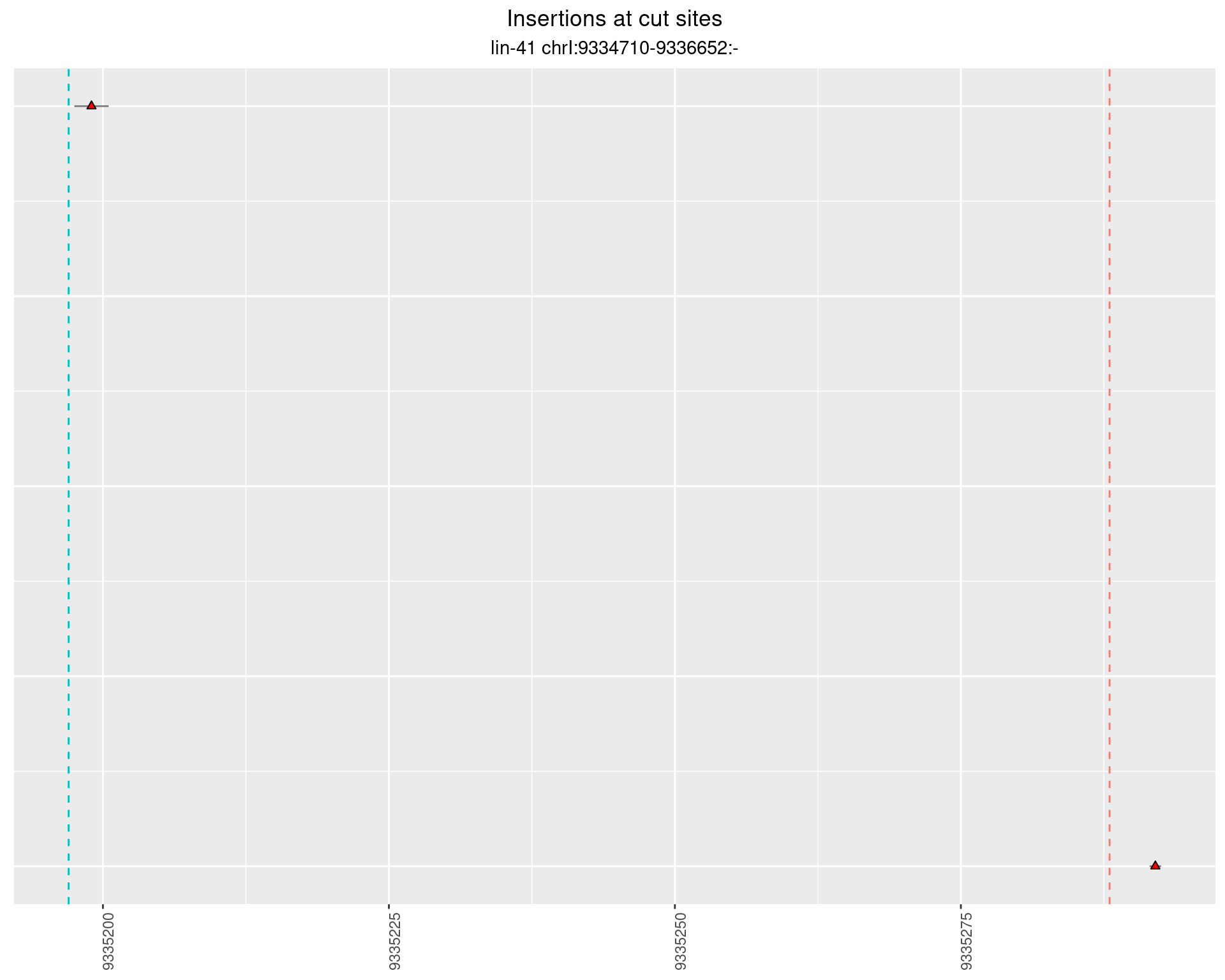
2.103.3 Freq: 1e-04 - 0.001
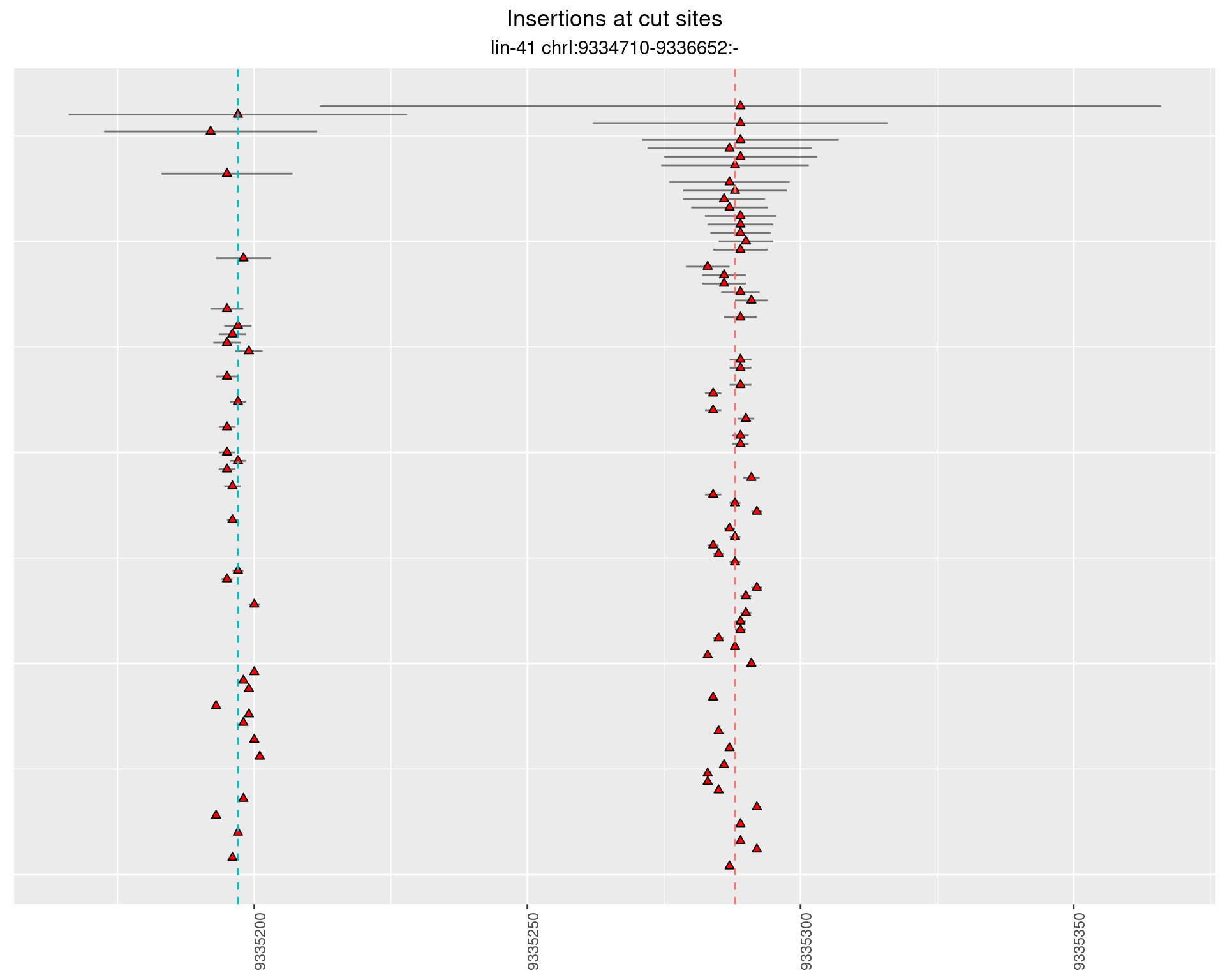
2.103.4 Freq: 1e-05 - 1e-04
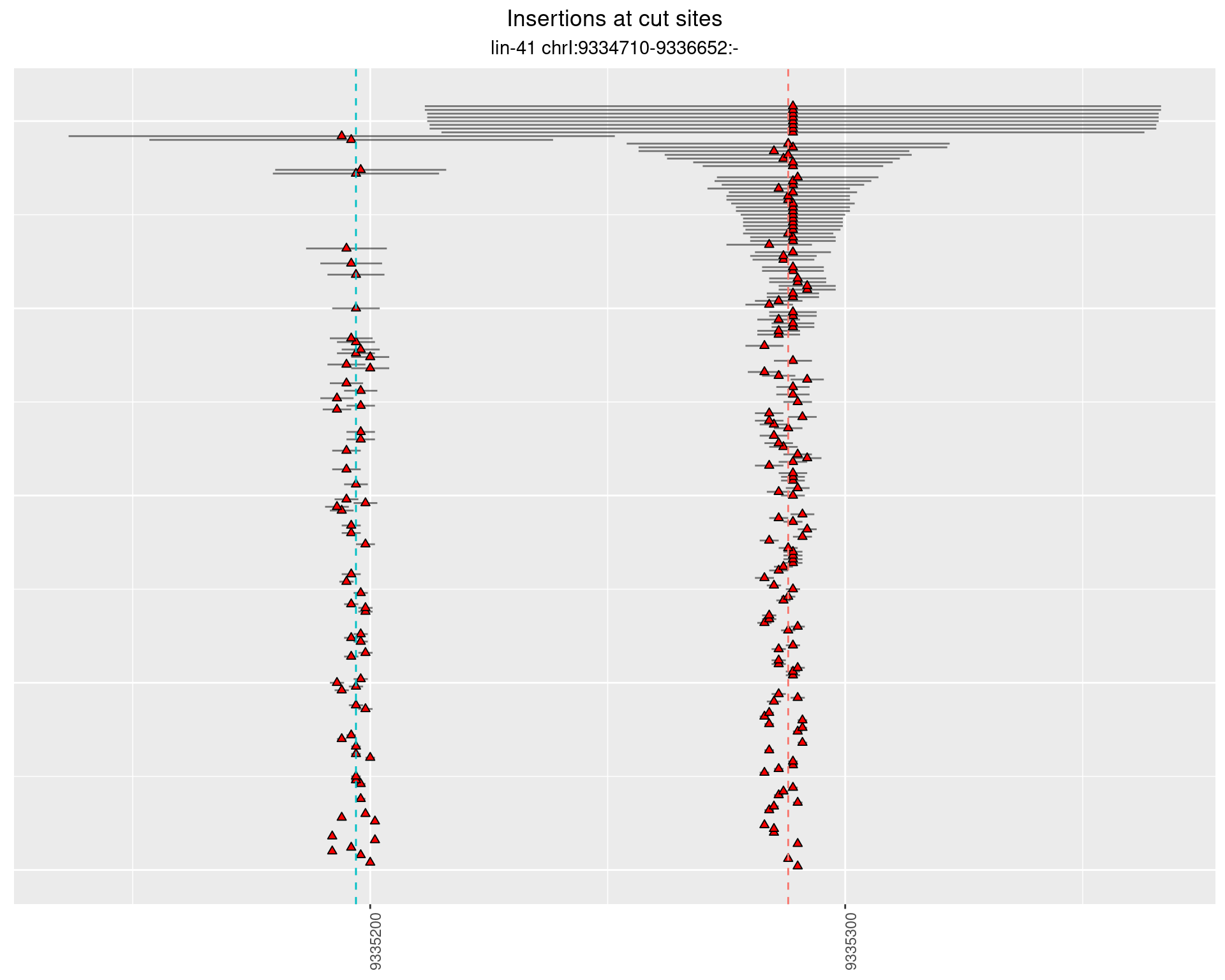
2.104 lin41_RNApacbio_L1_pool3
2.104.1 Freq: 1e-05 - 1e-04
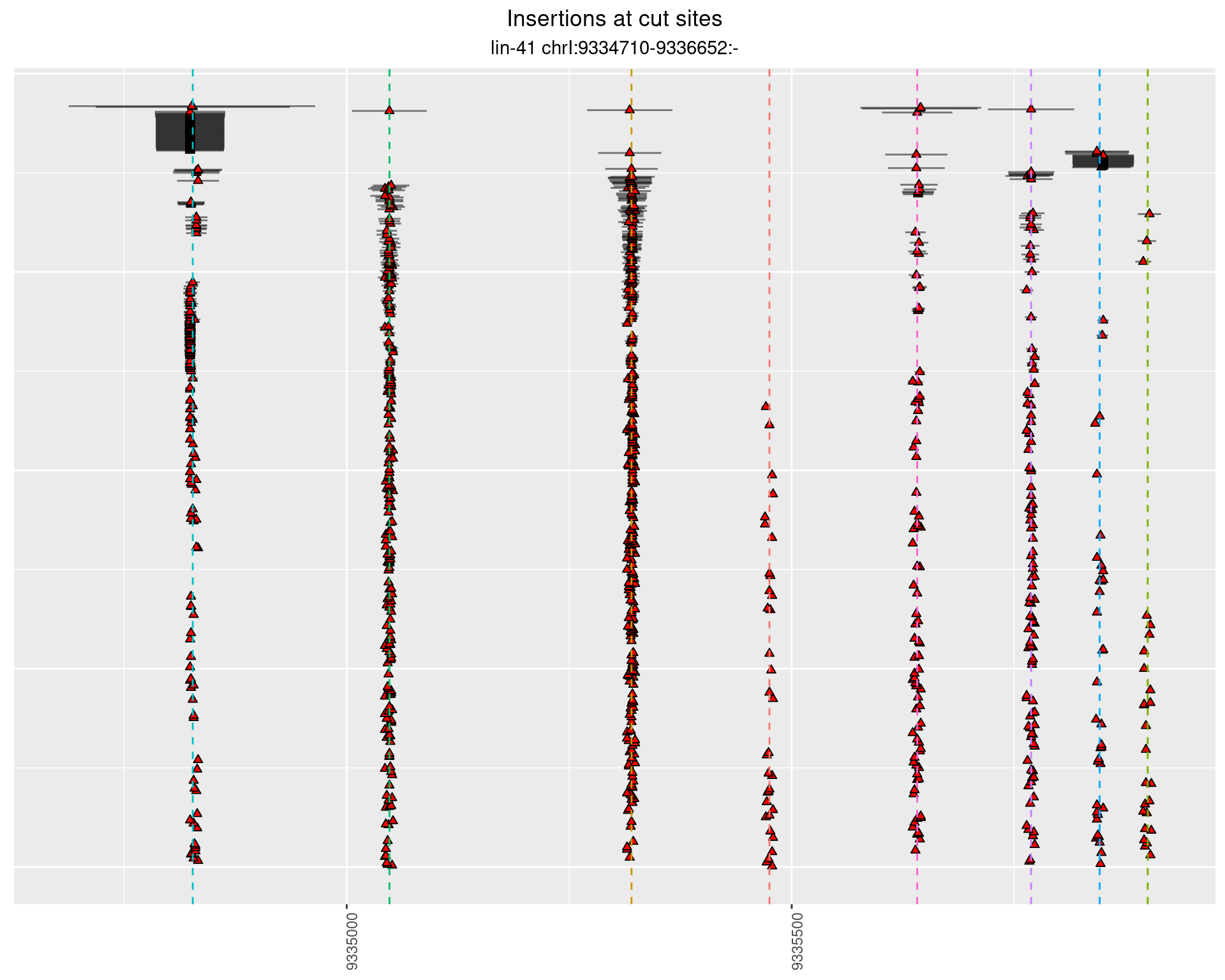
2.104.2 Freq: 0.01 - 0.1
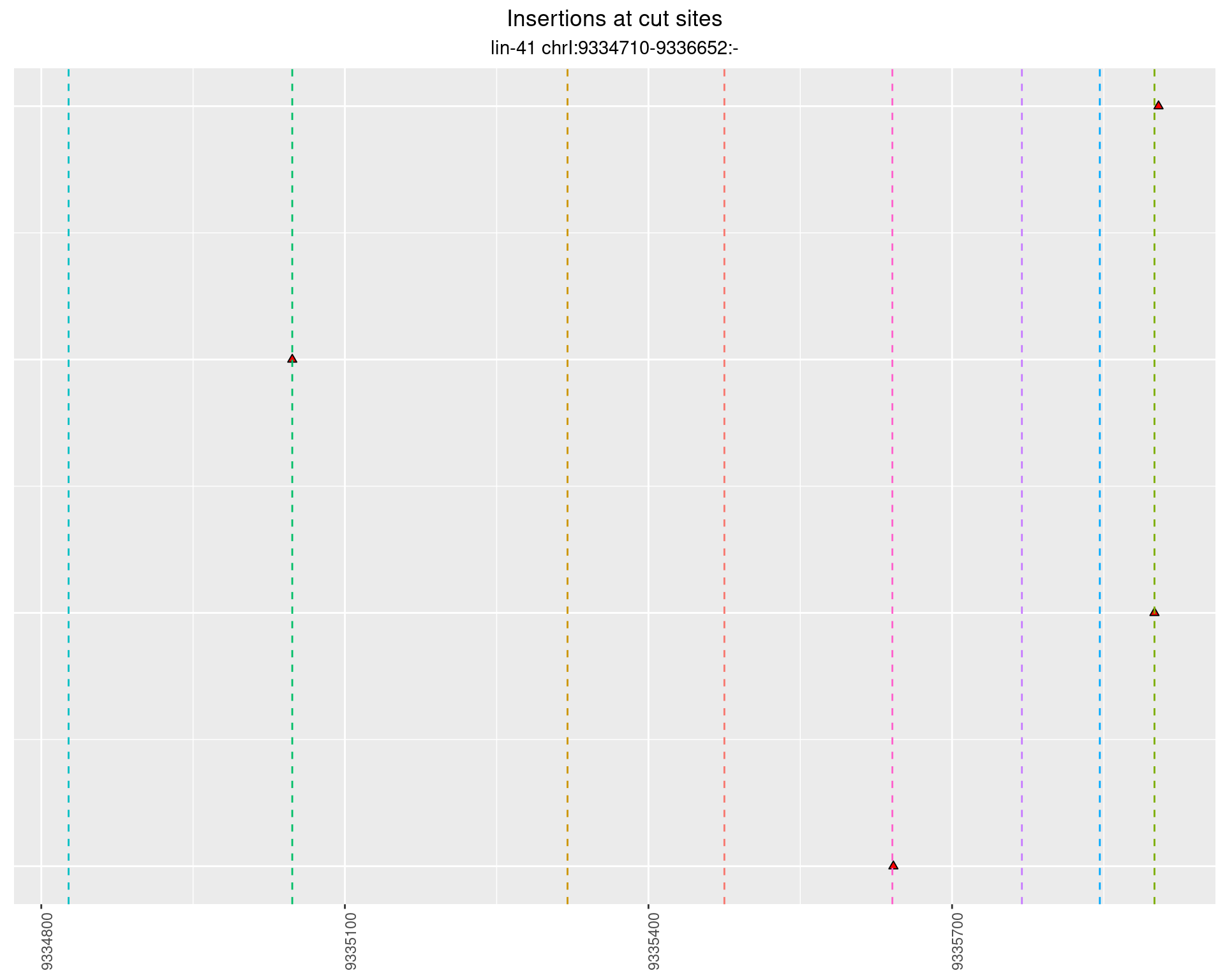
2.104.3 Freq: 0.001 - 0.01
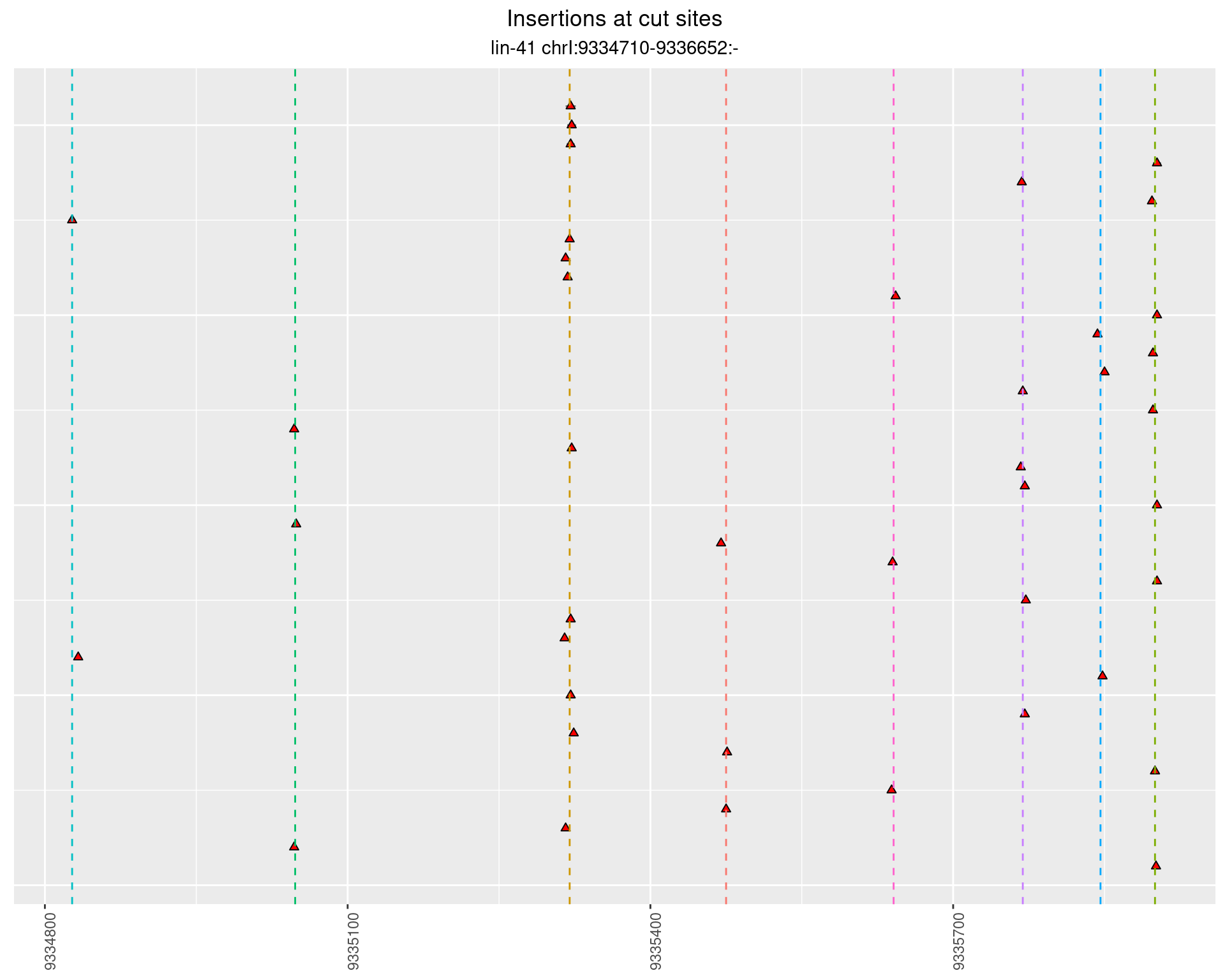
2.104.4 Freq: 1e-04 - 0.001
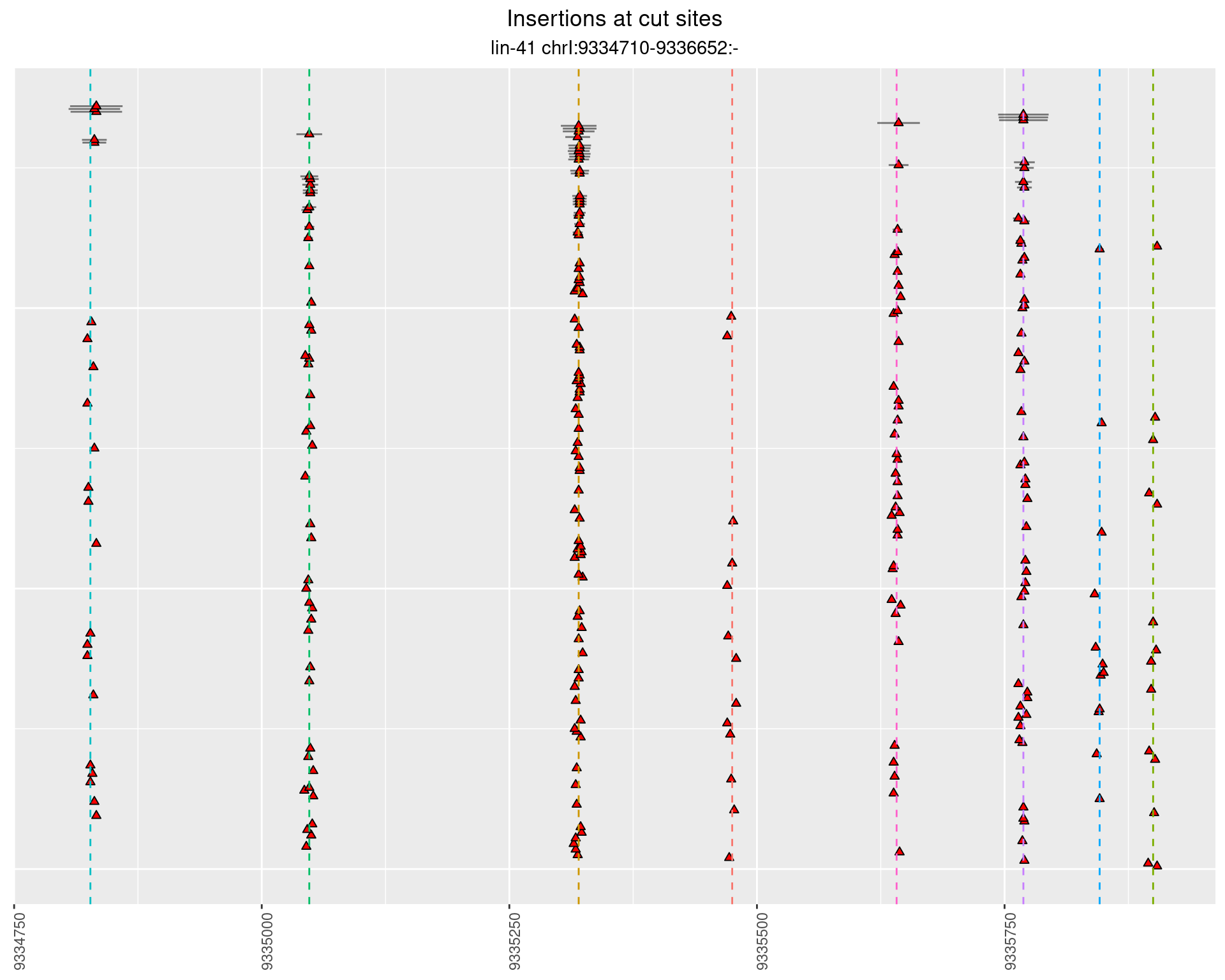
2.105 lin41_RNApacbio_L1_N2
2.105.1 Freq: 1e-04 - 0.001
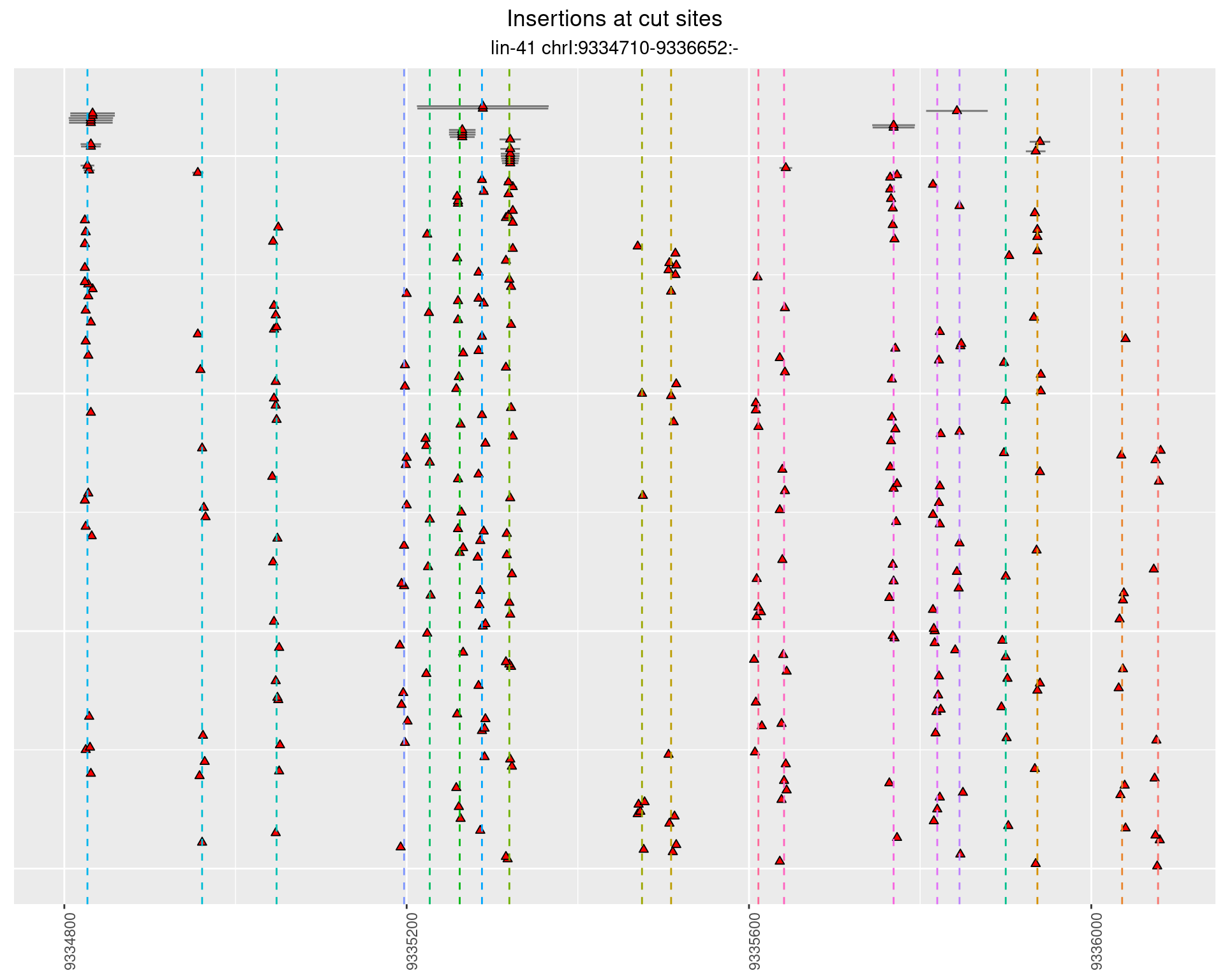
2.105.2 Freq: 0.01 - 0.1
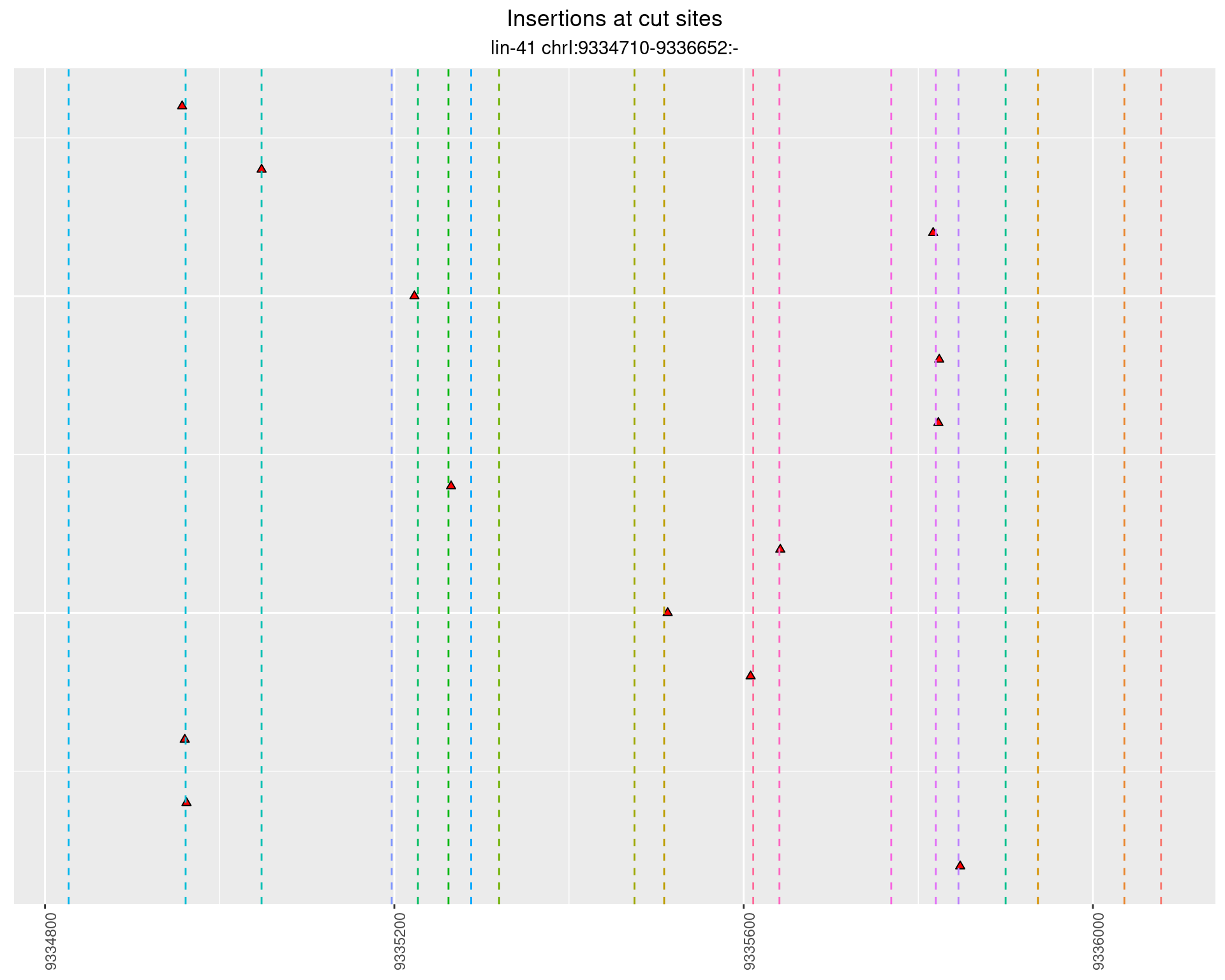
2.105.3 Freq: 0.001 - 0.01
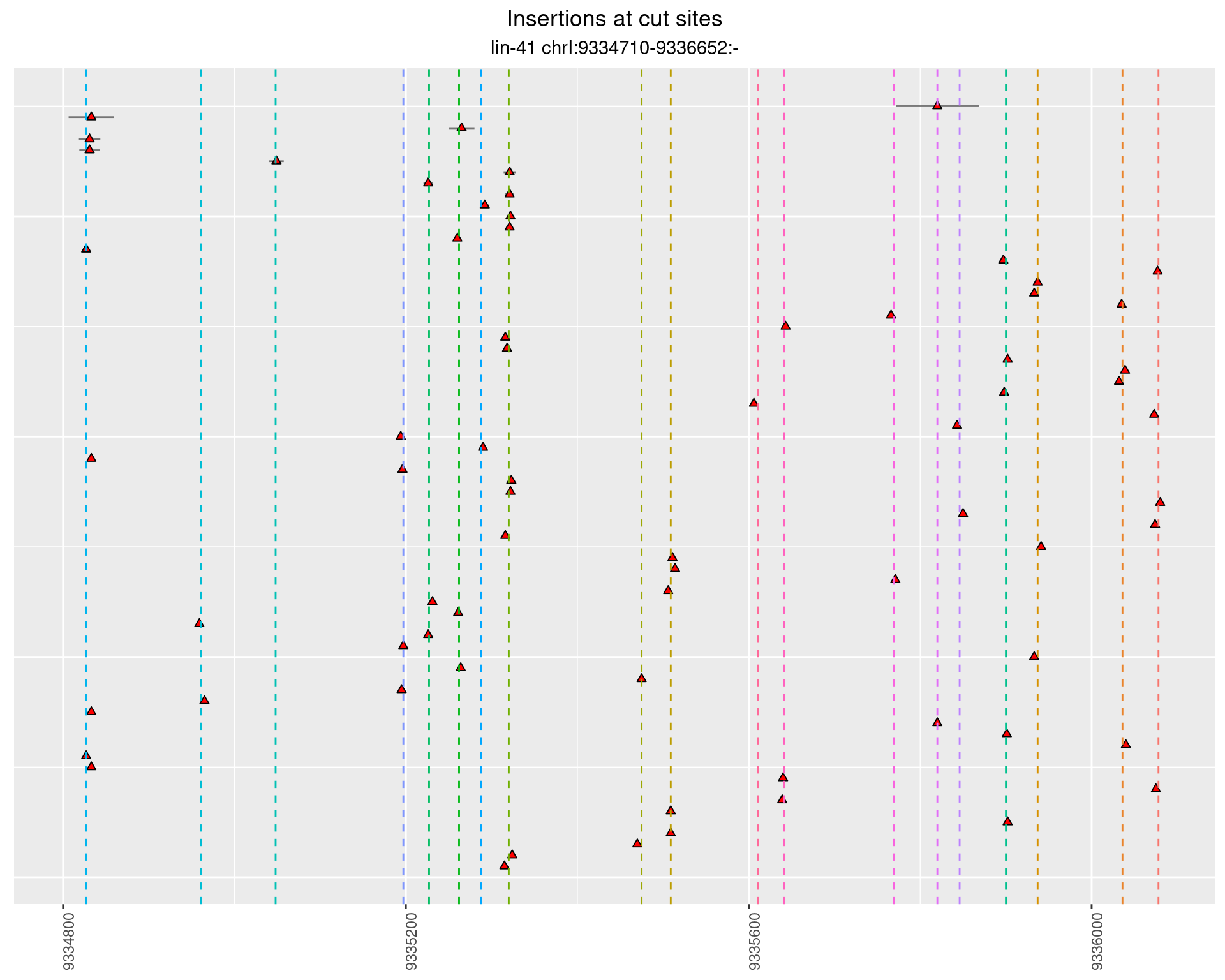
2.105.4 Freq: 1e-05 - 1e-04
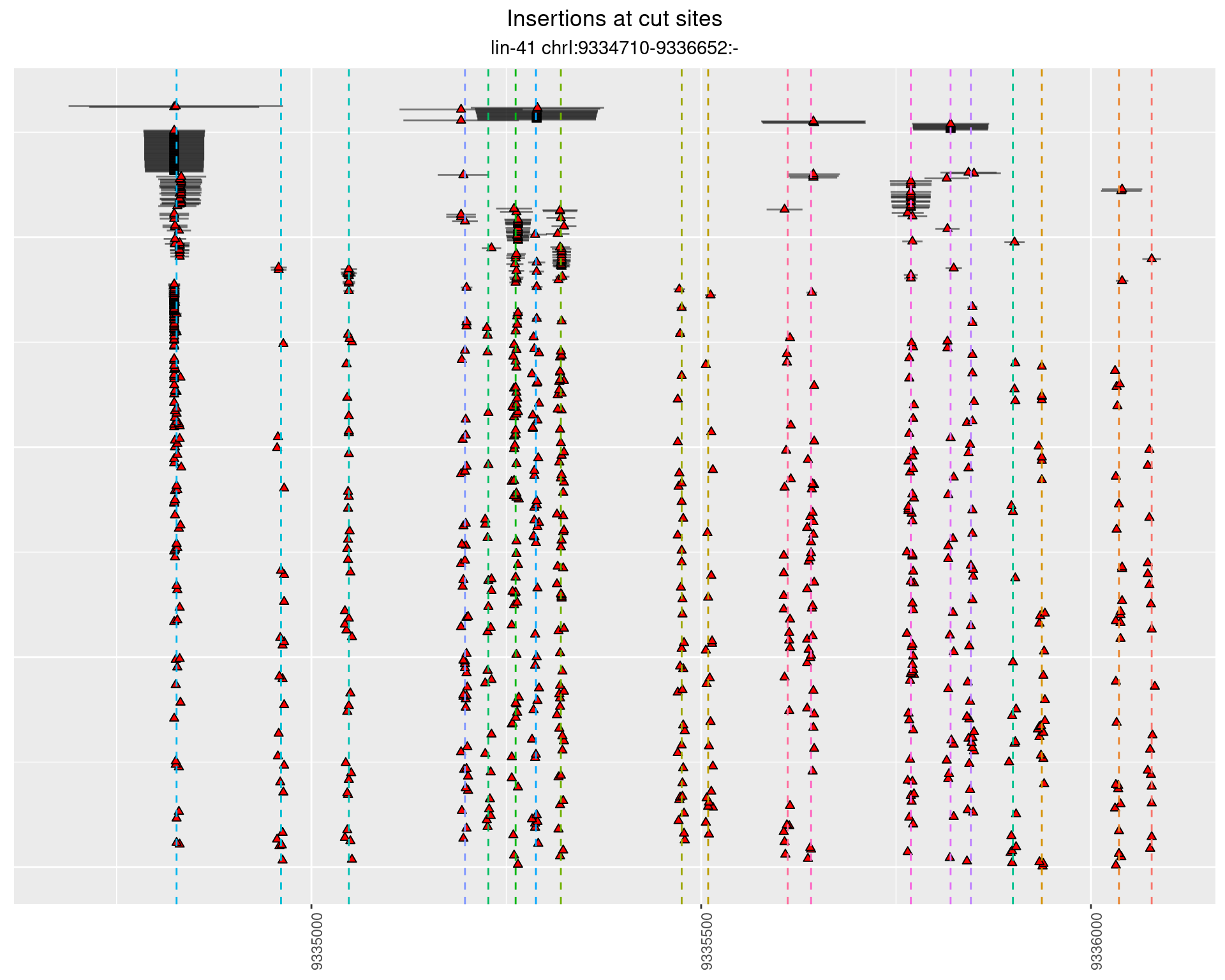
2.106 lin41_RNApacbio_L4_1516
2.106.1 Freq: 0.001 - 0.01
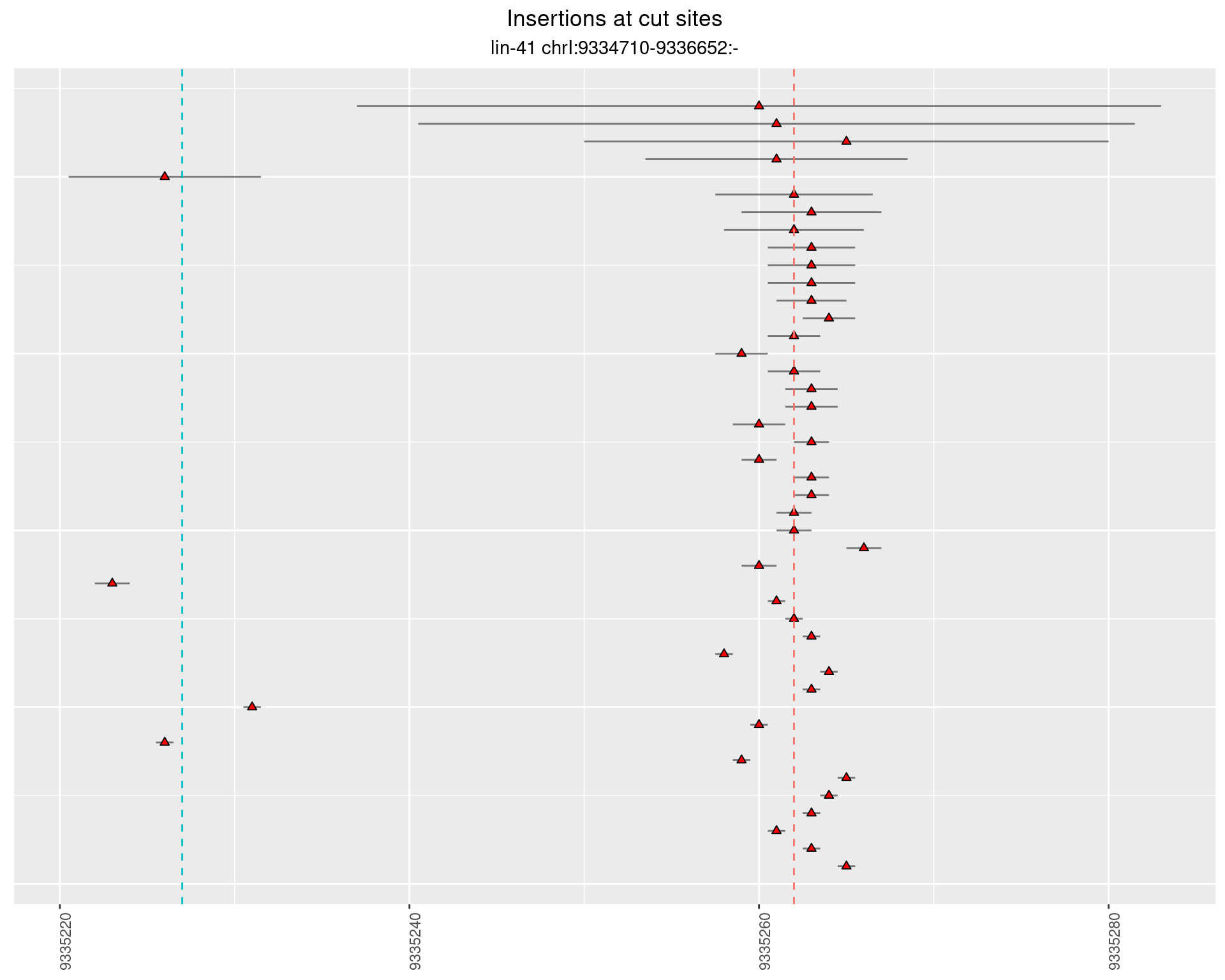
2.106.2 Freq: 1e-04 - 0.001
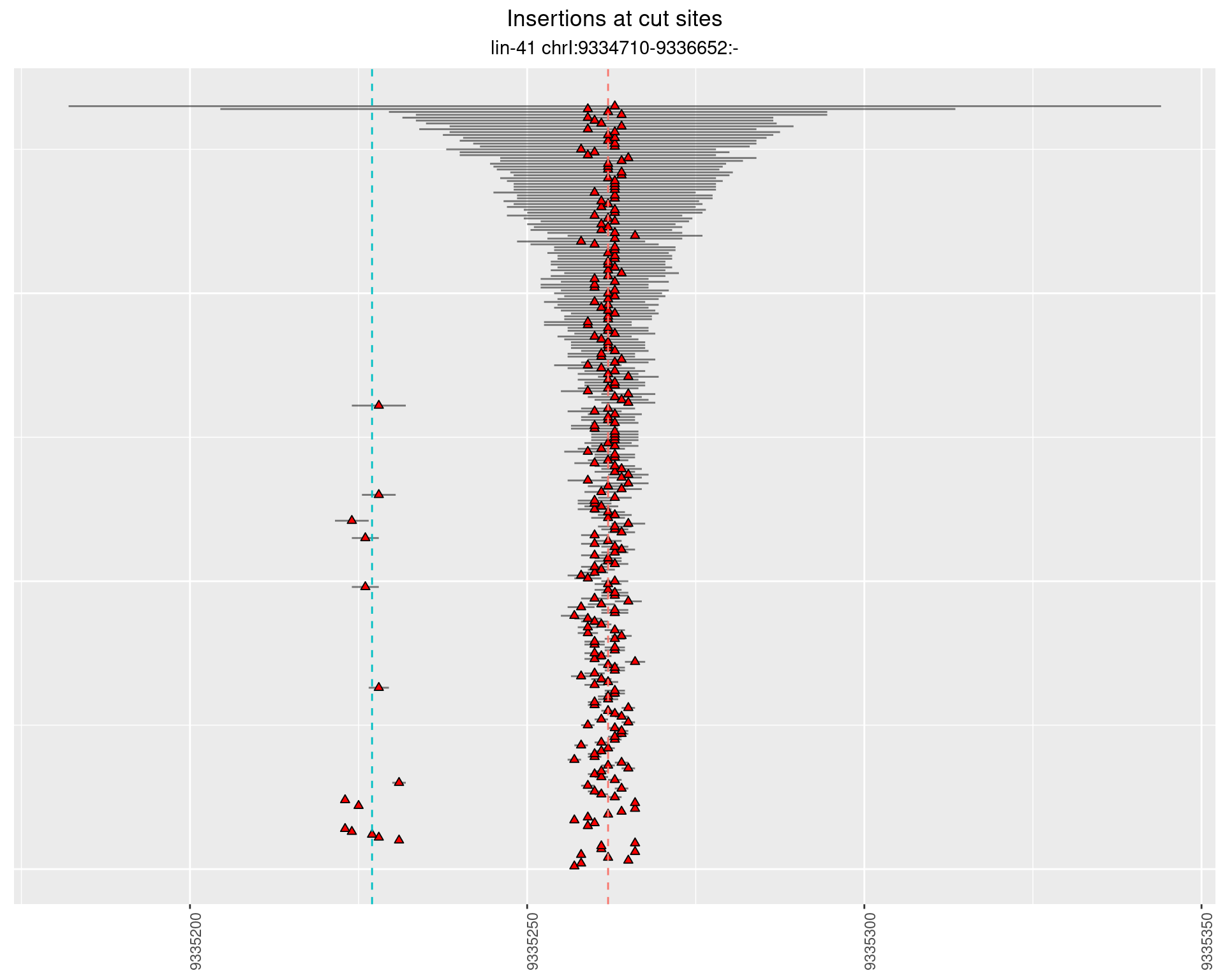
2.106.3 Freq: 1e-05 - 1e-04
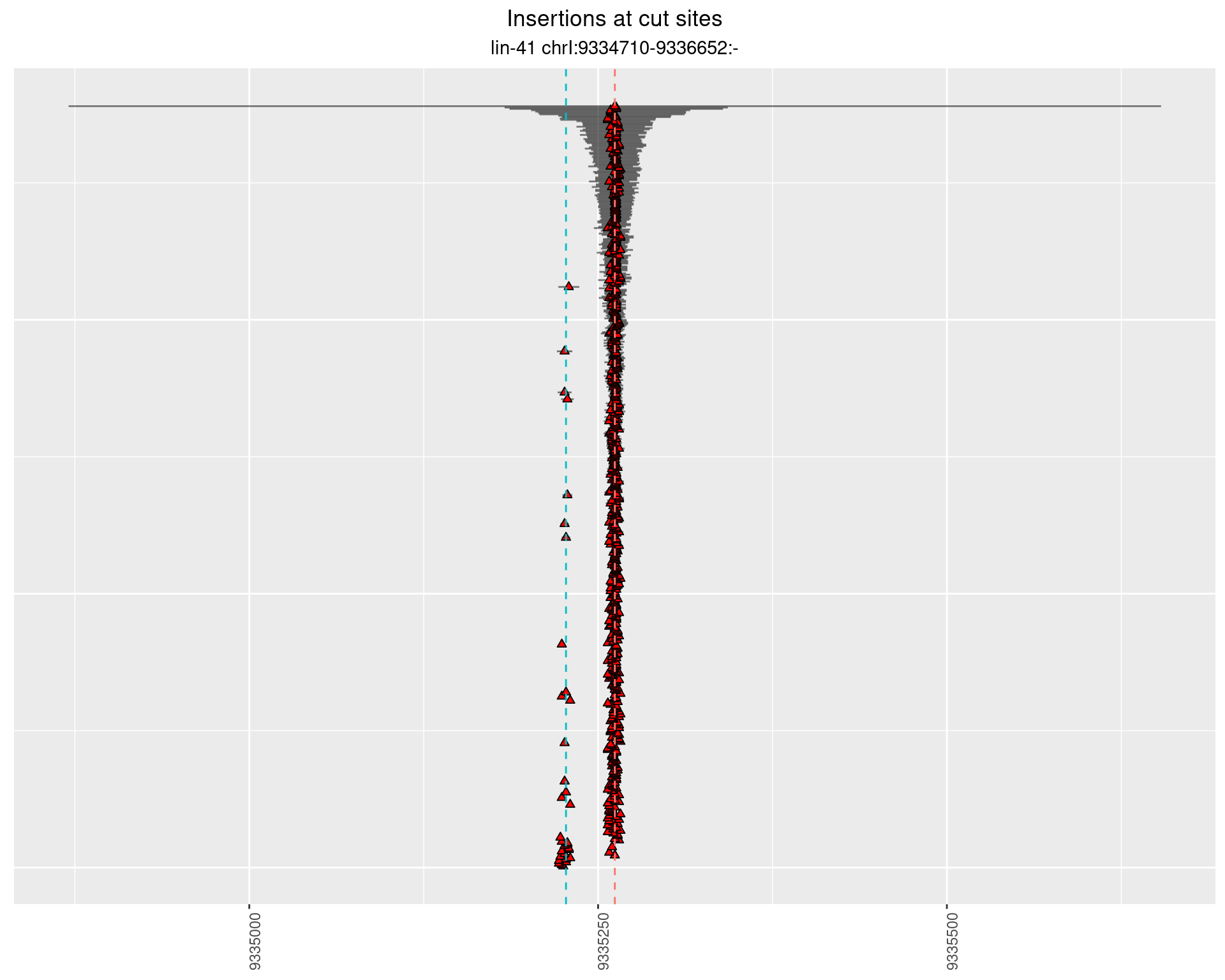
2.107 lin41_RNApacbio_L4_2627
2.107.1 Freq: 1e-04 - 0.001
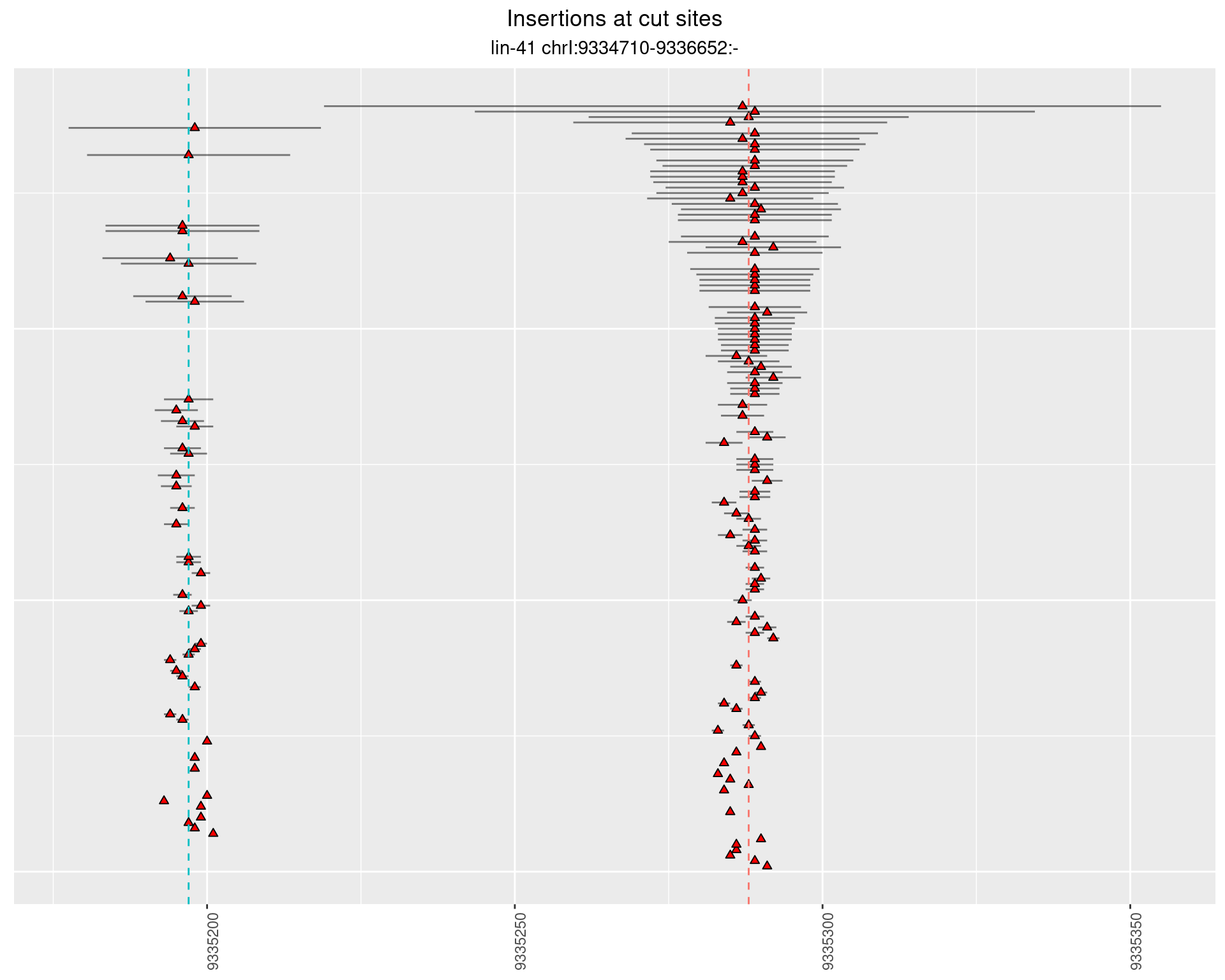
2.107.2 Freq: 0.001 - 0.01
2.107.3 Freq: 1e-05 - 1e-04
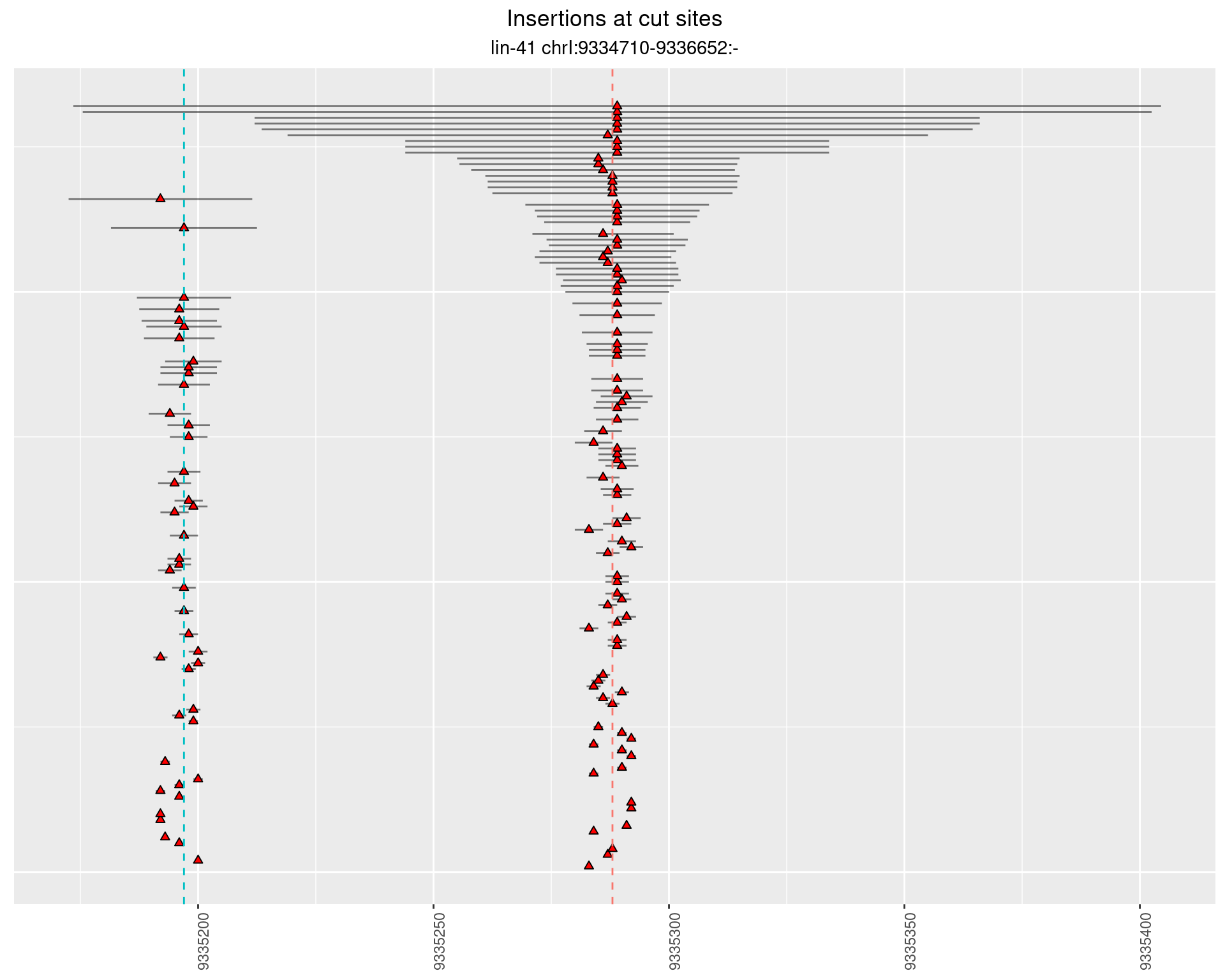
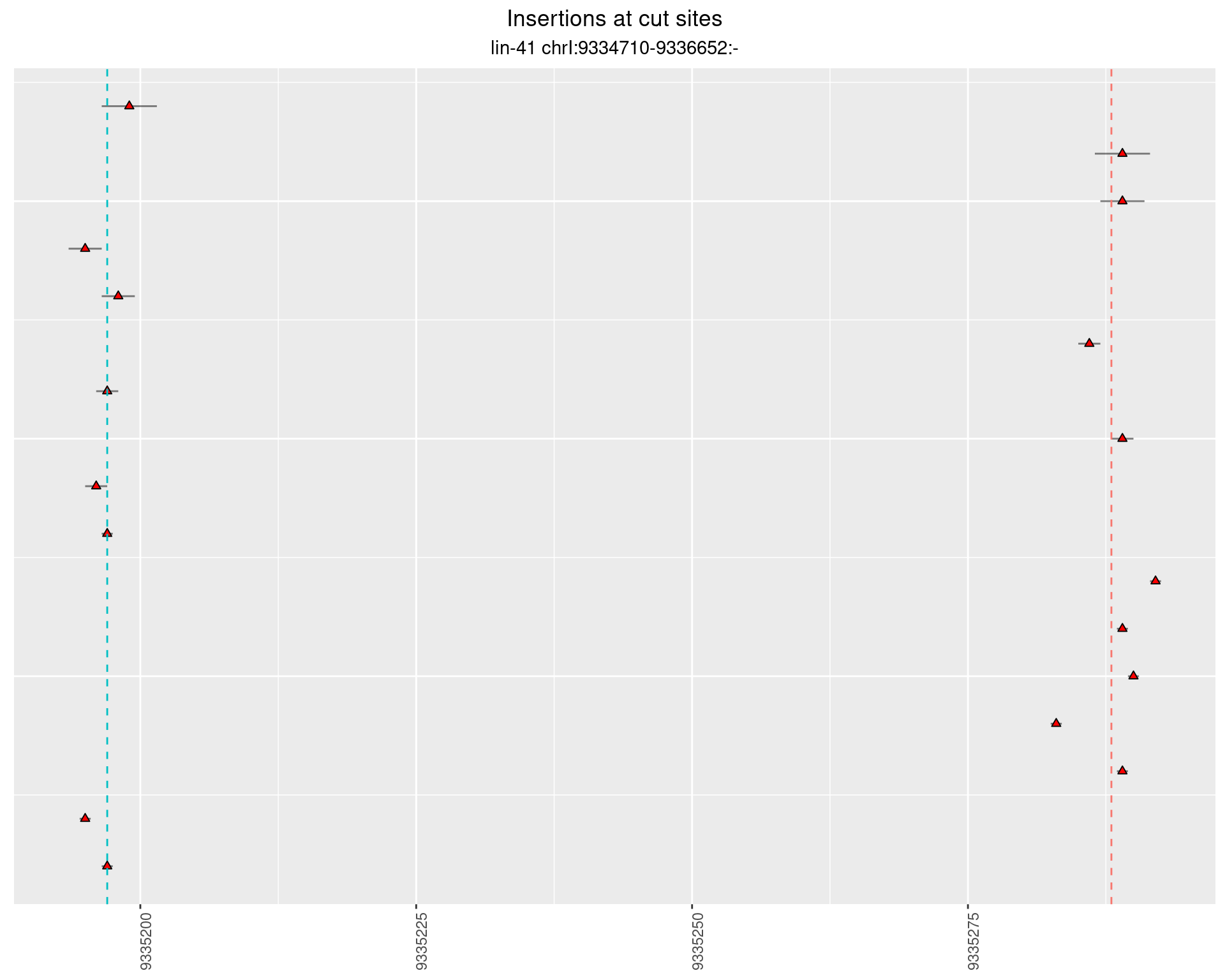
2.108 lin41_RNApacbio_L4_pool3
2.108.1 Freq: 0.001 - 0.01
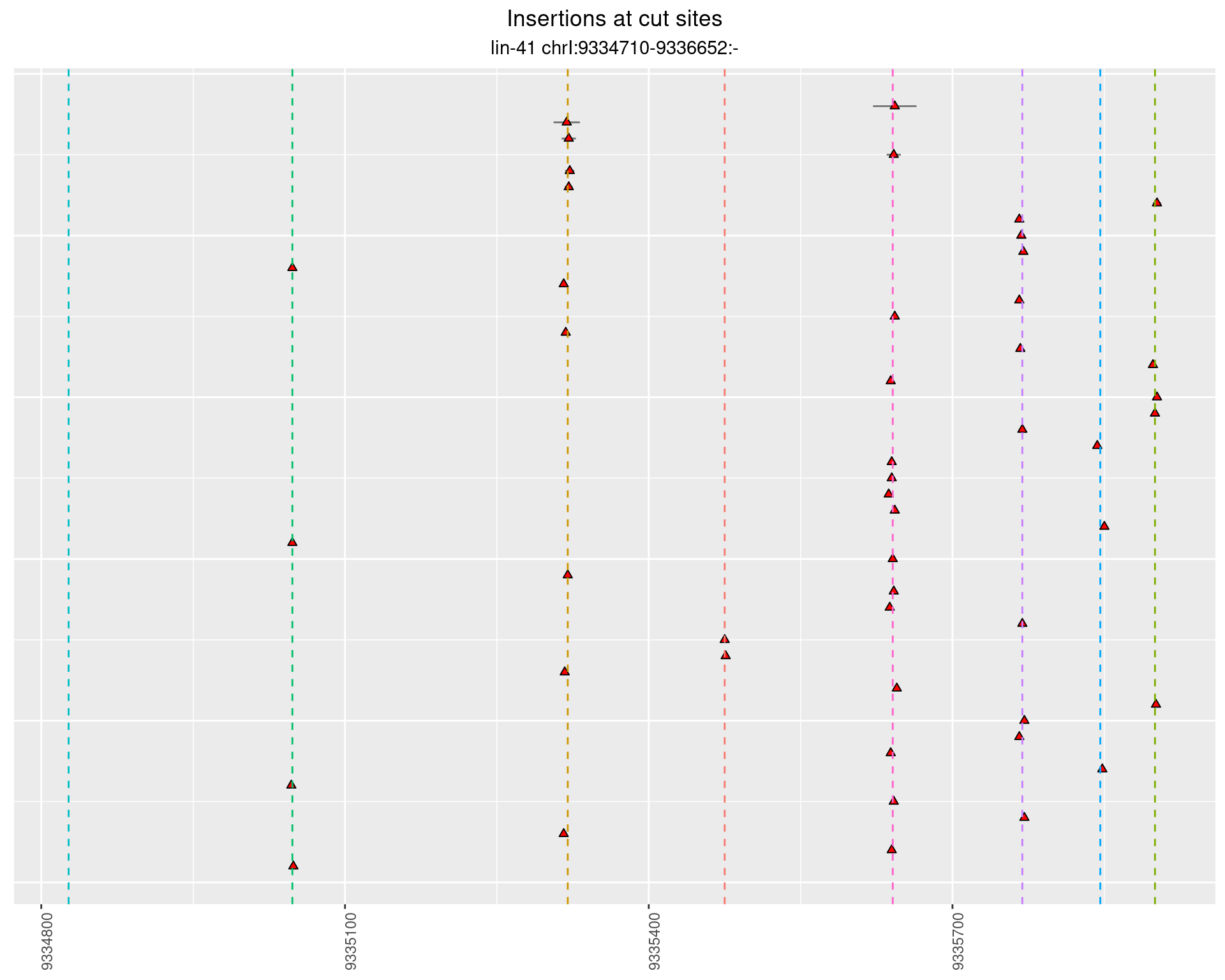
2.108.2 Freq: 1e-04 - 0.001
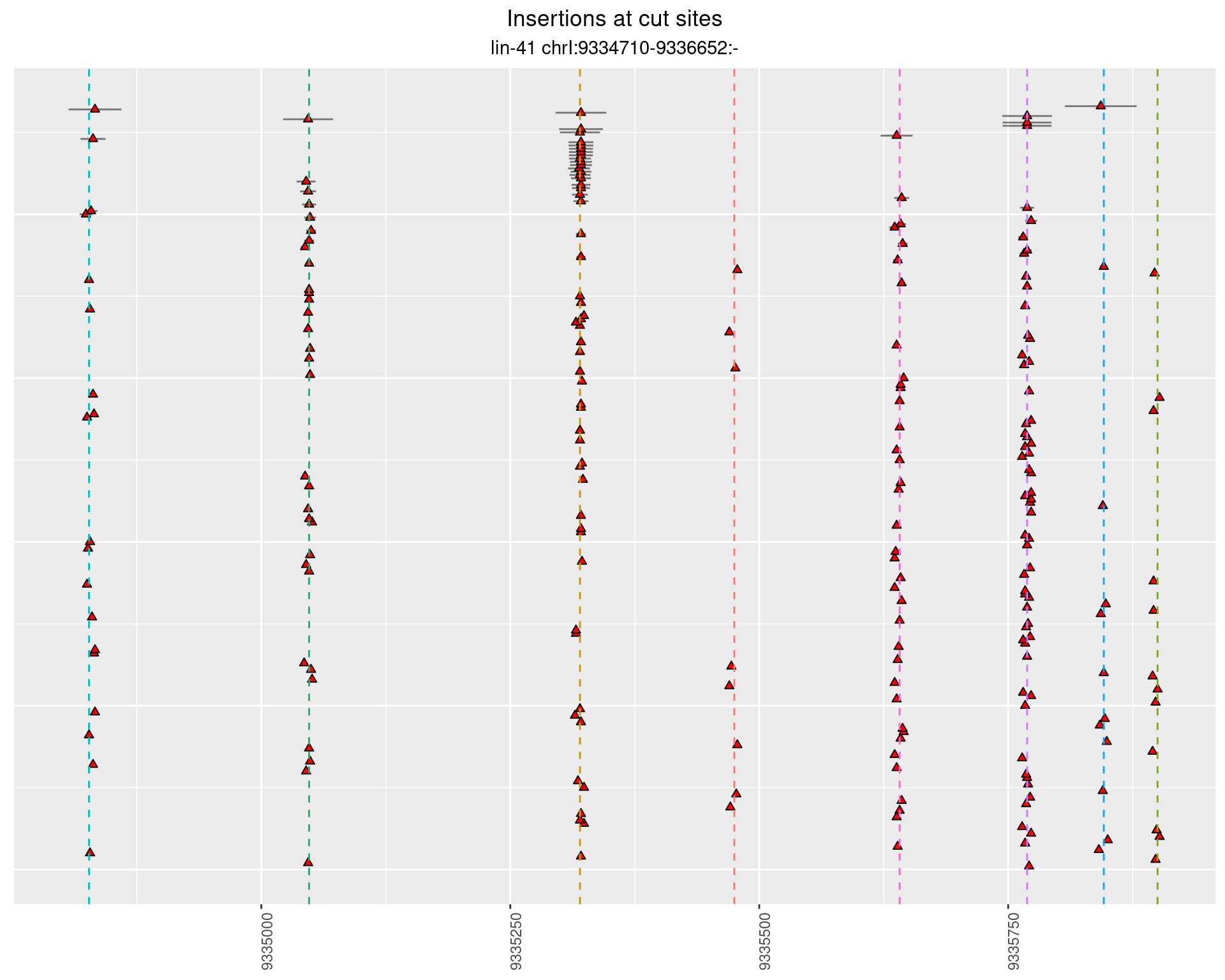
2.108.3 Freq: 0.01 - 0.1
2.108.4 Freq: 1e-05 - 1e-04
2.109 lin41_RNApacbio_L4_N2
2.109.1 Freq: 0.01 - 0.1
2.109.2 Freq: 1e-04 - 0.001
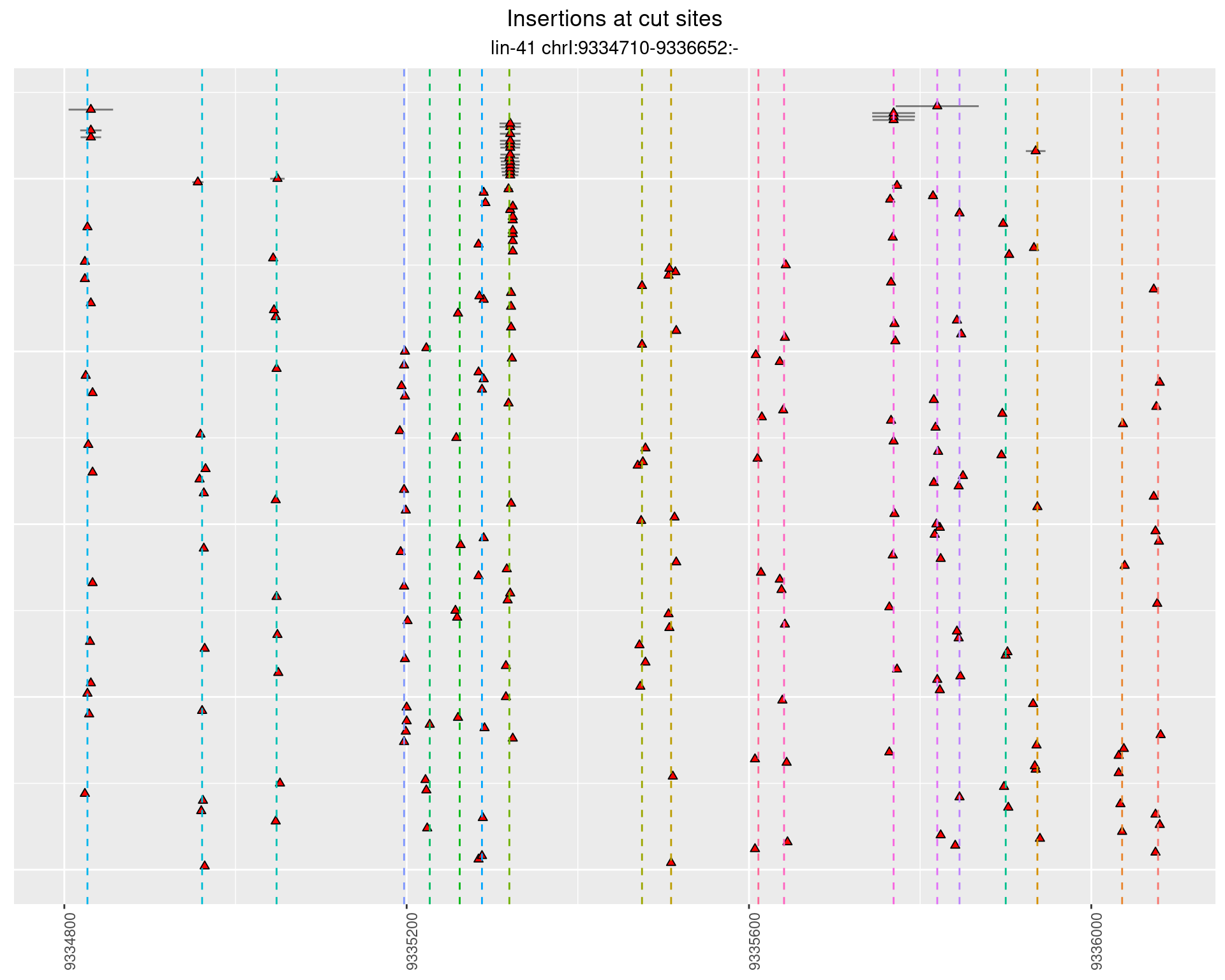
2.109.3 Freq: 0.001 - 0.01
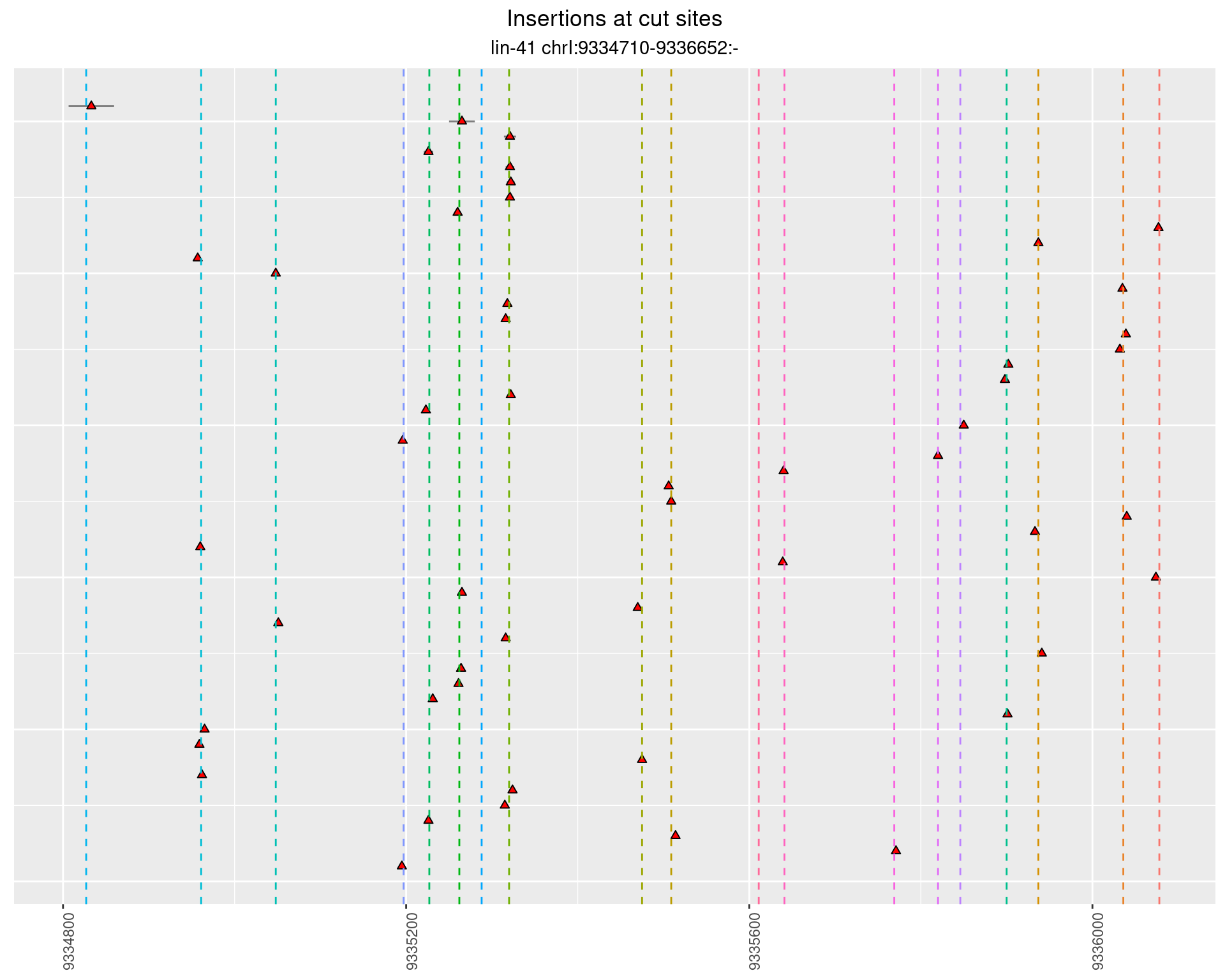
2.109.4 Freq: 1e-05 - 1e-04
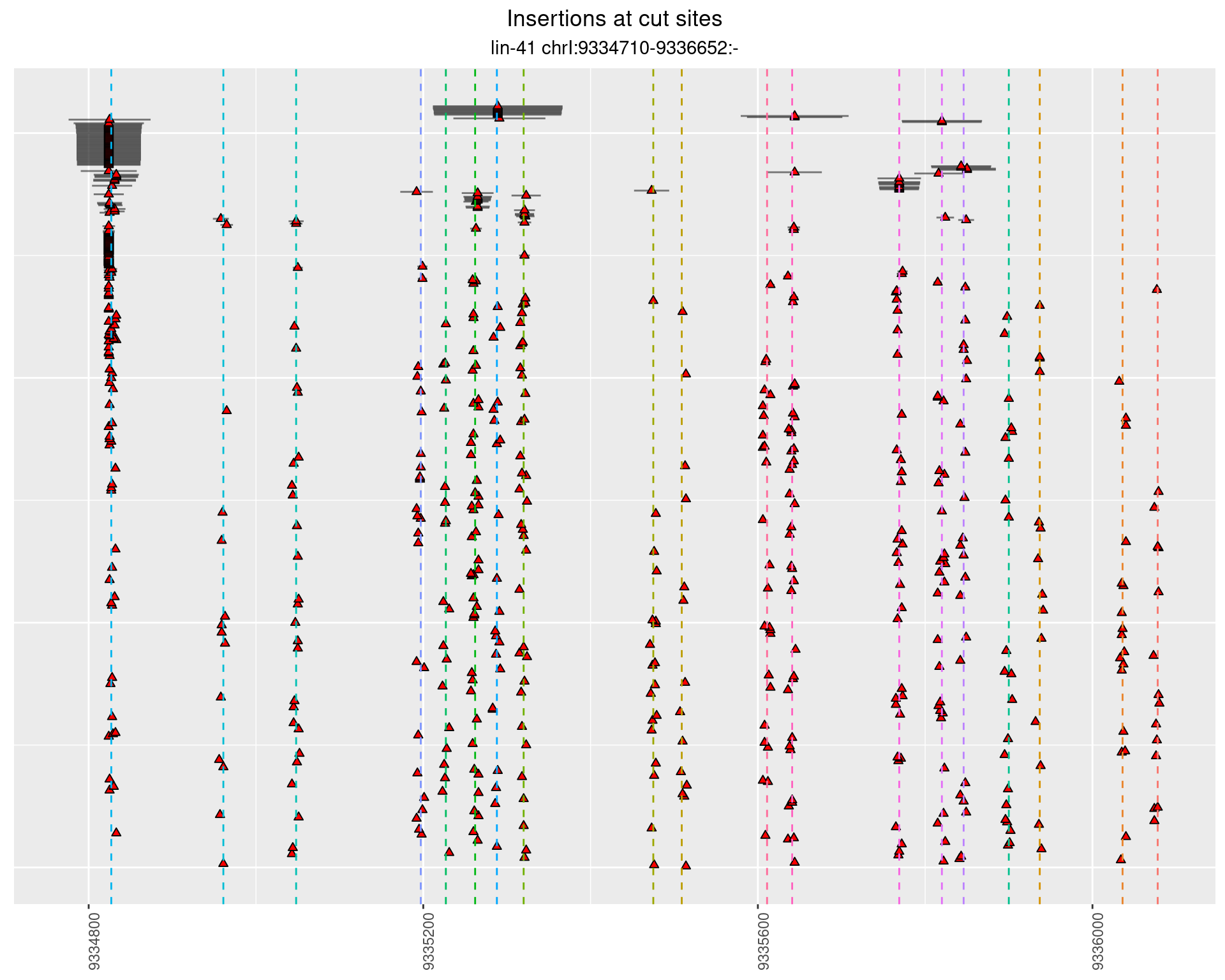
2.110 lin41_RNApacbio_L1_all
2.110.1 Freq: 1e-05 - 1e-04
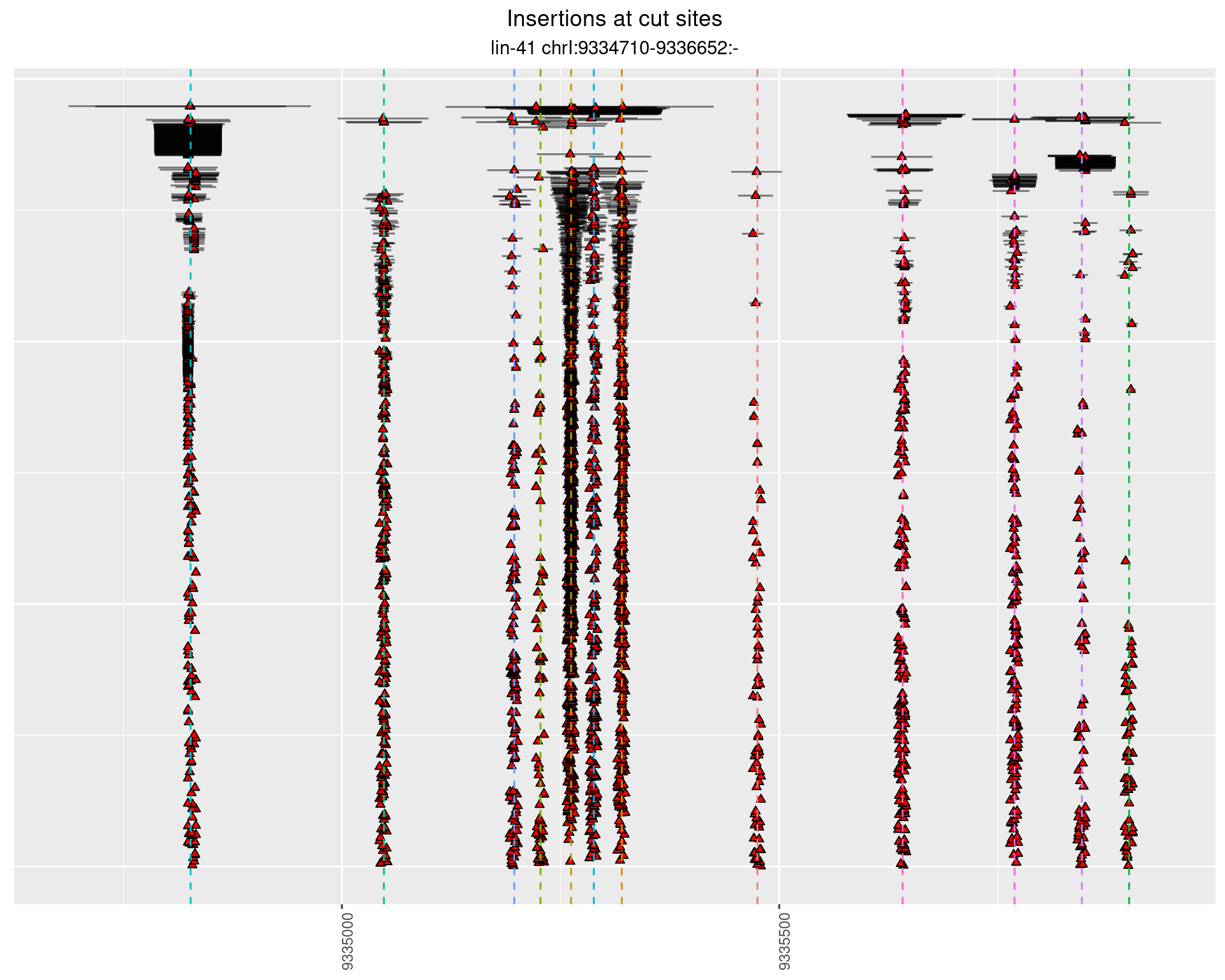
2.110.2 Freq: 0.01 - 0.1
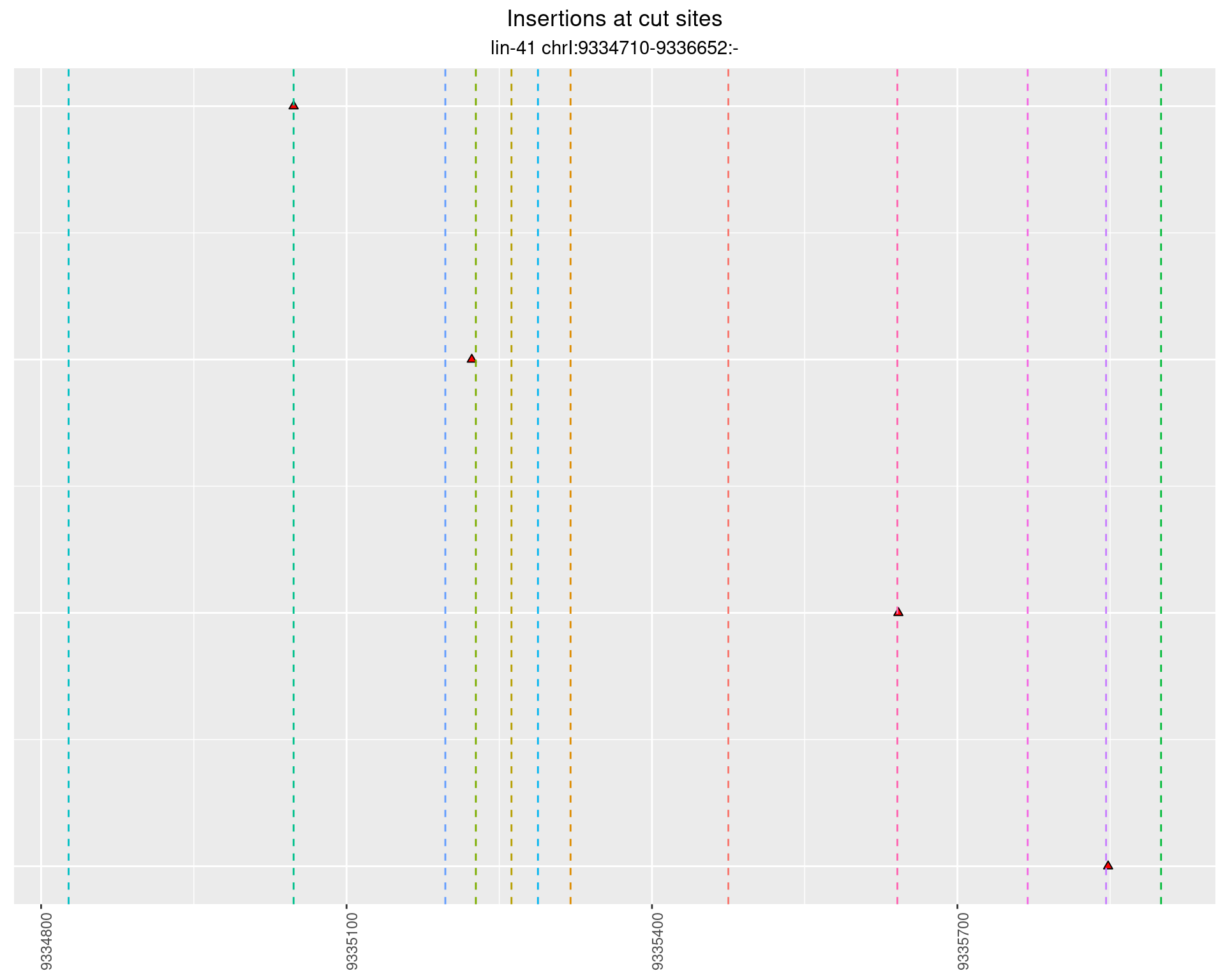
2.110.3 Freq: 0.001 - 0.01
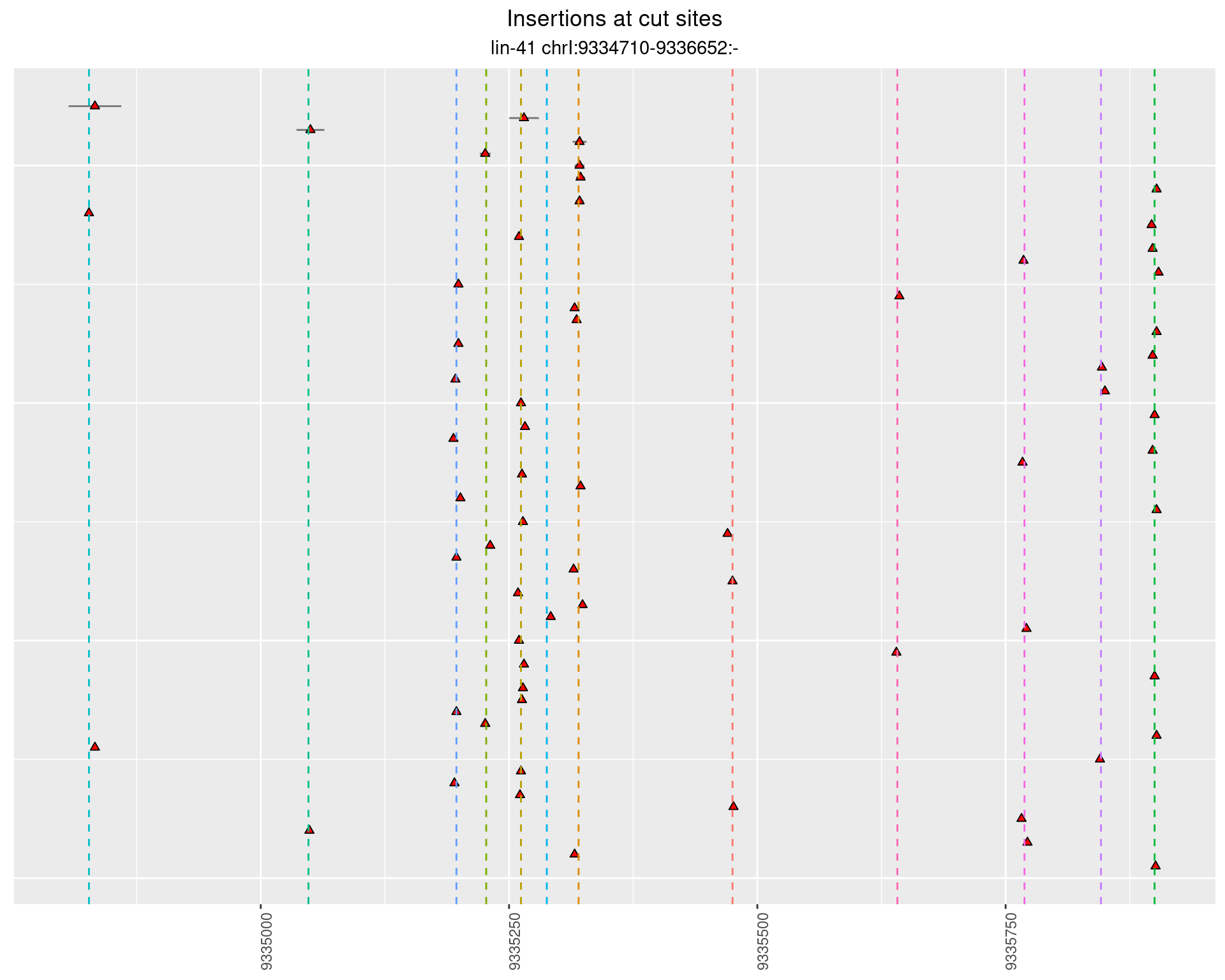
2.110.4 Freq: 1e-04 - 0.001
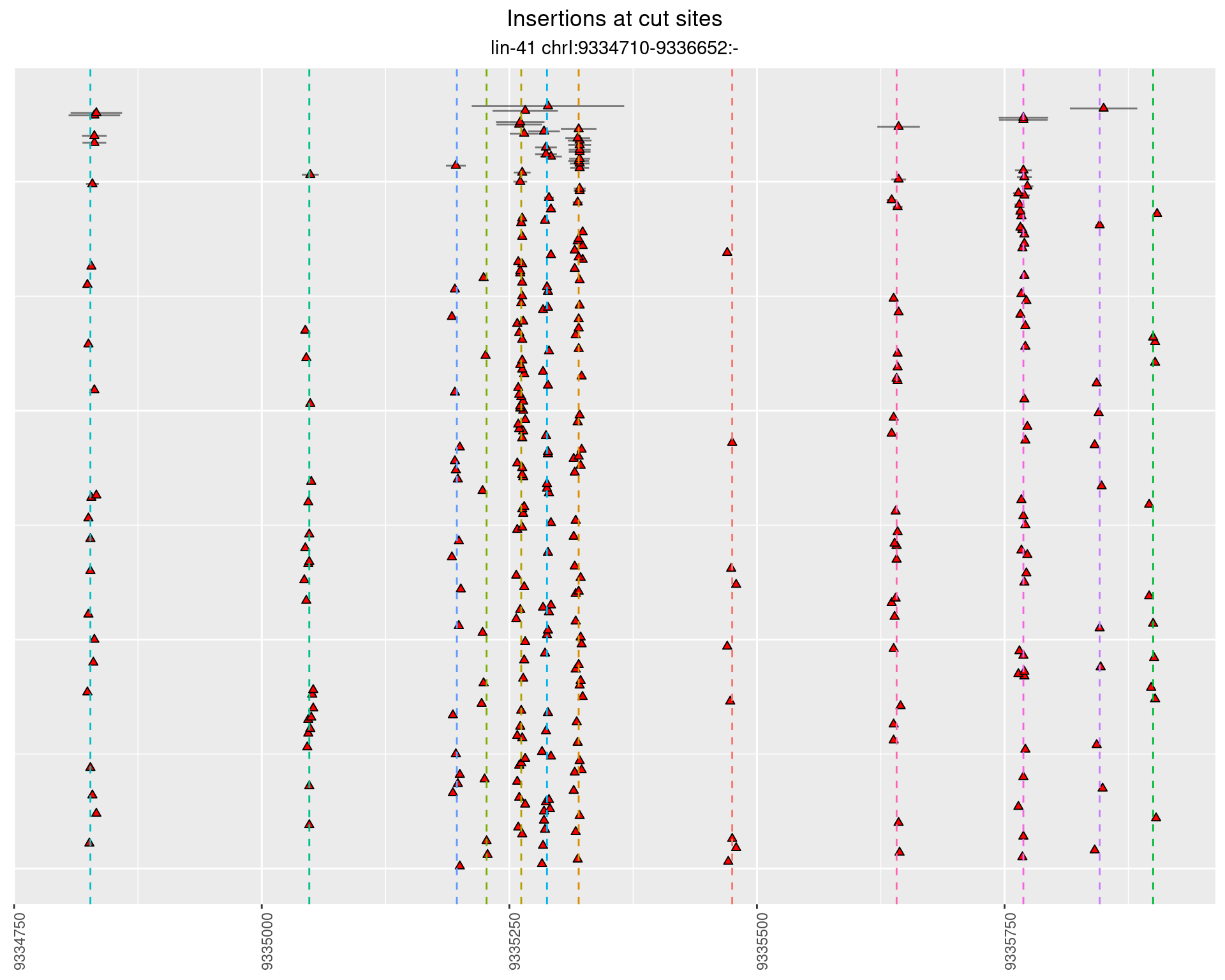
2.111 lin41_RNApacbio_L4_all
2.111.1 Freq: 0.001 - 0.01
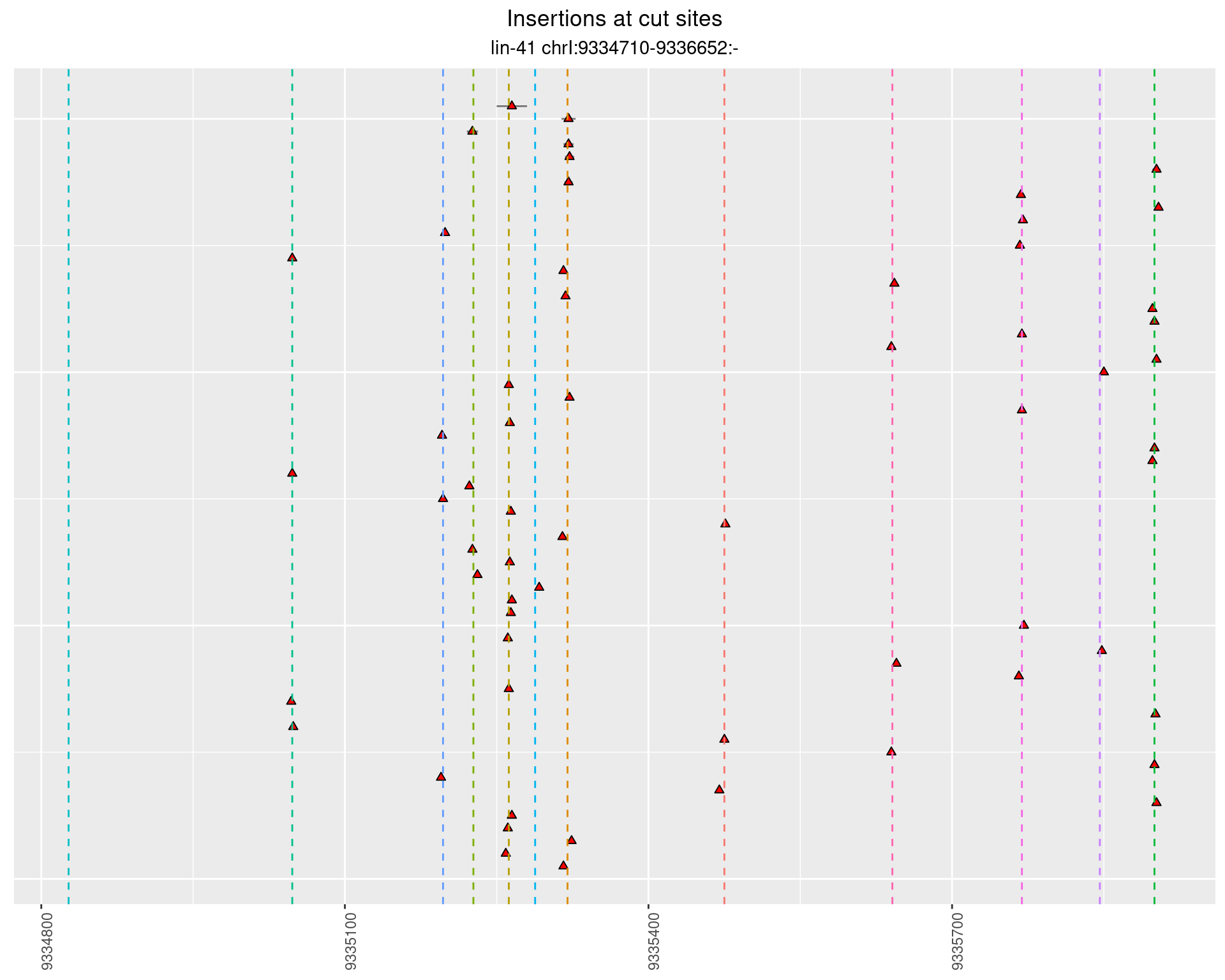
2.111.2 Freq: 1e-04 - 0.001
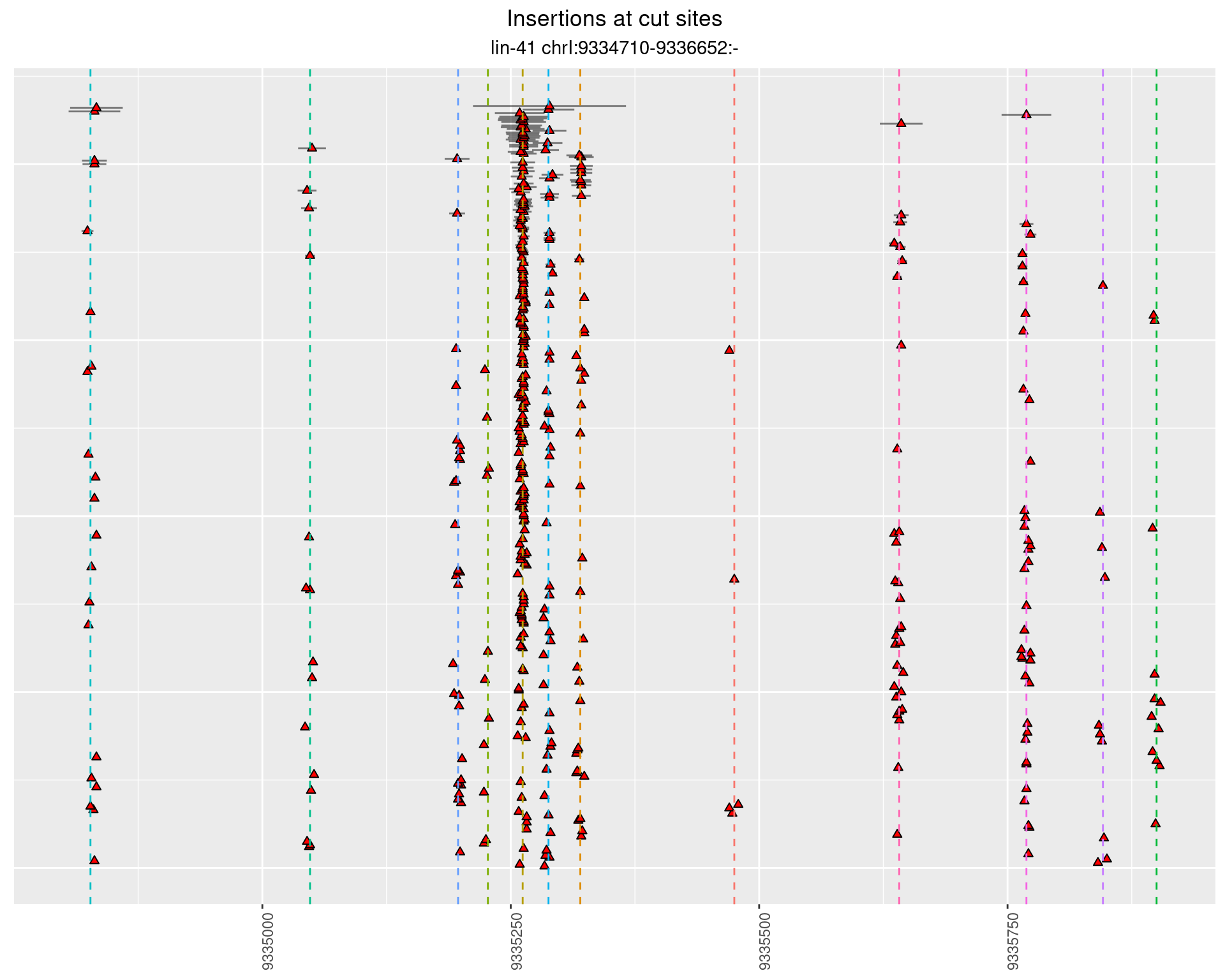
2.111.3 Freq: 0.01 - 0.1
2.111.4 Freq: 1e-05 - 1e-04
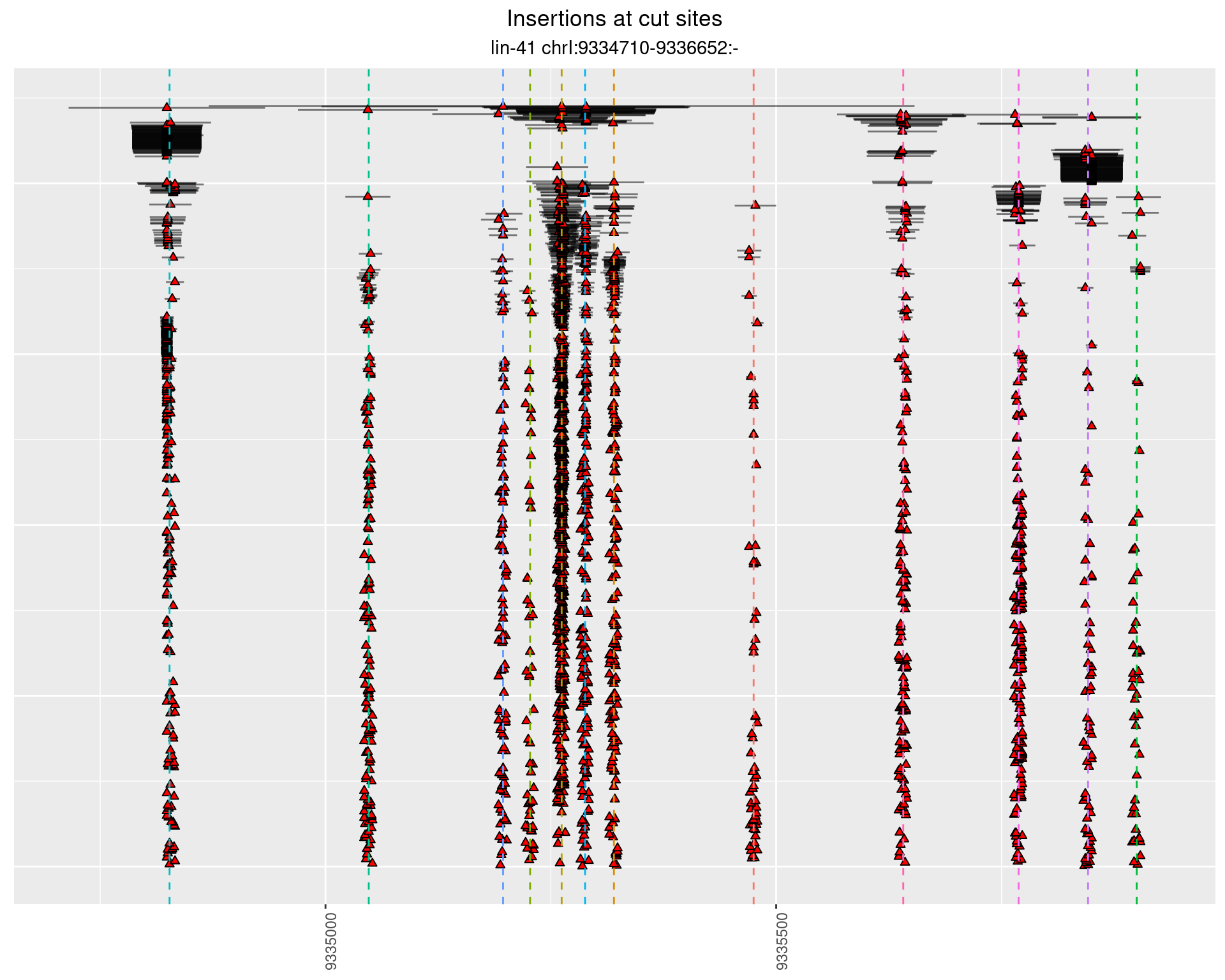
2.112 lin41_DNA_sg1516_2627_pool3
2.112.1 Freq: 1e-05 - 1e-04
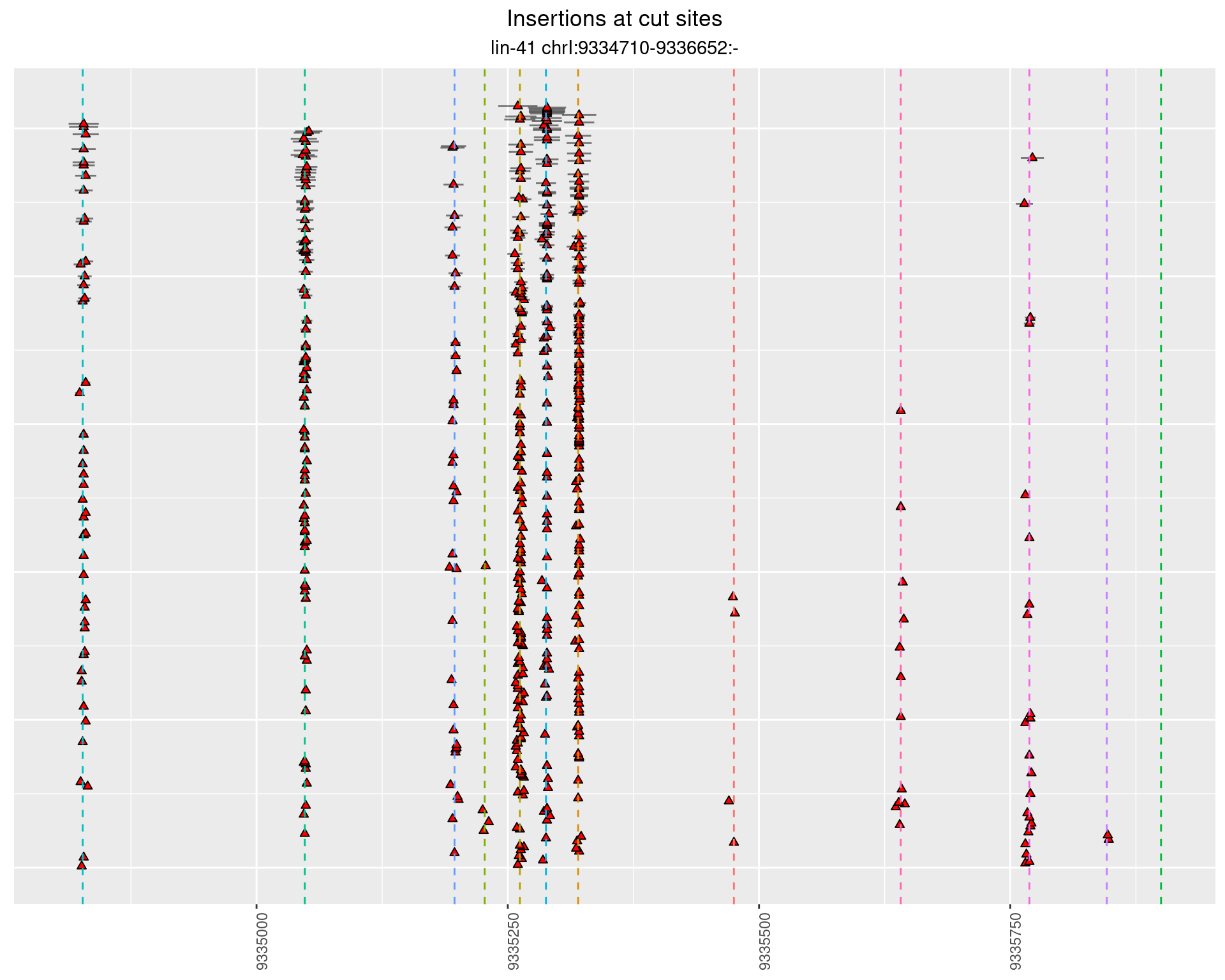
2.112.2 Freq: 1e-06 - 1e-05
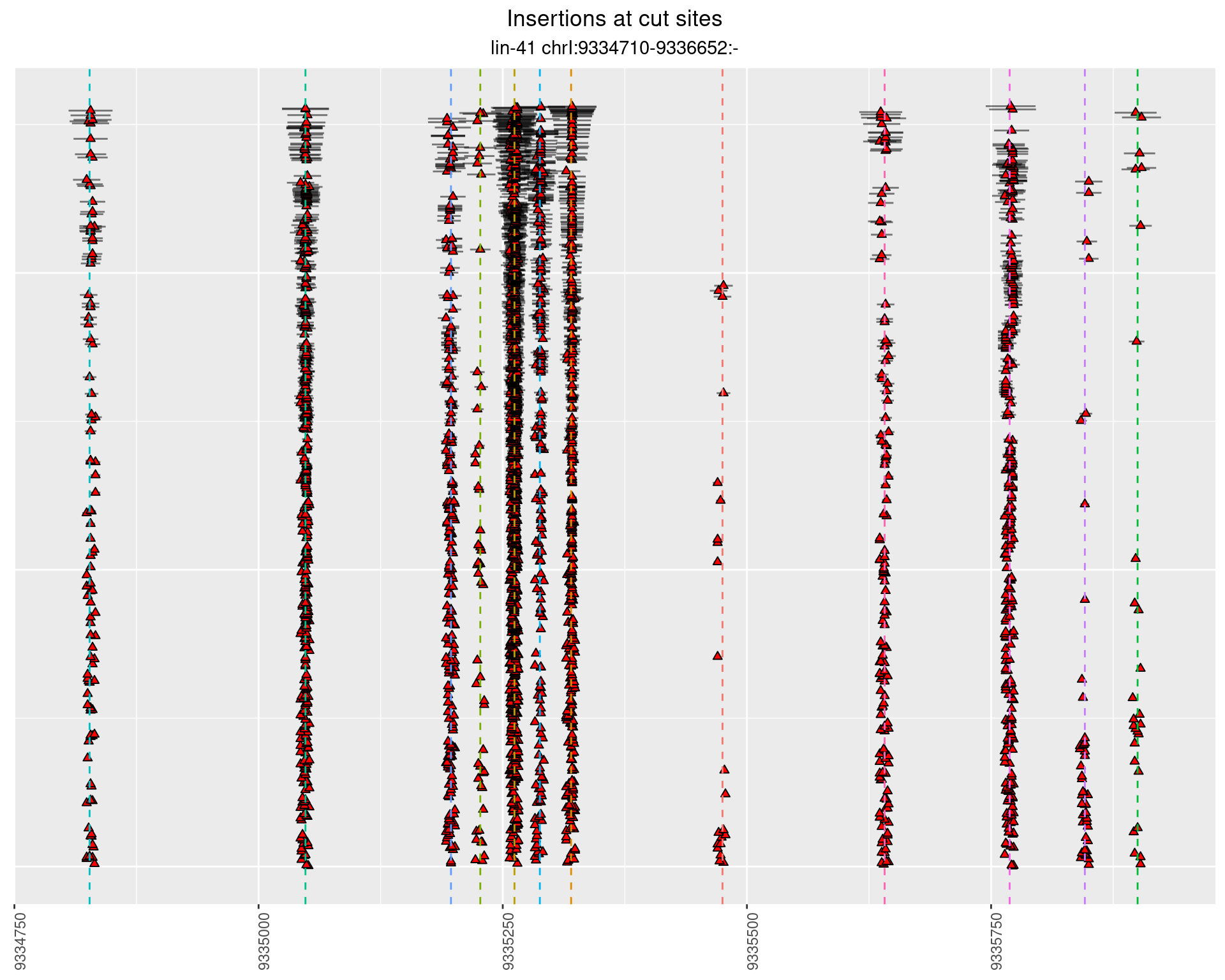
2.112.3 Freq: 1e-04 - 0.001